Staphylococcus aureus infections associated with the formation of biofilms on medical implants or host tissue play a critical role in the persistence of chronic infections. One critical mechanism of biofilm infection that leads to persistent infection lies in the capacity of biofilms to evade the macrophage-mediated innate immune response.
KEYWORDS: KLF2, S. aureus biofilm, macrophages
ABSTRACT
Staphylococcus aureus infections associated with the formation of biofilms on medical implants or host tissue play a critical role in the persistence of chronic infections. One critical mechanism of biofilm infection that leads to persistent infection lies in the capacity of biofilms to evade the macrophage-mediated innate immune response. It is now increasingly apparent that microorganisms exploit the negative regulatory mechanisms of the pattern recognition receptor (PRR)-mediated inflammatory response to subvert host cell functions by using various virulence factors. However, the detailed molecular mechanism, along with the identity of a target molecule, underlying the evasion of the macrophage-mediated innate immune response against S. aureus infection associated with biofilm formation remains to be elucidated. Here, using an in vitro culture model of murine macrophage-like RAW 264.7 cells, we demonstrate that S. aureus biofilm-conditioned medium significantly attenuated the capacity for macrophage bactericidal and proinflammatory responses. Importantly, the responses were associated with attenuated activation of NF-κB and increased expression of Kruppel-like factor 2 (KLF2) in RAW 264.7 cells. Small interfering RNA (siRNA)-mediated silencing of KLF2 in RAW 264.7 cells could restore the activation of NF-κB toward the bactericidal activity and generation of proinflammatory cytokines in the presence of S. aureus biofilm-conditioned medium. Collectively, our results suggest that factors secreted from S. aureus biofilms might exploit the KLF2-dependent negative regulatory mechanism to subvert macrophage-mediated innate immune defense against S. aureus biofilms.
INTRODUCTION
Staphylococcus aureus is a major human pathogen that causes a wide range of clinical infections, including nosocomial, osteoarticular, skin and soft tissue, and device-related infections (1, 2). In particular, the establishment of a mature biofilm by the attachment of S. aureus to medical implants or host tissue plays a critical role in the persistence of chronic infections (3). In response to invading S. aureus, both tissue-resident and tissue-infiltrating macrophages act as key regulators of the antibacterial and proinflammatory response for innate immune defense (4). In an acute infection associated with the planktonic phase of S. aureus, the macrophage-mediated innate immune response can be induced by the ligation of pathogen-associated molecular patterns (PAMPs) to pattern recognition receptors (PRRs) expressed on macrophages (5). In particular, the recognition of S. aureus-derived PAMPs by PRRs, including Toll-like receptor 2 (TLR2), TLR9, and nucleotide-binding oligomerization domain-containing protein 2 (NOD2), triggers distinct signaling pathways that converge to activate NF-κB (6–9), which is critical for the production of numerous cytokines, chemokines, and reactive oxygen intermediates to combat S. aureus. However, S. aureus infections involving the formation of biofilms were shown to be associated with decreased production of proinflammatory cytokines, which contributed to diminished bactericidal activity (10, 11).
One potential mechanism by which S. aureus biofilm interferes with antimicrobial proinflammatory responses has been attributed to the enhanced recruitment of alternatively polarized macrophages that exhibit an anti-inflammatory phenotype, characterized by increased arginase-1 (Arg-1) and decreased inducible nitric oxide synthase (iNOS) expression (11, 12). This alternatively activated response in macrophages was associated with attenuation of macrophage invasion and phagocytosis of biofilm-associated bacteria (13–15). However, the detailed molecular mechanism underlying the evasion of the macrophage-mediated innate immune response against S. aureus biofilms remains to be elucidated.
It is now increasingly apparent that microorganisms exploit the negative regulatory mechanisms of the PRR-mediated inflammatory response to subvert host cell functions by using various virulence strategies (8, 16, 17). In particular, a series of recent reports have implicated members of the Kruppel-like factor (KLF) family of transcriptional regulators as targets for bacterial pathogens as a means to subvert host cell functions (18–20). S. aureus exotoxins were shown to induce sustained expression of KLF2 in J774A.1 macrophage cells (18). Accumulating evidence demonstrates that KLF2 acts as a potent negative regulator of the proinflammatory response by interfering with the NF-κB activation pathway in monocytes and macrophages (19).
In this study, we have reasoned that factors secreted from S. aureus biofilms might alter the extent of macrophage polarization and activation toward the attenuation of antibacterial activities by aberrantly activating the negative regulatory cascade to modulate NF-κB pathways. To address this, we used an in vitro culture model of murine macrophage-like RAW 264.7 cells to determine whether the impaired functional activities of macrophages in the presence of factors secreted from S. aureus biofilms are associated with deregulated NF-κB activation. The results of this study have revealed the functional significance of KLF2 in regulating macrophage-mediated immune responses against S. aureus biofilms.
RESULTS
Conditioned medium from S. aureus biofilm skews the macrophage phenotype toward attenuation of phagocytic and bactericidal activities.
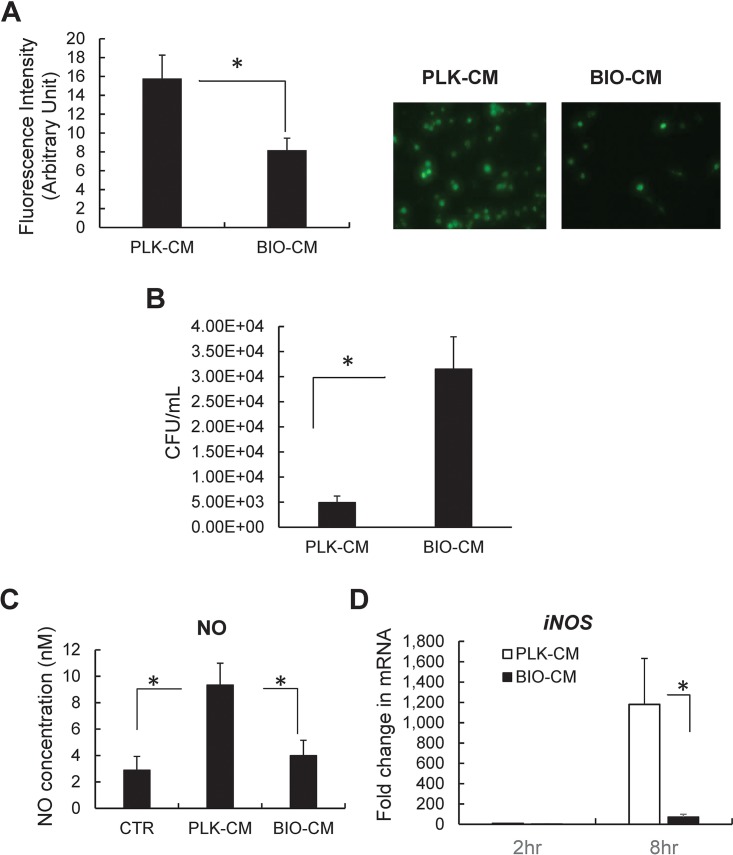
To evaluate the role of the S. aureus biofilm environment in macrophage functions, RAW 264.7 macrophage-like cells were treated with conditioned medium (CM) collected from cultures of mature S. aureus biofilms (BIO-CM), and the phagocytic and bactericidal activities of the cells were examined. The extent of phagocytosis in RAW 264.7 cells exposed to BIO-CM was significantly attenuated, compared with that in cells exposed to CM from a planktonic culture of S. aureus (PLK-CM) (Fig. 1A). This finding is consistent with data from a recent study showing that conditioned medium from mature S. aureus biofilms inhibits macrophage phagocytosis and induces cytotoxicity (21). S. aureus has been reported to survive within macrophages, which contributes to the persistence of S. aureus during infections (22). By observing the impaired phagocytic function of macrophages treated with BIO-CM, we subsequently assessed whether BIO-CM can also impair the capacity for bactericidal activities in macrophages. This was examined using an antibiotic protection assay, which enables the quantification of the number of bacterial colonies of intracellular S. aureus that survived within macrophages. The number of CFU counted from RAW 264.7 cells exposed to BIO-CM was significantly higher than that in RAW 264.7 cells exposed to PLK-CM (Fig. 1B). Since the capacity of macrophages to produce nitric oxide (NO) is functionally important in conferring bactericidal activity (23), we subsequently examined whether culture from the S. aureus biofilm environment was associated with an alteration in NO production. The secretion of NO from RAW 264.7 macrophages exposed to BIO-CM was significantly decreased by 50%, compared with cells exposed to PLK-CM (Fig. 1C). The expression of the iNOS gene, a gene involved in NO production, was also substantially diminished in RAW 264.7 cells exposed to BIO-CM, compared with cells exposed to PLK-CM (Fig. 1D).
FIG 1.
Effects of S. aureus biofilm-conditioned medium on the phagocytic and bactericidal capacities of RAW 264.7 macrophages. (A) Effect of S. aureus biofilm on the phagocytic activity of macrophages. RAW 264.7 cells were treated with conditioned medium from either a planktonic culture (PLK-CM) or a biofilm culture (BIO-CM) of S. aureus for 2 h. Next, the extent of bacterial phagocytosis by RAW 264.7 cells was assessed by measuring the fluorescence intensity from the phagocytized bacteria (opsonized Alexa Fluor 488-conjugated E. coli). (Left) Quantification of phagocytosis (n = 8). (Right) Representative fluorescence images of RAW 264.7 cells. (B) Effect of S. aureus biofilm on the bactericidal activity of macrophages. RAW 264.7 cells were exposed to PLK-CM or BIO-CM for 2 h and then incubated with live S. aureus bacteria for 1 h, followed by an antibiotic protection assay to count the number of intracellular S. aureus bacteria that survived within macrophages (n = 5 per group). (C) Effect of S. aureus biofilm on secretion of nitric oxide from RAW 264.7 macrophages. RAW 264.7 cells were treated with normal culture medium (control [CTR]), PLK-CM, or BIO-CM for 8 h, and nitric oxide concentrations in the supernatant of the cell culture were measured. (D) qPCR analysis of iNOS mRNA from RAW 264.7 cells 2 and 8 h following treatment with PLK-CM or BIO-CM (n = 8). *, P < 0.05.
Conditioned medium from S. aureus biofilm attenuates NF-κB activation necessary for a proinflammatory response.
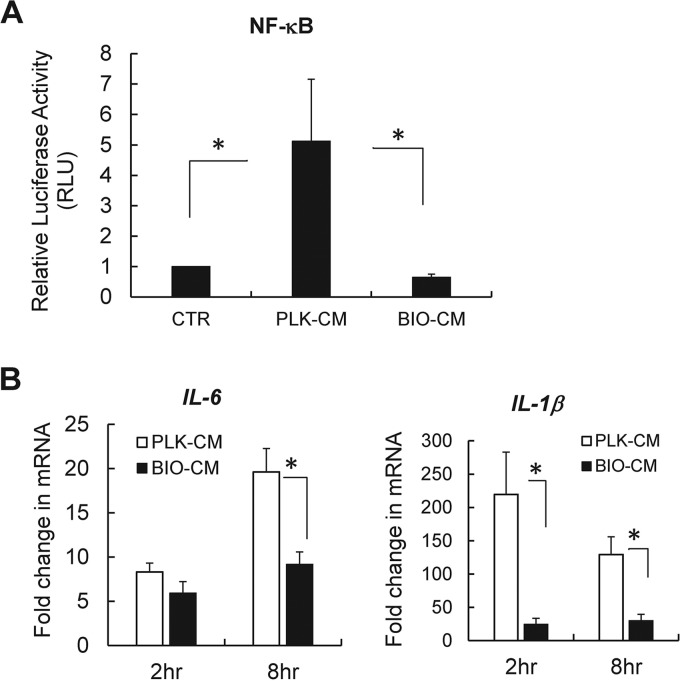
The response of macrophages to planktonic S. aureus involves the activation of key transcriptional mediators, including NF-κB, which in turn regulate the production of proinflammatory cytokines and bactericidal activity (21). Thus, we next examined whether the deregulated macrophage activation in response to BIO-CM is associated with the alteration in the NF-κB-mediated proinflammatory response. For this, we employed an NF-κB luciferase reporter assay, in which RAW 264.7 cells were transfected with an NF-κB luciferase reporter plasmid and the transfected cells were then exposed to either PLK-CM or BIO-CM. The extent of NF-κB activation was assessed by quantifying the luciferase expression of the NF-κB reporter (Fig. 2A). The NF-κB transcriptional activity was strongly reduced by ∼5-fold in RAW 264.7 cells exposed to BIO-CM, compared with cells treated with PLK-CM, which supports that factors secreted from cultures of S. aureus biofilms might act on macrophages to interfere with the NF-κB activation pathway.
FIG 2.
Effects of S. aureus biofilm-conditioned medium on activation of NF-κB and expression of proinflammatory cytokines in RAW 264.7 macrophages. (A) Transcriptional activation of NF-κB quantified by NF-κB luciferase expression in RAW 264.7 cells transfected with the NF-κB luciferase reporter following treatment with normal culture medium (CTR), PLK-CM, or BIO-CM for 8 h (n = 4 per group). RLU, relative luciferase units. (B) qPCR analysis of mRNA expression of the proinflammatory cytokines IL-1β and IL-6 from RAW 264.7 cells following treatment with PLK-CM or BIO-CM for 2 or 8 h (n = 6 to 8). *, P < 0.05.
The central role played by NF-κB in macrophage-mediated innate immunity has been revealed by the coordinated expression of multiple genes essential for the immune response, including those for proinflammatory cytokines, such as interleukin-1β (IL-1β) and IL-6, whose expression and production are critical for the recruitment of phagocytes, including neutrophils and macrophages, to the site of infection toward bactericidal activity (24, 25). Thus, we next examined whether the S. aureus biofilm environment was associated with alteration in the expression of proinflammatory cytokines, including IL-1β and IL-6. RAW 264.7 cells treated with BIO-CM exhibited substantially diminished expression of IL-1β and IL-6 compared to cells treated with PLK-CM (Fig. 2B).
Conditioned medium from S. aureus biofilm inhibits PRR-dependent expression of iNOS mRNA.
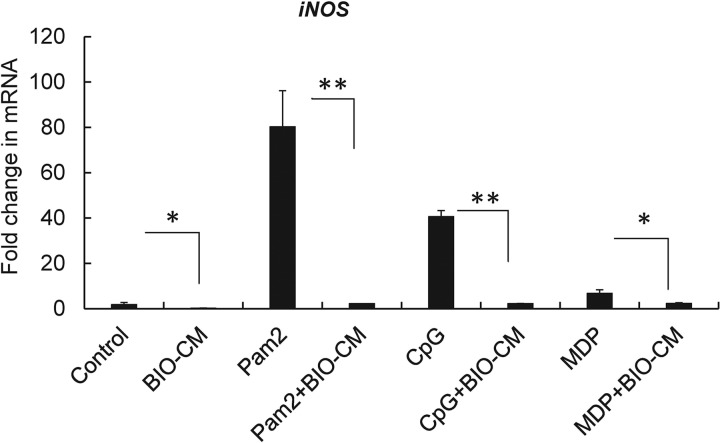
By observing that S. aureus biofilm-conditioned medium could significantly diminish the expression of proinflammatory cytokines, along with an attenuated activation of NF-κB, we reasoned that factors secreted from the S. aureus biofilm environment might interfere with either the expression of PRRs or PRR-mediated signaling, which can alter downstream signaling for NF-κB activation. However, treatment with BIO-CM did not alter the mRNA expression of PRRs, including TLR2, TLR9, and NOD2, in RAW 264.7 cells, compared with treatment with PLK-CM (see Fig. S1 in the supplemental material). We next sought to test whether the S. aureus biofilm-conditioned-medium-induced attenuation of the expression of proinflammatory cytokines is associated with an alteration of PRR-mediated signaling necessary to stimulate the response. This was assessed by quantifying the expression of iNOS mRNA in RAW 264.7 macrophages following the stimulation of the cells with an agonist of TLR2 (Pam2CSK4 [2 ng/ml]), TLR9 (CpG [3 μM]), or NOD2 (muramyl dipeptide [MDP] [10 ng/ml]) in the presence and absence of BIO-CM, since NF-κB-mediated expression of iNOS mRNA in response to PRR-mediated signaling is critical for promoting bactericidal activities of phagocytes. Treatment with the PRR agonist significantly increased the expression of iNOS mRNA in RAW 264.7 cells for all three agonists in control medium (P < 0.01 versus the control), in which the extent of iNOS mRNA expression was greater in RAW 264.7 cells stimulated with the TLR2 agonist than in cells stimulated with the TLR9 or NOD2 agonist. However, the responses were substantially attenuated in the presence of BIO-CM for all the agonists of PRRs (Fig. 3). Taken together, these results indicate that factors secreted from S. aureus biofilms might interfere with PRR-dependent signaling for the induction of a proinflammatory immune response in macrophages.
FIG 3.
Effects of S. aureus biofilm-conditioned medium on PRR-mediated expression of iNOS mRNA in RAW 264.7 macrophages. RAW 264.7 cells were treated with normal culture medium (Control) or BIO-CM in the presence or absence of the PRR agonists Pam2CSK4 (Pam2) (agonist for TLR2), CpG (agonist for TLR9), and MDP (agonist for NOD2). Treatment was conducted for 8 h, and the expression of iNOS mRNA was quantified by qPCR analysis (n = 4). *, P < 0.05; **, P < 0.01.
Conditioned medium from S. aureus biofilm increases the expression of KLF2.
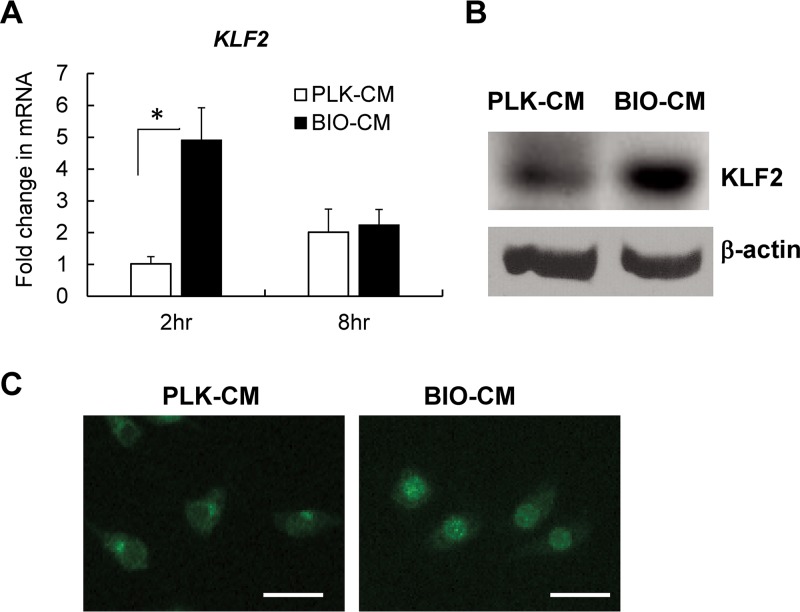
Based on previous understanding of the role of KLF2 as a negative regulator of NF-κB activation (19, 26), along with the observation that exotoxins from S. aureus could induce sustained expression of KLF2 in macrophages (18), we next sought to determine whether attenuated bactericidal and proinflammatory responses of RAW 264.7 cells in the S. aureus biofilm environment are associated with altered expression of KLF2. Our data demonstrate a significantly increased expression of KLF2 mRNA, by ∼3-fold, in RAW 264.7 cells exposed to BIO-CM compared to cells exposed to PLK-CM (Fig. 4A). The increased expression of KLF2 was also evident at the protein level, as revealed by Western blot analysis (Fig. 4B) and immunofluorescence staining (Fig. 4C) of KLF2 protein from RAW 264.7 cells following treatment with BIO-CM. Interestingly, the expression of KLF2 in RAW 264.7 cells exposed to BIO-CM was localized in the nucleus, while its expression in PLK-CM-treated cells was localized to the nuclear periphery (Fig. 4C). Taken together, our results suggest that factors secreted from cultures of S. aureus biofilms might act on macrophages to result in the increased expression of KLF2 and its translocation to the nucleus.
FIG 4.
Effects of S. aureus biofilm-conditioned medium on expression of KLF2 in RAW 264.7 macrophages. (A) qPCR analysis of expression of KLF2 mRNA from RAW 264.7 cells treated with PLK-CM or BIO-CM for 2 or 8 h (n = 6 per group). *, P < 0.05. (B) Representative Western blot images of expression of the KLF2 protein from RAW 264.7 cells treated with PLK-CM or BIO-CM for 2 h. β-Actin expression was used as an internal control. (C) Fluorescence images of RAW 264.7 cells expressing KLF2 protein. Cells were treated with PLK-CM or BIO-CM for 30 min and then immunostained with anti-KLF2 antibody (green). Bar = 40 μm.
siRNA-mediated silencing of KLF2 in macrophages restores bactericidal activity in response to biofilm-conditioned medium.
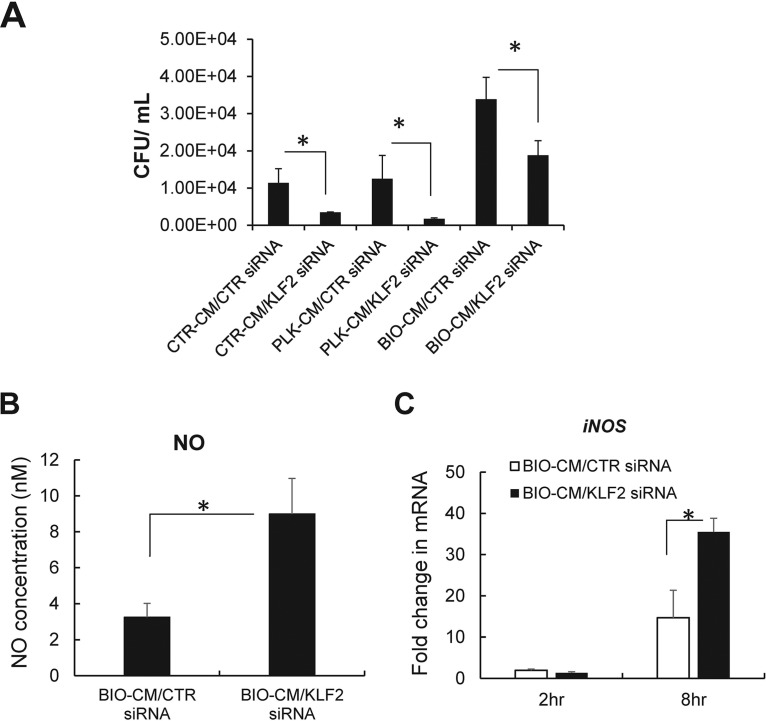
By observing an increased expression of KLF2 along with a diminished capacity for bactericidal activity in RAW 264.7 cells following exposure to BIO-CM, we next examined whether silencing of the KLF2 gene can functionally restore the bactericidal activity of macrophages toward S. aureus biofilms. For this, small interfering RNA (siRNA)-mediated knockdown studies were undertaken in RAW 264.7 cells. The efficiency of siRNA-mediated knockdown of KLF2 in RAW 264.7 cells was measured to be ∼60%, as determined by quantitative PCR (qPCR) analysis (Fig. S2A), which was also confirmed at the protein level by Western blot analysis (Fig. S2B). We next determined the effect of KLF2 knockdown on the bactericidal function of macrophages pretreated with BIO-CM using an antibiotic protection assay, in which live S. aureus bacteria were added to a monolayer of RAW 264.7 cells transfected with KLF2 siRNA or scrambled siRNA to assess intracellular bacterial killing. RAW 264.7 cells with siRNA-mediated knockdown of KLF2 exhibited a significantly enhanced bactericidal capacity compared with the control group (P < 0.05) (Fig. 5A).
FIG 5.
Effects of siRNA-mediated knockdown of the KLF2 gene on bactericidal activity and nitric oxide secretion in RAW 264.7 macrophages. siRNA-mediated knockdown of the KLF2 gene was induced in RAW 264.7 cells, and cells were exposed to normal culture medium (CTR-CM), PLK-CM, or BIO-CM. (A) Bactericidal activity of RAW 264.7 cells with siRNA knockdown of KLF2 (KLF2 siRNA) or scrambled control siRNA (CTR siRNA) following treatment with CTR-CM, PLK-CM, or BIO-CM for 2 h (n = 4 to 5 per group). (B) Nitric oxide concentrations measured from the supernatant of RAW 264.7 cells transfected with CTR siRNA or KLF2 siRNA following treatment with PLK-CM or BIO-CM for 8 h (n = 5). (C) qPCR analysis of iNOS mRNA from RAW 264.7 cells transfected with CTR siRNA or KLF2 siRNA following treatment with PLK-CM or BIO-CM for 2 or 8 h (n = 8). *, P < 0.05.
We next examined whether the silencing of KLF2 can enhance the secretion of nitric oxide, a major effector molecule for bactericidal activity in macrophages. The knockdown of KLF2 in RAW 264.7 cells treated with BIO-CM for 8 h significantly increased the secretion of NO by 3-fold compared with cells treated with control siRNA (Fig. 5B). Consistent with this, parallel effects were observed for iNOS mRNA encoding NO production. The siRNA-mediated knockdown of KLF2 in macrophages exhibited significantly high expression levels of iNOS mRNA in RAW 264.7 cells, compared with control macrophages, in response to exposure to BIO-CM (Fig. 5C). Taken together, our results support that the silencing of KLF2 with siRNA in macrophages could tip the balance in favor of an antibiofilm and proinflammatory phenotype, which could result in enhanced bactericidal activity against S. aureus biofilms.
siRNA-mediated silencing of KLF2 in macrophages restores the activation of NF-κB and the expression of proinflammatory genes in response to biofilm-conditioned medium.
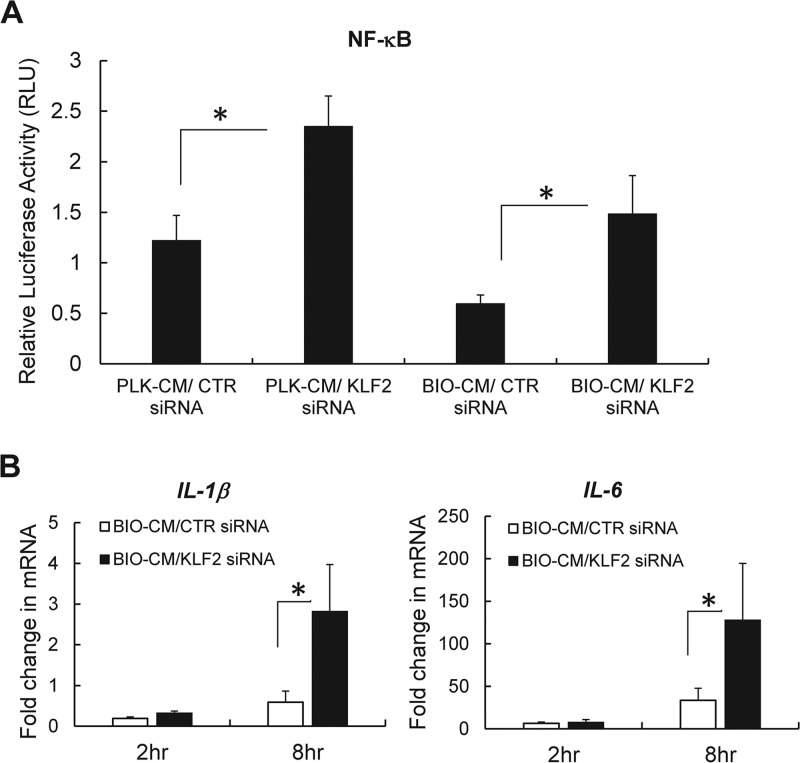
The overexpression of KLF2 in monocytes was shown to inhibit their proinflammatory activation through the inhibition of the transcriptional activity of NF-κB (27). By observing that the exposure of BIO-CM to macrophages significantly increases the expression of KLF2 (Fig. 4), along with attenuated NF-κB activation, we reasoned that the antibiofilm effects induced by KLF2 silencing might be mediated by restoring NF-κB transcriptional activation. To assess this, RAW 264.7 cells were double transfected with KLF2 siRNA (or scrambled siRNA) and an NF-κB luciferase reporter plasmid. The cells were then exposed to BIO-CM, and the extent of NF-κB activation was assessed by quantifying the luciferase expression of the NF-κB reporter. The induction of NF-κB luciferase activity was significantly increased in RAW 264.7 cells with KLF2 siRNA knockdown by ∼2-fold compared with cells transfected with scrambled siRNA (Fig. 6A). The enhanced NF-κB activation with KLF2 siRNA knockdown was associated with increased expression of genes for the proinflammatory cytokines IL-1β and IL-6 in response to BIO-CM. The expression levels of IL-1β and IL-6 were significantly increased by ∼4-fold and ∼3-fold, respectively, in macrophages with KLF2 siRNA knockdown, compared with cells transfected with control siRNA (Fig. 6B). Taken together, our results support that the S. aureus biofilm environment increased KLF2 expression in macrophages, which in turn attenuated NF-κB-dependent proinflammatory responses.
FIG 6.
Effects of siRNA-mediated knockdown of the KLF2 gene on activation of NF-κB and expression of proinflammatory cytokines in RAW 264.7 macrophages. (A) Transcriptional activation of NF-κB quantified by NF-κB luciferase expression in RAW 264.7 cells transfected with the NF-κB luciferase reporter and KLF2 siRNA (or scrambled siRNA [CTR siRNA]), following treatment with PLK-CM or BIO-CM for 8 h (n = 4 to 6 per group). (B) Effect of siRNA-mediated knockdown of the KLF2 gene on expression of proinflammatory cytokines in RAW 264.7 cells. siRNA-mediated knockdown of the KLF2 gene was induced in RAW 264.7 cells, and the cells were treated with BIO-CM for 2 or 8 h. The expression levels of IL-1β and IL-6 mRNAs were then quantified by qPCR analysis. Results were compared with those for RAW 264.7 cells transfected with scrambled siRNA (CTR siRNA) (n = 6 to 8 per group). *, P < 0.05.
DISCUSSION
A series of recent studies demonstrated that S. aureus biofilms evade host immune defense by skewing macrophage polarization toward an anti-inflammatory phenotype that promotes persistent bacterial survival (11–15). However, the detailed molecular mechanism underlying the evasion of macrophage-mediated innate immune responses against S. aureus biofilms remains to be elucidated. The major finding of this study is that S. aureus biofilm-conditioned-medium-induced impairment of bactericidal and proinflammatory activities of macrophages is mediated via KLF2-dependent attenuation of NF-κB activation. Our study identifies KLF2 in macrophages as a molecular target of S. aureus biofilms to evade innate immune attack.
In response to an infectious challenge involving planktonic S. aureus, the recognition of PAMPs by macrophages is mediated by PRRs, including TLR2, TLR9, and NOD2, which trigger NF-κB-dependent signaling cascades that induce the production of proinflammatory cytokines, nitric oxide, and reactive oxygen intermediates to combat S. aureus (6–9). It has been shown that various virulence factors secreted from S. aureus biofilms have the ability to interfere with various steps in the PRR-dependent NF-κB activation pathway to dampen antibacterial and proinflammatory responses in host cells (8, 17). Consistent with this understanding, our results show that factors secreted from S. aureus biofilms significantly attenuate the activation of NF-κB and deregulate PRR-mediated signaling for the expression of proinflammatory cytokines. This was evidenced by the result that treatment with BIO-CM could significantly inhibit TLR2-, TLR9-, and NOD2-mediated expression of iNOS mRNA in RAW 264.7 macrophages. Since PRR-mediated signaling preferentially activates NF-κB and the subsequent expression of iNOS (28), the attenuation of iNOS mRNA expression may indicate that factors secreted from S. aureus biofilms might interfere with or negatively regulate PRR-dependent NF-κB activation for the innate immune response of macrophages.
Our subsequent analysis of RAW 264.7 cells cultured in the presence of BIO-CM revealed that the BIO-CM-induced attenuation of NF-κB activation of innate immune responses might be mediated by upregulation of the expression of KLF2. The functional role of KLF2 in this response was evidenced by our finding that siRNA-mediated silencing of KLF2 in RAW 264.7 cells could restore the activation of NF-κB and the capacities for bactericidal activity and generation of proinflammatory cytokines. KLF2 is a subclass of the zinc finger family of transcription factors, and accumulating evidence supports the functional role of KLF2 in modulating the immune response (29). In particular, it has been shown that KLF2 can act as a negative regulator of proinflammatory activation in monocytes and macrophages, as evidenced by the finding that a deficiency of KLF2 in monocytes and macrophages results in augmented proinflammatory gene expression, while the overexpression of KLF2 could significantly attenuate the expression of proinflammatory cytokines (19).
The molecular mechanism by which S. aureus biofilm-conditioned-medium-induced KLF2 signaling modulates the activation of NF-κB in RAW 264.7 cells remains to be elucidated. One possible explanation is that KLF2 may suppress NF-κB transcriptional activity by inhibiting the recruitment of coactivators to NF-κB. It has been shown that KLF2 can directly interact with p300 and p300/CBP-associated factor (PCAF), critical coactivators for NF-κB-mediated transcriptional activity, which results in diminished recruitment of the coactivators to NF-κB in monocytes (27). Collectively, our results suggest that factors secreted from S. aureus biofilms exploit the KLF2-dependent negative regulatory mechanism to subvert macrophage-mediated innate immune responses against S. aureus biofilms.
What is the identity of the effector molecules secreted from S. aureus biofilms that act on macrophages to increase the expression of KLF2? It has been shown that S. aureus utilizes numerous secreted virulence factors, such as alpha-hemolysin (30) and leukocidins (31), to evade the host immune system. One plausible explanation is the role played by alpha-hemolysin, which is required for biofilm formation by S. aureus (32) and induces the apoptosis of macrophages (21). The S. aureus strain used in our study (ATCC 6538) was shown to express alpha-hemolysin (33). Mice infected with an alpha-hemolysin mutant strain of S. aureus exhibited a robust immune response, resulting in the expression of proinflammatory cytokines and chemokines involved in immune cell infiltration and increased bacterial clearance (34). Importantly, other studies have shown that KLF2 in host cells is considered a target of S. aureus exotoxins (20, 35). The factors secreted from S. aureus biofilms that act on macrophages along with molecular mechanisms for the increase in KLF2 expression in macrophages remain to be elucidated.
In summary, we have demonstrated that the S. aureus biofilm environment skews macrophage polarization toward the inhibition of bactericidal and proinflammatory activities by increasing the expression of KLF2 in macrophages, which in turn negatively regulates the activation of NF-κB pathways for innate immune responses. Our study is the first to underscore the functional role of KLF2 in regulating the macrophage-mediated immune response against S. aureus biofilms and provides an important insight into the mechanism by which biofilm-associated infections evade innate immune defense functions.
MATERIALS AND METHODS
Cell culture.
RAW 264.7 cells (American Type Culture Collection [ATCC], Manassas, VA) were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with glutamine (2 mM), penicillin (100 U/ml), streptomycin sulfate (100 mg/ml), and 10% heat-inactivated, lot-selected fetal bovine serum (FBS). Cells were incubated in cell culture flasks at a defined density at 37°C with 5% CO2.
Preparation of conditioned medium from planktonic and biofilm cultures of S. aureus.
S. aureus (ATCC 6538 strain), purchased from the ATCC (Manassas, VA), was streaked onto tryptic soy agar. Next, a single colony of S. aureus was incubated in 5 ml of sterile RPMI 1640 supplemented with 10% heat-inactivated FBS at 37°C for 18 h in a shaking incubator (180 rpm). Following culture, bacteria were pelleted by centrifugation at 4,000 rpm at 4°C for 15 min. Planktonic conditioned medium (PLK-CM) was collected from the supernatant of the pellet, filtered using a 0.2-μm filter, and stored at −80°C for further experimental use. Biofilm-conditioned medium (BIO-CM) was collected from the supernatant of a static biofilm culture of S. aureus. Briefly, two-well glass chamber slides (Thermo Scientific, Rochester, NY) coated with 20% human plasma in sterile carbonate-bicarbonate buffer were used for static biofilm culture. After coating, each chamber was inoculated with 2 ml of a culture of S. aureus (1 × 106 CFU/ml) grown overnight and cultured at 37°C for 6 days. The culture medium was replaced every 24 h with 2 ml of sterile RPMI 1640 supplemented with 10% heat-inactivated FBS. BIO-CM was then collected from the supernatant of the culture 24 h following the last medium change from the 6-day-old biofilm. The conditioned medium was filtered using a 0.2-μm filter after centrifugation for 15 min at 4,000 rpm and stored at −80°C for further use in the experiment.
Bacterial phagocytosis assay.
RAW 264.7 cells were seeded in a 24-well plate at a density of 1 × 106 cells per well and then incubated with PLK-CM or BIO-CM for 2 h at 37°C. After incubation, 50 μl of fluorescent Escherichia coli bioparticles (opsonized Alexa Fluor 488-conjugated E. coli; Thermo Fisher Scientific, Waltham, MA) in RPMI 1640 supplemented with 10% heat-inactivated FBS was added, and the cells were incubated at 37°C for 2 h. After incubation, the excess bioparticles were removed, and cells were washed twice with phosphate-buffered saline (PBS). Cells were then immediately observed under a fluorescence microscope (IX81; Olympus, Tokyo, Japan) to evaluate the presence of intracellular bioparticles. The fluorescence intensity of each well was also quantified with a plate reader (SpectraMax M4; Molecular Devices, Sunnyvale, CA) at an excitation wavelength of 480 nm and an emission wavelength of 520 nm.
Real-time quantitative PCR assay.
Total RNAs were isolated from RAW 264.7 cells using an RNA extraction kit (Omega Bio-Tek, Norcross, GA). The purity and concentration of RNA were measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, MA). A cDNA template was then prepared using a high-capacity cDNA reverse transcription kit (Quantabio, Germantown, MD) in a Bio-Rad (Hercules, CA) T100 thermal cycler. The qPCR analysis was performed with a real-time PCR system (Realplex2 master cycler; Eppendorf, Hauppauge, NY) in 20-μl volumes per reaction. In brief, each reaction mixture contained 5 ng cDNA, 100 nmol/liter primers, and 10 μl 2× SYBR green PCR master mix (Quantabio). The qPCR cycles were composed of an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s and an elongation step at 72°C for 20 s. qPCR results were normalized against cycle thresholds for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and are presented as fold changes in mRNA levels. The primer sequences of genes, including GAPDH, inducible nitric oxide synthase (iNOS), interleukin-6 (IL-6), interleukin-1β (IL-1β), KLF2, TLR2, TLR9, and NOD2, are listed in Table S1 in the supplemental material.
Western blot analysis.
RAW 264.7 macrophages were seeded into 24-well plates at a density of 1 × 106 cells/well and then incubated for the indicated times with either PLK-CM or BIO-CM for 2 h. After treatment, the supernatant was discarded, and total protein was isolated from RAW 264.7 cells with cell-lytic radioimmunoprecipitation assay (RIPA) buffer with a phosphatase inhibitor cocktail (Pierce, Rockford, IL). The total protein concentration was estimated by a Bradford assay, and protein samples (35 μg) were subjected to separation on a 10% SDS-PAGE gel (Bio-Rad, Hercules, CA). After electrophoresis, the protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA) by using the Trans-blot Turbo system (Bio-Rad). The membrane was blocked for 1 h in 5% milk in Tris-buffered saline–Tween (TBST) buffer. Subsequently, the membrane was blotted overnight using mouse anti-KLF2 (1:500; Abcam, Cambridge, MA) or rabbit anti-β-actin (1:1,000; Cell Signaling, Danvers, MA), followed by incubation with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibodies, respectively, for 1 h at room temperature. Proteins were visualized on a Kodak image station.
Treatment of RAW 264.7 macrophages with PRR agonists.
The effect of S. aureus biofilm-conditioned medium on PRR-mediated signaling in macrophages was assessed by stimulating RAW 264.7 cells with Pam2CSK4 (2 ng/ml) for TLR2 activation, CpG oligonucleotide (3 μM) for TLR9 activation, or muramyl dipeptide (MDP) (10 ng/ml) for NOD2 activation. RAW 264.7 cells were treated with the indicated PRR agonist solution for 8 h in the presence or absence of BIO-CM and then subjected to a qPCR assay for iNOS mRNA expression. All the PRR agonists were obtained from InvivoGen (San Diego, CA) and prepared using sterile RPMI supplemented with 10% heat-inactivated FBS.
Immunofluorescence imaging of KLF2 in RAW 264.7 cells.
RAW 264.7 macrophages were seeded into 24-well plates at a density of 2.5 × 105 cells/well and then incubated with either PLK-CM or BIO-CM for 30 min. After treatment, cells were fixed in PBS with 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min. Cells were washed with PBS and blocked with antibody diluent (Dako, Santa Clara, CA) at room temperature for 1 h. The cells were then washed with PBS, incubated with primary antibody against KLF2 (1:100; Abcam, Cambridge, MA) at 4°C overnight, and incubated with Alexa Fluor 488-labeled donkey anti-mouse IgG (Thermo Fisher Scientific, Waltham, MA) at room temperature for 1 h. Finally, the cells were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and observed under a fluorescence microscope (IX81; Olympus, Tokyo, Japan) to evaluate the expression and localization of KLF2 protein.
Nitric oxide measurement.
RAW 264.7 cells were treated with PLK-CM or BIO-CM for 8 h, and the supernatant of each sample was collected for nitric oxide (NO) measurement. NO levels were quantified by measuring the levels of the stable end product, nitrite, using the Griess reagent (Thermo Fisher Scientific, Waltham, MA).
NF-κB luciferase reporter assay.
The effect of BIO-CM on NF-κB transcriptional activity in macrophages was assessed using the Cignal NF-κB pathway reporter assay kit (catalog number CCS-013L; Qiagen, Germantown, MD), which comprises an inducible NF-κB-responsive construct that carries the firefly luciferase reporter gene and a constitutively expressing Renilla luciferase construct. The NF-κB reporter gene was transfected into RAW 264.7 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with 100 ng of each plasmid (reporter or negative-control plasmid). The cells were then treated with control sterile RPMI 1640 supplemented with 10% FBS, PLK-CM, or BIO-CM for 8 h. The activation of NF-κB was quantified using firefly luciferase and Renilla luciferase substrates in a dual-luciferase reporter assay system (Promega, Madison, WI). The luciferase signal was read in a plate reader (SpectraMax M4; Molecular Devices, Sunnyvale, CA). The luciferase values were determined by normalization to Renilla values (as a control) to correct for variation in transfection efficiencies.
siRNA-mediated knockdown of KLF2 in RAW 264.7 cells.
RAW 264.7 cells were seeded at 1 × 106 cells/well in 12-well plates to achieve 70 to 80% confluence on the day of transfection. The cells were then transfected with scrambled or KLF2 siRNA (Thermo Fisher Scientific, Waltham, MA) at a final concentration of 50 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocol. At 48 h posttransfection, the cells were subjected to subsequent assays. The efficiency of KLF2 knockdown was confirmed by qPCR and Western blot assays.
Double transfection of the NF-κB luciferase reporter and KLF2 siRNA in RAW 264.7 macrophages.
The activation of NF-κB from macrophages with siRNA-mediated knockdown of KLF2 was quantified by an NF-κB luciferase reporter assay as described above. RAW 264.7 cells were plated at 4 × 104 cells per well in 96-well plates and then transfected with the NF-κB reporter and KLF2 siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 48 h of transfection, cells were collected and assayed with firefly luciferase and Renilla luciferase substrates using a dual-luciferase reporter assay system (Promega, Madison, WI).
Antibiotic protection assay for quantifying bactericidal activity of macrophages.
RAW 264.7 cells were seeded at 2.5 × 105 cells/well in a 24-well plate and then incubated with either PLK-CM or BIO-CM for 2 h. After treatment, cells were incubated with planktonic S. aureus bacteria at a multiplicity of infection (MOI) of 1:10 at 37°C for 1 h. The cells were washed with PBS to remove nonadherent bacteria. The cells were treated with serum-free DMEM with 25 μg/ml gentamicin (Sigma-Aldrich, St. Louis, MO) for 30 min and washed with PBS to kill and remove the extracellular bacteria. Cells were lysed with PBS containing 0.1% Triton X-100 at room temperature for 10 min. The cell lysates were then serially diluted with sterile PBS, and 50 μl of each dilution was plated onto triplicate tryptic soy agar plates. The plates were incubated overnight at 37°C. The number of internalized colonies of bacteria was enumerated, and CFU per milliliter were determined.
Statistical analysis.
Data analysis was performed using Origin 2015 software (Origin Lab, Northampton, MA). Statistical significance between two groups was determined by two-tailed unpaired t tests. Statistical tests among multiple groups were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s posttest for secondary analysis for comparison. For all analyses, P values of less than 0.05 were considered statistically significant. Data are presented as means ± standard errors (SE).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH R01 NR015674) and the Kent State University startup fund.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00643-18.
REFERENCES
- 1.Noskin GA, Rubin RJ, Schentag JJ, Kluytmans J, Hedblom EC, Smulders M, Lapetina E, Gemmen E. 2005. The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Arch Intern Med 165:1756–1761. doi: 10.1001/archinte.165.15.1756. [DOI] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister JL, Horswill AR. 2014. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 4:178. doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier B, Philpott DJ. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat Rev Immunol 4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 7.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johannessen M, Askarian F, Sangvik M, Sollid JE. 2013. Bacterial interference with canonical NFkappaB signalling. Microbiology 159:2001–2013. doi: 10.1099/mic.0.069369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanneganti TD, Lamkanfi M, Nunez G. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity 27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Secor PR, James GA, Fleckman P, Olerud JE, McInnerney K, Stewart PS. 2011. Staphylococcus aureus biofilm and planktonic cultures differentially impact gene expression, MAPK phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol 11:143. doi: 10.1186/1471-2180-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol 186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snowden JN, Beaver M, Beenken K, Smeltzer M, Horswill AR, Kielian T. 2013. Staphylococcus aureus sarA regulates inflammation and colonization during central nervous system biofilm formation. PLoS One 8:e84089. doi: 10.1371/journal.pone.0084089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gries CM, Kielian T. 2017. Staphylococcal biofilms and immune polarization during prosthetic joint infection. J Am Acad Orthop Surg 25:S20–S24. doi: 10.5435/JAAOS-D-16-00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanke ML, Angle A, Kielian T. 2012. MyD88-dependent signaling influences fibrosis and alternative macrophage activation during Staphylococcus aureus biofilm infection. PLoS One 7:e42476. doi: 10.1371/journal.pone.0042476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanke ML, Heim CE, Angle A, Sanderson SD, Kielian T. 2013. Targeting macrophage activation for the prevention and treatment of Staphylococcus aureus biofilm infections. J Immunol 190:2159–2168. doi: 10.4049/jimmunol.1202348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddick LE, Alto NM. 2014. Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell 54:321–328. doi: 10.1016/j.molcel.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zecconi A, Scali F. 2013. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol Lett 150:12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Dach K, Zovko J, Hogardt M, Koch I, van Erp K, Heesemann J, Hoffmann R. 2009. Bacterial toxins induce sustained mRNA expression of the silencing transcription factor klf2 via inactivation of RhoA and Rhophilin 1. Infect Immun 77:5583–5592. doi: 10.1128/IAI.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, McManus R, Ryan T, Leahy P, Lin Z, Haldar SM, Atkins GB, Wong HR, Lingrel JB, Jain MK. 2011. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity 34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Grady E, Mulcahy H, Adams C, Morrissey JP, O’Gara F. 2007. Manipulation of host Kruppel-like factor (KLF) function by exotoxins from diverse bacterial pathogens. Nat Rev Microbiol 5:337–341. doi: 10.1038/nrmicro1641. [DOI] [PubMed] [Google Scholar]
- 21.Scherr TD, Hanke ML, Huang O, James DB, Horswill AR, Bayles KW, Fey PD, Torres VJ, Kielian T. 2015. Staphylococcus aureus biofilms induce macrophage dysfunction through leukocidin AB and alpha-toxin. mBio 6:e01021-15. doi: 10.1128/mBio.01021-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. 2008. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One 3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathan CF, Hibbs JB Jr. 1991. Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol 3:65–70. doi: 10.1016/0952-7915(91)90079-G. [DOI] [PubMed] [Google Scholar]
- 24.Dev A, Iyer S, Razani B, Cheng G. 2011. NF-kappaB and innate immunity. Curr Top Microbiol Immunol 349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- 25.Newton K, Dixit VM. 2012. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4:a006049. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munitz A, Cole ET, Beichler A, Groschwitz K, Ahrens R, Steinbrecher K, Willson T, Han X, Denson L, Rothenberg ME, Hogan SP. 2010. Paired immunoglobulin-like receptor B (PIR-B) negatively regulates macrophage activation in experimental colitis. Gastroenterology 139:530–541. doi: 10.1053/j.gastro.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. 2006. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A 103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie QW, Kashiwabara Y, Nathan C. 1994. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J Biol Chem 269:4705–4708. [PubMed] [Google Scholar]
- 29.Bieker JJ. 1996. Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol 15:347–352. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 30.Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, Jania C, Doerschuk CM, Tilley SL, Duncan JA. 2012. Staphylococcus aureus alpha-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis 205:807–817. doi: 10.1093/infdis/jir846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonzo F III, Torres VJ. 2013. Bacterial survival amidst an immune onslaught: the contribution of the Staphylococcus aureus leukotoxins. PLoS Pathog 9:e1003143. doi: 10.1371/journal.ppat.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caiazza NC, O’Toole GA. 2003. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J Bacteriol 185:3214–3217. doi: 10.1128/JB.185.10.3214-3217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YG, Lee JH, Raorane CJ, Oh ST, Park JG, Lee J. 2018. Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front Microbiol 9:1241. doi: 10.3389/fmicb.2018.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tkaczyk C, Hamilton MM, Datta V, Yang XP, Hilliard JJ, Stephens GL, Sadowska A, Hua L, O’Day T, Suzich J, Stover CK, Sellman BR. 2013. Staphylococcus aureus alpha toxin suppresses effective innate and adaptive immune responses in a murine dermonecrosis model. PLoS One 8:e75103. doi: 10.1371/journal.pone.0075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama M, Kurokawa K, Nakamura K, Lee BL, Sekimizu K, Kubagawa H, Hiramatsu K, Yagita H, Okumura K, Takai T, Underhill DM, Aderem A, Ogasawara K. 2012. Inhibitory receptor paired Ig-like receptor B is exploited by Staphylococcus aureus for virulence. J Immunol 189:5903–5911. doi: 10.4049/jimmunol.1201940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.