Abstract
Risk metabolites of postmenopausal osteoporosis (PO) were explored to offer a theoretical basis for future therapy. The data E-GEOD-7429 were downloaded from ArrayExpress database. In total 20 samples deprived from postmenopausal women having low or high bone mineral density (BMD) were covered in this expression profile. After screening of differentially expressed genes (DEGs), gene-gene network was constructed taking the intersection between the DEGs and genes in the seed protein-protein interaction network. Then, the other five networks were established, including metabolite, phenotype, gene-metabolite, phenotype-gene, and phenotype-metabolite networks. Next, these 6 networks were integrated into one weighted multi-omics network to further identify the candidate metabolites using random walk with restart based on the PO-related seed genes, seed metabolites and phenotype. Using the score among nodes of the weighted composite network, the top 50 metabolites, and the top 100 co-expressed genes interacting with the top 50 metabolites were detected. A set of 601 DEGs between low BMD and high BMD samples were selected. Significantly, the top 5 metabolites were respectively glucosylgalactosyl hydroxylysine, all-trans-5,6-epoxyretinoic acid, tretinoin, colecalciferol, and rocaltrol. Moreover, 3 metabolites (estraderm, triphosadenine, and tretinoin) had a degree >50 in the co-expression network. Tretinoin was the member of the top 5 metabolites, and estraderm was a metabolite with the seventh interaction score. A series of metabolites, tretinoin and estraderm might be closely associated with the onset and progression of PO.
Keywords: postmenopausal osteoporosis, metabolites, bone mineral density, multi-omics composite network, differentially expressed genes
Introduction
Postmenopausal osteoporosis (PO), one type of osteoporosis, is believed to result directly from the decreased endogenous estrogen in menopausal women (1,2). The measurement of bone mineral density (BMD) has been regarded as the ‘gold standard’ in osteoporosis diagnosis (3). Therefore, extraction of genes predisposing to BMD will be beneficial for the understanding of genetic mechanisms and aid in developing novel treatment of PO and PO-related fractures.
An increasing number of genes has been identified to have independent effects on BMD in osteoporosis in recent years. For example, POSTN, a regulator of osteoblast differentiation and bone formation, has been demonstrated to influence the susceptibility to low BMD and osteoporosis (4). Moreover, genes VDR, ESR1, NRIP1 in B cells have been reported to be closely associated with BMD in patients with osteoporosis (5). Furthermore, Jemtland et al (6) measured the changes in 84 bone biopsies related to BMD variations in postmenopausal women to extract osteoporosis-associated candidate genes and detected that SOX4, MMP13, and MEPE were all under-expressed. Nevertheless, these studies paid main attention on the gene levels related to the BMD, not an intensive analysis of the metabolites in PO.
In 2008, Xiao et al (7) provided the gene profile data of E-GEOD-7429, analyzing the gene expression profile in B cells of PO patients and identified that downregulation of ESR1 and MAPK3 in B cells led to the increased osteoclastogenesis or decreased osteoblastogenesis. Moreover, in 2016, Ma et al (8) used this microarray data (E-GEOD-7429) to identify several crucial genes related to PO, and found that the interactions, for example, CSTA/TYROBP, CCNE1/REL and TUBA1B/ESR1 may play an important role in the development of PO. However, these analyses are mainly focused on identifying altered genes between disease and control groups, and do not perform the corresponding analysis for identifying metabolites. Metabolites, the end products of biological regulatory process, are regarded as the final response of biological systems to the changes of environment or inheritance (9). Significantly, detecting and prioritizing disease-associated metabolites is crucial for our understanding of metabolite processes to improve medicine (10). As reported, metabolites rarely play an important role in isolation, but they exert a key function in the connection of phenotypes as well as genotypes, which are frequently influenced by genome and phenome (11). With the development of various ‘omics’ information, genomic, metabolic, as well as phenomic data, offer valuable information for disease risk candidate metabolites prioritization. Fortunately, Yao et al have prioritized several candidate metabolites using multi-omics composite network (12). Complementarily, we used multi-omics composite network to screen the metabolite in PO. Our efforts aimed to characterize new metabolite biomarkers as tools allowing for better diagnosis and preventing the progression of PO in the future.
Materials and methods
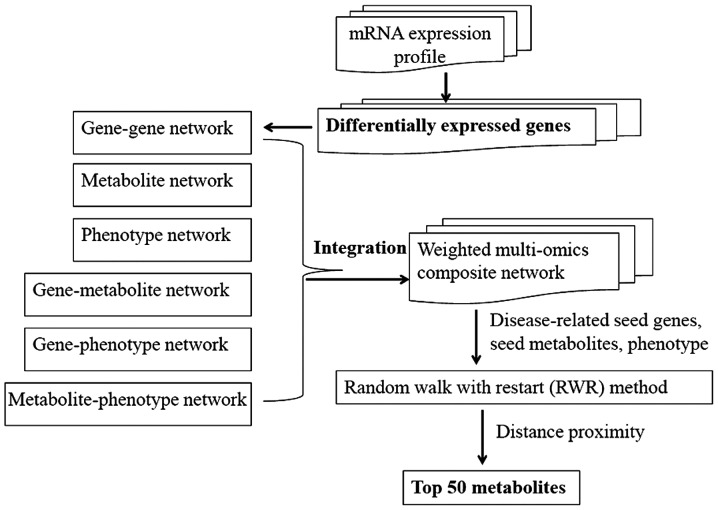
The present analysis was comprised of the following steps: microarray data selection (E-GEOD-7429) and identification of differentially expressed genes (DEGs), construction of a weighted composite network through integrating six data sets, and prioritization of candidate metabolites. The detailed information of each step is described in Fig. 1.
Figure 1.
Flow diagram of the method.
Data set
With the goal of revealing the molecular mechanisms of PO, microarray analyses of monocytes were performed using microarray data (accession number: E-GEOD-7429) (7) which were downloaded from the ArrayExpress database. A total of 20 samples were available. B cells were isolated from the whole blood of 20 unrelated postmenopausal women 54–60 years of age, including 10 with high BMD and 10 with low BMD. The platform of E-GEOD-7429 was A-AFFY-33-Affymetrix GeneChip Human Genome HG-U133A [HG-U133A].
Data pretreatment and DEGs identification
Probe IDs having concentrated expression level were transformed into human gene symbols. Duplicated genes of expression value in matrix were then eliminated. Overall, 12,437 genes were obtained. As known, the difference of gene expression levels reflected the disease characteristics. In our study, LIMMA package and t-test were used to identify DEGs between the two groups, following by the multiple correction test using Benjamini & Hochberg (13) method based on false discovery rate (FDR). DEGs were subsequently extracted using the criteria of FDR <0.05. These identified DEGs were used to build a gene network.
Establishment of multi-omics composite network
Six data sets (denoted by six networks) were combined to build a composite network, and six networks were called gene, phenotype, phenotype-gene, metabolite, gene-metabolite, and phenotype-metabolite networks.
Construction of gene network
In our study, all human protein-protein interactions (PPIs) having combine-scores (1,048,576 interactions) were obtained from STRING to establish the background PPI network. After we removed the duplicated PPIs, and transformed proteins into human gene symbols, 1,515,370 highly correlated gene-gene interactions covering 16,785 genes were extracted to construct a seed PPI network (combine-score not less than 0.8 was used herein). Next, we took the intersection between the 16,785 genes in the seed PPI network and DEGs to establish the informative gene-gene network.
Construction of metabolite network
To begin with, there were 4,994 human metabolites obtained from the KEGG, HMDB, Reactome, MSEA (14), as well as SMPDB (15). Afterwards, the metabolite-metabolite interactions of human as well as their corresponding confidence scores were extracted from STITCH (16). At the end, overall 3,764 human metabolites as well as 74,667 human metabolite-metabolite interactions were acquired.
Establishment of phenotype network
A total of 5,080 phenotypes as well as the scores exist in the phenotype-phenotype similarity associations (17). Relying on the phenotype-phenotype similarity associations, a phenotype network was constructed.
Construction of gene-metabolite association network
In order to obtain gene-metabolite interactions, the chemical and gene associations of human and the corresponding confidence scores were first extracted from the STITCH. After that, based on the 4,994 human metabolites, we gained human metabolite and gene associations. When removing the metabolites not included in the metabolite network mentioned above and eliminating the genes not covered in the gene network described above, a total of 192,763 gene-metabolite interactions involving in 12,342 genes as well as 3,278 metabolites were gained.
Construction of phenotype-gene association network
Phenotype-gene associations were picked out relying on the curated Morbid Map file of the OMIM database. After discarding the phenotypes that were not included in the phenotype network and the genes that were not involved in the gene network, there were 2,603 gene-phenotype associations (covering 1,715 genes as well as 1,886 phenotypes). The weighted score was determined as 1 for each phenotype-gene association.
Establishment of phenotype-metabolite association network
After filtration, there were 664 associations between 388 metabolites and 149 phenotypes. In addition, we defined the weighted value as 1 for each phenotype-metabolite interaction.
Establishment of a weighted composite network
To extract the potential metabolites, we merged the six networks mentioned above into a weighted composite network. Specific steps were described in Yao et al (12).
Prioritizing candidate metabolites on the basis of the weighted multi-omics composite network
The known PO-related metabolites (CID: 5460164, and 123986) were extracted from the Human Metabolome Database (HMDB) (18) which gathers specific information of small-molecule metabolites of human as well as the disease phenotype data. The known PO-related genes were collected from the Morbid Map file of Online Mendelian Inheritance in Man (OMIM) (19). The corresponding seed genes of PO were CALCR, LRP5, VDR, COL1A1, ESR1, NPPB, CNR1, CNR2, COL1A2, which were deposited in the database of OMIM. The phenotype of PO was 166710 in OMIN. Next, we mapped these seed genes, the phenotype, and the known metabolites to the weighted multi-omics composite network.
In an attempt to obtain the candidate metabolite prioritization from the weighted composite network involved in the seed genes, the phenotype, and the known metabolites, RWR method was expanded to the multi-omics composite network (20). Based on the distance proximity of every candidate metabolite, candidate metabolites were scored and ranked. According to the interaction score, the top 50 metabolites were identified which were determined as the PO-prioritized metabolites.
Subsequently, co-expressed genes interacting with the top 50 metabolites were identified, and then, we ranked these co-expressed genes relying on the score distribution. Finally, we extracted the top 100 co-expressed genes.
The subnetwork of the top 50 metabolites obtained from the composite network, and the co-expressed network between the top 100 co-expressed genes and the top 50 metabolites were constructed. Significantly, degree analyses were performed for these two networks to further identify several important AF-related metabolites.
Results
Identification of DEGs and establishment of multi-omics composite network
In the present study, we applied DEGs to construct the gene network, hence, DEGs between the two groups were first extracted. Using the criteria of the FDR <0.05, overall 601 genes were extracted as differential expression. The top 40 DEGs are listed in Table I. In this study, the PO-related metabolites were identified and prioritized by combining multi-omics data. Thus, in our study, a multi-omics composite network was first established through combining the information of genome, metabolome, and phenome. A total of 6 kinds of interactions were included in the multi-omics composite network. In this network, there were 9,360 nodes and 10,224,741 edges (Table II).
Table I.
The top 40 (DEGs).
| Gene symbols | FDR | Gene symbols | FDR |
|---|---|---|---|
| MAPK3 | 8.52E-09 | UBQLN4 | 9.85E-05 |
| HAO2 | 8.21E-09 | KIRREL | 9.73E-05 |
| TMEM8B | 7.38E-08 | POMT1 | 9.04E-05 |
| PSTPIP1 | 5.52E-08 | SNAI2 | 8.78E-05 |
| HGD | 5.37E-08 | CYB5R4 | 7.92E-05 |
| STK11 | 4.55E-08 | ODF2 | 7.22E-05 |
| ARMCX4 | 1.61E-08 | LOC728392 | 7.21E-05 |
| SETD3 | 7.71E-07 | ZNHIT3 | 8.36E-04 |
| NEO1 | 7.39E-07 | XDH | 8.12E-04 |
| ACKR1 | 7.12E-07 | GALR1 | 7.49E-04 |
| PRG3 | 6.53E-07 | PIGK | 7.12E-04 |
| BEGAIN | 5.69E-07 | PRX | 7.05E-04 |
| NRXN1 | 4.27E-07 | PTOV1-AS2 | 7.01E-04 |
| ZNF446 | 3.95E-07 | SULT1A2 | 6.82E-04 |
| C7 | 3.20E-07 | ABT1 | 7.31E-04 |
| NBR2 | 3.18E-07 | PAQR4 | 7.19E-04 |
| ISYNA1 | 4.39E-06 | PAM16 | 6.97E-04 |
| RGSL1 | 4.15E-06 | TMEM92-AS1 | 6.02E-04 |
| ODF1 | 1.85E-06 | EVX1 | 3.18E-04 |
| SLC26A3 | 1.56E-06 | CD1A | 1.71E-04 |
DEGs, differentially expressed genes; FDR, false discovery rate.
Table II.
Statistical information of the composite network.
| Statistics of the composite network | Nodes | Edges |
|---|---|---|
| Gene-gene network | 516 | 3325 |
| Metabolite-metabolite network | 3764 | 74,667 |
| Phenotype-phenotype network | 5080 | 10,140,046 |
| Gene-metabolite association network | 516 | 3023 |
| Phenotype-gene association network | 5080 | 2510 |
| Phenotype-metabolite association network | 537 | 664 |
| Total | 9360 | 10,224,741 |
Prioritization of the PO-related metabolites
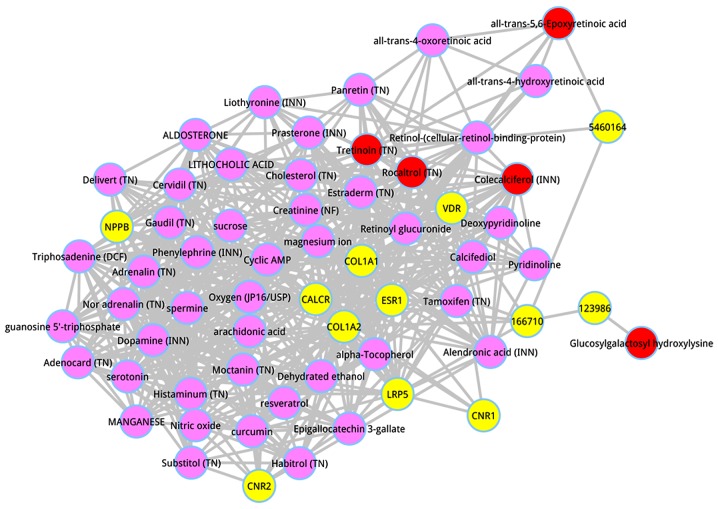
A total of 10 disease-related genes of PO are deposited in OMIM database, including CALCR, LRP5, VDR, COL1A1, ESR1, NPPB, CNR1, CNR2, and COL1A2, which were downloaded and determined as seed genes. There were 2 known disease metabolites (5460164, and 123986) data on PO in HMDB, which were defined as the seed metabolites. In our analysis, the 2 seed metabolites, and the 10 seed genes were utilized as seeds to identify the candidate metabolites of PO. To illustrate the intrinsic mode of this computational approach, the metabolites of the composite network were ranked in descending order on the basis of the interaction scores. The top 50 metabolites were dissected, and shown in Table III. The top 5 metabolites were respectively glucosylgalactosyl hydroxylysine (score = 0.027662), all-trans-5,6-Epoxyretinoic acid (score = 0.008912), tretinoin (TN) (score = 0.004934), colecalciferol (INN) (score = 0.000686), and rocaltrol (TN) (score = 0.000675). A subnetwork of the top 50 metabolites was extracted from the whole composite network, as shown in Fig. 2.
Table III.
The top 50 metabolites.
| Metabolite ID | Metabolite name | Score |
|---|---|---|
| 122304 | Glucosylgalactosyl hydroxylysine | 0.027662 |
| 5363137 | All-trans-5,6-Epoxyretinoic acid | 0.008912 |
| 444795 | Tretinoin (TN) | 0.004934 |
| 5280795 | Colecalciferol (INN) | 0.000686 |
| 5280453 | Rocaltrol (TN) | 0.000675 |
| 445354 | Retinol-(cellular-retinol-binding-protein) | 0.000487 |
| 5757 | Estraderm (TN) | 0.000473 |
| 6076 | Cyclic AMP | 0.000439 |
| 5283731 | Calcifediol | 0.000407 |
| 5280360 | Cervidil (TN) | 0.0004 |
| 439260 | Nor adrenalin (TN) | 0.000369 |
| 774 | Histaminum (TN) | 0.000357 |
| 945 | Nitric oxide | 0.000347 |
| 5988 | sucrose | 0.000323 |
| 5839 | Aldosterone | 0.00032 |
| 6437063 | all-trans-4-oxoretinoic acid | 0.000311 |
| 9903 | Lithocholic acid | 0.000305 |
| 105071 | Deoxypyridinoline | 0.000304 |
| 5288826 | Substitol (TN) | 0.000299 |
| 2088 | Alendronic acid (INN) | 0.000298 |
| 172198 | Delivert (TN) | 0.000296 |
| 702 | Dehydrated ethanol | 0.000277 |
| 588 | Creatinine (NF) | 0.000257 |
| 888 | magnesium ion | 0.000256 |
| 6438629 | all-trans-4-hydroxyretinoic acid | 0.000255 |
| 5202 | serotonin | 0.000254 |
| 65064 | Epigallocatechin 3-gallate | 0.000252 |
| 681 | Dopamine (INN) | 0.00025 |
| 5281877 | Retinoyl glucuronide | 0.000246 |
| 969516 | curcumin | 0.00024 |
| 449171 | Panretin (TN) | 0.000232 |
| 5957 | Triphosadenine (DCF) | 0.000231 |
| 105068 | Pyridinoline | 0.000227 |
| 5881 | Prasterone (INN) | 0.000225 |
| 753 | Moctanin (TN) | 0.000224 |
| 445154 | resveratrol | 0.000222 |
| 14985 | α-Tocopherol | 0.000221 |
| 2733526 | Tamoxifen (TN) | 0.00022 |
| 5816 | Adrenalin (TN) | 0.000218 |
| 977 | Oxygen (JP16/USP) | 0.000213 |
| 60961 | Adenocard (TN) | 0.00021 |
| 89594 | Habitrol (TN) | 0.000209 |
| 23930 | Manganese | 0.000203 |
| 444899 | arachidonic acid | 0.000202 |
| 5997 | Cholesterol (TN) | 0.000201 |
| 77999 | Gaudil (TN) | 0.0002 |
| 1103 | spermine | 0.000198 |
| 6830 | guanosine 5′-triphosphate | 0.000195 |
| 6041 | Phenylephrine (INN) | 0.000195 |
| 5920 | Liothyronine (INN) | 0.000192 |
Figure 2.
Multi-omics composite network involved in the top 50 metabolites and the seed genes. Yellow nodes are the seed genes. Pink nodes are the metabolites. Red nodes stand for the top 5 metabolites.
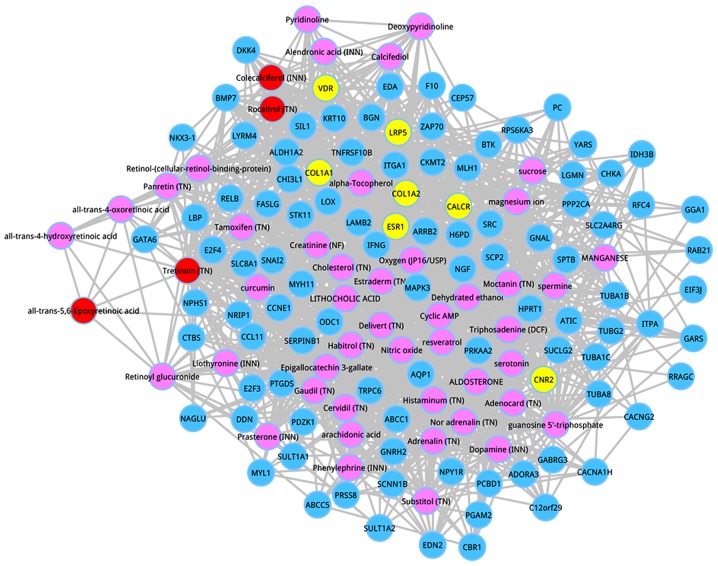
Afterwards, the co-expressed genes were extracted, which had interactions with the top 50 metabolites according to the score ranking. On the basis of the pre-defined criteria, the top 100 co-expressed genes were identified, and the co-expressed network of the top 100 genes are shown in Fig. 3. Following degree analysis for the co-expressed network, 3 metabolites had a degree >50, including estraderm (degree = 64), triphosadenine (DCF) (degree = 57), and tretinoin (TN) (degree = 55). Significantly, tretinoin (TN) was the member of the top 5 metabolites, and estraderm was a metabolite with the seventh interaction score.
Figure 3.
Co-expressed network. Blue nodes represent the top 100 co-expressed genes. Yellow nodes are the seed genes. Pink nodes are the metabolites. Red nodes stand for the members of the top 5 metabolites.
Discussion
Postmenopausal women have a high incidence of osteoporosis because of simultaneous existence of many independent predisposing factors, including estrogen deficiency, calcium loss, as well as aging (21). Osteoporosis is a main public health concern globally, particularly in countries with large aging populations, for example, China (22). Thus, it is urgently needed to seek for novel effective therapy strategy as well as etiologic explanations for PO. For a better understanding of the potential molecular mechanisms of PO, we used a computational method to extract metabolites signatures in PO by integrating the information among genes, metabolites, and phenotype related to pathogenesis. Our study identified several important metabolites between PO and normal samples, including tretinoin (TN) and estraderm.
Ahmed et al (23) demonstrated that tretinoin can suppress osteoblast proliferation, and inhibit the production of alkaline phosphatase, osteocalcin and IL-6 in normal human osteoblasts. In addition, suppression of IL-6 generation in normal osteoblasts might contribute to bone abnormalities. Moreover, in another study, tretinoin has been implicated to be able to activate the differentiation and proliferation of osteoclasts, thereby enhancing bone absorption (24). The imbalance in bone resorption and formation results in bone loss during aging and osteoporosis (25). Furthermore, many studies have used tretinoin to construct the model of osteoporosis (26–28). Thus, we infer that tretinoin is important for bone absorption and may be a target to suppress bone loss related to PO.
PO is believed to result directly from the decreased endogenous estrogen in menopausal women (2). Estraderm is a kind of steroid hormone that is mainly secreted by ovaries, placenta or testicles, and it is the most biologically active estrogen. Low concentration estraderm has been implicated to influence the proliferation and differentiation of osteoblast in a positive way (29). Estrogens may enhance fractional gastrointestinal calcium absorption, and an increased calcium intake might play a role in promoting bone mass (30). Recently, researchers have demonstrated that the combination of calcium and estrogen results in a significant increase in bone mass of the femoral neck, relative to estrogen alone (31). In addition, increased calcium intake decreases bone loss of premenopausal women (32,33).
In conclusion, we successfully extracted several important metabolites in PO on the basis of the combination of multi-omics data. Significantly, our identified metabolites (tretinoin and estraderm) are available as biomarkers to diagnose PO, and our study has established available therapeutic options (tretinoin and estraderm) for the treatment of PO. However, we must take several limitations into consideration. Firstly, there are many gene expression profiles on PO, but we only used E-GEOD-7429 dataset in our analysis. Hence, we will utilize other datasets on this disease to confirm our results. In addition, we used existing data to extract candidate metabolite biomarkers based on the bioinformatics method. Yet, our results have not been verified by experiments and this remains the main weak point of the present study. Consequently, future investigations are required to reveal the changes of these metabolites in the understanding of the progression procedure in PO based on animal experiments or patient tissues. Despite these disadvantages, we are satisfied as our findings could offer some preliminary evidence to reveal the potential candidate therapeutic strategies for PO. The application of specific regulation-related metabolites in PO may provide new insights for preventive and therapeutic strategies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CZ conceived the study and drafted the manuscript. YW and CLZ acquired and analyzed the data. HRW conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Marcus R. Post-menopausal osteoporosis. Best Pract Res Clin Obstet Gynaecol. 2002;16:309–327. doi: 10.1053/beog.2002.0284. [DOI] [PubMed] [Google Scholar]
- 2.Seibel MJ, Dunstan CR, Zhou H, Allan CM, Handelsman DJ. Sex steroids, not FSH, influence bone mass. Cell. 2006;127(1079):1080–1081. doi: 10.1016/j.cell.2006.12.002. author reply. [DOI] [PubMed] [Google Scholar]
- 3.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao SM, Gao Y, Cheung CL, Bow CH, Lau KS, Sham PC, Tan KCB, Kung AWC. Association of CDX1 binding site of periostin gene with bone mineral density and vertebral fracture risk. Osteoporos Int. 2012;23:1877–1887. doi: 10.1007/s00198-011-1861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Xia RG. Screening and functional microarray analysis of differentially expressed genes related to osteoporosis. Genet Mol Res. 2014;13:3228–3236. doi: 10.4238/2014.April.25.8. [DOI] [PubMed] [Google Scholar]
- 6.Jemtland R, Holden M, Reppe S, Olstad OK, Reinholt FP, Gautvik VT, Refvem H, Frigessi A, Houston B, Gautvik KM. Molecular disease map of bone characterizing the postmenopausal osteoporosis phenotype. J Bone Miner Res. 2011;26:1793–1801. doi: 10.1002/jbmr.396. [DOI] [PubMed] [Google Scholar]
- 7.Xiao P, Chen Y, Jiang H, Liu YZ, Pan F, Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, et al. In vivo genome-wide expression study on human circulating B cells suggests a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J Bone Miner Res. 2008;23:644–654. doi: 10.1359/jbmr.080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S, Yin F, Cai J. Identification of crucial genes related to postmenopausal osteoporosis using gene expression profiling. Aging Clin Exp Res. 2016;28:1067–1074. doi: 10.1007/s40520-015-0509-y. [DOI] [PubMed] [Google Scholar]
- 9.Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson JK, Wilson ID. Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 12.Yao Q, Xu Y, Yang H, Shang D, Zhang C, Zhang Y, Sun Z, Shi X, Feng L, Han J, et al. Global prioritization of disease candidate metabolites based on a multi-omics composite network. Sci Rep. 2015;5:17201. doi: 10.1038/srep17201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Wishart DS. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 38 (Web Server) 2010:W71–7. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al. SMPDB 2.0: Big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(D1):D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, Beyer A, Bork P. STITCH 2: An interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38(Suppl_1):D552–D556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res 37 (Database) 2009:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul TV, Thomas N, Seshadri MS, Oommen R, Jose A, Mahendri NV. Prevalence of osteoporosis in ambulatory postmenopausal women from a semiurban region in Southern India: relationship to calcium nutrition and vitamin D status. Endocr Pract. 2008;14:665–671. doi: 10.4158/EP.14.6.665. [DOI] [PubMed] [Google Scholar]
- 22.Feng Z, Liu C, Guan X, Mor V. China's rapidly aging population creates policy challenges in shaping a viable long-term care system. Health Aff (Millwood) 2012;31:2764–2773. doi: 10.1377/hlthaff.2012.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed N, Sammons J, Khokher MA, Hassan HT. Retinoic acid suppresses interleukin 6 production in normal human osteoblasts. Cytokine. 2000;12:289–293. doi: 10.1006/cyto.1999.0588. [DOI] [PubMed] [Google Scholar]
- 24.Xu P, Guo X, Yao JF, Cai QK, Li YF. The effect and the mechanism of osteoporotic model female rats induced by retinoic acid. Orthopedic J China. 2001;10:995–998. (In Chinese) [Google Scholar]
- 25.Shiraishi A, Takeda S, Masaki T, Higuchi Y, Uchiyama Y, Kubodera N, Sato K, Ikeda K, Nakamura T, Matsumoto T, et al. Alfacalcidol inhibits bone resorption and stimulates formation in an ovariectomized rat model of osteoporosis: Distinct actions from estrogen. J Bone Miner Res. 2000;15:770–779. doi: 10.1359/jbmr.2000.15.4.770. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Xu Y, Qian Z. The changes of hepcidin in tretinoin-induced osteoporosis in rats. Jiangsu Med J. 2008;8:027. [Google Scholar]
- 27.Chen Q, Xiong Y, Wang Z, Xue L, Wang L. Osteoporosis model of rats caused by tretinoin acid and experimental study on effects of gallium salts on apoptosis, contents of DNA, molecular weight of DNA and lipid peroxidation. Zhong Guo Di Fang Bing Fang Zhi Za Zhi. 2003;19:68–70. (In Chinese) [Google Scholar]
- 28.Dai X, Xiong Y, Wang Z, Lei Y, Zuo H, Xue L. Experimental study on effect of gallium salt and estradiol on bone metabolism in tretinoin-induced osteoporosis of rats. J Xi'an Med University. 2001;3:215–217. (In Chinese) [Google Scholar]
- 29.Yu S-J, Huang XY, Wang CH, Shan WL, Zhu GX, Liu HC. Effect of low-concentration estradiol on proliferation and differentiation of osteoblasts from jaws of osteoporotic rats. Chin J Prosthodontics. 2016;1:17–21. (In Chinese) [Google Scholar]
- 30.Lindsay R, Bush TL, Grady D, Speroff L, Lobo RA. Therapeutic controversy: Estrogen replacement in menopause. J Clin Endocrinol Metab. 1996;81:3829–3838. doi: 10.1210/jcem.81.11.8923821. [DOI] [PubMed] [Google Scholar]
- 31.Haines CJ, Chung TK, Leung PC, Hsu SY, Leung DH. Calcium supplementation and bone mineral density in postmenopausal women using estrogen replacement therapy. Bone. 1995;16:529–531. doi: 10.1016/8756-3282(95)00084-Q. [DOI] [PubMed] [Google Scholar]
- 32.Nowson CA, Green RM, Hopper JL, Sherwin AJ, Young D, Kaymakci B, Guest CS, Smid M, Larkins RG, Wark JD. A co-twin study of the effect of calcium supplementation on bone density during adolescence. Osteoporos Int. 1997;7:219–225. doi: 10.1007/BF01622292. [DOI] [PubMed] [Google Scholar]
- 33.Nieves JW, Golden AL, Siris E, Kelsey JL, Lindsay R. Teenage and current calcium intake are related to bone mineral density of the hip and forearm in women aged 30–39 years. Am J Epidemiol. 1995;141:342–351. doi: 10.1093/aje/141.4.342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.