During Salmonella enterica serovar Typhimurium infection, host inflammation alters the metabolic environment of the gut lumen to favor the outgrowth of the pathogen at the expense of the microbiota. Inflammation-driven changes in host cell metabolism lead to the release of l-lactate and molecular oxygen from the tissue into the gut lumen.
KEYWORDS: Salmonella, gut inflammation, microbiome
ABSTRACT
During Salmonella enterica serovar Typhimurium infection, host inflammation alters the metabolic environment of the gut lumen to favor the outgrowth of the pathogen at the expense of the microbiota. Inflammation-driven changes in host cell metabolism lead to the release of l-lactate and molecular oxygen from the tissue into the gut lumen. Salmonella utilizes lactate as an electron donor in conjunction with oxygen as the terminal electron acceptor to support gut colonization. Here, we investigated transcriptional regulation of the respiratory l-lactate dehydrogenase LldD in vitro and in mouse models of Salmonella infection. The two-component system ArcAB repressed transcription of l-lactate utilization genes under anaerobic conditions in vitro. The ArcAB-mediated repression of lldD transcription was relieved under microaerobic conditions. Transcription of lldD was induced by l-lactate but not d-lactate. A mutant lacking the regulatory protein LldR failed to induce lldD transcription in response to l-lactate. Furthermore, the lldR mutant exhibited reduced transcription of l-lactate utilization genes and impaired fitness in murine models of infection. These data provide evidence that the host-derived metabolites oxygen and l-lactate serve as cues for Salmonella to regulate lactate oxidation metabolism on a transcriptional level.
INTRODUCTION
Salmonella enterica serovar Typhimurium is an enteric pathogen that causes subacute, self-limiting gastroenteritis in the immunocompetent host (1). S. Typhimurium virulence is primarily mediated by two distinct type 3 secretion systems. The type 3 secretion system encoded by Salmonella pathogenicity island 1 (2) enables S. Typhimurium to invade cultured epithelial cells and the mucosa of infected animals (3, 4). A second type 3 secretion system, encoded by Salmonella pathogenicity island 2, enables intracellular replication inside of macrophages (5, 6) and is required for full virulence in murine and bovine models of infection (7–9). Both type 3 secretion systems are required for efficient induction of host inflammatory responses. Development of inflammation drives changes in the microbiota composition (dysbiosis), including an expansion of the pathogen population (10, 11). Increased colonization of the intestinal tract by Salmonella enhances fecal shedding and host transmission through the fecal-oral route (12, 13).
The onset of intestinal inflammation changes the metabolic environment of the gut. Recent studies have shown that inflammation leads to the production of the terminal electron acceptors nitrate (14, 15) and tetrathionate (16), as well as leakage of oxygen from host tissue into the gut lumen (13). The availability of electron acceptors supports the outgrowth of S. Typhimurium, which is capable of anaerobic and aerobic respiration. In contrast, the microbiota mostly consists of obligate anaerobes that cannot use these electron acceptors. Exogenous electron acceptors allow S. Typhimurium to run a complete, oxidative tricarboxylic acid cycle (17). This in turn facilitates the catabolism of poorly fermentable carbon sources, such as ethanolamine and succinate (17, 18). Recently, we have shown that host-derived l-lactate serves as a nutrient source during S. Typhimurium expansion in the inflamed gut (19).
Under homeostatic conditions, nondigestible polysaccharides are fermented into the short-chain fatty acid butyrate by members of the class Clostridia. Butyrate is oxidized as the primary carbon and energy source for intestinal epithelial cells, a process which consumes oxygen and which keeps the gut lumen anaerobic (20, 21). Butyrate is also a cue for the transcription factor PPARγ, which controls the central metabolism of epithelial cells and drives their metabolism toward oxidative metabolism (13, 19, 22). During S. Typhimurium infection, inflammation results in a depletion of butyrate-producing Clostridia from the gut as a part of gut microbiota dysbiosis (10, 13). Butyrate deprivation alters host cell metabolism, leading to leakage of oxygen and l-lactate into the gut lumen (13, 19). The respiratory l-lactate dehydrogenase LldD facilitates degradation of host-derived l-lactate in a stereospecific manner (19). The terminal electron acceptor for l-lactate utilization is host-derived oxygen, as lactate oxidation is entirely dependent on the cytochrome bd oxidase CydAB (19). Since l-lactate and oxygen become available in the gut through the same mechanism, i.e., inflammation-associated changes in host metabolism, lactate oxidation by the LldD-CydAB electron transport chain constitutes a disease-specific metabolic module that contributes to S. Typhimurium outgrowth through manipulation of host metabolism (19).
Here, we investigated how l-lactate utilization is regulated during S. Typhimurium infection in the murine gut. LldD is encoded in an operon that contains other genes with putative functions in l-lactate utilization. The first gene in the operon, lldP, encodes a putative l-lactate permease. The second gene, lldR, encodes a putative regulatory protein. The gene products of lldPRD in S. Typhimurium 14028 exhibit considerable sequence identity at the amino acid level to the orthologues in Escherichia coli K-12 MG1665 (94%, 86%, and 94% for LldP, LldR, and LldD, respectively). While some work has been done to understand the regulation of the homologous operon in E. coli under in vitro conditions (23, 25, 26), the regulation of the l-lactate utilization operon in S. Typhimurium, especially in vivo, remains largely uncharacterized. Our results from in vitro experiments and murine models of colitis indicate that host-derived oxygen and l-lactate induce transcription of l-lactate utilization genes, suggesting that sensing of these host-derived metabolites governs transcriptional control of the lldPRD operon.
RESULTS
Oxygen, nitrate, and l-lactate induce lactate utilization genes.
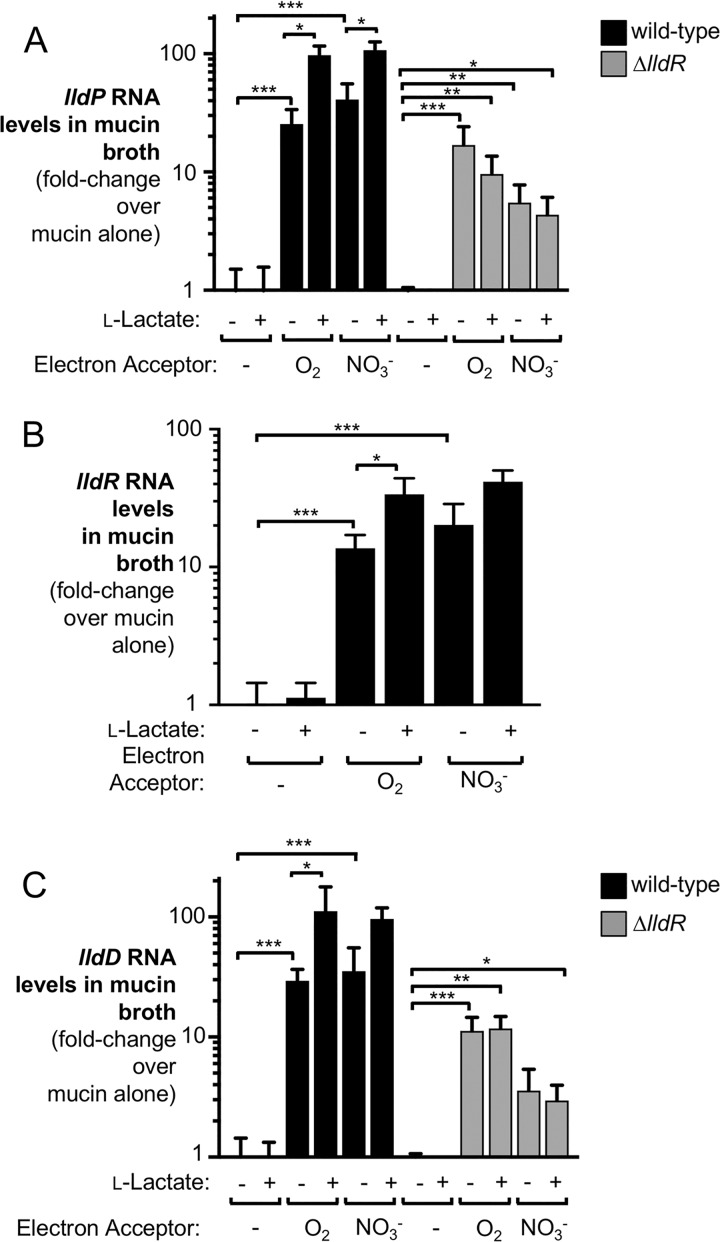
To investigate expression of the S. Typhimurium lldPRD operon, we first analyzed transcription of individual genes in response to key host-derived metabolites in vitro. Mucin broth containing l-lactate or the electron acceptor nitrate was inoculated with an S. Typhimurium wild-type strain (IR715). After 3 h of growth under microaerobic or anaerobic conditions, we extracted bacterial RNA and evaluated transcription of lldP, lldR, and lldD by reverse transcription (RT)-quantitative PCR (qPCR) (Fig. 1). In the absence of exogenous electron acceptors, addition of l-lactate alone was insufficient to induce transcription of lldP, lldR, and lldD. In contrast, addition of the electron acceptors oxygen and nitrate was sufficient to significantly upregulate expression by more than 10-fold. Exposure to l-lactate in the presence of oxygen further increased transcription beyond that induced by the electron acceptor alone, up to about 100-fold over that under uninduced conditions (Fig. 1). In addition to LldD, S. Typhimurium expresses a second respiratory lactate dehydrogenase termed Dld (27). Unlike LldD, Dld is specific for d-lactate (19). Under the conditions tested, dld was not inducible by oxygen, nitrate, or l-lactate (see Fig. S1 in the supplemental material).
FIG 1.
Analysis of lldPRD transcription in response to various stimuli in vitro. The S. Typhimurium wild-type strain and a ΔlldR mutant strain were grown in mucin broth. Nitrate (NO3−) and l-lactate were added as indicated at concentrations of 40 mM and 20 mM, respectively. Cultures were grown for 3 h anaerobically (no electron acceptor and nitrate conditions) or in the presence of 1% oxygen. RNA was extracted, and lldP (A), lldR (B), and lldD (C) mRNA levels were assessed by RT-qPCR. Transcription was normalized to that of the housekeeping gene gmk. All experiments were conducted with at least 3 biological replicates. To determine differences between groups, a two-tailed, unpaired Student's t test on ln-transformed data was used. Bars indicate the geometric mean ± standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
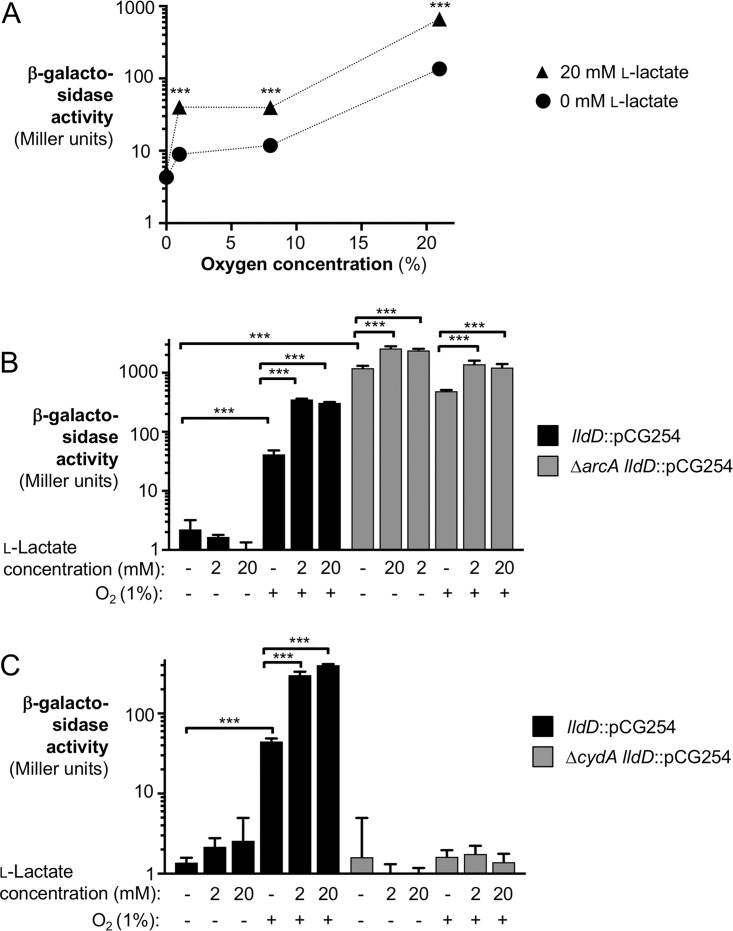
Previous reports suggest that under inflammatory conditions, the gut environment becomes microaerobic (13, 22). We thus analyzed how various levels of oxygenation affected lldD transcription with no l-lactate and 20 mM l-lactate in lysogeny broth (LB) (Fig. 2A). For this experiment, we created a transcriptional fusion of the lldD gene and lacZ (CG254), leaving the coding sequence of lldD intact to avoid any changes in expression due to the absence of LldD. We exposed cultures of S. Typhimurium CG254 to 0% (anaerobic), 1% (microaerobic), 8% (tissue oxygenation), and 21% (atmospheric) oxygen and assessed lldD transcription in a standard β-galactosidase assay. Increasing levels of oxygen correlated with increased lldD expression, with 21% oxygen inducing the strongest response. Addition of 20 mM l-lactate further increased expression in the presence of oxygen.
FIG 2.
Influence of oxygen levels, ArcAB regulation, and CydAB-mediated respiration on lldD transcription. (A) The lldD-lacZ transcriptional reporter strain CG254 (lldD::pCG254) was grown in LB in the presence of various concentrations of oxygen. The medium was supplemented with 20 mM l-lactate or mock treated. After 135 min, β-galactosidase activity was quantified. Note that the size of the error bar is smaller than the symbol representing the geometric mean. (B and C) S. Typhimurium strains CG254 (lldD::pCG254), CG267 (ΔarcA lldD::pCG254), and CG270 (ΔcydA lldD::pCG254) were grown anaerobically (no-oxygen conditions) or with 1% oxygen in LB supplemented with l-lactate, as indicated. After 5 h, β-galactosidase activity was determined. (B) β-Galactosidase activity in the wild-type background and a ΔarcA mutant background. (C) β-Galactosidase activity in the wild-type background and a ΔcydA mutant background. All experiments were conducted with at least 3 biological replicates. To determine differences between groups, a two-tailed, unpaired Student's t test was applied to the ln-transformed data. Bars indicate the geometric mean ± standard error. ***, P < 0.001.
The two-component system ArcAB represses lldD expression in the absence of oxygen.
ArcB is a sensor kinase that detects the redox state of the quinone pool and phosphorylates the response regulator ArcA. Phosphorylated ArcA then regulates the transcription of a wide variety of genes (28). In E. coli, the ArcAB two-component system represses lactate utilization genes (25, 26, 29, 30). In S. Typhimurium, lldP and lldR were also found to be repressed under anaerobic conditions in a broad survey of the ArcA regulatory program (28). We thus hypothesized that ArcAB would also be involved in the regulation of lldD. To test this idea, we compared lldD expression in the wild-type background (CG254) with that in strains which lack ArcA (CG267) or ArcB (CG268). LB supplemented with and without l-lactate was inoculated with the indicated strains and incubated anaerobically or microaerobically. Since the ΔarcA and ΔarcB mutants displayed a slight growth defect under aerobic conditions, the cultures were incubated for 5 h. lldD expression was then evaluated by β-galactosidase assay (Fig. 2B and S2). Under anaerobic conditions, the wild-type strain had minimal lldD expression, even in the presence of l-lactate, while transcription was induced under the oxygenated condition as well as in the presence of oxygen and l-lactate. In contrast, the transcription of lldD in the ΔarcA and ΔarcB mutants was derepressed, and induction by oxygen was lost entirely (Fig. 2B and S2). Of note, these strains still exhibited significant induction by the addition of l-lactate both in the presence and in the absence of oxygen. Taken together, these experiments suggest that ArcAB is responsible for transcriptional repression of S. Typhimurium lldD under anaerobic conditions.
The terminal oxidase CydAB is necessary for lldD expression under microaerobic conditions.
In our previous study, we found that l-lactate utilization was dependent on the activity of the terminal oxidase CydAB in vivo (19). ArcB senses electron flux via the redox state of the quinone pool (31), and thus, we hypothesized that aerobic respiration would contribute to derepression of lldD. To test this, we performed β-galactosidase assays as described above using a mutant strain which lacks the terminal oxidase CydAB (CG270) (Fig. 2C). While the wild-type strain had increased lldD transcription with the addition of oxygen and oxygen with l-lactate, the ΔcydA mutant exhibited no increase in lldD transcription in the presence of oxygen. Our data suggest that under microaerobic conditions, the electron flux mediated by CydAB is sensed by ArcAB, which in turn enables lldD expression.
Induction of lldD transcription occurs at physiological levels of lactate.
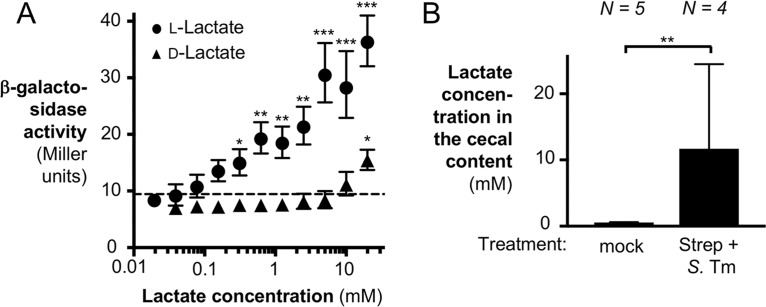
In our previous work, we showed that lactate levels in the murine gut vary considerably (19). The experiments described thus far have used an initial concentration of 20 mM l-lactate. To test if lactate utilization genes were still induced in vitro at physiologically relevant concentrations of l-lactate, we performed a dose-response experiment with 1% oxygen and various concentrations of l-lactate (Fig. 3A). There was a significant, dose-dependent increase in the level of lldD transcription over that of the oxygen-only condition (dashed line) for a large number of l-lactate concentrations ranging from 0.3125 mM to 20 mM l-lactate.
FIG 3.
Analysis of lldPRD transcription in response to various concentrations of l-lactate and d-lactate. (A) The lldD-lacZ transcriptional reporter strain CG254 (lldD::pCG254) was grown in LB for 135 min in the presence of 1% oxygen with various concentrations of l-lactate or d-lactate. lldD transcription was assessed by determination of β-galactosidase activity. The dashed line indicates the β-galactosidase activity in the absence of lactate. All experiments were conducted with at least 3 biological replicates. Note that in some instances, the size of the error bar is smaller than the symbol representing the geometric mean. (B) C57BL/6 mice were treated intragastrically with streptomycin, followed by infection with the S. Typhimurium ΔlldD mutant (CG6) 1 day later (Strep + S. Tm). Mock-treated mice (mock) received water, followed by LB 1 day later. The cecal content was collected 5 days after infection for lactate quantification by GC-MS. To determine differences between groups, a two-tailed, unpaired Student's t test on ln-transformed data was used. Bars and symbols indicate the geometric mean ± standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we analyzed the lactate concentrations in the murine gut lumen during S. Typhimurium infection. Streptomycin-pretreated C57BL/6 mice were intragastrically inoculated with the S. Typhimurium ΔlldD mutant, and the lactate concentrations in the cecal content were quantified by gas chromatography-mass spectrometry (GC-MS). Consistent with our previous results (19), the lactate concentration in mock-treated animals (no streptomycin or S. Typhimurium treatment) was approximately 0.6 mM (Fig. 3B). Lactate levels in the Salmonella-infected animals rose to about 11.7 mM (Fig. 3B). As such, the responsiveness of lldD transcription to lactate in vitro (Fig. 3A) encompasses the relevant in vivo concentrations of lactate (Fig. 3B).
Host cells produce the l-enantiomer of lactate. To determine the potential contribution of d-lactate to the induction of lldD expression, we repeated this experiment with various concentrations of the d-enantiomer (Fig. 3A). In contrast to our previous results with l-lactate, exposure to d-lactate did not increase lldD expression significantly, with the exception of exposure to d-lactate at 20 mM, the highest concentration tested. The commercially available d-lactate compound is 98% enantiopure, and thus, the 20 mM d-lactate condition likely contains 0.4 mM l-lactate, which would be sufficient to induce expression. As such, the observed increase in lldD expression with 20 mM d-lactate was likely a reflection of this impurity (Fig. 3A).
The transcriptional regulator LldR enables the response to l-lactate in vitro.
The l-lactate utilization operon includes the putative transcriptional regulator LldR. In E. coli, LldR has been shown to increase transcription of l-lactate utilization genes (23, 25). Given that the l-lactate utilization genes in S. Typhimurium are also inducible by l-lactate in the presence of electron acceptors, we hypothesized that LldR could also be responsible for the increase in lldPD expression in the presence of lactate. To test this, we evaluated transcription of lldP and lldD in a ΔlldR mutant (Fig. 1A and C, gray bars). Unlike the wild-type strain, the ΔlldR mutant was unable to increase transcription of lldP and lldD when l-lactate was added to the medium, even in the presence of oxygen and nitrate. These data strongly support the idea that the increase in lldPD expression in the presence of l-lactate is mediated by LldR.
LldR mediates lldPD transcription in the murine gut.
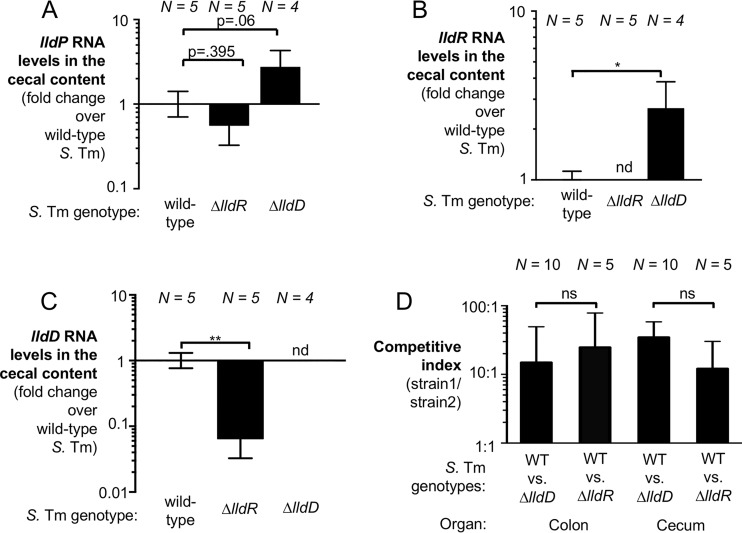
We next sought to examine transcription of lldPRD in vivo in a mouse model of Salmonella infection. One of the pathological features of nontyphoidal salmonellosis is the development of a neutrophilic infiltrate into the intestinal mucosa (32, 33). Transmigrated neutrophils and other leukocytes are frequently detected in the feces of patients (34, 35). The streptomycin-treated C57BL/6 mouse model has been used extensively by numerous groups as it allows the reproducible colonization of C57BL/6 mice and neutrophils infiltrate the intestinal mucosa upon oral S. Typhimurium infection (36). Perturbation of the microbiota by streptomycin prior to S. Typhimurium infection ensures the depletion of Clostridia and leads to lactate production by the host (Fig. 3B) and leakage of oxygen into the gut lumen (13, 19, 22). Therefore, we initially chose to assess transcription of genes required for lactate utilization in the streptomycin-treated C57BL/6 mouse model. To assess how lldP transcription occurs in the murine cecum, we treated wild-type C57BL/6 mice with streptomycin sulfate by oral gavage. One day later, we intragastrically infected mice with an S. Typhimurium wild type (IR715), a ΔlldR mutant (CG200), or a ΔlldD mutant (CG6). Five days after infection, the mice were sacrificed and the cecal contents were collected for bacterial RNA extraction and RT-qPCR (Fig. 4A to C). We were initially concerned that members of the gut microbiota express genes similar to S. Typhimurium lldPRD, thus possibly interfering with our RT-qPCR assay. However, no lldR or lldD mRNA was detected in mice infected with the S. Typhimurium ΔlldR or ΔlldD mutant (Fig. 4A to C), respectively, demonstrating that the RT-qPCR assay is specific for the intended S. Typhimurium mRNA targets. Similarly, no lldP transcription was detected in mice infected with a ΔlldP mutant (Fig. S3). mRNA for all three genes of the lldPRD operon was detected during S. Typhimurium colonization of the cecal lumen. Consistent with our in vitro data showing that LldR is required for full expression of l-lactate utilization genes in the presence of l-lactate, expression of lldD was decreased in the ΔlldR mutant (P < 0.01). A similar trend was observed for lldP; however, this decrease was not statistically significant. Intriguingly, lldP and lldR expression increased in the ΔlldD mutant (Fig. 4A and B). We speculate that this was due to an accumulation of lactate within the cell, as the ΔlldD mutant cannot degrade lactate. To ensure that differences in lldPRD expression were not due to attenuation of the mutant strains, we analyzed inflammatory cytokines in cecal tissue, which revealed significant inflammation in the streptomycin-treated, S. Typhimurium-infected cecum. Inflammation levels were similar across all groups, with the exception of mice infected with the ΔlldD mutant, which exhibited somewhat lower Nos2 and Cxcl1 expression (Fig. S4).
FIG 4.
Effect of LldR on S. Typhimurium (S. Tm) lldPRD transcription and fitness in vivo. (A to C) Groups of wild-type C57BL/6 mice were treated with streptomycin by oral gavage. One day later, animals were intragastrically infected with the S. Typhimurium wild-type strain (IR715), a ΔlldD mutant (CG6), or a ΔlldR mutant (CG200). Cecal content was collected for RNA extraction 5 days after infection. S. Typhimurium RNA levels were assessed for lldP (A), lldR (B), and lldD (C). Transcription was normalized to that of S. Typhimurium 16S rRNA. (D) Swiss Webster mice were inoculated with a 1:1 ratio of an S. Typhimurium wild-type (WT) strain (AJB715) and a ΔlldD mutant (CG6) or the wild-type strain (AJB715) and a ΔlldR mutant (CG200). Colonic and cecal contents were collected 8 days after infection. Wild-type and mutant populations were enumerated on selective agar, and the competitive index was calculated. The number of mice per group (N) is indicated for each bar. To determine differences between groups, a two-tailed, unpaired Student's t test was applied to the ln-transformed data. Bars are the geometric mean ± standard error. *, P < 0.05; **, P < 0.01; nd, not detected; ns, not significant.
LldR is required for optimal fitness in the intestinal tract.
We next wanted to explore the contribution of LldR to the fitness of S. Typhimurium in vivo. To do this, we assessed the role of LldR in Swiss Webster mice. Swiss Webster mice do not require antibiotic pretreatment to develop neutrophilic inflammation, and thus, the perturbation of the microbiota that occurs is dependent on S. Typhimurium virulence factors and not direct killing of the microbiota by antibiotics (19). To determine if LldR provides a fitness advantage in the murine gut, we performed competitive colonization experiments (Fig. 4D). Groups of Swiss Webster mice were intragastrically inoculated with an equal ratio of the S. Typhimurium wild type (AJB715) and a ΔlldD mutant (CG6) or the S. Typhimurium wild-type (AJB715) and a ΔlldR mutant (CG200), and the abundance of these strains in the cecal and colonic contents was determined 8 days after infection. As we have shown before, the ΔlldD mutant was recovered in lower numbers than the wild-type strain. Similarly, LldR also provided a significant fitness advantage in both the colon and the cecum contents. The competitive fitnesses for the ΔlldD mutant and the ΔlldR mutant were not significantly different from one another. We also attempted to analyze the transcription of lldD in this mouse model. S. Typhimurium gut colonization was considerably lower in Swiss Webster mice than in streptomycin-treated C57BL/6 mice, which prevented us from reproducibly quantifying lldD transcription in the Swiss Webster mouse model. Collectively, these data suggest that LldR regulates l-lactate utilization on the transcriptional level in vivo and that LldR-mediated regulation contributes to optimal gut colonization.
DISCUSSION
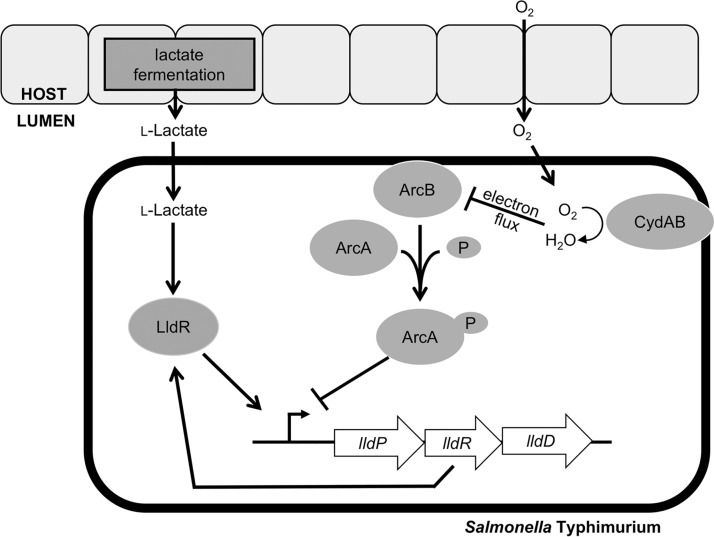
While the majority of Salmonella enterica serovars are associated with gastroenteritis, a subset of serovars causes disseminated disease, such as typhoid and paratyphoid fever. The genomes of extraintestinal serovars typically exhibit an increased accumulation of pseudogenes compared to the genomes of intestinal serovars (37, 38), which may reflect adaptations to different niches within the host (39). We have shown previously that S. Typhimurium exploits changes in host metabolism, such as the release of oxygen and l-lactate into the gut lumen (Fig. 5) (13, 19). Our data suggest that the inducibility of the lldPRD operon by l-lactate may be a specific adaptation to changes in mammalian l-lactate production during infection with the intestinal Salmonella serovar Typhimurium. Consistent with this idea, lldP and lldR were identified to be pseudogenes in the extraintestinal serovars Salmonella enterica serovar Gallinarum and Salmonella enterica serovar Choleraesuis, respectively (37, 40, 41). Furthermore, during experimental S. Typhimurium infection of mice, LldD contributes gut colonization but is dispensable for fitness at systemic sites (19).
FIG 5.
Central model of regulation of lactate utilization during S. Typhimurium infection. Refer to the text for details.
Our results indicate that the regulation of the lldPRD operon is stereospecific to l-lactate. Mammalian cells produce l-lactate (42), whereas members of the microbiota can produce both isomers (43). Little is known about changes in short-chain fatty acids and lactate during human salmonellosis. Of note, patients with noninfectious diarrhea exhibit increased l-lactate levels in the feces (44). Increases in fecal l-lactate in ulcerative colitis are due to augmented excretion by the mucosa (45). In patients with ulcerative colitis suffering from pancolitis and active Crohn’s disease, the average fecal l-lactate concentration exceeds 10 mM, whereas it is less than 4 mM in quiescent patients (44).
Here, we report that induction of the S. Typhimurium l-lactate dehydrogenase LldD in vitro occurred at l-lactate concentrations that span the experimentally determined in vivo lactate concentrations. In contrast, exposure to d-lactate had little effect on lldD transcription. Consistent with the idea that l-lactate utilization is an adaptation to the environment of the inflamed gut, the d-lactate dehydrogenase Dld was not inducible by oxygen, nitrate, or l-lactate. Furthermore, Dld activity does not provide a fitness advantage in the murine gut (19).
Unlike the cytoplasmic lactate dehydrogenase LdhA used during fermentation, LldD is membrane bound and donates electrons directly to the quinone pool (46). In the murine gut, oxygen likely is the terminal electron acceptor for S. Typhimurium l-lactate utilization (19). Our data showed that oxygen availability and aerobic respiration modulated l-lactate utilization genes in vitro. Signaling through the respiration-responsive regulatory system ArcAB was required for repression under anaerobic conditions, when electron acceptors are not available for coupling with l-lactate conversion to pyruvate. Similarly, the activity of the terminal oxidase CydAB was essential for expression of l-lactate utilization genes under microaerobic conditions. This shows a dual function for CydAB during lactate oxidation, which serves as the terminal electron acceptor and modulates the electron flux that relieves ArcAB-mediated repression of l-lactate utilization genes. Taken together, our study demonstrates that sensing of host-derived metabolites provides an important cue for a dynamic transcriptional program that benefits S. Typhimurium in the murine gut through lactate oxidation and, thus, ensures successful S. Typhimurium outgrowth.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
All bacterial strains and plasmids used in this study are listed in Table 1. All primers are listed in Tables 2 and 3. S. Typhimurium and E. coli strains were routinely grown in lysogeny broth (LB; 10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter sodium chloride) and on LB plates (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter sodium chloride, 15 g/liter agar) under aerobic conditions at 37°C. Nalidixic acid (Nal), kanamycin (Kan), and chloramphenicol (Cm) were added to the media as needed at a concentration of 50 mg/liter, 100 mg/liter, and 15 mg/liter, respectively. 5-Bromo-4-chloro-3-indolylphosphate (X-phos; Chem-Impex) was added to the plates to detect the activity of the acidic phosphatase PhoN. All plasmids used were created by use of a Gibson Assembly cloning kit (New England Biolabs). To generate pCG200, pCG226, pMW195, and pMW89, the upstream and downstream regions of the gene of interest were amplified using Q5 HotStart high-fidelity DNA polymerase (New England Biolabs). The upstream and downstream regions were then inserted into SphI-digested pRDH10 using the Gibson Assembly reaction. All plasmids were sequenced to check for point mutations prior to mutagenesis. For cloning experiments, the host for suicide plasmids was DH5α λpir. S17-1 λpir served as the donor strain for conjugation into S. Typhimurium. After conjugation, single crossovers were selected with LB plates supplemented with Cm. Counterselection was performed by plating on sucrose plates (5% sucrose, 15 g/liter agar, 8 g/liter nutrient broth base) to check for second crossover events. This cloning strategy generates clean, unmarked deletions and was used to create strains CG200, CG226, MW118, and MW119. To generate pCG254, 379 nucleotides of the 3′ end of the lldD coding sequence (including the stop codon) and 31 nucleotides immediately downstream of the stop codon were amplified and cloned into SmaI-digested pFUSE. pCG254 was conjugated into IR715, CG124, MW119, and MW118 as described above to generate the lldD transcriptional fusion in strains CG254, CG270, CG267, and CG268, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid identifier | Description | Source or reference |
|---|---|---|
| S. Typhimurium strains | ||
| IR715 | Nalidixic acid-resistant strain of Salmonella enterica serovar Typhimurium 14028S | 53 |
| AJB715 | IR715 phoN::Kanr | 54 |
| CG6 | IR715 ΔlldD | 19 |
| CG124 | IR715 ΔcydA | 19 |
| CG200 | IR715 ΔlldR | This study |
| CG226 | IR715 ΔlldP | This study |
| CG254 | IR715 lldD::pCG254 | This study |
| CG267 | IR715 ΔarcA lldD::pCG254 | This study |
| CG268 | IR715 ΔarcB lldD::pCG254 | This study |
| CG270 | IR715 ΔcydA lldD::pCG254 | This study |
| MW118 | IR715 ΔarcB | This study |
| MW119 | IR715 ΔarcA | This study |
| E. coli strains | ||
| DH5α λpir | F− endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 φ80dlacZΔM15 λpir | 55 |
| S17-1 λpir | zxx::RP4 2-(Tetr::Mu) (Kanr::Tn7) λpir | 56 |
| Plasmids | ||
| pRDH10 | ori(R6K) mobRP4 Cmr Tetr sacRB | 54 |
| pFUSE | ori(R6K) mobRP4 Cmr lacZYA | 57 |
| pCG226 | Upstream and downstream regions of S. Typhimurium lldP in pRDH10 | This study |
| pCG254 | Fragment of lldD in pFUSE | This study |
| pCG200 | Upstream and downstream regions of S. Typhimurium lldR in pRDH10 | This study |
| pMW195 | Upstream and downstream regions of S. Typhimurium arcA in pRDH10 | This study |
| pMW89 | Upstream and downstream regions of S. Typhimurium arcB in pRDH10 | This study |
TABLE 2.
Primers and RT-qPCR parameters
| Target | Sequences | Limit of quantification (CT value) | Linear dynamic range (no. of copies/μl) | Source or reference |
|---|---|---|---|---|
| lldP | 5′-TGCTGGCGTTCGCGTTTATC-3′, 5′-CTCTCTACAGGCTACCGCGG-3′ | 38 | 2.69 × 108–2.7 | This study |
| lldR | 5′-ACCGCTGCCGACAAAGAGAA-3′, 5′-CTTTCATCTGGCGATCGCGG-3′ | 37 | 2.69 × 108–2.7 | This study |
| lldD | 5′-TCCGTAACGGGCTGGATGTC-3′, 5′-GAAGGTCGCCATGACCCTGA-3′ | 33 | 2.69 × 108–2.7 | This study |
| dld | 5′-CCGGAGCAGATCCTGAGCAA-3′, 5′-TTTGAATCTTCCGGCTGCGC-3′ | 37 | 2.10 × 108–2.1 | This study |
| gmk | 5′-TTGGCAGGGAGGCGTTT-3′, 5′-GCGCGAAGTGCCGTAGTAAT-3′ | 34 | 2.27 × 109–22.7 | 58 |
| 16S rRNA | 5′-CAGAAGAAGCACCGGCTAACTC-3′, 5′-GCGCTTTACGCCCAGTAATT-3′ | 33 | 7.21 × 107–72.1 | 58 |
| Cxcl1 | 5′-TGCACCCAAACCGAAGTCAT-3′, 5′-TTGTCAGAAGCCAGCGTTCAC-3′ | 32 | 4.86 × 107–48.6 | 59 |
| Nos2 | 5′-TTGGGTCTTGTTCACTCCACGG-3′, 5′-CCTCTTTCAGGTCACTTTGGTAGG-3′ | 37 | 1.04 × 108–1.04 × 102 | 60 |
| Tnf | 5′-AGCCAGGAGGGAGAACAGAAAC-3′, 5′-CCAGTGAGTGAAAGGGACAGAACC-3′ | 40 | 3.2 × 107–32 | 61 |
| Gapdh | 5′-TGTAGACCATGTAGTTGAGGTCA-3′, 5′-AGGTCGGTGTGAACGGATTTG-3′ | 40 | 2.31 × 108–2.31 × 102 | 59 |
TABLE 3.
Primers for mutagenesis
| Target/purpose | Sequences | Source or reference |
|---|---|---|
| Deletion of S. Typhimurium lldR | 5′-GCCATCTCCTTGCATGTTAACGTTCCGGTACCGTATC-3′, 5′-GTTTTAACCTGGATGATTCCGTGAATGATTATTTCA-3′, 5′-TTCCGTGAATGATTATTTCAGCAGCCAGCGATTATC-3′, 5′-GTCGGCCTGAACGGTCGCCATGCACCATTCCTTG-3′ | This study |
| Deletion of S. Typhimurium arcA | 5′-GCCATCTCCTTGCATGCACCGTGCTTAACGAGCGCG-3′, 5′-TTTGGCGCCTGGGCCGAAAAATTGCCA-3′, 5′-GGGCCGAAAAATTGCCAACTAAATCGAAAC-3′, 5′-CGTATTGTCGACAGCCAGCATGCACCATTCCTTG-3′ | This study |
| Deletion of S. Typhimurium arcB | 5′-GCCATCTCCTTGCATGCTCCACAACGATATCATCAACCGGG-3′, 5′-ACCCCGGTCAAACCGGGGTTCCTTCAC-3′, 5′-AACCGGGGTTCCTTCACCACAACTTC-3′, 5′-GAAATAGGCCAGATAGCGTTGCATGCACCATTCCTTG-3′ | This study |
| Deletion of S. Typhimurium lldP | 5′-GGGCGCCATCTCCTTGCATGTCGAAGAAGCAAACACTTATAC-3′, 5′-GTTCTCAGGAGACCTGCATTGTGATGCC-3′, 5′-GAGACCTGCATTGTGATGCCAAAACGCC-3′, 5′-CGGAACAACACCAGGCAGCATGCACCATTCCTTGCGGC-3′ | This study |
| Creation of pCG254 | 5′-GCCGCTCTAGAACTAGTGGATCCCCGGTGTCGAATCACGGCGG-3′, 5′-CTTTATGTACTGCGTGACCGGGAATTCCGATCCGACAAC-3′ | This study |
Quantification of mRNA during growth in mucin broth.
About 100 mg of mucin (porcine stomach type II; Sigma) was suspended in 1 ml ethanol, and the mixture was incubated at room temperature overnight. The ethanol was evaporated in a vacuum centrifuge at 30°C. Dried mucin was resuspended in no-carbon E medium (0.2 g/liter MgSO4·7H2O, 3.9 g/liter KH2PO4, 5.0 g/liter anhydrous K2HPO4, 3.5 g/liter NaNH4HPO4·4H2O) (47, 48) at a final concentration of 0.5% (wt/vol) mucin. Other compounds were dissolved in water and filter sterilized (pore size, 0.2 μm) prior to addition to the complete mucin broth. Sodium l-lactate (Sigma) and sodium nitrate (Sigma) were added to the media as specified for a final concentration of 20 mM and 40 mM, respectively. All media were preincubated in an anaerobic chamber (5% hydrogen, 5% CO2, 90% nitrogen; Sheldon Manufacturing) 1 day prior to inoculation. Five milliliters of medium was inoculated with 100 μl of an overnight culture of the strains of interest. The cultures were grown for 3 h in the anaerobic chamber (no electron acceptor condition and nitrate condition) or a hypoxic cabinet (1% oxygen, 99% nitrogen; Coy Laboratory Products), as indicated in each figure legend. RNA was extracted using with an Aurum total RNA minikit (Bio-Rad) according to the manufacturer’s protocol. The RNA was then treated with DNase I (Thermo Fisher) twice according to the manufacturer’s instructions prior to analysis by RT-qPCR. RNA samples were stored at −80°C.
Preparation of cDNA and qPCR.
cDNA was prepared using TaqMan reverse transcription reagents (Thermo Fisher) as described by the manufacturer. Briefly, the reaction mixture was prepared using 2.5 μl of 10× RT-PCR buffer, 5.5 μl MgCl2 (25 mM), 5 μl of deoxynucleoside triphosphates (2.5 mM each), 1.25 μl of random hexamers (50 μM), 1 μl RNase inhibitor, 0.625 μl of reverse transcriptase, and 9.6 μl of template RNA. The RNA concentration and the A260/A280 ratio were evaluated. For bacterial RNA samples, a reaction with RT-PCR buffer, water, and template RNA (no reverse transcriptase) was performed to quantify contamination with DNA. Samples with more than 5% DNA contamination were excluded from analysis. The reverse transcription reaction was performed using the following protocol: 10 min at 25°C, 30 min at 48°C, 5 min at 95°C, and 4°C indefinitely. cDNA and no-reverse-transcription controls were stored at −20°C prior to analysis.
qPCR was performed using SYBR Green (Thermo Fisher) with 2 μl of template DNA and 250 nM each primer in a final reaction volume of 11 μl. The primer sequences and important experimental parameters are listed in Table 2. qPCR was performed using a QuantStudio 6 Flex instrument (Thermo Fisher) with the vendor-supplied, standard SYBR green qPCR protocol. Unless indicated otherwise, the ramp speed was 1.6°C/s. The hold stage was at 50°C for 2 min and 95°C for 10 min. The amplification stage consisted of two steps, 95°C for 15 s and 60°C for 1 min, which were repeated for a total of 40 times. The melt curve was determined by increasing the temperature from 60°C to 95°C at a speed of 0.05°C/s. A nontemplate control (water) was run in addition to the samples. Two technical replicates per biological replicate were assayed. Melt curves for each reaction were evaluated prior to analysis. Data were analyzed using QuantStudio real-time PCR software (v1.2). Baselines were determined using the baseline threshold algorithm. Data were further analyzed using the comparative threshold cycle (CT) method for Fig. 1 and 4 and for Fig. S1 and S3 in the supplemental material in Microsoft Excel software (49). PCR fragments containing known concentrations of the qPCR target were used to determine the limit of quantification (the CT value for the lowest value in the linear dynamic range), the linear dynamic range, and the efficiency of the qPCR for bacterial targets. Serial dilutions of plasmids containing the gene of interest were used to quantify the absolute counts of the target gene in Fig. S4 and to perform quality control analysis for the qPCR assay (50).
β-Galactosidase assays.
Five milliliters of LB supplemented with various concentrations of l- or d-lactate (Sigma) was preincubated in the anaerobic chamber 1 day prior to inoculation. Overnight cultures were grown in the anaerobic chamber (Fig. 2B and C and S2) or aerobically with shaking (Fig. 2A and 3A). The prereduced medium was inoculated with 100 μl of overnight culture and incubated in the anaerobic chamber (no electron acceptor conditions) or hypoxic cabinet (1% or 8% oxygen conditions) as indicated for 135 min (Fig. 2A and 3A) or 5 h (Fig. 2B and C and S2). At the end of incubation, cultures were placed on ice for 20 min and β-galactosidase assays were performed based on the protocol described by Miller (51). Briefly, the optical density at 600 nm (OD600) of the chilled cultures was taken. One hundred microliters or 500 μl of chilled culture was added to 900 μl or 500 μl of phosphate-buffered saline (PBS), respectively. Bacterial cells were lysed with 20 μl of 0.1% SDS and 40 μl of chloroform, vortexed for 10 s, and incubated at room temperature for 5 min. Two hundred microliters of 4 mg/ml o-nitrophenyl-β-d-galactoside (ONPG; Thermo Scientific) was then added to the reaction mixture. After sufficient yellow color had developed, the reaction was stopped with 500 μl of 1 M Na2CO3 and the time was recorded. The A420 and A550 were taken, and the Miller units were calculated according to the following equation: relative activity (in Miller units) = 1,000 × {[A420 − (1.75 × A550)]/(t × v × OD600)}, where t is time in minutes and v is volume in milliliters.
Mouse experiments.
Conventional C57BL/6 and Swiss Webster mice, originally obtained from The Jackson Laboratory, were bred under specific-pathogen-free conditions in a barrier facility at UT Southwestern. Both male and female mice (age, 6 to 8 weeks) were used for the experiments, and no marked differences between male and female mice were observed. All mice were on a 12-h light/12-h dark cycle and consumed food (Envigo 2919, Teklad Global 16% protein diet, irradiated) and water ad libitum. All experiments were conducted in accordance with the policies of the Institutional Animal Care and Use Committee at UT Southwestern.
Streptomycin-treated mouse model of salmonellosis.
Wild-type C57BL/6 mice were intragastrically treated with 20 mg of sterile streptomycin sulfate (lot number 1796C493; VWR) in water or mock treated with water. One day later, the mice were infected intragastrically with 1 × 105 CFU of S. Typhimurium strains or mock treated with LB. Mice were euthanized 5 days after infection. The cecal content and cecal tissue were flash frozen in liquid nitrogen for RNA extraction and stored at −80°C. For analysis of the cecal content by GC-MS, the cecal content was placed in sterile PBS for further analysis.
Swiss Webster mouse model of salmonellosis.
Wild-type Swiss Webster mice were intragastrically inoculated with 1 × 109 CFU of S. Typhimurium by using 5 × 108 CFU of each strain. Mice were sacrificed at 8 days after infection, and colonic and cecal contents and tissues were collected for analysis. Colonic and cecal contents were placed in sterile PBS, vortexed, and serially diluted on selective agar to determine the number of CFU per gram of each strain. Competitive indices were calculated by dividing the number of CFU per gram of the wild-type strain by the number of CFU per gram of the mutant strain, corrected by the same ratio in the inoculum. Cecal tissue was flash frozen for later analysis and stored at −80°C.
RNA extraction from cecal content.
Cecal content was collected, flash frozen in liquid nitrogen, and stored at −80°C. RNA was extracted with the TRI reagent (Molecular Research Center). Briefly, frozen samples were resuspended in 0.5 ml of TRI reagent and homogenized in a Mini-BeadBeater (BioSpec Products) twice for 30 s each time. Chloroform (0.1 ml) was then added to the tube, and the tube was shaken and incubated for 5 min at room temperature. The samples were then centrifuged at 12,000 × g for 15 min at 4°C. The aqueous-phase supernatant (0.2 ml) was transferred to a new tube. RNA was precipitated by adding 0.25 ml of isopropanol, followed by incubation at room temperature for 10 min. Samples were then centrifuged at 12,000 × g for 8 min at 25°C to pellet the RNA. The pellet was then washed with 0.5 ml of 75% ethanol and centrifuged at 12,000 × g for 5 min at 25°C. The pellet was then air dried and resuspended in DNase/RNase-free water. RNA preparations were then treated twice with DNase I (Ambion) prior to use. cDNA preparation and qPCR were performed as described above.
GC-MS quantification of lactate.
Lactate measurements were performed as described previously (19, 52). Cecal contents were collected and placed into sterile PBS. Samples were vortexed for 2 min and centrifuged at 6,000 × g at 4°C for 15 min. Supernatant was then aliquoted and 50 μM deuterated lactate (sodium l-lactate-3,3,3-d3; CDN Isotopes) was added as the internal standard. Samples were then evaporated to dryness in a vacuum centrifuge and stored at −80°C prior to analysis. Standards were prepared in the same way as the samples with set concentrations of deuterated and nondeuterated lactate. Samples were then resuspended in pyridine (Sigma), sonicated for 1 min, and incubated at 80°C for 20 min. Samples were derivatized with N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane (Sigma) and incubated for 1 h at 80°C. Samples were then centrifuged at 16,000 × g for 1 min to remove the debris, and 80 μl of supernatant was transferred to autosampler vials. Analysis was performed using a Shimadzu TQ8040 triple-quadrupole GC-MS. The injection temperature was 250°C with a split ratio of 1:100 and a volume of 1 μl. An Rtx-5 SilMS fused-silica capillary column was used with helium as the carrier gas (velocity, 50 cm/s). The oven temperature began at 50°C for 2 min and rose to 100°C in increments of 20°C per minute, with a hold at 100°C for 3 min. The oven temperature was then increased to a final temperature of 330°C, rising in increments of 40°C per minute with a final hold for 3 min. The ion source was used in electron ionization mode (70 V, 150 μA, 200°C). Selected ion monitoring (SIM) and multiple-reaction monitoring (MRM) were used, with the event time being 50 ms. The following mass spectrometry parameters were used: for lactate-d3, an MRM of 264 > 236 and a collision energy (CE) of 6 V or an MRM of 264 > 189 and a CE of 8 V, and for lactate, an MRM of 261 > 233 and a CE of 6 V or an MRM of 261 > 189 and a CE of 8 V. The m/z used for quantification is italicized. The recovery of each sample was calculated using the recovery of internal deuterated standards.
Cytokine mRNA quantification from cecal tissue.
Inflammatory cytokines were assessed as previously described (19). Flash-frozen tissue was homogenized with a Mini-BeadBeater (BioSpec Products). RNA was extracted with the TRI reagent (Molecular Research Center) according to the manufacturer’s instructions. RNA was treated with DNase I (Thermo Fisher) following extraction. cDNA preparation and qPCR were then performed as described above.
Statistical analysis.
Data analysis was done in Microsoft Excel (v16.15.5) and GraphPad Prism (v7.0c) software. All data were transformed with the natural logarithm (ln) prior to analysis. The normality of all transformed data was tested using the D’Agostino-Pearson test (for sample sizes of 8 or more) or the Shapiro-Wilk test (for less than 8 samples). Data for mice that were euthanized for health reasons prior to the end of the experiment were excluded from analysis. To determine statistical differences between groups of mice, a two-tailed, unpaired Student's t test was applied to the logarithmically transformed data. To determine statistical differences between wild-type and mutant bacterial populations within the same animal (competition experiments), a two-tailed, paired Student's t test was used. P values of less than 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
Work in S.E.W.’s lab was funded by the NIH (AI118807, AI128151), the Welch Foundation (I-1858), the Burroughs Wellcome Fund (1017880), and a research scholar grant (RSG-17-048-01-MPC) from the American Cancer Society. C.C.G. received an NSF graduate research fellowship (1000194723). R.B.C. was funded by an NIH training grant (GM109776). W.Z. was supported by a research fellows award from the Crohn’s and Colitis Foundation of America (454921).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
We thank Julie Pfeiffer, David Hendrixson, Ezra Burstein, and Vanessa Sperandio for their advice and comments.
S.E.W. is listed as an inventor on patent application WO2014200929A1, which describes a treatment to prevent the inflammation-associated expansion of Enterobacteriaceae. The other authors have no additional financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00773-18.
REFERENCES
- 1.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, Baumler AJ. 2009. Life in the inflamed intestine, Salmonella style. Trends Microbiol 17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills DM, Bajaj V, Lee CA. 2006. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol 15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 3.Galan JE, Curtiss R. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med 180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, Dinauer MC, Mastroeni P, Fang FC. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658. [DOI] [PubMed] [Google Scholar]
- 7.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsolis RM, Adams LG, Ficht TA, Baumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67:4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. 2008. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez CA, Rivera-Chávez F, Byndloss MX, Bäumler AJ. 2015. The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar Typhimurium during colitis. Infect Immun 83:3470–3478. doi: 10.1128/IAI.00351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio 3:e00143-12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, Beiting DP, Santos RL, Hooper LV, Winter SE. 2017. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe 22:291–301.e6. doi: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado D, Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23:54–64.e6. doi: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. 2012. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One 7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilera L, Campos E, Gimenez R, Badia J, Aguilar J, Baldoma L. 2008. Dual role of LldR in regulation of the lldPRD operon, involved in l-lactate metabolism in Escherichia coli. J Bacteriol 190:2997–3005. doi: 10.1128/JB.02013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin EC. 1993. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol 175:6671–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iuchi S, Aristarkhov A, Dong JM, Taylor JS, Lin EC. 1994. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked l-lactate dehydrogenase. J Bacteriol 176:1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong JS, Kaback HR. 1972. Mutants of Salmonella typhimurium and Escherichia coli pleiotropically defective in active transport. Proc Natl Acad Sci U S A 69:3336–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, Hassan HM. 2011. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol 11:58. doi: 10.1186/1471-2180-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch AS, Lin EC. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol 178:6238–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iuchi S, Lin EC. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A 85:1888–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgellis D, Kwon O, Lin EC. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 32.Day DW, Mandal BK, Morson BC. 1978. The rectal biopsy appearances in Salmonella colitis. Histopathology 2:117–131. [DOI] [PubMed] [Google Scholar]
- 33.McGovern VJ, Slavutin LJ. 1979. Pathology of salmonella colitis. Am J Surg Pathol 3:483–490. [DOI] [PubMed] [Google Scholar]
- 34.Harris JC, Dupont HL, Hornick RB. 1972. Fecal leukocytes in diarrheal illness. Ann Intern Med 76:697–703. [DOI] [PubMed] [Google Scholar]
- 35.Alvarado T. 1983. Faecal leucocytes in patients with infectious diarrhoea. Trans R Soc Trop Med Hyg 77:316–320. [DOI] [PubMed] [Google Scholar]
- 36.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuccio SP, Baumler AJ. 2014. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5:e00929-14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang C, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du F, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 39.Tanner JR, Kingsley RA. 2018. Evolution of Salmonella within hosts. Trends Microbiol 26:986–998. doi: 10.1016/j.tim.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, Barron A, Layton A, Pickard D, Kingsley RA, Bignell A, Clark L, Harris B, Ormond D, Abdellah Z, Brooks K, Cherevach I, Chillingworth T, Woodward J, Norberczak H, Lord A, Arrowsmith C, Jagels K, Moule S, Mungall K, Sanders M, Whitehead S, Chabalgoity JA, Maskell D, Humphrey T, Roberts M, Barrow PA, Dougan G, Parkhill J. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res 18:1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu CH, Tang P, Chu C, Hu S, Bao Q, Yu J, Chou YY, Wang HS, Lee YS. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res 33:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adeva-Andany M, López-Ojén M, Funcasta-Calderón R, Ameneiros-Rodríguez E, Donapetry-García C, Vila-Altesor M, Rodríguez-Seijas J. 2014. Comprehensive review on lactate metabolism in human health. Mitochondrion 17:76–100. doi: 10.1016/j.mito.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. 2007. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol 73:6526–6533. doi: 10.1128/AEM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hove H, Mortensen PB. 1995. Influence of intestinal inflammation (IBD) and small and large bowel length on fecal short-chain fatty acids and lactate. Dig Dis Sci 40:1372–1380. [DOI] [PubMed] [Google Scholar]
- 45.Hove H, Holtug K, Jeppesen PB, Mortensen PB. 1995. Butyrate absorption and lactate secretion in ulcerative colitis. Dis Colon Rectum 38:519–525. [DOI] [PubMed] [Google Scholar]
- 46.Futai M, Kimura H. 1977. Inducible membrane-bound l-lactate dehydrogenase from Escherichia coli. Purification and properties. J Biol Chem 252:5820–5827. [PubMed] [Google Scholar]
- 47.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 48.Berkowitz D, Hushon JM, Whitfield HJ Jr, Roth J, Ames BN. 1968. Procedure for identifying nonsense mutations. J Bacteriol 96:215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 50.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 51.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 52.Luethy PM, Huynh S, Ribardo DA, Winter SE, Parker CT, Hendrixson DR. 2017. Microbiota-derived short-chain fatty acids modulate expression of Campylobacter jejuni determinants required for commensalism and virulence. mBio 8:e00407-17. doi: 10.1128/mBio.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Bäumler AJ. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. 2005. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J Bacteriol 187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 57.Baumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207–213. [DOI] [PubMed] [Google Scholar]
- 58.Bohez L, Ducatelle R, Pasmans F, Botteldoorn N, Haesebrouck F, Van Immerseel F. 2006. Salmonella enterica serovar Enteritidis colonization of the chicken caecum requires the HilA regulatory protein. Vet Microbiol 116:202–210. doi: 10.1016/j.vetmic.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 59.Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. 2003. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech 14:33–43. [PMC free article] [PubMed] [Google Scholar]
- 60.Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, Baumler AJ. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun 76:2008–2017. doi: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Buttner L, Smoot MP, Behrendt CL, Cherry S, Santos RL, Hooper LV, Winter SE. 2017. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.