Cryptosporidium, a protozoan parasite that infects the gastrointestinal epithelium and other mucosal surfaces in humans and animals, is an important opportunistic pathogen in AIDS patients and one of the most common enteric pathogens affecting young children in developing regions. This parasite is referred to as a “minimally invasive” mucosal pathogen, and epithelial cells play a central role in activating and orchestrating host immune responses.
KEYWORDS: Cryptosporidium, NF-κB, cryptosporidiosis, exosomes, intestinal epithelium, parasitic infection, splenocytes
ABSTRACT
Cryptosporidium, a protozoan parasite that infects the gastrointestinal epithelium and other mucosal surfaces in humans and animals, is an important opportunistic pathogen in AIDS patients and one of the most common enteric pathogens affecting young children in developing regions. This parasite is referred to as a “minimally invasive” mucosal pathogen, and epithelial cells play a central role in activating and orchestrating host immune responses. We previously demonstrated that Cryptosporidium parvum infection stimulates host epithelial cells to release exosomes, and these released exosomes shuttle several antimicrobial peptides to carry out anti-C. parvum activity. In this study, we detected the upregulation of inflammatory genes in the liver and spleen following C. parvum intestinal infection in neonatal mice. Interestingly, exosomes released from intestinal epithelial cells following C. parvum infection could activate the nuclear factor kappa B signaling pathway and trigger inflammatory gene transcription in isolated primary splenocytes. Several epithelial cell-derived proteins and a subset of parasite RNAs were detected in the exosomes released from C. parvum-infected intestinal epithelial cells. Shuttling of these effector molecules, including the high mobility group box 1 protein, was involved in the induction of inflammatory responses in splenocytes induced by the exosomes released from infected cells. Our data indicate that exosomes released from intestinal epithelial cells upon C. parvum infection can activate immune cells by shuttling various effector molecules, a process that may be relevant to host systemic responses to Cryptosporidium infection.
INTRODUCTION
Cryptosporidium is an obligate intracellular protozoan parasite that infects the gastrointestinal epithelium and other mucosal surfaces in humans and animals (1, 2). Immunosuppressed individuals, particularly AIDS patients with a late diagnosis or without access to highly active antiretroviral therapy, are extremely susceptible to Cryptosporidium infection (3, 4). Cryptosporidium is also a common cause of diarrhea in young children in developing countries (5). Cryptosporidium attaches to the apical membrane surface of epithelial cells and forms an intracellular but extracytoplasmic vacuole in which the organism remains and develops (4). Due to this minimally invasive feature of Cryptosporidium infection, epithelial cells play a central role in activating and orchestrating host immune responses (6).
The invasion of intestinal epithelial cells by Cryptosporidium parvum in vitro activates Toll-like receptor 4 (TLR4) and nuclear factor kappa B (NF-ĸB) signaling, resulting in the production and secretion of various cytokines, chemokines, and antimicrobial peptides (7, 8). Interferon gamma (IFN-γ) is key to the mucosal innate anti-C. parvum defense (9), but acquired resistance requires T cells with the α/β-type T-cell receptor (10). Infiltration of natural killer cells, macrophages, and other immune cells has been identified at the site of infection. Activation of these immune cells may facilitate the epithelial anti-C. parvum defense through direct and/or indirect effects on the production of IFN-γ (6, 7). In addition, although Cryptosporidium infection is limited to the intestinal mucosal surface, a mild systemic inflammatory response, evident by the induction of inflammatory genes in extraintestinal tissues, has been reported in previous studies (11, 12). Therefore, cell-cell communication between infected epithelial cells (mainly enterocytes) and immune cells is essential to mucosal antimicrobial immunity. Nevertheless, current understanding of the underlying molecular mechanisms is very limited.
Intestinal epithelial cells can sense the environment of the intestinal lumen and transmit essential signals to cells in the underlying gastrointestinal mucosa. One characteristic of mucosal immunity is that infection at the primary infection site can initialize the activation of a defense response at all the mucosal surface sites (13). Upon C. parvum infection, intestinal epithelial cells quickly initiate a series of innate immune reactions, including production of antimicrobial molecules and release of inflammatory chemokines/cytokines (14, 15). These chemokines/cytokines of epithelial cell origin may mobilize and activate immune effector cells to the infection sites (6). Exosomes are extracellular vesicles, about 100 nm in diameter, derived from all types of cells (16). Exosomes can shuttle various molecules and function as bioactive vesicles to promote cell-cell communication and immunoregulation (17–20). Exosomes derived from gastrointestinal epithelial cells can modulate lymphocyte immune responses in vitro (21–23).
We previously demonstrated that C. parvum infection stimulates host epithelial cells to release exosomes through activation of TLR4/NF-ĸB signaling (24). Released exosomes shuttle several antimicrobial peptides, can bind to C. parvum sporozoites, and exhibit anti-C. parvum activity (24). In this study, we further investigated the involvement of epithelial cell-derived exosomes in host immune responses to Cryptosporidium infection. Our data indicate that exosomes released from intestinal epithelial cells upon C. parvum infection can activate immune cells by shuttling various effector molecules, a process that may be relevant to the activation of mucosal and systemic immunity against Cryptosporidium infection.
RESULTS
Induction of inflammatory genes in the liver and spleen following C. parvum intestinal infection in neonatal mice.
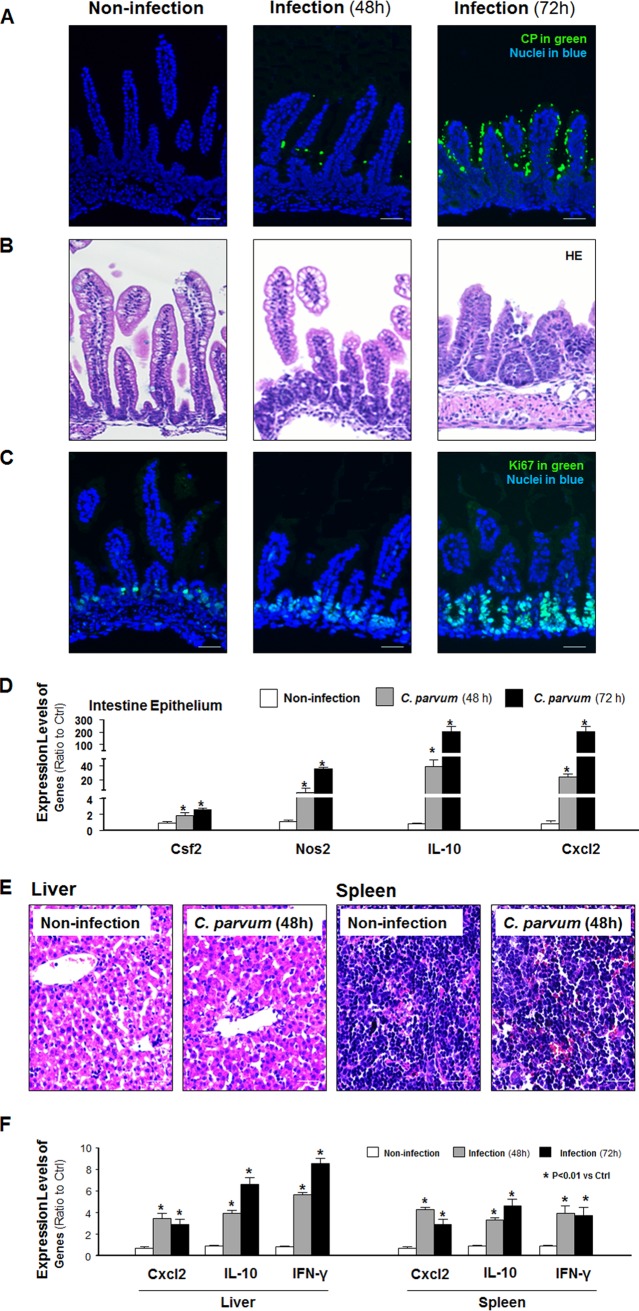
Because adult immunocompetent mice are resistant to infection (25), a well-developed infection model of cryptosporidiosis in neonatal mice (26, 27) was used for our in vivo experiments. We measured the expression levels of several inflammatory genes in the liver and spleen of the animals following oral gavage of C. parvum oocysts. C. parvum parasites were detected using immunofluorescent staining in the villus regions of the ileum of infected animals at 48 and 72 h after C. parvum oocyst administration (Fig. 1A). A mild inflammatory infiltration with a shorter height of the villi and an expansion of the crypt region was observed in the ileal epithelium of the mice after C. parvum infection (Fig. 1A to C). These cells in the crypt regions of infected animals showed a strong straining for Ki67 (a marker of proliferating cells) (Fig. 1C). Consistent with results from previous studies (14, 28), the upregulation of inflammatory genes, such as those for colony-stimulating factor 2 (Csf2), inducible nitric oxide synthase 2 (Nos2), interleukin 10 (Il-10), and C-X-C motif chemokine ligand 2 (Cxcl2), in the intestinal epithelium of infected animals compared with their expression in the intestinal epithelium of the noninfected control mice was detected (Fig. 1D). No parasite was detected in the liver or spleen of the infected animals using immunofluorescent staining (data not shown). Hematoxylin and eosin (HE) staining revealed no obvious morphological changes in the liver or spleen tissues from the infected animals compared with those from the noninfected controls (Fig. 1E). Nevertheless, we detected a significant induction of the Cxcl2, Il-10, and Ifn-γ genes in the liver and spleen tissues of infected animals (Fig. 1F), although the levels of Cxcl2 and Il-10 induced in the liver and spleen tissue were much less than those induced in the infected ileal epithelium.
FIG 1.
Inflammatory responses in the liver and spleen following C. parvum intestinal infection in neonatal mice. (A) C. parvum (CP) parasites were detected, using immunofluorescent staining, in the villus regions of the ileum of infected animals at 48 and 72 h after C. parvum oocyst administration. (B) A shorter height of the villi, an expansion of the crypt region, and a mild inflammatory infiltration were observed in the ileal epithelium of mice after C. parvum infection. (C) Strong straining for Ki67 (a marker of proliferating cells) was detected in these cells in the crypt regions of infected animals. (D) Upregulation of the Csf2, Nos2, Il-10, and Cxcl2 genes was detected in the intestinal epithelium of infected animals compared with their expression in the intestinal epithelium of the noninfected control by using real-time PCR analysis. (E) No obvious morphological changes of liver and spleen tissues from the infected animals compared with those from the noninfected controls were observed by HE staining. (F) Induction of Cxcl2, Il-10, and Ifn-γ genes in the liver and spleen tissues of infected animals compared with that in the tissues of the noninfected controls determined by using real-time PCR analysis. Data represent the means ± SEs from 3 independent experiments. *, P < 0.01 by ANOVA versus noninfected control. Bars = 50 μm.
Induction of inflammatory genes in splenocytes after incubation with supernatants of C. parvum-infected intestinal epithelial cell cultures.
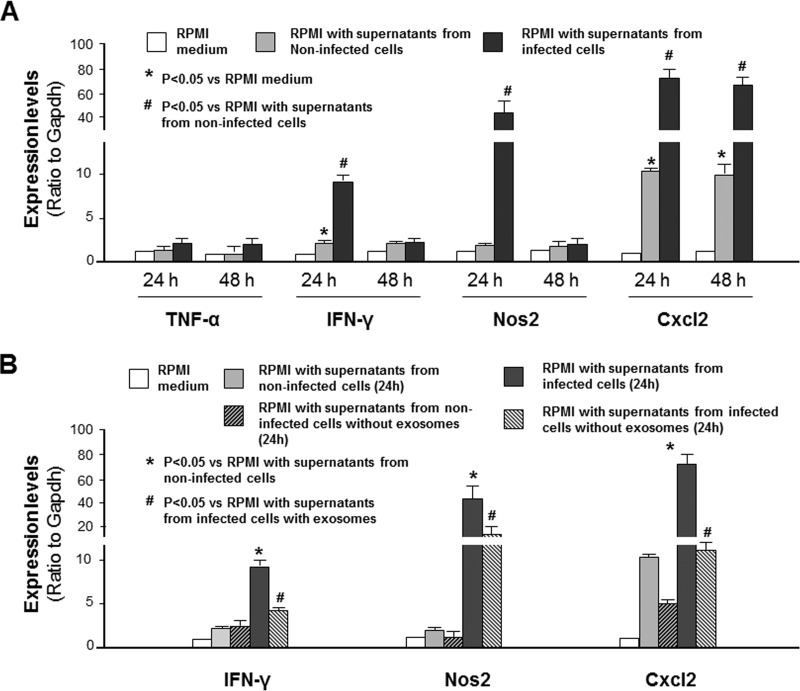
To explore the underlying mechanisms of extraintestinal inflammatory responses during Cryptosporidium infection, we asked whether the supernatants of infected cells could trigger inflammatory gene expression in splenocytes. Nonmalignant murine intestinal epithelial (IEC4.1) cells (29) were exposed to C. parvum infection for 24 h, and supernatants were collected. Primary splenocytes were isolated from mice, and they were highly responsive to lipopolysaccharide (LPS) stimulation, showing a significant increase in the expression levels of the Cxcl2 and Nos2 genes following LPS stimulation (see Fig. S1 in the supplemental material). A significant increase in the expression levels of Ifn-γ, Nos2, and Cxcl2 (Fig. 2A), as well as those of Il-10, the gene for interleukin 25 (Il-25), and the gene for interleukin 33 (Il-33) (Fig. S2), was detected in splenocytes after incubation in RPMI medium with the addition of supernatant medium from infected IEC4.1 cell cultures compared with that in splenocytes incubated with RPMI medium with the addition of supernatants from noninfected IEC4.1 cells. Of note, an increase in the expression levels of the Ifn-γ and Cxcl2 genes was also detected in cells after incubation in RPMI medium with the addition of supernatant from noninfected IEC4.1 cells compared with that after incubation in the RPMI medium control (Fig. 2A), presumably due to stimulation by potential effector molecules, such as cytokines, released from the noninfected cells into the supernatants. No significant increase in the expression level of the gene for tumor necrosis factor alpha (TNF-α) was detected between cells after incubation with the addition of supernatants from noninfected cell cultures and after incubation with the addition of supernatants from infected IEC4.1 cell cultures (Fig. 2A).
FIG 2.
Induction of inflammatory responses in splenocytes after incubation with supernatants of C. parvum-infected intestinal epithelial cell cultures and its association with exosome release. (A) Expression levels of inflammatory genes in splenocytes following incubation with supernatants from infected cell cultures. *, P < 0.05 by ANOVA versus incubation with RPMI medium; #, P < 0.05 by ANOVA versus RPMI incubation with supernatant from noninfected cells. (B) Expression levels of inflammatory genes in splenocytes following incubation with supernatants from infected cell cultures after removal of exosomes. *, P < 0.05 by ANOVA versus RPMI incubation with supernatants from noninfected cells; #, P < 0.05 by ANOVA versus RPMI incubation with supernatants from infected cells with exosomes. Data represent the means ± SEs from 3 independent experiments.
Following infection, a panel of cytokines and chemokines was induced and secreted into the supernatants (7, 8). In our previous studies (24), we demonstrated an increase in exosome release from intestinal epithelial cells following Cryptosporidium infection. We then asked whether removal of epithelial cell-derived exosomes from the conditional medium could block the associated inflammatory response in splenocytes. Indeed, a significant inhibition of the inflammatory responses in the splenocytes, reflected by the expression levels of the Ifn-γ, Nos2, and Cxcl2 genes, was detected after incubation with the conditional medium following removal of exosomes by ultracentrifugation (Fig. 2B).
Release of exosomes to the supernatants from cultured host epithelial cell cultures following C. parvum infection.
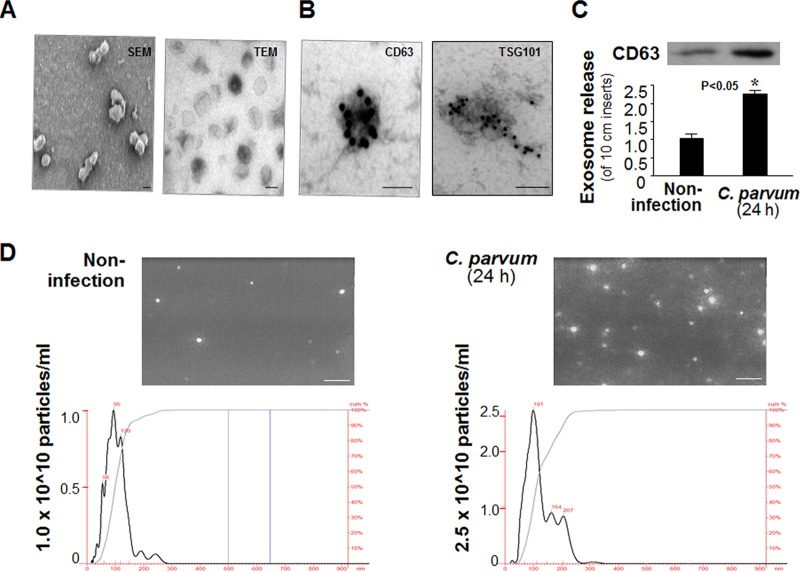
Due to their small size (30 to 90 nm), exosomes are neither visible by confocal microscopy nor reliably detectable by flow cytometry (17, 18). Using an immunomagnetic isolation approach (21), we isolated highly purified exosomes from epithelial cell cultures with and without C. parvum infection. These exosomes were cup-shape membrane vesicles with a diameter of 40 to 100 nm under scanning and transmission electron microscopy (Fig. 3A) and positive for exosome markers CD63 antigen (CD63) and tumor susceptibility gene 101 (Tgs101) (30, 31) by immunogold staining (Fig. 3B) or Western blotting for CD63 (Fig. 3C). No obvious difference in the morphological characteristics was observed in exosomes isolated from the cell cultures with and without C. parvum infection (data not shown). Nanoparticle tracking analysis of these isolated exosomes showed a peak size of about 100 nm, characteristic of exosomes (24), and revealed a higher concentration of exosomes from IEC4.1 cell cultures after C. parvum infection (Fig. 3D). Of note, the concentration scores for exosomes from noninfected and infected monolayers in Fig. 3D are different.
FIG 3.
Release of exosomes to the supernatants from host epithelial cell cultures following C. parvum infection. (A) Scanning electron micrographs (SEM) and transmission electron micrographs (TEM) of exosomes isolated from the supernatants of host epithelial cell cultures. (B) Isolated exosomes are positive by immunogold staining for exosome markers CD63 and Tsg101. (C) Increased exosome release from cultured host epithelial cells following C. parvum infection, as assessed by Western blotting for CD63. Data represent the means ± SEs from 3 independent experiments. *, P < 0.05 by t test versus the noninfected control. (D) Nanoparticle tracking analysis of exosomes from cell cultures following infection. Size and particle distribution plots of exosomes isolated from noninfected and C. parvum-infected (24 h) IEC4.1 cells determined by nanoparticle tracking analysis. Data represent the results of an analysis of isolated exosomes in a 1-ml volume from a total of 12 ml of supernatants from one infected or noninfected 10-cm dish. (Insets) Representative screenshots of exosomes isolated by nanoparticle tracking analysis. Bars = 100 nm (A and B) and 1,000 nm (D).
Induction of inflammatory genes in splenocytes following incubation with exosomes released from C. parvum-infected intestinal epithelial cells.
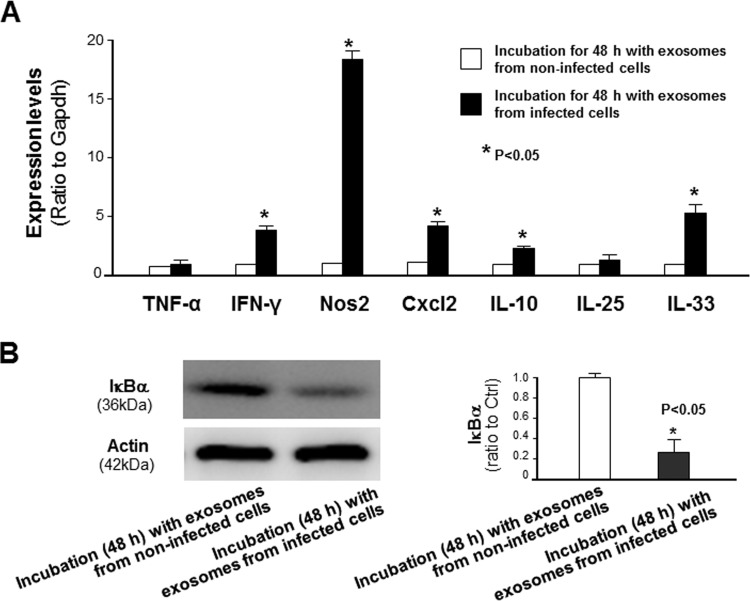
We then asked whether exosomes released from C. parvum-infected intestinal epithelial cells can activate inflammatory responses in splenocytes. We isolated splenocytes and incubated them for 48 h with medium containing exosomes released from IEC4.1 cells with or without C. parvum infection. The expression levels of selected inflammatory genes were significantly higher in splenocytes after incubation with the exosomes released from C. parvum-infected IEC4.1 cells than in cells after incubation with the exosomes released from noninfected IEC4.1 cells (Fig. 4A). We also observed the activation of the NF-κB signaling pathway, reflected by the degradation of IкBα, in splenocytes after incubation with the exosomes from C. parvum-infected IEC4.1 cell cultures (Fig. 4B). Complementarily, an increase in inflammatory gene expression and activation of NF-κB signaling were also detected in murine monocyte macrophages (RAW 264.7 cells) after incubation with the exosomes from C. parvum-infected IEC4.1 cell cultures (Fig. S3).
FIG 4.
Induction of inflammatory responses in splenocytes following incubation with exosomes released from C. parvum-infected intestinal epithelial cells. (A) Upregulation of selected inflammatory genes in primary splenocytes following incubation with exosomes isolated from C. parvum-infected or noninfected IEC4.1 cell cultures. (B) Degradation of IкBα in splenocytes following incubation with exosomes isolated from infected or noninfected IEC4.1 cell cultures. Data represent the means ± SEs from 3 independent experiments. *, P < 0.05 by t test versus incubation with exosomes from noninfected cells.
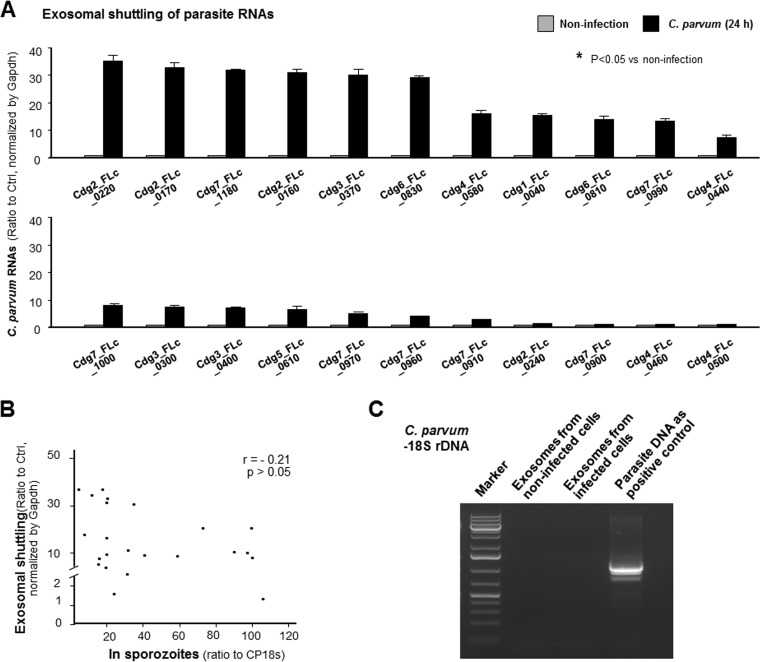
Exosomes released from C. parvum-infected intestinal epithelial cells shuttle a panel of parasite RNAs but not parasite 18S rDNA.
A recent report of an analysis of a full-length cDNA library constructed from C. parvum identified 118 RNAs of low protein-coding potential (32). In our previous studies (33, 34), we demonstrated that several C. parvum RNA transcripts of low protein-coding potential are selectively delivered into epithelial cells during host-parasite interactions. Here, we detected a certain amount of several C. parvum RNA transcripts of low protein-coding potential in the exosomes released from IEC4.1 cells following C. parvum infection (Fig. 5A). Exosomal shuttling of parasite RNA transcripts was not at random, as the parasite RNA contents in the exosomes were not corelated to their expression levels in the parasite (Fig. 5B). In addition, we failed to detect the parasite 18S ribosomal DNA (rDNA) in the exosomes released from IEC4.1 cells following C. parvum infection (Fig. 5C), using the nested PCR-restriction fragment length polymorphism (PCR-RFLP) of the 18S rDNA, as previously reported (35).
FIG 5.
Exosomes released from C. parvum-infected intestinal epithelial cells shuttle parasite RNAs but not parasite 18S rDNA. (A) Increased shuttling of several parasite RNAs in exosomes released from intestinal epithelial cells following infection. Exosomes from noninfected cell cultures were used as the control. (B) Exosomal shuttling of parasite RNAs is not correlated to the expression levels of parasite RNAs in the infective sporozoites. Expression levels of selected parasite RNAs in exosomes and in freshly exocysted C. parvum sporozoites were measured by real-time PCR. (C) C. parvum 18S rDNA was not detected in the exosomes released from intestinal epithelial cells following C. parvum infection. C. parvum DNA was used as the positive control, and a representative gel image is shown. Data represent the means ± SEs from 3 independent experiments. *, P < 0.05 by t test versus the noninfected control.
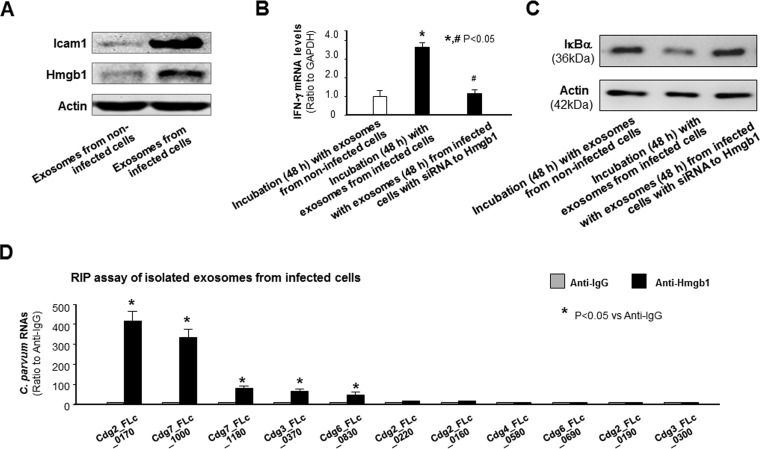
Shuttling of Hmgb1 is involved in the induction of inflammatory genes in splenocytes induced by exosomes released from C. parvum-infected intestinal epithelial cells.
Besides CD63 (Fig. 3B), we also detected by Western blotting intercellular adhesion molecule 1 (Icam1) and high mobility group box 1 (Hmgb1) proteins in the released exosomes, with higher levels of Icam1 and Hmgb1 proteins being detected in the exosomes released from infected IEC4.1 cells than in the exosomes released from noninfected cells (Fig. 6A). To explore how exosomes released from infected intestinal epithelial cells may activate inflammatory responses in splenocytes, we tested a potential role for Hmgb1, because extracellular Hmgb1 has been demonstrated to activate the NF-ĸB pathway in immune cells by binding and signaling through the receptor for advanced glycation end products (RAGE) (36). We used an RNA interference approach to knock down Hmgb1 in IEC4.1 cells and then exposed the cells to C. parvum infection. The exosomes were then collected and incubated with splenocytes. Cells displayed a much lower increase in Ifn-γ gene induction after incubation with exosomes released from infected IEC4.1 cells treated with small interfering RNA (siRNA) to Hmgb1 (with >60% silencing; data not shown) than after incubation with exosomes from infected cells not treated with siRNA (Fig. 6B). Accordingly, the degradation of IкBα in the cells was also partially inhibited in cells incubated with exosomes released from infected IEC4.1 cells treated with siRNA to Hmgb1 (Fig. 6C). HMGB1 is an evolutionarily highly conserved protein in mammals and can interact with RNA molecules through its zinc finger domains (37, 38). To investigate a potential physiological association of Hmgb1 with these RNAs in the exosomes, we performed an RNA immunoprecipitation (RIP) analysis. A significant amount of several parasite RNAs was detected in the immunoprecipitates from exosomes from infected cell cultures using an anti-Hmgb1 antibody (Fig. 6D). Parasite RNAs associated with Hmgb1 were not correlated with the parasite RNA contents in the exosomes (data not shown).
FIG 6.
Shuttling of Hmgb1 is involved in the induction of inflammatory responses in splenocytes induced by exosomes released from C. parvum-infected intestinal epithelial cells. (A) Increased content of Icam1 and Hmgb1 in exosomes released from intestinal epithelial cells following C. parvum infection. The content of Icam1 and Hmgb1 in the isolated exosomes was measured by Western blotting, and actin was also blotted as a control. (B) Knockdown of Hmgb1 in IEC4.1 cells attenuated Ifn-γ gene expression in splenocytes following incubation of exosomes released from infected cells. *, P < 0.05 by ANOVA versus incubation with exosomes from noninfected cells; #, P < 0.05 by ANOVA versus incubation with exosomes from infected cells. (C) Attenuation of IкBα degradation in splenocytes following incubation with exosomes isolated from infected IEC4.1 cell cultures treated with siRNA to Hmgb1. Representative gel images are shown. (D) Assembly of RNAs to the Hmgb1 complex in exosomes released from IEC4.1 cells following C. parvum infection. IEC4.1 cells were exposed to C. parvum infection for 24 h, and exosomes were isolated from the supernatants, followed by RIP analysis using anti-Hmgb1. Assembly of RNAs in the immunoprecipitates was measured by real-time PCR analysis. Data represent the means ± SEs from 3 independent experiments. *, P < 0.05 by t test versus anti-IgG.
DISCUSSION
One feature of the human mucosal immune system is that stimulation at one mucosal surface often results in mucosal immunity at many, if not all, mucosae (13). This process is finely controlled so that systemic immune responses are activated for an appropriate immune response to pathogen infection, but it maintains a tolerance toward nonharmful commensal microbes and benign environmental substances. The life cycle of Cryptosporidium infection in the intestine is well-known, but the systemic impact of infection is less studied. Upon C. parvum infection, intestinal epithelial cells initiate a series of innate defense responses, resulting in infiltration and activation of immune effector cells to the infection sites (6). We previously demonstrated that C. parvum infection stimulates the release of exosomes from infected epithelial cells (24). Data from this study support the notion that exosomes released from intestinal epithelial cells upon C. parvum infection can activate immune cells by shuttling various effector molecules of both host epithelial cell and parasite origin, a process that may be relevant to the systemic host immune responses against Cryptosporidium infection.
One of the key findings of this study is the activation of splenocytes by exosomes released from C. parvum-infected intestinal epithelial cells. Although other effector molecules released from infected epithelial cells, such as cytokines and chemokines, may also be involved in the activation of splenocytes after incubation with the supernatants from infected cell cultures, the released exosomes may be key players in splenocyte activation because removal of the exosomes from the supernatants significantly, if not totally, blocked the associated inflammatory response in splenocytes. Of note, exosomes from infected cell cultures were more potent than those from the noninfected control cultures in activating splenocytes. We speculated that the difference in the exosomal content of the two exosome populations may account for this phenomenon. These molecules shuttled in the released exosomes may activate NF-ĸB signaling in the recipient cells.
Increasing evidence supports the notion that exosomes shuttle proteins, lipids, and RNA molecules and function as bioactive vesicles to promote cell-cell communication and immunoregulation (17–20). We detected a much higher level of Hmgb1 and Icam1 proteins in the exosomes released from infected cells than in exosomes released from the noninfected controls. HMGB1 is a ubiquitous DNA-binding protein (37), but extracellular HMGB1 can activate the NF-ĸB pathway in immune cells by binding and signaling through RAGE (36). While HMGB1 release in most cell types is mediated by inflammasome activation during pyroptosis (a form of proinflammatory programmed cell death), the release of HMGB1 from gastrointestinal epithelial cells is through exosomes (39). Indeed, knockdown of Hmgb1 in host cells resulted in a decrease of Hmgb1 in the released exosomes and, consequently, a significant inhibition of NF-ĸB activation in the recipient splenocytes.
Interestingly, besides these molecules of host epithelial cell origin, we detected some parasite RNA transcripts of low protein-coding potential in the released exosomes. The C. parvum genome does not encode the key components of the small RNA-mediated posttranscriptional gene silencing system (40). Nevertheless, 118 orphan candidate genes with little homology to known annotated protein-coding genes were identified in C. parvum (32). These orphan genes are of low protein-coding potential, as their RNA transcripts predict no complete open reading frames (32, 41). We recently demonstrated that several RNA transcripts of these C. parvum orphan genes can be delivered into epithelial cells and modulate gene transcription in infected host cells (34). Interestingly, some of these C. parvum RNA transcripts are present in exosomes released from infected intestinal epithelial cells. It is unclear whether these parasite RNAs are involved in the activation of splenocytes triggered by exosomes released from infected cells. There are several ubiquitous cytoplasmic nucleic acid-recognizing receptors and sensors (such as RAGE for HMGB1) that can recognize foreign RNA molecules and trigger the activation of intracellular signaling pathways, such as NF-κB signaling (42, 43). Interestingly, we detected a direct physiological association between several parasite RNAs and the Hmgb1 protein in the released exosomes.
In a recent report (35), Cryptosporidium 18S rDNA was detected in stool and tissue samples, as well as in blood and cerebrospinal fluid samples, from HIV-infected patients with intestinal cryptosporidiosis. DNA molecules are usually not detectable in exosomes from various types of cells (19, 20). Accordingly, we failed to detect the parasite 18S rDNA in these exosomes released from infected cells, suggesting that exosome release from infected epithelial cells may not contribute to the systemic circulation of parasite DNA, as reported in HIV-infected patients with intestinal cryptosporidiosis (35).
From a pathophysiological viewpoint, exosomes released from the intestinal epithelium following Cryptosporidium infection may enter the blood and be circulated to multiple organs. It was not a surprise that induction of inflammatory genes was detected in the liver and spleen tissues from the mice following C. parvum infection. Given the limitation of the neonatal model of intestinal cryptosporidiosis (28, 44), the pathophysiological significance for circulating exosomes in intestinal cryptosporidiosis requires further investigation with additional animal models, such as the piglet model of cryptosporidiosis (45), and evaluation of plasma-derived exosomes from patients with intestinal cryptosporidiosis. Cryptosporidium is generally referred as a “minimally invasive” mucosal pathogen, and infection is limited to the mucosal surfaces (1, 2, 4). Given the potential systemic impact of exosomes from infected epithelium following Cryptosporidium infection, coupled with the recent detection of Cryptosporidium DNA in blood and cerebrospinal fluid samples from HIV-infected patients with intestinal cryptosporidiosis, our data suggest that intestinal cryptosporidiosis may involve inflammatory responses in multiple organs not limited to the intestinal infection site.
MATERIALS AND METHODS
C. parvum and cell lines.
C. parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm, Deary, ID). RAW 264.7 cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured according to the instructions. Cells of the IEC4.1 cell line were a kind gift from Pingchang Yang (McMaster University, Hamilton, ON, Canada) and were cultured as previously described (33).
Infection models and infection assays.
Models of intestinal cryptosporidiosis using IEC4.1 cells were employed as previously described; infection was done with a 1:1 ratio between C. parvum oocysts and host cells (33, 46). All parasite preparations were tested using the Limulus amebocyte lysate gel formation test as previously described to exclude the possibility of contamination with lipopolysaccharides (24). For Cryptosporidium infection in vivo, C57BL/6J neonatal mice (both male and female mice at 6 days after birth) received C. parvum oocysts by oral gavage (105 oocysts per mice) (26, 27). Mice receiving phosphate-buffered saline (PBS) by oral gavage were used as controls. The mice were from The Jackson Laboratory, and the study was approved by the Creighton University Institutional Animal Care and Use Committee. At least five animals from each group were sacrificed at 24 and 48 h after C. parvum or PBS administration; liver, spleen, and ileal tissues were obtained for biochemical analysis, and paraffin sections were used for HE staining and immunohistochemistry. Immunofluorescence microscopy was used to assay for C. parvum infection as previously reported (7, 47).
Quantitative real-time PCR and Western blotting.
For quantitative analysis of mRNA and C. parvum RNA expression, comparative real-time PCR was performed as previously reported (34, 46) using SYBR Green PCR master mix (Applied Biosystems, Carlsbad, CA). Briefly, RNA was extracted using the TRI Reagent and treated with a DNA-free kit (Ambion) to remove any remaining DNA. Quantified (500 ng) RNA was reverse transcribed using T100 thermal cyclers (Bio-Rad). Real-time PCR was then performed using 25 ng of template cDNA for each RNA gene of interest. Each sample was run in triplicate. The relative abundance of each RNA was calculated using the ΔΔCT threshold cycle (CT) method and normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA (for measurement of whole-cell lysates) or U2 small nuclear (RNU2-1) RNA (for measurement of nuclear extracts). PCR-RFLP was carried out to detect C. parvum 18S rDNA as previously reported (35). The sequences for all the primers are listed in Table S1 in the supplemental material. For Western blotting, whole-cell extracts were prepared using the mammalian protein extraction reagent (Fisher) supplemented with a protease inhibitor cocktail. The cell pellet was incubated in the mammalian protein extraction reagent and centrifuged at 16,100 × g for 20 min, and the supernatants were saved for use as whole-cell extracts as previously reported (48). Antibodies to Icam1 (0.4 μg/ml; Santa Cruz), Hmgb1 (0.4 μg/ml; Santa Cruz), CD63 (0.4 μg/ml; Santa Cruz), IкBα (0.4 μg/ml; Santa Cruz), and β-actin (0.2 μg/ml; Sigma-Aldrich) were used. The details for Western blot analysis are described in our previous studies (34, 46).
siRNAs and plasmids.
The siRNA duplexes for mouse Hmgb1 were purchased from Santa Cruz. A nonspecific scrambled sequence, 5′-UUCUCCGAACGUGUCACGUUU-3′, synthesized by Integrated DNA Technologies (Coralville, IA), was used as a control. siRNA oligonucleotides were transfected into IEC4.1 cells for 24 h with the Lipofectamine RNAiMAX reagent (Invitrogen). The efficiency of knockdown was verified by real-time PCR and Western blotting.
Exosome purification and nanoparticle tracking analysis.
Supernatant medium from IEC4.1 cell cultures (12 ml per 10-cm culture dish) was collected at the time points after infection indicated above. Supernatant medium from uninfected cell cultures was used as a control. The collected medium was then centrifuged at 200 × g for 10 min to eliminate cells and again spun at 10,000 × g for 30 min, followed by filtration through a 0.22-μm-pore-size filter to remove the cell debris. Exosomes were pelleted by ultracentrifugation at 120,000 × g for 70 min and precipitated using exosome precipitation solution (Exo-Quick; System Biosciences) following the manufacturer’s instructions. Exosomes were further quantified by nanoparticle tracking analysis, as we previously reported (24).
Splenocyte isolation and incubation with exosomes.
Splenocytes were isolated from mice of 6 to 8 weeks of age, as previously reported (49). C57BL/6J mice (both male and female; The Jackson Laboratory) were used, and their use was approved by the Creighton University Biosafety and Institutional Animal Care and Use Committees. Freshly isolated splenocytes were first cultured with RPMI medium for 4 h. Cells were then cultured for an additional 24 to 72 h with the conditional medium (RPMI medium with the addition of supernatants or exosomes from IEC4.1 cell cultures). For supernatants or exosomes from noninfected IEC4.1 cells, the conditional medium was conditioned with a 10:1 ratio, by volume, of RPMI and the supernatants or exosomes (in RPMI). For supernatants or exosomes from infected IEC4.1 cells, the volume of supernatants or exosomes added to the conditional medium was adjusted with the same number of exosomes as that from the uninfected cell cultures. The number of exosomes isolated or in the supernatants was quantified by nanoparticle tracking analysis. Some splenocytes were cultured with RPMI medium with LPS (from Escherichia coli strain K-12; 1 to 20 μg/ml; InvivoGen) for 2 h. Cells were then collected for real-time PCR or Western blot analysis.
Electron microscopy.
Electron microscopy was performed as previously described (24). Briefly, for transmission electron microscopy, exosome pellets were resuspended in 2.5% glutaraldehyde, embedded in a mixture of 4% uranyl acetate and 2% methylcellulose (1:9 ratio), and observed with a JEOL 1400 electron microscope (JEOL USA). For scanning electron microscopy, isolated exosomes were fixed immediately in 2.5% glutaraldehyde, dehydrated, dried in a critical point drying device, sputter coated, and examined with a Hitachi S-4700 microscope (Hitachi). For immunogold analysis, exosomes were fixed in 4% paraformaldehyde, blocked with 10% fetal calf serum in phosphate-buffered saline for 20 min, and incubated overnight at 4°C with antibodies to CD63 (1 μg/ml; Santa Cruz) and Tgs101 (1 μg/ml; Santa Cruz). After incubation with the gold-conjugated secondary antibody (goat anti-mouse IgG, 2 μg/ml; Abcam), samples were embedded and observed with a JEOL 1400 electron microscope.
RIP analysis.
RIP was performed as we previously reported (24). Briefly, IEC4.1 cells were collected, and lysates were precleared with 20 μl of PBS-washed Magna protein A+G magnetic beads (Millipore, MA). The precleared lysate (1,000 μg protein) was then mixed with beads coated with anti-Hmgb1 (0.2 μg/ml; Santa Cruz) or anti-IgG (0.2 μg/ml; Santa Cruz) as a control. The collected immunoprecipitated RNA-protein complexes and inputs were digested with proteinase K. RNA was extracted from these samples using the TRI Reagent (Invitrogen) and treated with a DNA-free DNase treatment and removal I kit according to the manufacturer’s protocol (Ambion Inc., Austin, TX). The presence of RNA was measured by quantitative real-time PCR using a CFX Connect real-time system (Bio-Rad). The gene-specific PCR primer pairs are listed in Table S1.
Statistical analyses.
All values are given as the means ± standard errors (SEs). The means for groups were from at least three independent experiments. Multiple comparisons were carried out by analysis of variance (ANOVA). Differences between two groups were evaluated with the unpaired t test. Statistical analyses were performed using Prism software (GraphPad, Inc., San Diego, CA, USA). P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Guoku Hu (University of Nebraska Medical Center, USA) and Shibin Ma (Creighton University, USA) for helpful and stimulating discussions, Bing Q. Huang and Nicholas F. LaRusso (Mayo Clinic) for electronic microscopy, and Barbara L. Bittner (Creighton University) for her assistance in writing the manuscript.
This work was supported by the National Institutes of Health (AI095532, AI116323, and AI136877 to X.-M.C.) and the Nebraska Department of Health & Human Services (LB595 Cancer and Smoking Disease Research Program Development Grant to X.-M.C.).
We disclose that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00705-18.
REFERENCES
- 1.Striepen B. 2013. Parasitic infections: time to tackle cryptosporidiosis. Nature 503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 2.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manabe YC, Clark DP, Moore RD, Lumadue JA, Dahlman HR, Belitsos PC, Chaisson RE, Sears CL. 1998. Cryptosporidiosis in patients with AIDS: correlates of disease and survival. Clin Infect Dis 27:536–542. doi: 10.1086/514701. [DOI] [PubMed] [Google Scholar]
- 4.Chen XM, Keithly JS, Paya CV, LaRusso NF. 2002. Cryptosporidiosis. N Engl J Med 346:1723–1731. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- 5.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 6.Hu G, Feng Y, O’Hara SP, Chen XM. 2014. Immunology of cryptosporidiosis, p 423–454. In Cryptosporidium: parasite and disease. Springer, New York, NY. [Google Scholar]
- 7.Zhou R, Gong AY, Eischeid AN, Chen XM. 2012. miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PLoS Pathog 8:e1002702. doi: 10.1371/journal.ppat.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurent F, Kagnoff MF, Savidge TC, Naciri M, Eckmann L. 1998. Human intestinal epithelial cells respond to Cryptosporidium parvum infection with increased prostaglandin H synthase 2 expression and prostaglandin E2 and F2alpha production. Infect Immun 66:1787–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollok RC, Farthing MJ, Bajaj-Elliott M, Sanderson IR, McDonald V. 2001. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology 120:99–107. doi: 10.1053/gast.2001.20907. [DOI] [PubMed] [Google Scholar]
- 10.Pantenburg B, Castellanos-Gonzalez A, Dann SM, Connelly RL, Lewis DE, Ward HD, White AC Jr. 2010. Human CD8(+) T cells clear Cryptosporidium parvum from infected intestinal epithelial cells. Am J Trop Med Hyg 82:600–607. doi: 10.4269/ajtmh.2010.09-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anatskaya OV, Sidorenko NV, Vinogradov AE, Beyer TV. 2007. Impact of neonatal cryptosporidial gastroenteritis on epigenetic programming of rat hepatocytes. Cell Biol Int 31:420–427. doi: 10.1016/j.cellbi.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 12.Filatova NA, Knyazev NA, Skarlato SO, Anatskaya OV, Vinogradov AE. 2018. Natural killer cell activity irreversibly decreases after Cryptosporidium gastroenteritis in neonatal mice. Parasite Immunol 40:e12524. doi: 10.1111/pim.12524. [DOI] [PubMed] [Google Scholar]
- 13.Otczyk DC, Cripps AW. 2010. Mucosal immunization: a realistic alternative. Hum Vaccin 6:978–1006. doi: 10.4161/hv.6.12.13142. [DOI] [PubMed] [Google Scholar]
- 14.Deng M, Lancto CA, Abrahamsen MS. 2004. Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol 34:73–82. doi: 10.1016/j.ijpara.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Alcantara CS, Yang CH, Steiner TS, Barrett LJ, Lima AA, Chappell CL, Okhuysen PC, White AC Jr, Guerrant RL. 2003. Interleukin-8, tumor necrosis factor-alpha, and lactoferrin in immunocompetent hosts with experimental and Brazilian children with acquired cryptosporidiosis. Am J Trop Med Hyg 68:325–328. [PubMed] [Google Scholar]
- 16.Cocucci E, Racchetti G, Meldolesi J. 2009. Shedding microvesicles: artefacts no more. Trends Cell Biol 19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Théry C, Ostrowski M, Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 18.Leone DA, Rees AJ, Kain R. 2018. Dendritic cells and routing cargo into exosomes. Immunol Cell Biol 96:683–693. doi: 10.1111/imcb.12170. [DOI] [PubMed] [Google Scholar]
- 19.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atay S, Gercel-Taylor C, Suttles J, Mor G, Taylor DD. 2011. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am J Reprod Immunol 65:65–77. doi: 10.1111/j.1600-0897.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 21.van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. 2001. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 22.Büning J, von Smolinski D, Tafazzoli K, Zimmer KP, Strobel S, Apostolaki M, Kollias G, Heath JK, Ludwig D, Gebert A. 2008. Multivesicular bodies in intestinal epithelial cells: responsible for MHC class II-restricted antigen processing and origin of exosomes. Immunology 125:510–521. doi: 10.1111/j.1365-2567.2008.02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallegol J, Van Niel G, Lebreton C, Lepelletier Y, Candalh C, Dugave C, Heath JK, Raposo G, Cerf-Bensussan N, Heyman M. 2007. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology 132:1866–1876. doi: 10.1053/j.gastro.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 24.Hu G, Gong AY, Roth AL, Huang BQ, Ward HD, Zhu G, LaRusso NF, Hanson ND, Chen XM. 2013. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog 9:e1003261. doi: 10.1371/journal.ppat.1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungar BL, Kao TC, Burris JA, Finkelman FD. 1991. Cryptosporidium infection in an adult mouse model. Independent roles for IFN-gamma and CD4+ T lymphocytes in protective immunity. J Immunol 147:1014–1022. [PubMed] [Google Scholar]
- 26.Kapel N, Benhamou Y, Buraud M, Magne D, Opolon P, Gobert J-G, Magne D. 1996. Kinetics of mucosal ileal gamma-interferon response during cryptosporidiosis in immunocompetent neonatal mice. Parasitol Res 82:664–667. doi: 10.1007/s004360050182. [DOI] [PubMed] [Google Scholar]
- 27.Lacroix S, Mancassola R, Naciri M, Laurent F. 2001. Cryptosporidium parvum specific mucosal immune response in C57BL/6 neonatal and gamma interferon-deficient mice: role of tumor necrosis factor alpha in protection. Infect Immun 69:1635–1642. doi: 10.1128/IAI.69.3.1635-1642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitch GJ, He Q. 1999. Reactive nitrogen and oxygen species ameliorate experimental cryptosporidiosis in the neonatal BALB/c mouse model. Infect Immun 67:5885–5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bharhani MS, Borojevic R, Basak S, Ho E, Zhou P, Croitoru K. 2006. IL-10 protects mouse intestinal epithelial cells from Fas-induced apoptosis via modulating Fas expression and altering caspase-8 and FLIP expression. Am J Physiol Gastrointest Liver Physiol 291:G820–G829. doi: 10.1152/ajpgi.00438.2005. [DOI] [PubMed] [Google Scholar]
- 30.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem 273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 31.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. 2012. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A 109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagishi J, Wakaguri H, Sugano S, Kawano S, Fujisaki K, Sugimoto C, Watanabe J, Suzuki Y, Kimata I, Xuan X. 2011. Construction and analysis of full-length cDNA library of Cryptosporidium parvum. Parasitol Int 60:199–202. doi: 10.1016/j.parint.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Ming Z, Wang Y, Gong AY, Zhang XT, Li M, Chen T, Mathy NW, Strauss-Soukup JK, Chen XM. 2018. Attenuation of intestinal epithelial cell migration during Cryptosporidium parvum infection involves parasite Cdg7_FLc_1030 RNA-mediated induction and release of Dickkopf-1. J Infect Dis 218:1336–1347. doi: 10.1093/infdis/jiy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Gong AY, Ma S, Chen X, Li Y, Su CJ, Norall D, Chen J, Strauss-Soukup JK, Chen XM. 2017. Delivery of parasite RNA transcripts into infected epithelial cells during Cryptosporidium infection and its potential impact on host gene transcription. J Infect Dis 215:636–643. doi: 10.1093/infdis/jiw607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velásquez JN, Pantano ML, Vittar N, Nigro MG, Figueiras O, Astudillo OG, Ricart J, Della Paolera D, Carnevale S. 2018. First detection of Cryptosporidium DNA in blood and cerebrospinal fluid of HIV-infected patients. Parasitol Res 117:875–881. doi: 10.1007/s00436-018-5766-1. [DOI] [PubMed] [Google Scholar]
- 36.Kokkola R, Andersson A, Mullins G, Ostberg T, Treutiger CJ, Arnold B, Nawroth P, Andersson U, Harris RA, Harris HE. 2005. RAGE is the major receptor for the proinflammatory activity of HMGB1 in rodent macrophages. Scand J Immunol 61:1–9. doi: 10.1111/j.0300-9475.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 37.Andersson U, Tracey KJ. 2011. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol 29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianchi ME, Manfredi AA. 2007. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev 220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Stolz DB, Sappington PL, Macias CA, Killeen ME, Tenhunen JJ, Delude RL, Fink MP. 2006. HMGB1 is secreted by immunostimulated enterocytes and contributes to cytomix-induced hyperpermeability of Caco-2 monolayers. Am J Physiol Cell Physiol 290:C990–C999. doi: 10.1152/ajpcell.00308.2005. [DOI] [PubMed] [Google Scholar]
- 40.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 41.Puiu D, Enomoto S, Buck GA, Abrahamsen MS, Kissinger JC. 2004. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res 32:D329–D331. doi: 10.1093/nar/gkh050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brencicova E, Diebold SS. 2013. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front Cell Infect Microbiol 3:37. doi: 10.3389/fcimb.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawasaki T, Kawai T, Akira S. 2011. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev 243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Favennec L. 1997. Physiopathologic and therapeutic studies in in vitro and in vivo models of Cryptosporidium parvum infection. J Eukaryot Microbiol 44:69S–70S. [DOI] [PubMed] [Google Scholar]
- 45.Gookin JL, Chiang S, Allen J, Armstrong MU, Stauffer SH, Finnegan C, Murtaugh MP. 2006. NF-kappaB-mediated expression of iNOS promotes epithelial defense against infection by Cryptosporidium parvum in neonatal piglets. Am J Physiol Gastrointest Liver Physiol 290:G164–G174. doi: 10.1152/ajpgi.00460.2004. [DOI] [PubMed] [Google Scholar]
- 46.Ming Z, Gong AY, Wang Y, Zhang XT, Li M, Mathy NW, Strauss-Soukup JK, Chen XM. 2018. Involvement of Cryptosporidium parvum Cdg7_FLc_1000 RNA in the attenuation of intestinal epithelial cell migration via trans-suppression of host cell SMPD3 gene. J Infect Dis 217:122–133. doi: 10.1093/infdis/jix392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasahara T, Maruyama H, Aoki M, Kikuno R, Sekiguchi T, Takahashi A, Satoh Y, Kitasato H, Takayama Y, Inoue M. 2003. Apoptosis of intestinal crypt epithelium after Cryptosporidium parvum infection. J Infect Chemother 9:278–281. doi: 10.1007/s10156-003-0259-1. [DOI] [PubMed] [Google Scholar]
- 48.Abmayr SM, Yao T, Parmely T, Workman JL. 2006. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Pharmacol Chapter 12:Unit12.3. doi: 10.1002/0471142727.mb1201s75. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura A, Nagai K, Ando K, Tamura G. 1986. Selective suppression by prodigiosin of the mitogenic response of murine splenocytes. J Antibiot 39:1155–1159. doi: 10.7164/antibiotics.39.1155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.