Abstract
Objective
Ubiquitination of proteins leads to their degradation by the proteasome, and is regulated by ubiquitin ligases and substrate-specific ubiquitin-specific peptidases (USPs). The ubiquitination process also plays important roles in the regulation of cell metabolism and cell cycle. Here, we found that the expression of several USPs is increased in SSc tenosynovial and skin biopsies, and we demonstrated that USP inhibition decreases TGF-β signalling in primary fibroblast cell lines.
Methods
High-density transcriptomic studies were performed using total RNA obtained from SSc tenosynovial samples. Confirmatory immunostaining experiments were performed on tenosynovial and skin samples. In vitro experiments were conducted in order to study the influence of USP modulation on responses to TGF-β stimulation.
Results
Tenosynovial biopsies from SSc patients overexpressed known disease-associated gene pathways: fibrosis, cytokines and chemokines, and Wnt/TGF-β signalling, but also several USPs. Immunohistochemistry experiments confirmed the detection of USPs in the same samples, and in SSc skin biopsies. Exposure of primary fibroblast cell lines to TGF-β induced USP gene expression. The use of a pan-USP inhibitor decreased SMAD3 phosphorylation, and expression of COL1A1, COL3A1 and fibronectin gene expression in TGF-β-stimulated fibroblasts. The effect of the USP inhibitor resulted in increased SMAD3 ubiquitination, and was blocked by a proteasome inhibitor, thereby confirming the specificity of its action.
Conclusion
Overexpression of several USPs, including USP15, amplifies fibrotic responses induced by TGF-β, and is a potential therapeutic target in SSc.
Keywords: systemic sclerosis, TGF-β, SMAD3, ubiquitin-specific peptidase
Rheumatology key messages
Ubiquitin-specific peptidases are overexpressed in SSc tenosynovial and skin biopsies.
Ubiquitin-specific peptidases 15 amplifies TGF-β stimulation of fibroblasts through inhibition of SMAD3 ubiquitination and degradation.
Ubiquitin-specific peptidases inhibition decreases collagen production by TGF-β-stimulated fibroblasts.
Introduction
SSc is a connective tissue disease affecting skin and internal organs, characterized by a triad of inflammation, vasculopathy and progressive fibrosis, due to deposition of mainly type I collagen. Out of the intricate mechanisms involved in the pathogenesis of the disease, evidence indicates that TGF-β signalling plays a central role in mediating the effects of several pro-fibrotic effectors (tribbles homologue 3, IL-6, Ubiquitin-conjugating enzyme 9 (Ubc9), thrombospondin 1, integrins) [1–9]. In addition, TGF-β is induced by hypoxia in cultured fibroblasts, an observation suggesting a role for this cytokine in linking vasculopathy and fibrosis in the disease [10, 11]. Not surprisingly, TGF-β and wingless mouse mammary tumour virus integration (Wnt) signalling are among the most prevalent pathways found in global gene expression studies performed on SSc skin biopsies [12–16]. In this perspective, modulation of TGF-β activity remains a top therapeutic target in SSc drug development [17].
We recently performed whole-body MRI studies in SSc patients, and evidenced deep connective tissue infiltrates surrounding tendons in patients with active disease, and tendon friction rubs [18]. Tenosynovitis and arthritis were also found by MRI in one-third of the patients. We performed tenosynovial biopsies in patients with clinically active tenosynovitis, in order to evaluate whether such samples would provide additional information on disease mechanisms. Here, we report that these samples are characterized by the overexpression of genes involved in fibrosis, TGF-β/Wnt signalling, and chemokines and cytokines, but also by the concurrent overexpression of several ubiquitin-specific peptidases (USPs).
Ubiquitinated proteins are addressed to proteasomes where they are degraded. The ubiquitination process is controlled by the concerted action of three (E1, E2 and E3) ubiquitin ligases, and a large number of substrate-specific USPs. In normal cells, regulation of the ubiquitination/deubiquitination balance plays a role in numerous cellular pathways, such as cell cycle, regulation of transcription and response to stress [19]. In tumours, overexpression of specific USPs is associated with increased proliferation, and USP inhibitors are currently developed for oncological purposes [20–24]. Among the USPs overexpressed in SSc tenosynovial biopsies, USP15 is known to specifically deubiquitinate SMAD3, and TGF-β Receptor 1 (TGF-βR1) [24–30]. We therefore wanted to test the hypothesis that USP15 overexpression plays a role in the pathogenesis of SSc via decreased ubiquitin-mediated degradation of proteins involved in TGF-β signalling.
Methods
Tenosynovial and skin biopsy samples
Five tenosynovial biopsies were obtained from diffuse SSc patients undergoing surgical procedures due to refractory pain and/or loss of function caused by tenosynovitis. These samples were used in high-density transcriptomic and immunohistochemistry experiments. Paired cutaneous biopsies were obtained from clinically affected and clinically normal skin from 20 patients with diffuse SSc. All patients fulfilled the LeRoy and Medsger criteria [31], and were in the active phase of the disease. Ten normal cutaneous biopsies were used as negative control (from breast and abdominal wall reduction specimens). The study was approved by the ethics committee of the Université catholique de Louvain. All subjects provided written informed consent.
High-density transcriptomic analyses
RNA was extracted from the tenosynovial biopsy samples using a NucleoSpin RNA II extraction kit (Macherey-Nagel, Düren, Germany), including DNase treatment of the samples. RNA quality was assessed using an Agilent 2100 Bioanalyzer and RNA nanochips. All samples had an RNA integrity number >7. Complementary RNA was synthetized from 1 μg total RNA, and biotin-labelled according to a standard Affymetrix procedure (GeneChip® 3’ IVT express kit, ThermoFisher Scientific, Waltham, MA, USA). GeneChip Human Genome U133 Plus 2.0 arrays were hybridized overnight at 45°C with 10 μg fragmented biotinylated complementary RNA. The slides were then washed and stained using a EukGE-WS2v5 fluidics protocol on a GeneChip Fluidics Station 450, before being scanned on a GeneChip Scanner 3000 (Affymetrix, ThermoFisher Scientific). The Affymetrix .CEL and .CHP files were deposited in the Gene Expression Omnibus of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo) and are accessible through Gene Expression Omnibus accession number GSE93698.
USP15, USP16, USP37 and USP48 immunostaining
Fresh tissue samples were fixed overnight in 10% formalin buffer at pH 7.0 and embedded in paraffin for histological and immunohistochemical analyses. Formalin-fixed, paraffin-embedded 5 μm sections were incubated for 30 min in citrate buffer (pH 5.8) at 95°C for antigen retrieval, next incubated overnight at 4°C with the following primary antibodies: anti-USP15 (1/800, NovusBio, Centennial, CO, USA), anti-USP16 (1/1500, Proteintech, Chicago, IL, USA), anti-USP37 (1/500, NovusBio) and anti-USP48 (1/1000, Proteintech). Bound antibodies were labelled with EnVision + system horseradish peroxidase-labelled polymer (Dako, Santa Clara, CA, USA). Serous ovarian tumour sections were used as positive control for USP15 and USP16, prostatic adenocarcinoma for USP37 and pancreatic adenocarcinoma for USP48.
Evaluation of optical parameters (density of collagen bundles and inflammatory cell infiltrates) was performed using a semi-quantitative score on a 0–3 scale, where 0 indicates absence, 1 weak and very focal staining, 2 moderate and focal staining, and 3 moderate in several foci. Perivascular fibroblastic densification was assessed as present or not. USP15-, USP16-, USP37- and USP48-stained slides were digitalized on a SCN400 slide scanner (Leica), and quantification of the stains was performed using FRIDA software v1.1.0 (average of the quantification of 6 pictures/slide at a 40× magnification, normalized for the surface of the nuclei).
Fibroblast cell cultures
Short-term human fibroblasts cell lines were isolated from synovial biopsies obtained in patients with OA. Cells were maintained at 37°C in a 8% CO2 atmosphere, in DMEM (ThermoFisher Scientific), supplemented with 10% fetal bovine serum, 1% sodium pyruvate (Gibco) and 1% antibiotic-antimycotic (Gibco), and used between passages 4 and 12. For the experiments themselves, cells were washed, then plated in X-VIVO 10 medium (Lonza, Basel, Switzerland) in order to avoid interferences from fetal bovine serum with responses to TGF-β.
In some experiments, cells were plated in 12-well plates, at 120 000 cells/well and cultured in duplicates in the presence or the absence of TGF-β (4 ng/ml, R&D Systems, Minneapolis, MN, USA), or dimethyloxalylglycine (DMOG) (500 μM, Sigma-Aldrich, Darmstadt, Germany). After 16 h, cells were harvested and pelleted for RNA extraction and quantitative PCR (qPCR) evaluation of USP gene expression. In other experiments, cells were incubated for 16 h with or without TGF-β (4 ng/ml), in the presence or absence of USP inhibitor NSC632839 (10 μM, LifeSensors, Malvern, PA, USA) for qPCR analyses of COL1A1, COL3A1 and fibronectin gene expression.
In order to evaluate the effects of USP inhibition on SMAD3 phosphorylation in response to TGF-β stimulation, cells were plated in 24-well plates at 30 000 cells/well in X-VIVO 10, in the presence or absence of 10 μM NSC632839, and left to adhere overnight, then stimulated with TGF-β (4 ng/ml) for 15 min before being harvested in 50 μl RIPA buffer (ThermoFisher Scientific) with 1% Halt protease and phosphatase inhibitor cocktail (ThermoFisher Scientific). In this experimental setting, SMAD3 ubiquitination was evaluated by adding 125 nM epoxomicin (Sigma-Aldrich), an inhibitor of proteasome degradation, to the cell cultures 1 h before the overnight incubation step with NSC632839, and subsequent TGF-β stimulation. Whole-cell lysates were prepared by lysing cells in cold lysis buffer (Tris-HCl 50 mM, pH 8; NaCl 150 mM; NP40 1%; EDTA 2 mM) supplemented with protease and phosphatase inhibitor cocktail tablets (PhosSTOP, Roche).
Finally, for the small interfering RNA (siRNA) experiments, cells were plated at 30 000 cells/well in 24-well plates, and transfected in duplicate wells with 50 nM USP15 or 50 nM non-targeting siRNA pools (On-Target Plus siRNA, Dharmacon, Lafayette, CO, USA), using the DharmaFECT1 siRNA Transfection Reagent, according the to the manufacturer’s protocol. After 24 h, the culture medium was switched to serum-free X-VIVO 10. After 30 h, cells were stimulated with 4 ng/ml TGF-β for 15 min, and then harvested in 50 μl RIPA buffer with 1% Halt protease and phosphatase inhibitor cocktail. All experiments were repeated three times. Due to low transfection efficiency, USP15 gene expression was never down-regulated by more than 60% after transfection of USP15 siRNA compared with non-targeting siRNA pools.
Plasmids and transfection of human embryonic kidney cells
In order to test the effects of selected USPs on SMAD3 expression and phosphorylation in response to TGF-β, we constructed USP15, USP37, USP48 and USP16 expression plasmids, as described in the supplementary materials and methods, available at Rheumatology online. Because of the low transfection efficiency of short-term human fibroblasts cell lines in the previous experiments, human embryonic kidney (HEK293) cells were used in these experiments. HEK293 cells were maintained in X-VIVO 10 serum-free medium for 24 h, then transfected in triplicate wells with 1 μg of each plasmid separately, using Lipofectamin 2000 (Invitrogen, Carlsbad, CA, USA). Negative controls were transfected with the corresponding empty plasmids. After 24 of 48 h, cells were stimulated using 4 ng/ml TGF-β for 15 min, and then harvested in 50 μl RIPA buffer (Pierce) with 1% Halt protease and phosphatase inhibitor cocktail (ThermoFisher Scientific). All experiments were repeated two or three times.
Real-time qRT-PCR
Total RNA was extracted from the fibroblast cell cultures using the NucleoSpin RNAII kit. cDNA was synthesized from 500 ng total RNA using RevertAid Moloney murine leukaemia virus RT (Fermentas, Waltham, MA, USA) and oligo(dT) primers. qPCRs and analyses were carried out on a Light Cycler 480 real-time PCR system (Roche, Basel, Switzerland) using SYBR Green PCR Master mix (BioRad, Hercules, CA, USA), as further described in the supplementary materials and methods, available at Rheumatology online.
Western blot and immunoprecipitation experiments
SMAD3, phospho-SMAD3, USP15, USP16, USP37 and USP48 western blots were performed as described in the supplementary materials and methods, available at Rheumatology online. In order to evaluate SMAD3 ubiquitination, SMAD3 was immunoprecipitated from whole-cell lysates using an anti-SMAD3 antibody (Rockland, Limerick, PA, USA) with Protein A/G UltraLink Resin (ThermoFisher Scientific). The immunoprecipitates were dissolved in 20 μl protein loading buffer (ThermoFisher Scientific), loaded on 10% Tris-glycine gels and transferred to nitrocellulose membranes as above. An anti-polyubiquitinylated conjugate (Enzo Life Sciences, diluted 1/2000 in Tris-buffered saline, 0.1% Tween 20, 5% non-fat milk) was used as primary antibody, and an anti-mouse-horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX, USA, diluted at 1/2000) as secondary antibody. After quantification of the ubiquitin signal, membranes were stripped for 5 min at room temperature in Restore PLUS Western Blot Stripping Buffer (ThermoFisher Scientific), and re-stained with an anti-SMAD3 antibody as above, in order to normalize the amount of ubiquitinated proteins to total SMAD3.
Statistical analyses
Analysis of gene expression data was performed on GeneSpring (Agilent, Santa Clara, CA, USA) after normalization by robust multi-array analysis [32]. SSc tenosynovial samples were compared with synovial biopsy samples harvested previously by our group in patients with several rheumatic conditions [33, 34]. ANOVA and t-tests were performed, using OA samples as the comparator in the latter case. GOTERM pathway analyses were done (on all transcripts overexpressed in SSc compared with OA samples, with a fold-change threshold >2) using DAVID [35, 36].
Other statistical tests were run on Prism (GraphPad, San Diego, CA, USA), using paired or unpaired Student’s t-tests, as indicated in the figure legends.
Results
In our first experiments, we performed global transcriptomic studies in tenosynovial biopsy samples (n = 5) from patients with SSc. Gene expression in these specimens was compared with synovial biopsy samples from patients with several rheumatic conditions known to induce fibroblast activation and/or proliferation (RA, OA, PsA, gout).
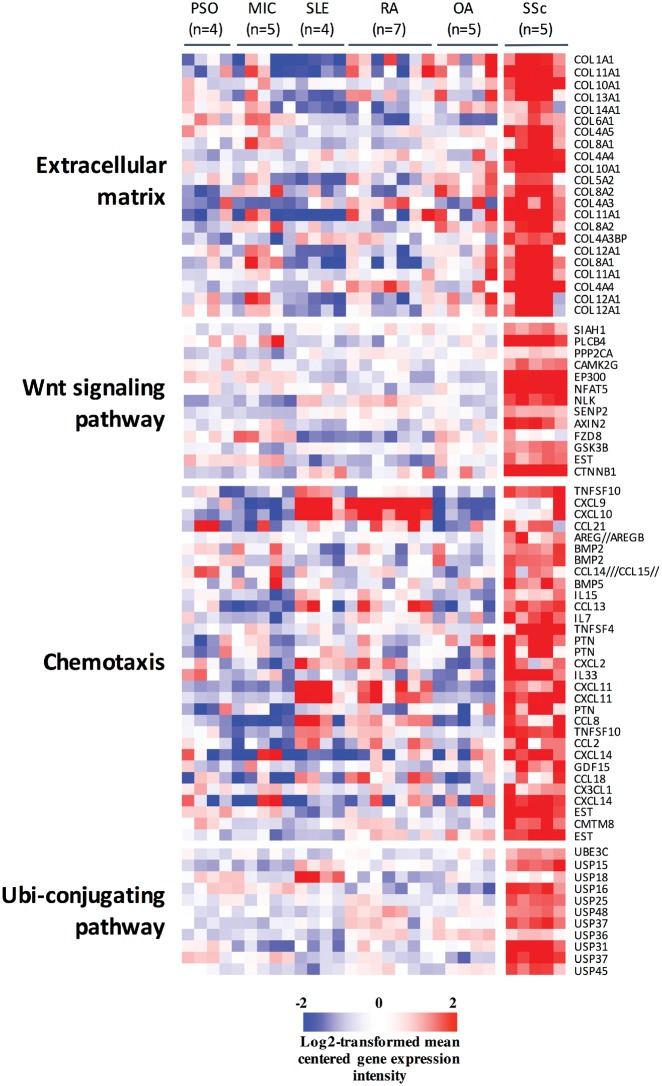
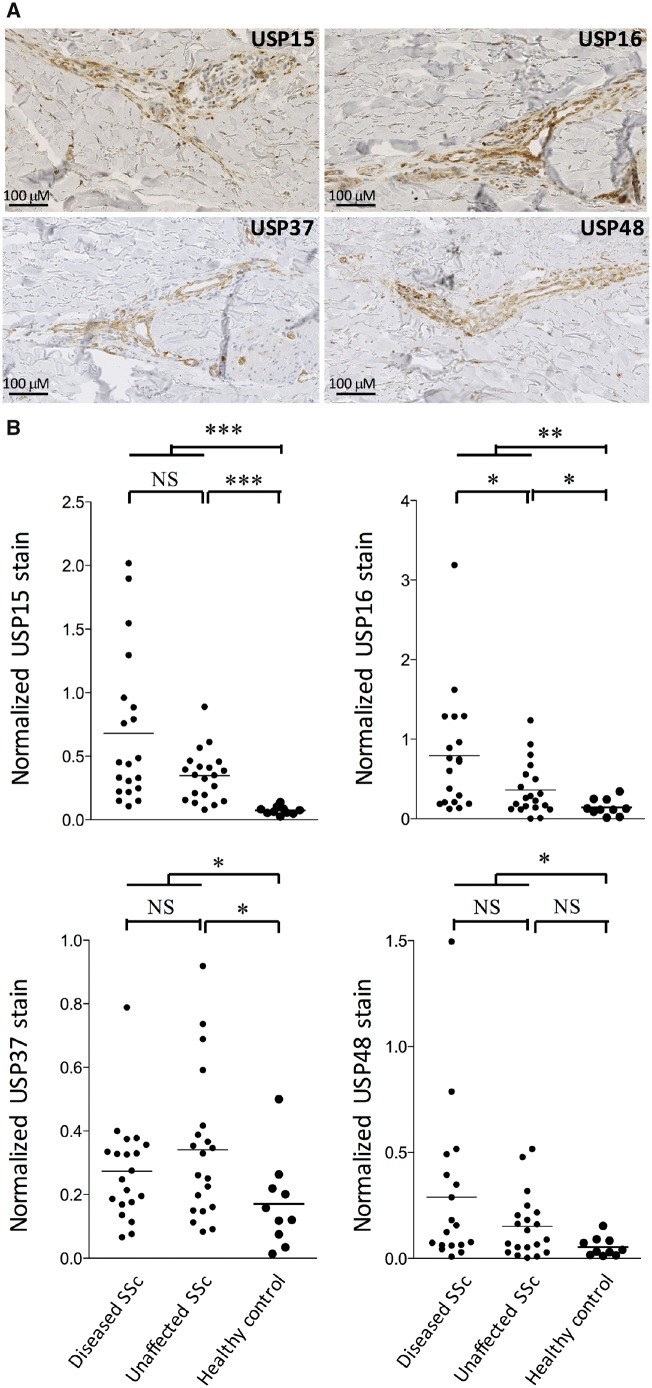
As expected, tenosynovial SSc samples were characterized by a strong overexpression of collagen transcripts, transcripts encoding chemokines and cytokines, and transcripts involved in Wnt/TGF-β signalling. Intriguingly, we also found concurrent overexpression of numerous transcripts encoding USPs in these samples: USP15, -16, -18, -25, -31, -36, -37, -45 and -48 (Fig. 1, supplementary Table S1, available at Rheumatology online). Immunohistochemistry experiments, performed on the same tenosynovial samples, confirmed the presence of USP15 in all samples, located in parts of the biopsy showing abundant collagen deposits, or around small vessels (supplementary Fig. S1, available at Rheumatology online). USP15, USP16, USP37 and USP48 stains were also performed in independent superficial paired (clinically affected and unaffected) skin biopsies obtained from patients with diffuse SSc, compared with normal skin biopsies. Of note, collagen bundles, peri-vascular fibroblasts and inflammatory cell aggregates were still observed in clinically unaffected skin samples from diffuse SSc patients, albeit significantly less than in their clinically affected counterparts (supplementary Fig. S2, available at Rheumatology online). Although the USP15, USP16, USP37 and USP48 stains were patchy (Fig. 2A), they were significantly increased in SSC affected and unaffected skin compared with control biopsies, with a gradient between affected and unaffected SSc skin specimens (Fig. 2B).
Fig. 1.
Overexpression of USPs in SSc tenosynovial biopsies
Genes differentially expressed in SSc tenosynovial compared with synovial tissue samples from patients with inflammatory joint disorders. Student’s t-tests indicated that 2549 transcripts were significantly overexpressed in SSc compared with OA samples, with a fold-change >2. Pathway analyses showed that these transcripts were significantly enriched in the indicated GOTERM pathways (all pathways and enrichment scores are listed in supplementary Table S1, available at Rheumatology online). The data are displayed as log2-transformed, mean-centered values. MIC: microcrystalline arthritis; PSO: psoriatic arthritis; USP: ubiquitin-specific peptidase.
Fig. 2.
Overexpression of USP15, USP16, USP37 and USP48 in SSc skin biopsies
Biopsies were obtained from clinically affected and unaffected skin regions in 20 patients with diffuse SSc and 10 healthy controls. (A) Characteristic images of USP15, USP16, USP37 and USP48 stains, showing positive fibroblasts, endothelial and some inflammatory cells, mainly in perivascular areas. (B) Immunoquantification show significant overexpression of the stained USPs in SSc compared with healthy skin samples. *P < 0.05; **P < 0.005; ***P < 0.0005 using Mann–Whiney tests. USP: ubiquitin-specific peptidase.
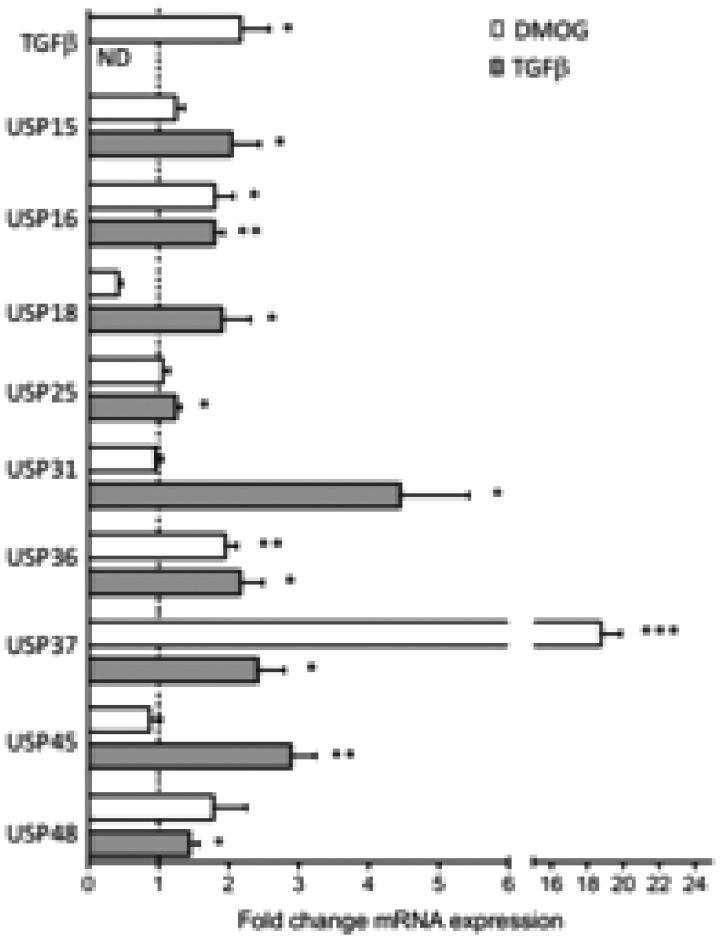
We wondered whether hypoxia or TGF-β were involved in the induction of USP gene expression. Synovial fibroblasts (from non-SSc individuals) were cultured overnight in the presence of DMOG, a hypoxia-inducible factor 1α-hydroxylase inhibitor that mimics the effects of hypoxia, or TGF-β. Our results confirmed the stimulatory effects of DMOG on TGF-β gene expression, but we did not observe consistent in vitro effects on overall USP gene expression. By contrast, TGF-β itself induced a significant overexpression of most USP transcripts tested in the panel (Fig. 3).
Fig. 3.
Regulation of USP expression by DMOG and TGF-β in fibroblast cell lines
Synovial fibroblasts (n = 6 cell lines), obtained from the knee of patients with OA, were cultured for 24 h in duplicate wells with DMOG, a hypoxia-inducible factor 1α hydroylase inhibitor, or TGF-β. Expression of the indicated USP was evaluated by quantitative PCR. Results are displayed as mean (± s.e.m.) fold-changes compared with unstimulated cells from three different experiments. *P < 0.01; **P < 0.005; ***P < 0.0001 using unpaired Student’s t-tests. Mean value for USP37 induction by DMOG, 19.1. DMOG: dimethyloxalylglycine; ND: not done; USP: ubiquitin-specific peptidase.
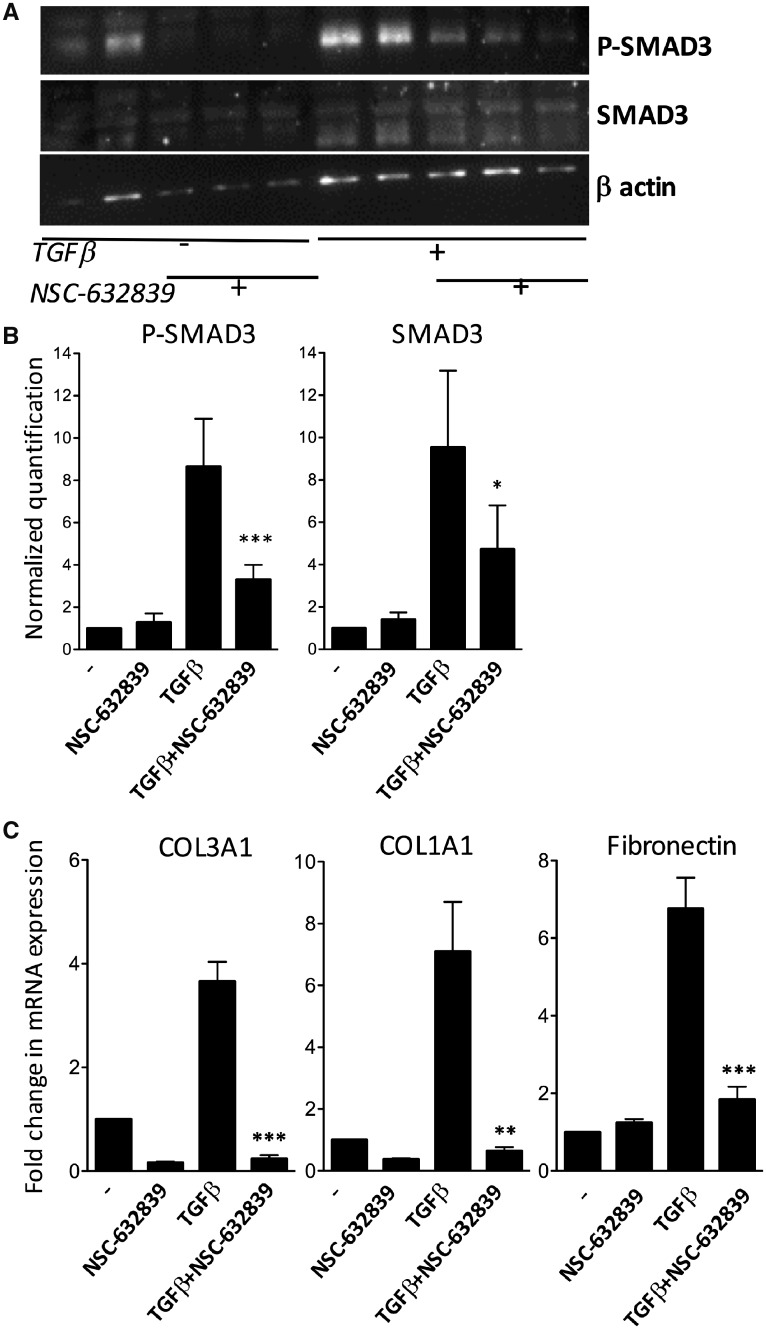
USP15 specifically deubiquitinates SMAD3 and TGF-βR1, which suggests that USP15 overexpression in SSc tissues could play a pathogenic role through increased responses to TGF-β stimulation via inhibition of ubiquitin-mediated degradation of these proteins. In order to support this hypothesis, we evaluated the in vitro effects of NSC632839, a pan-USP inhibitor, on SMAD3 phosphorylation and collagen gene expression in synovial fibroblasts exposed to TGF-β. Western blot experiments showed that NSC632839 significantly inhibited expression of total SMAD3 protein and SMAD3 phosphorylation in TGF-β-stimulated cells (Fig. 4A and B). As shown by qPCR experiments, this was paralleled by a significant decrease in COL1A1, COL3A1 and fibronectin gene expression in response to TGF-β stimulation (Fig. 4C). In order to verify that the effects of NSC632839 were mediated by modulation of the ubiquitination process, we immunoprecipitated SMAD3 from treated vs untreated cells. We found that the use of NSC632839 in TGF-β-stimulated fibroblasts is associated with increased SMAD3 ubiquitination (Fig. 5A and B). Additional experiments indicated that the inhibitory effects of NSC632839 on SMAD3 phosphorylation were not observed in the presence of epoxomicin, a proteasome inhibitor (Fig. 5C and D), thereby indicating that the mode of action of NSC632839 involves increased ubiquitination and proteasomal degradation of SMAD3.
Fig. 4.
Effects of USP inhibition on SMAD3 phosphorylation and collagen gene expression in TGF-β-stimulated fibroblasts
(A, B) Total and phospho-SMAD3 western blots in unstimulated or TGF-β-stimulated synovial fibroblasts, cultured in multiplicate wells in the presence or the absence of NSC632839, a pan-USP inhibitor. (A) Representative experiment. (B) β-actin-normalized SMAD3 and phospho-SMAD3 quantification in four different experiments. P-values were calculated using paired Student’s t-tests. *P < 0.01; ***P < 0.0001. (C) Quantitative PCR evaluation of COL1A1, COL3A1 and fibronectin (FN1) gene expression in unstimulated or TGF-β-stimulated synovial fibroblasts, cultured in duplicate wells in the presence or the absence of NSC632839. Results are displayed as mean (± s.e.m.) fold-changes in β2-microglobulin-normalized gene expression compared with unstimulated cells, from four different experiments. P-values were calculated using paired Student’s t-tests. **P < 0.005; ***P < 0.0001. USP: ubiquitin-specific peptidase.
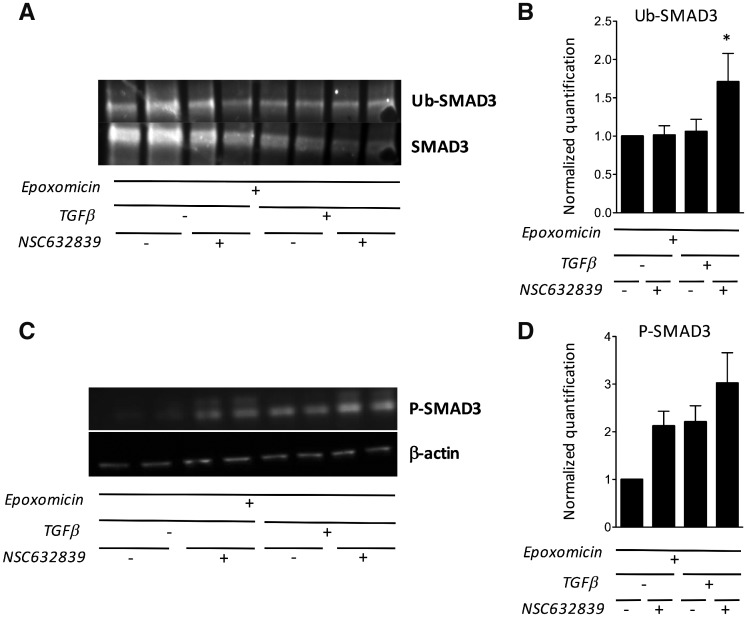
Fig. 5.
NSC632839 induces SMAD3 ubiquitination, and its effects are proteasome-dependent
(A, B) Total SMAD3 was immunoprecipitated from unstimulated or TGF-β-stimulated synovial fibroblasts, cultured in duplicate wells in the presence or the absence of NSC632839. All cells were pre-incubated with epoxomicin, in order to inhibit proteosomal degradation of ubiquitinated SMAD3. The presence of ubiquitinated chains was studied by western blot (Ub-SMAD3) and normalized for total SMAD3 in the immunoprecipitates. (A) Representative experiment. (B) Total SMAD3-normalized quantification of SMAD3 ubiquitination. The results are displayed as the mean (± s.e.m.) from three different experiments. *P < 0.05 using unpaired Student’s t-tests. (C, D) Total and phospho-SMAD3 western blots (C) and β-actin-normalized quantification (D) in unstimulated or TGF-β-stimulated synovial fibroblasts, pre-incubated with epoxomicin, and cultured in duplicate wells in the presence or the absence of NSC632839. Results are displayed as the mean (± s.e.m.) values from three different experiments.
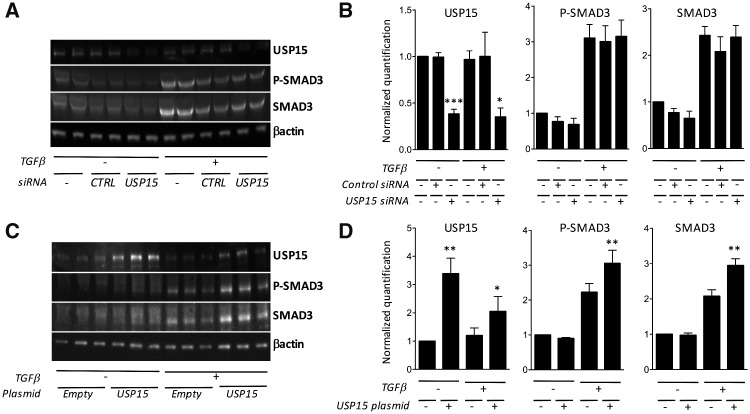
We finally transfected USP15 siRNAs in synovial fibroblasts. In these cells, USP15 inhibition using siRNAs was not able to down-regulate SMAD3 phosphorylation in response to TGF-β stimulation (Fig. 6A and B). Although the absence of noticeable effects in the presence of USP15 siRNAs is probably due to resistance of fibroblasts to transfection and incomplete USP15 inhibition, these results prompted us to test the possibility that other USPs, in particular those overexpressed in SSc tenosynovial biopsies, are involved in regulation of TGF-β responses. In order to address this question, we transfected expression plasmids in HEK293 cells (rather than in synovial fibroblasts, given their resistance to transfection). As expected, USP15 overexpression in HEK293 cells induced a significant increase in TGF-β-induced SMAD3 phosphorylation and total SMAD3 (Fig. 6C and D). However, we could not demonstrate any significant effect of USP16, USP37, USP45 or USP48 on SMAD3 phosphorylation in HEK293 cells exposed to TGF-β (data not shown).
Fig. 6.
Effects of USP15 overexpression or inhibition on TGF-β-induced SMAD3 phosphorylation in vitro
(A, B) Human synovial fibroblasts were left non-transfected, or transfected with control or USP15 siRNA, and then stimulated with TGF-β. (A) Representative USP15, phospho-SMAD3 and total SMAD3 western blot. (B) β-actin-normalized USP15, total SMAD3 and phospho-SMAD3 quantification in two different experiments. P-values were calculated using unpaired Student’s t-tests. *P < 0.01, ***P < 0.0001. (C, D) Human embryonic kidney (HEK293) cells were transfected with a control or USP15-encoding plasmid, and stimulated in duplicate wells with TGF-β. (C) Representative USP15, phospho-SMAD3 and total SMAD3 western blot. (D) β-actin-normalized USP15, total SMAD3 and phospho-SMAD3 quantification in three different experiments. P-values were calculated using unpaired Student’s t-tests. **P < 0.005. USP: ubiquitin-specific peptidase.
Discussion
We found that SSc tenosynovial biopsies are characterized by the overexpression of transcripts involved in fibrosis, TGF-β/Wnt signalling, and chemokines and cytokine, but also transcripts encoding several USPs. In particular, USP15 is overexpressed in SSc tenosynovial, but also in skin biopsy specimens. USP15, a TGF-β-induced gene, amplifies TGF-β signalling, and in vitro USP inhibition using a pharmacological inhibitor decreases collagen and fibronectin gene expression by TGF-β-stimulated fibroblasts.
Several reports described gene expression profiles in SSc skin biopsies and found consistent results, i.e. overexpression of transcripts involved in fibrosis, inflammation and proliferation, with a high level of sample-to-sample variations in the amplitude of the changes for each of these pathways [12–16]. Overexpression of fibrosis- and TGF-β/Wnt signalling-associated transcripts in our samples is in line with these previous observations, and is correlative to the significant levels of fibrosis observed on histological examination of the biopsies. Our samples were not characterized by the overexpression of numerous transcripts found in inflammatory or immune cells, but this is because the comparators we used were synovial biopsies from patients with rheumatic disorders, themselves characterized by fluctuating levels of expression of these transcripts. Yet, overexpression of several chemokines (CXCL9, CXCL10, CCL2, CCL13, CCL18) and cytokines (IL-7, IL-15, IL-33, TNFSF4/OX40L) was observed in the SSc samples, pointing at specific patterns of immune cell recruitment and activation, known to be involved in the pathogenesis of the disease [37, 38].
Increased expression of several USP-encoding transcripts in our samples is a novel finding, of potential interest for the pathogenesis of the disease in view of the known role of USP15 on SMAD3 and TGF-βR1 deubiquitination. Immunohistochemistry experiments confirmed overexpression of USP15 not only in tenosynovial biopsy fragments, but also in superficial biopsies obtained from diseased skin areas of patients with diffuse SSc. In these samples, the patchy staining pattern probably explains why overexpression of USP15 fails to be detected in global gene expression approaches performed on superficial SSc skin biopsies. Of note, the control skin samples used in the latter confirmatory experiments were not obtained from the arm but from breast and abdominal reduction specimens. Gene expression profiles in normal fibroblasts is influenced by their anatomical location (changes in expression of transcripts involved in positional identity, cell fate determination, cell migration, extracellular matrix remodelling). Yet, there is no indication that ubiquitin-related transcripts belong to these pathways in normal skin [39]. In SSc skin samples, it should also be noted that the predominant perivascular stain opens the possibility that USP15 is overexpressed in heterogeneous cell populations, including endothelial cells.
It is not obvious why USP overexpression in SSc tissues displays a gradient of expression from skin to tenosynovium. Differences in tissue oxygenation between skin and tendon sheets is a plausible feature of the disease, in view of the dense fibrotic changes observed in the latter samples, and it was therefore tempting to look at the effects of hypoxia on induction of USP gene expression. However, we could not evidence any consistent effect of hypoxia-inducible factor 1α inhibition on USP expression by cultured fibroblasts. In contrast, TGF-β did display significant effects on USP expression in fibroblast cell cultures, and the presence of active TGF-β in SSc teguments is well documented. Yet, there is no formal evidence of locoregional variations in bioactive TGF-β concentrations in the affected skin of patients with the disease. Our group did show the presence of focal deep connective tissue infiltrates by whole-body MRI imaging of SSc patients [18], which might translate into local biological variations, involving TGF-β or other cytokines with potential effects on USP expression, but additional work is needed in order to confirm this hypothesis.
Our experiments indicate that USP15 expression in SSc fibroblasts is part of an amplification loop leading to increased responses to TGF-β through inhibition of SMAD3 phosphorylation. As such, these results were expected, given the known effects of USP15 on SMAD3 and TGF-βR1 deubiquitination. Yet, our work, by demonstrating the significant effect of pharmacological USP inhibition on the production of collagen genes by TFGβ-stimulated fibroblasts, points at potential therapeutic developments in the modulation of fibrosis, at least in tenosynovial structures (considering that our ex vivo experiments were performed on synovial fibroblasts). Additional experiments in pre-clinical models of the disease will be needed in order to further substantiate the role of USP15 in dermal or lung fibrosis. Of note, USP15 is not the only USP overexpressed in SSc samples, and it is possible that several of the other overexpressed USPs also play a role in disease-associated mechanisms. According to in silico prediction algorithms, USP45 would have the ability to deubiquinate SMAD3 as well. However, we could not find any significant effect of USP45 overexpression on SMAD3 phosphorylation in response to TGF-β stimulation in HEK293 cells, and this was also the case for other USPs tested: USP16, USP37 and USP48. Whether not only TGF-β signalling but also additional cellular processes are impacted by overexpression of USPs in SSc tissues needs to be further studied using proteome ubiquitination (‘ubiquitome’) assessment tools.
Our observations are not the first documentation of the presence of amplificatory mechanisms leading to increased response to TGF-β in SSc tissues. Thus, Khodzhigorova et al. [6] evidenced a significant increase in sumoylated proteins in SSc skin compared with control biopsies. Addition of Small Ubiquitin-like MOdifyer-1 tags is a post-translational modification of proteins mediated by the Small Ubiquitin-like MOdifyer E2-conjugating enzyme Ubc9, and the authors found that in vivo Ubc9 inhibition using siRNA led to decreased fibrosis and decreased nuclear accumulation of phosphorylated SMAD3 in two mouse models of the disease (bleomycin-induced dermal fibrosis, and fibrosis induced by overexpression of a constitutively active TGF-βR1). Overall, their results indicate that increased sumoylation in SSc may contribute to the pathogenesis of SSc through mechanisms involving TGF-β/SMAD signalling. Similarly, Bhattacharyya et al. [40] modulated the expression of A20 in cultured skin fibroblasts. A20 is a ubiquitin-editing enzyme with both E3 ubiquitin ligase and deubiquitinating functions, and its expression is down-regulated by TGF-β in fibroblast cell lines. In vitro A20 inhibition using siRNA led to increased TGF-β-induced COL1A1 and COL1A2 gene expression, and myofibroblast differentiation. Conversely, overexpression of A20 led to decreased collagen and α-smooth muscle actin gene expression, and decreased myofibroblast differentiation in response to TGF-β stimulation, together with decreased TGF-β-induced SMAD2 phosphorylation and nuclear localization.
In conclusion, the strong in vitro decrease in collagen and fibronectin gene expression by TGF-β-stimulated fibroblasts after exposure to an USP inhibitor illustrates the regulatory role of protein ubiquitination/deubiquitination on TGF-β signalling and fibroblast effector functions. Overexpression of several USPs in SSc tenosynovial biopsies suggests that this mechanism also operates in vivo, and further investigations will indicate whether USP modulation has the potential to translate in new therapeutic options in this severe disease.
Funding: This work was supported in part by grants from FRSR (Fonds de Recherche Scientifique en Rhumatologie), the Fonds National de la Recherche Scientifique (Communauté française de Belgique) and the Association sclérodermique de France (ASF).
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Lafyatis R. Transforming growth factor β—at the centre of systemic sclerosis. Nat Rev Rheumatol 2014;10:706–19. [DOI] [PubMed] [Google Scholar]
- 2. Varga J, Abraham D.. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest 2007;117:557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hua F, Mu R, Liu J. et al. TRB3 interacts with SMAD3 promoting tumor cell migration and invasion. J Cell Sci 2011;124:3235–46. [DOI] [PubMed] [Google Scholar]
- 4. Zhang L, Zhang J, Liu X, Liu S, Tian J.. Tribbles 3 regulates the fibrosis cytokine TGF-β1 through ERK1/2-MAPK signaling pathway in diabetic nephropathy. J Immunol Res 2014;2014:240496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mimura Y, Ihn H, Jinnin M. et al. Constitutive thrombospondin-1 overexpression contributes to autocrine transforming growth factor-beta signaling in cultured scleroderma fibroblasts. Am J Pathol 2005;166:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khodzhigorova A, Distler A, Lang V. et al. Inhibition of sumoylation prevents experimental fibrosis. Ann Rheum Dis 2012;71:1904–8. [DOI] [PubMed] [Google Scholar]
- 7. Asano Y, Ihn H, Yamane K. et al. Involvement of alphavbeta5 integrin-mediated activation of latent transforming growth factor beta1 in autocrine transforming growth factor beta signaling in systemic sclerosis fibroblasts. Arthritis Rheum 2005;168:3088–98. [DOI] [PubMed] [Google Scholar]
- 8. Asano Y, Ihn H, Yamane K. et al. Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-β signaling in scleroderma fibroblasts. J Immunol 2005;52:2897–905. [DOI] [PubMed] [Google Scholar]
- 9. Kawaguchi Y, Hara M, Wright TM.. Endogenous IL-1alpha from systemic sclerosis fibroblasts induces IL-6 and PDGF-A. J Clin Invest 1999;103:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beyer C, Schett G, Gay S, Distler O, Distler JHW.. Hypoxia in the pathogenesis of systemic sclerosis. Arthritis Res Ther 2009;11:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Distler JH, Jüngel A, Pileckyte M, et al. Hypoxia-induced increase in the production of extracellular matrix proteins in systemic sclerosis. Arthritis Rheum 2007;56:4203–15. [DOI] [PubMed] [Google Scholar]
- 12. Wei J, Fang F, Lam AP. et al. Wnt/β-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells. Arthritis Rheum 2012;64:2734–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Milano A, Pendergrass SA, Sargent JL. et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One 2008;3:e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pendergrass SA, Lemaire R, Francis I. et al. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol 2012;132:1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sargent JL, Milano A, Bhattacharyya S. et al. A TGFβ-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol 2010;130:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rice LM, Ziemek J, Stratton EA. et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol 2015;67:3004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rice LM, Padilla CM, McLaughlin SR. et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest 2015;125:2795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stoenoiu MS, Houssiau F, Lecouvet F.. Tendon friction rubs in systemic sclerosis: a possible explanation – an ultrasound and magnetic resonance imaging study. Rheumatology (Oxford) 2013;52:529–33. [DOI] [PubMed] [Google Scholar]
- 19. Nijman SMB, Luna-Vargas MPA, Velds A. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005;123:772–86. [DOI] [PubMed] [Google Scholar]
- 20. Ramakrishna S, Suresh B, Baek KH.. The role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci 2011;68:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luise C, Capra M, Donzelli M. et al. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLos One 2011;6:e15891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allende-Vega N, Saville MK.. Targeting the ubiquitin-proteasome system to activate wild-type p53 for cancer therapy. Semin Cancer Biol 2010;20:29–39. [DOI] [PubMed] [Google Scholar]
- 23. Hoeller D, Dikic I.. Targeting the ubiquitin system in cancer therapy. Nature 2009;456:438–44. [DOI] [PubMed] [Google Scholar]
- 24. Eichhorn PJ, Rodón L, Gonzàlez-Juncà A. et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med 2012;18:429–35. [DOI] [PubMed] [Google Scholar]
- 25. Herhaus L, Sapkota GP.. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFβ and BMP pathways. Cell Signal 2014;26:2186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Izzi L, Attisano L.. Regulation of the TGFβ signalling pathway by ubiquitin-mediated degradation. Oncogene 2004;23:2071–8. [DOI] [PubMed] [Google Scholar]
- 27. Dupont S, Inui M, Newfeld J.. Regulation of TGFβ signal transduction by mono- and deubiquitylation of Smads. FEBS Lett 2012;586:1913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inoue Y, Imamura T.. Regulation of TGF-β family signaling by E3 ubiquitin ligases. Cancer Sci 2008;99:2107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aggarwal K, Massagué J.. Ubiquitin removal in the TGFβ pathway. Nat Cell Biol 2012;14:656–7. [DOI] [PubMed] [Google Scholar]
- 30. Inui M, Manfrin A, Mamidi A. et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 2011;13:1368–75. [DOI] [PubMed] [Google Scholar]
- 31. LeRoy EC, Medsger TA Jr.. Criteria for the classification of early systemic sclerosis. J Rheumatol 2001;8:1573–6. [PubMed] [Google Scholar]
- 32. Irizarry R, Hobbs B, Collin F. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 33. Nzeusseu Toukap A, Galant C, Theate I. et al. Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum 2007;56:1579–88. [DOI] [PubMed] [Google Scholar]
- 34. Lauwerys BR, Hernández-Lobato D, Gramme P. et al. Heterogeneity of synovial molecular patterns in patients with arthritis. PLoS One 2015;10:e0122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 36. Huang D. W, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acid Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Antonelli A, Fallahi P, Ferrari SM. et al. Systemic sclerosis fibroblasts show specific alterations of interferon-γ and tumor necrosis factor-α-induced modulation of interleukin 6 and chemokine ligand 2. J Rheumatol 2012;39:979–85. [DOI] [PubMed] [Google Scholar]
- 38. Chizzolini C, Brembilla NC, Montanari E, Truchetet ME.. Fibrosis and immune dysregulation in systemic sclerosis. Autoimmunity Rev 2011;10:276–81. [DOI] [PubMed] [Google Scholar]
- 39. Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY.. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2006;2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhattacharyya S, Wang W, Graham LV, Varga J.. A20 suppresses canonical Smad-dependent fibroblast activation: novel function for an endogenous inflammatory modulator. Arthritis Res Ther 2016;18:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.