Abstract
Chromosomal aneuploidy is a defining feature of carcinomas and results in tumor-entity specific genomic imbalances. For instance, most sporadic colorectal carcinomas carry extra copies of chromosome 7, an aneuploidy that emerges already in premalignant adenomas, and is maintained throughout tumor progression and in derived cell lines. A comprehensive understanding on how chromosomal aneuploidy affects nuclear organization and gene expression, i.e., the nucleome, remains elusive. We now analyzed a cell line established from healthy colon mucosa with a normal karyotype (46,XY) and its isogenic derived cell line that acquired an extra copy of chromosome 7 as its sole anomaly (47,XY,+7). We studied structure/function relationships consequent to aneuploidization using genome-wide chromosome conformation capture (Hi-C), RNA sequencing and protein profiling. The gain of chromosome 7 resulted in an increase of transcript levels of resident genes as well as genome-wide gene and protein expression changes. The Hi-C analysis showed that the extra copy of chromosome 7 is reflected in more interchromosomal contacts between the triploid chromosomes. Chromatin organization changes are observed genome-wide, as determined by changes in A/B compartmentalization and topologically associating domain (TAD) boundaries. Most notably, chromosome 4 shows a profound loss of chromatin organization, and chromosome 14 contains a large A/B compartment switch region, concurrent with resident gene expression changes. No changes to the nuclear position of the additional chromosome 7 territory were observed when measuring distances of chromosome painting probes by interphase FISH. Genome and protein data showed enrichment in signaling pathways crucial for malignant transformation, such as the HGF/MET-axis. We conclude that a specific chromosomal aneuploidy has profound impact on nuclear structure and function, both locally and genome-wide. Our study provides a benchmark for the analysis of cancer nucleomes with complex karyotypes.
Introduction
Abnormal numbers of whole chromosomes or chromosome arms, so called chromosomal aneuploidy, are a common feature of solid tumors [1], [2]. Complex karyotypes with multiple gains and losses of whole chromosomes and/or chromosome arms are commonly found in advanced malignancies [3], [4], [5], [6], [7], [8]. Chromosomal aneuploidies, and the ensuing genomic imbalances are tumor-type specific. Colorectal carcinomas (CRCs), for instance, are defined by gains of chromosomes and/or chromosome arms 1, 7, 8q, 13, and 20q as well as losses of 4, 8p, 17p and 18q [9]. Premalignant polyps frequently show whole chromosome 7 gains as one of the earliest genome mutation [9], [10], [11], [12]. However, the implication of this trisomy for subsequent cancer development remains elusive [13].
Roig et al. established a karyotypically normal (46, XY) human colonic epithelial cell line (HCEC) from non-cancerous colon tissue of a patient undergoing routine surveillance colonoscopy by immortalization with Cdk4 and hTERT transduction [14]. These cells maintain their diploid karyotype under long-term propagation in culture conditions with 2% serum and do not show a malignant phenotype. However, long-term propagation (~ 40 population doublings) of HCEC cells under serum-free culture conditions gave rise to cells with trisomy 7, which was the only cytogenetic aberration [15]. Trisomy 7 cells (HCEC+7) are impaired in cellular fitness, i.e., proliferation rate, colony formation and migration, compared to their normal diploid counterparts under optimal culture conditions. However, the HCEC+7 cells had a growth advantage under serum-deprived culture conditions. Whole exome sequencing of diploid HCEC and trisomic HCEC+7 cells identified 240 HCEC+7-specific mutated genes, suggesting that the gain of chromosome 7 induces the acquisition of mutations, which could potentially facilitate the initiation and/or progression of colorectal tumorigenesis [16].
A comprehensive understanding of complex biological systems depends on recognizing its structure–function relationships from molecules to the entire system. Such efforts form the basis for the NIH Common Fund Initiative, the 4D Nucleome (https://commonfund.nih.gov/4Dnucleome), which is aimed to elucidate how nuclear organization affects the cellular transcriptome and the phenotype of cells [17]. We have previously used Hi-C and RNA-seq to study structure/function relationships in the CRC cell line HT-29, a cell line that harbors multiple structural and numerical chromosomal aberrations which led to profound changes in genome structure and function. We could show that chromosome conformation capture identifies chromosomal aberrations at high resolution, and that these aberrations alter the relationship between structure and function [18]. We now use a model system that consists of a matched pair of isogenic HCEC and HCEC+7 cells to dissect the consequences of a single chromosomal aneuploidy on genome architecture and function. We analyzed the genome, transcriptome and proteome using Hi-C, RNAseq and multiplex fluorescent two-dimensional gel-electrophoresis (2-DIGE) with subsequent mass spectrometry. We chose trisomy of chromosome 7 as a model, because it occurs with high frequency in colorectal adenomas and must be considered as a “point of no return” in the progression towards malignancy.
Materials and Methods
Cell Lines and Tissue Culture
Cell lines HCEC 1CT and HCEC 1CT+ 7 were kindly provided by Dr. Jerry W. Shay (University of Texas Southwestern Medical Center, Dallas, TX) and cultured as reported elsewhere [15]. Briefly, cells were propagated in 4:1 high-glucose Dulbecco's Modified Eagle Medium (DMEM)/ Medium 199 (Life Technologies, Carlsbad, CA) uploidy: instigator and inhibitor of tumorigemented with 20 ng/ml epidermal growth factor (Life Technologies), 1 mg/ml hydrocortisone, 10 mg/ml insulin, 2 mg/ml transferrin and 5 nM sodium selenite (all Sigma, St Louis, MO) under 2% oxygen and 5% carbon dioxide. 2% cosmic calf serum (Hyclone, Logan, UT) were added if appropriate. Cells were routinely checked for mycoplasma contamination by PCR (MycoScope PCR Detection Kit, Genlantis, San Diego, CA). Cell line authentication was verified by short tandem repeat (STR) profiling using the AmpFLSTR™ Identifiler™ PCR Amplification Kit (Thermo Fisher, Waltham, MA) according to the manufacturer's instructions.
Array-CGH
DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) as instructed by the manufacturer. One microgram of sample DNA and sex-matched reference DNA was labeled with Cy3 or Cy5, respectively, using the SureTag DNA Labeling Kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions (protocol version 7.3). Labeled DNA was subsequently hybridized on SurePrint G3 human 4x 180 K CGH arrays (Agilent) for 24 h. After washing, slides were processed on an Agilent G2565BA scanner. Agilent Technologies' Feature Extraction (version 10.7.3.1) was used for data quality control and extraction. Data were visualized and analyzed using Nexus 7.5.
Spectral Karyotyping (SKY)
Preparation of metaphase chromosome suspension, spectral karyotyping (SKY), slide pretreatment, slide denaturation, detection and imaging were conducted as previously described [19].
Multiplex Fluorescence In Situ Hybridization (FISH)
Bacterial artificial chromosome contigs consisting of three to four overlapping clones were assembled targeting the following genes: COX2 (1q31.1), TERC (3q26), APC (5q22), EGFR (7p11), MYC (8q24.21), CCND1 (11q13.3), CDX2 (13q12), CDH1 (16q22.1), TP53 (17p13.1), HER2 (17q12), SMAD4 (18q21), ZNF217 (20q13.2). FISH probes were chosen based on tumor suppressors and oncogenes known to be involved in CRC. The probes were combined into three FISH panels (panel 1: TERC-COX2-APC-EGFR, panel 2: CDH1-HER2-TP53-ZNF217, panel 3: CDX2-CCND1-SMAD4-MYC). The miFISH procedure was performed as previously described [20]. A total of 500 nuclei were analyzed for each cell line. Images were automatically acquired with a fluorescence microscope and a 40x oil immersion objective (BX63, Olympus, Tokyo, Japan) equipped with custom optical filters (Chroma, Bellow Falls, VT, USA) with a motorized stage and custom scanning and analysis software (BioView, Rehovot, Israel).
3D-Fluorescence In Situ Hybridization (3D-FISH)
Whole chromosome painting probes (WCP) of chromosome 7 and 19 were prepared from flow-sorted chromosomes. After culturing, the cells were washed in PBS and fixed in 4% paraformaldehyde (PFA) for 10 min at RT. Then, cells were rinsed in 0.05% Triton X-100 and permeabilized in 0.5% Triton X-100 for 20 min, followed by incubation in 20% glycerol for at least 1.5 h before cycles of freezing and thawing in liquid nitrogen. The cells were incubated in HCl for 10–30 min, washed in 2xSSC and incubated in RNase A for 1 h at 37°C. Finally, cells were incubated in 50% formamide/2xSSC at RT at least 5 days before hybridization or stored in the same solution at 4°C. Cells and probes were co-denaturated at 72°C for 5 min and hybridized at 37°C for 48 h.
Images were acquired using a Delta Vision Elite High Resolution Microscope (GE Healthcare Life Sciences), with a 63× oil immersion objective. Images were taken in 896×896 size and 16-bit. Voxel size was 0 .2 μm × 0 .2 μm × 0.1 08 μm. Image deconvolution was done using the Delta Vision software.
FISH Image Analysis
Nuclei were extracted and analyzed semi-automatically through our MATLAB image processing pipeline. Nuclei boundaries were detected via Marker-Controlled Watershed Segmentation, following methods modified from (https://www.mathworks.com/help/images/marker-controlled-watershed-segmentation.html). The z-dimension provided little additional spatial resolution, so maximum projection of the images was analyzed. DAPI channel images were converted to grayscale and the image gradient magnitude was computed to determine nuclei borders. The original grayscale images were eroded then dilated to remove noise and create a flat regional maximum within nuclei. Gradient boundaries enclosing regional maxima determined where nuclei were found. The convex hull of the gradient boundaries defined the nuclei shape.
Each nucleus was analyzed individually, and chromosome territories (CTs) were automatically extracted using a method similar to that of nuclei extraction, but within each defined nucleus region. For each fluorescent channel (containing CT 19 and 7), the image was passed through a Gaussian filter and binarized. Images were then dilated and eroded to reduce noise. Connected components reflecting CTs were manually selected. The CT area, CT centroid distance to nuclear periphery and the CT centroid distance to other centroids were then calculated automatically. For HCEC+7, only nuclei that had 3 copies of chromosome 7 were kept. Forty-two cells were used for HCEC, 41 cells were used for HCEC+7.
RNA Sequencing and Data Analysis
RNA was extracted from three subsequent passages of each cell line using the RNeasy Mini Kit and on-column DNAse treatment (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA integrity was checked using the 2100 Bioanalyzer (Agilent, Santa Clara, CA). Ribosomal RNA (rRNA) was removed using biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads according to the Illumina total RNA sequencing protocol. The RNA was fragmented and the cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers, followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. The resulting double-strand cDNA was used as the input to a standard Illumina library prep with end-repair, adapter ligation and PCR. The final purified product was quantitated by qPCR. Samples were sequenced on a HiSeq2500 using Illumina TruSeq v4 chemistry with 125 bp paired-end.
Reads were aligned to reference genome hg19 using STAR [21]. Gene expression was quantified using RSEM [22]. Alignment parameters were set based on RSEM default parameters, “rsem-calculate-expression” “--star”. Transcripts per million (TPM) was used to quantify gene expression. TPM expression was binned into 100 kb regions for comparison with Hi-C matrix dimensions. To achieve this, the sum of all gene-level TPMs was calculated for all genes within each 100 kb region. Regions 100 kb that contain only a proportion of a gene (i.e. the gene spans multiple 100 kb regions) received a TPM value proportional to how much the gene overlaps that region.
Differential expression was determined following methods described in [23]. P values for differential expression were adjusted for false discovery following methods outlined in [24]. GSAASeqSP was used for Gene Set Analysis of RNA-seq data [25]. Raw counts were input to GSAASeqSP in the form of a .gct file, and the MSigDB hallmark gene sets were analyzed [26]. The following GSAASeqSP parameters were used: “Permutation type: gene_set”, “Metric for gene set analysis: Weighted_KS”, “Metric for differential expression analysis: Signal2Noise”, “Association statistic: weighted”, and “P value threshold: 0.05”.
Crosslinking of Cells for Hi-C
Cells were trypsinized, washed with PBS, re-suspended in 10 mL PBS containing 1% formaldehyde and incubated for 10 min at room temperature on a rocking platform. To quench the crosslinking reaction, glycine of 2.5 M was added to a final concentration of 0.2 M, and incubated for 5 min at room temperature on a rocking platform, then on ice for at least 15 min to stop crosslinking completely. Cells were collected by centrifugation at 800×g for 10 min at 4°C, washed in 1 ml ice-cold PBS, and spun down at 800×g for 10 min at 4°C. The supernatant was discarded, cells were snap-frozen in liquid nitrogen and stored at −80°C for Hi-C library construction.
Generation of Hi-C libraries for sequencing
The in situ Hi-C protocols from Rao et al. [27] were adapted with slight modifications. For each Hi-C library, approximate 3 x 106 cells were incubated in 250 μl of ice-cold Hi-C lysis buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% Igepal CA630) with 50 μl of protease inhibitors (Sigma) on ice for 30 min and washed with 250 μl lysis buffer. The nuclei were pelleted by centrifugation at 2500×g for 5 min at 4°C, re-suspended in 50 μl of 0.5% sodium dodecyl sulfate (SDS) and incubated at 62°C for 10 min. Afterwards 145 μl of water and 25 μl of 10% Triton X-100 (Sigma) were added and incubated at 37°C for 15 min.
Chromatin was digested with 100 units of restriction enzyme MboI (NEB) overnight at 37°C with rotation. Chromatin end overhangs were filled in and marked with biotin-14-dATP (Thermo Fisher Scientific) by adding the following components to the reaction: 37.5μl of 0 .4mM biotin-14-dATP (Life Technologies), 1 .5μl of 10 mM dCTP, 1 .5μl of 10 mM dGTP, 1 .5μl of 10 mM dTTP, and 8 μl of 5 U/μl DNA Polymerase I, Large (Klenow) Fragment (NEB). The marked chromatin ends were ligated by adding 900 μl of ligation master mix consisting of 663 μl of water, 120 μl of 10X NEB T4 DNA ligase buffer (NEB), 100 μl of 10% Triton X-100, 12 μl of 10 mg/ml BSA, 5 μl of 400 U/ μl T4 DNA Ligase (NEB), and incubated at room temperature for 4 h.
Chromatin decross-linking was performed by adding 50 μl of 20 mg/ml proteinase K (NEB) and 120 μl of 10% SDS and incubated at 55°C for 30 min, adding 130 μl of 5 M sodium chloride and incubate at 68°C overnight. DNA was precipitated with ethanol, washed with 70% ethanol, and dissolved in 105 μl of 10 mM Tris–HCl, pH 8. DNA was sheared on a Covaris S2 sonicator. Biotinylated DNA fragments were pulled with the MyOne Streptavidin C1 beads (Life Technologies). To repair the ends of sheared DNA and remove biotin from unligated ends, DNA-bound beads were re-suspended in 100 μl of mix containing 82 μl of 1X NEB T4 DNA ligase buffer with 10 mM ATP (NEB), 10 μl of 10 (2.5mM each) 25 mM dNTP mix, 5 μl of 10 U/μl NEB T4 PNK (NEB), 4 μl of 3 U/μl NEB T4 DNA polymerase (NEB), and 1 μl of 5 U/μl NEB DNA polymerase I, Large (Klenow) Fragment (NEB).
After end-repair, dATP attachment was carried out in in 100 μl of mix consisting of 90 μl of 1X NEBuffer 2, 5 μl of 10 mM dATP, 5 μl of 5 U/μl NEB Klenow exo minus (NEB), and incubated at 37°C for 30 min. The beads were then cleaned for Illumina sequencing adaptor ligation which was done in a mix containing 50 μl of 1X T4 ligase buffer, 3 μl T4 DNA ligases (NEB), and 2 μl of a 15 μM Illumina indexed adapter at room temperature for 1 h. DNA was dissociated from the bead by heating at 98°C for 10 min, separated on a magnet, and transferred to a clean tube.
Final amplification of the library was carried out in multiple PCRs using Illumina PCR primers. The reactions were performed in 25 μl scale consisting of 25 ng of DNA, 2 μl of 2 .5mM dNTPs, 0.35 μl of 10 μM each primer, 2.5 μl of 10X PfuUltra buffer, PfuUltra II Fusion DNA polymerase (Agilent). The PCR cycle conditions were set to 98°C for 30 seconds as the denaturing step, followed by 14 cycles of 98°C 10 seconds, 65°C for 30 seconds, 72°C for 30 seconds, then with an extension step at 72°C for 7 min.
After PCR amplification, the products from the same library were pooled and fragments ranging in sized of 300–500 bp were selected with AMPure XP beads (Beckman Coulter). The sized selected libraries were sequenced to produce paired-end Hi-C reads on the Illumina HiSeq 2500 platform with the V4 of 125 cycles.
Generation and Analysis of Hi-C Matrices
Paired end reads were processed using the juicer pipeline with default parameters [28]. Reads were mapped to reference genome hg19, with “-s” (site parameter) MboI. Reads with MAPQ >30 were kept for further analysis. Data was extracted and input to MATLAB using Juicebox tools command “dump”. Knight-Ruiz (KR) normalization was applied to all matrices, observed over expected (O/E) matrices were used for A/B compartmentalization and identification of topologically associating domains (TADs). Rows and columns for which more than 10% of entries had zeros were removed from the matrix. A/B compartmentalization was determined using the Fiedler vector, as previously described [29]. TAD structure was determined using previously described methods [30], with λthr = 0.8, and minimum TAD size defined as 300 kb. This technique is derived from spectral graph theory, and is used for graph segmentation. Here, the parameters are set so that 300 kb is the minimum domain size and 0.8 is the minimum Fiedler number of each domain. The Fiedler number is a measure of graph connectivity, with higher values implying stronger connectivity.
Multiplex Fluorescent Two-Dimensional Gel-Electrophoresis (2-DIGE)
Proteins for 2D-DIGE analysis were labeled with Refraction-D™ labeling kit (NH DyeAGNOSTIC, Halle, Germany). A total of 50 μg of each sample and a pooled internal standard for exact quantification was mixed with Tris–HCl (1.5 M, pH 8.8) and 1 μl of 50 nM G-100, G-200 or G-300, respectively, and incubated in darkness at 4°C for 30 min. Each reaction was terminated by adding 1 μl 10 mM lysine and incubating with the protein on ice for 10 min. Samples and internal standard were combined and diluted with rehydration sample buffer [7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 2% (v/v) ampholytes (pH 4–7, SERVA Electrophoresis, Heidelberg, Germany) and a trace of bromophenol blue] to a final volume of 450 μl. 24 cm Immobiline Dry Strips, pH 4–7 (GE Healthcare, Chicago, IL, USA) were used for isoelectric focusing which was carried out in a Protean® i12TM IEF cell (Bio-Rad Laboratories, Hercules, CA, USA) at 20°C reaching approximately 57,700 Vh. After equilibration (Buffer Kit for 2D HPETM Gels, SERVA Electrophoresis), horizontal second dimension was carried out using precast plastic-backed 12.5% acrylamide gels (2DHPETM Large Gel NF 12.5% Kit, 0.65 x 200 x 255 mm, SERVA Electrophoresis). DIGE images were acquired using a Typhoon FLA 9000 scanner (GE Healthcare). Detected spots were matched and analyzed using Progenesis SameSpots® (Nonlinear Dynamics, Newcastle upon Tyne, UK, v4.1). Spots that showed significant expression differences between groups were picked using a robotic spot picker (GE Healthcare). Gel plugs were washed in 25 mM ammonium bicarbonate in 50% (v/v) acetonitrile to remove inhibitory chemicals. Dried gel plugs were rehydrated in an ice-cold solution of 12.5 ng/μl sequencing grade trypsin (Promega, Fitchburg, WI, USA) in 10 mM NH4HCO3. Proteins were digested in-gel at 37°C for 4 h. Peptides were extracted for 30 min with 10 μl of 0.1% trifluoroacetic acid (TFA) and directly applied to a Prespotted AnchorChip MALDI target (BRUKER Daltonics, Billerica, MA, USA) according to the manufacturer's instructions. Subsequently, samples were analyzed in an Ultraflex MALDI-TOF/TOF mass spectrometer (BRUKER Daltonics). Acquired mass spectra were automatically calibrated and annotated using Compass 1.4 for flex software (BRUKER Daltonics). For protein identification, results from each individual protein spot were used to search a human subset in Swiss-Prot non-redundant database by means of Mascot search engine (Matrix Science Ltd., London, UK, v2.2). Not identified proteins were repeatedly evaluated using TripleTOF 5600+ mass spectrometer (ABSciex, Framingham, MA, USA). Dry peptide pellets were dissolved in 5 pmol sequencing grade trypsin (Promega, USA) in 50 mM Ammonium bicarbonate, 0.02% Protease Max and incubated overnight at 37°C. Peptides were extracted for 10 min at 37°C with 40–50 μl 1% formic acid, 66% acetonitrile and 33% 100 mM Ammonium bicarbonate prior subjection to LC–MS/MS analysis. Chromatographic separation of peptides was achieved using a C18 column, 5 cm (100 μm ID, Phenomenex, Torrance, CA) connected to the UltimateTM 3000 RSLCnano chromatography system (Thermo Scientific, USA). The survey MS spectrum was acquired in the range of m/z 350–1.250. MS/MS data for 10 most intense precursors were obtained with a higher-energy collisional dissociation for ions with charge 2 to 4. The mass spectrometric raw data were analyzed with the ProteinPilot software (version 5.0.1). A false discovery rate (FDR) of 0.01 for proteins and peptides was required. All mass spectrometric evaluations were carried out using the following settings: (i) enzyme “trypsin”, (ii) species “human”, (iii) fixed modifications “carbamidomethyl”, (iv) optional modifications “methionine oxidation” and (v) missed cleavages “1”.
Functional Analysis of Identified Proteins
Ingenuity pathway analysis (IPA, QIAGEN, USA) was used to determine the connectivity between identified proteins, their biofunction and localization. Only direct relationships were included in the analysis. Each protein symbol was mapped to its own object in the Ingenuity Pathways Knowledge Database. Interactive pathways were generated using all IPA available tissue/cell culture types as well as targeted tissue/cell cultures of the large intestine. A score> 5 was considered as significant.
Xenografting in Athymic Nude Mice
A total of 2 × 106 cells of each cell line were suspended in 1 ml PBS or Matrigel® Matrix (Corning, Tewksbury, MA) and injected subcutaneously in 6–7 weeks old narcotized (O2/isoflurane) athymic NCr-nu/nu mice (Charles River Laboratories). Cells were injected in male and female mice using either PBS or Matrigel® as a carrier. Six mice per cell line were analyzed.
Results
The karyotypically normal (46,XY) cell line HCEC has been derived from non-cancerous colon tissue of a patient undergoing routine surveillance colonoscopy [14]. Long-term propagation (~40 population doublings) of these cells under serum-free culture conditions gave rise to cells with trisomy 7 (HCEC+7), which was the only cytogenetic aberration [15]. We confirmed the karyotype by spectral karyotyping (SKY) and aCGH. HCEC showed a diploid normal karyotype (46,XY), whereas HCEC+7 showed a trisomy 7 as sole cytogenetic aberration (47,XY,+7) (Figure 1A). No additional chromosomal gains or losses were detected by aCGH (Supplementary Figure 1). Isogenicity was confirmed by STR profiling. The proportion of cells trisomic for chromosome 7 in the HCEC+7 cell line was determined by interphase FISH to be 60% to 70%. No morphological differences were observed between the HCEC and HCEC+7 cell lines. Trisomy 7 alone did not induce malignant transformation. HCEC and HCEC+7 cells were injected subcutaneously into athymic nude mice, however, neither cell line resulted in tumor growth after a follow up period of 6 months. This is consistent with previous results [15].
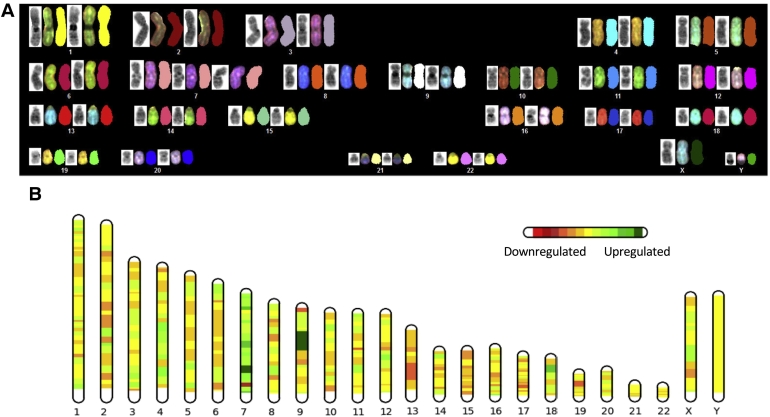
Figure 1.
(A) SKY analysis of the HCEC+7 cells. The trisomy of chromosome 7 is the sole cytogenetic anomaly. (B) Differential, global, chromosome-wide gene expression between HCEC and HCEC+7 cells. Upregulation (green) is most profound on chromosome 7, however, changes were not restricted to the aneuploid chromosome.
To decipher whether trisomy 7 facilitates the acquisition of additional numerical chromosomal aberrations, we used miFISH [20] to enumerate copy numbers of 12 gene-specific loci within each interphase nucleus analyzed (see Materials and Methods). The majority of HCEC cells were diploid (94.8%). Only 2.8% and 2.4% of cells were tetraploid or revealed copy number changes, respectively. 37.5% of the HCEC+7 cells were diploid, 58% were trisomic for chromosome 7, 0.4% were tetraploid, 2% were tetraploid with six copies of chromosome 7 and 2.9% showed additional copy number changes. We conclude that the acquired trisomy of chromosome 7 alone does not trigger additional chromosomal instability.
As shown by us and others before, chromosome wide average gene expression levels closely follow genomic copy number [31], [32], [33], [34]. This was also the case in our model system here. Average gene expression levels on chromosome 7 were higher compared to those on diploid chromosomes (Figure 1B).
Trisomy 7 Results in Specific Alterations to Nuclear Organization as Measured by Hi-C
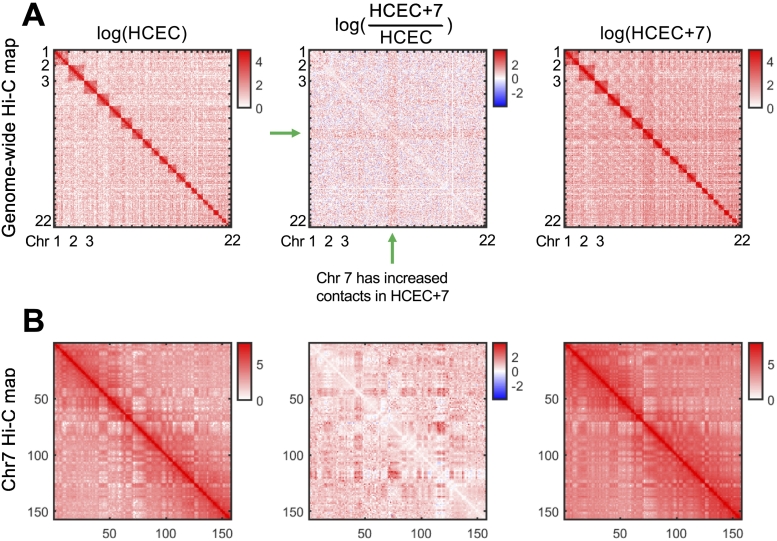
Changes to nuclear organization as a consequence of aneuploidy were determined by Hi-C. We show that three copies of chromosome 7 resulted in increased inter- and intrachromosomal contacts of the aneuploid chromosomes which is shown genome-wide in Figure 2A. Figure 2B displays chromosome 7 specific Hi-C maps, and confirms that extra copies of chromosome indeed results in increased intrachromosomal contacts. This is consistent with previous results from our group analyzing the colorectal cancer cell line HT-29, where we demonstrated that copy number changes resulted in increased contacts [18].
Figure 2.
Hi-C maps show an increase in chromosome 7 contacts genome-wide. (A) Genome-wide Hi-C contact maps for HCEC (left), HCEC+7 (right), and the difference between the two samples (center). Matrices are shown at 1 Mb resolution and log-scale. Red regions in the middle matrix are enriched in HCEC+7, blue regions in the middle matrix are enriched in HCEC. A clear increase in HCEC+7 contacts is observed in regions involving chromosome 7 (green arrow). (B) Chromosome 7 Hi-C contact maps for HCEC (left), HCEC+7 (right), and the difference between the 2 samples (center). Matrices are shown at 1 Mb resolution and log-scale. Red regions in the middle matrix are enriched in HCEC+7, blue regions in the middle matrix are enriched in HCEC. A clear increase in HCEC+7 contacts is observed for the majority of intra-chromosomal contacts in chromosome 7.
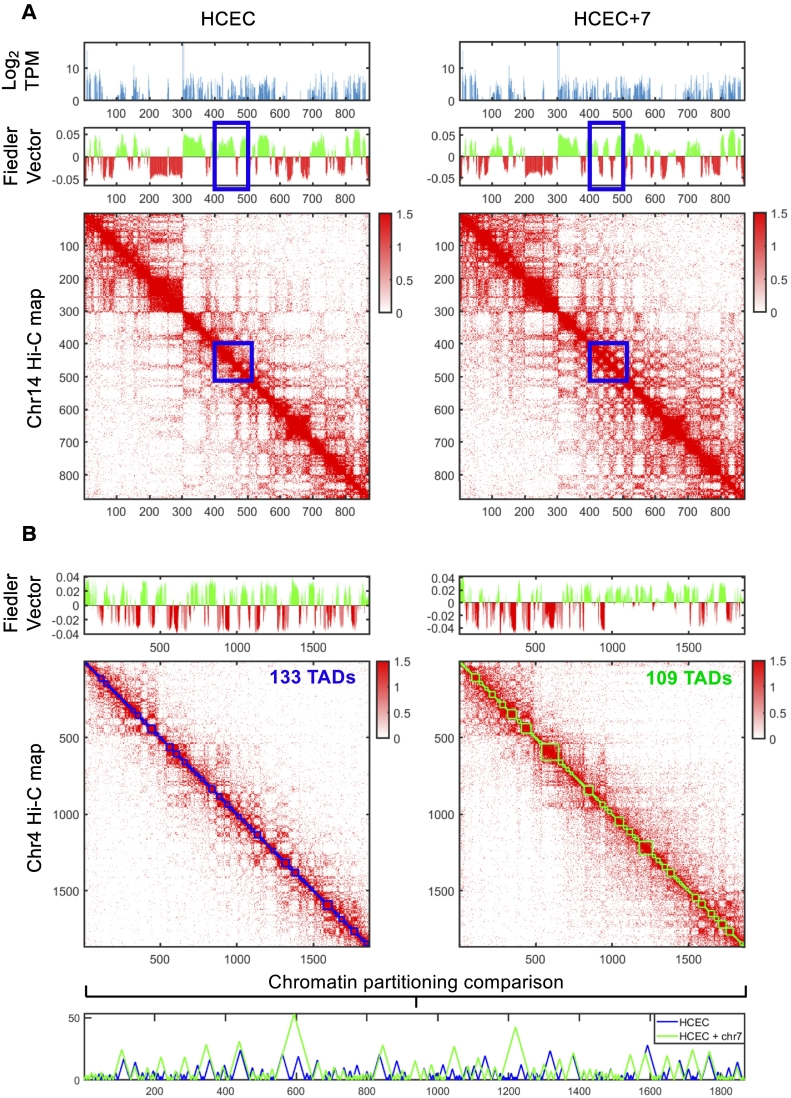
The global Hi-C maps revealed additional, aneuploidy specific alterations to nuclear organization. Chromatin is organized as active and inactive domains, also referred to as A and B compartments. We observed a clear change in this compartmentalization on chromosome 14. Here, we determined a clear switch in compartmentalization from A in HCEC to B in HCEC+7 in a region spanning chr14:62.4Mb–63.8Mb (Figure 3A and Supplementary Figure 2A). This region contains few proteins coding genes, many of which change significantly in expression. The most significant change in expression is observed in KCNH5 and RHOJ, both of which are down regulated and completely repressed in HCEC+7 (adjusted p-value 1.7824e-17 and 2.9776e-06, respectively), in line with their change from compartment A to B. KCNH5 hypermethylation has been observed in a number of cancers, which may explain the compartment switch from A to B [35].
Figure 3.
HCEC+7 results in genome-wide structural changes. (A) A change in chromatin partitioning is observed in chromosome 14. The left and right side show HCEC and HCEC+7, respectively. The top row shows the average gene expression for each 100 kb bin. The middle row shows the Fiedler vector partitioning. A clear change is observed between 425 and 450 (blue square). The bottom row is the Hi-C contact map for chromosome 14, shown at 100 kb resolution, log-scale. (B) HCEC+7 shows a clear change in chromosome 4 patterning. The left and right side show HCEC and HCEC+7, respectively. The top row shows the Fiedler vector partitioning. The middle row is the Hi-C contact map for chromosome 4, shown at 100 kb resolution, log-scale. Squares (green and blue) along the diagonal of the Hi-C matrix depict the TAD structure. A direct comparison of the TAD structure between samples is shown below.
Next, we observed a visible loss of chromosome 4 patterning, with clear changes in A/B compartmentalization as well as topologically associating domain (TAD) structure chromosome-wide (Figure 3B and Supplementary Figure 2B). Genes within regions that change from A in HCEC to B in HCEC+7 are generally down-regulated, as expected. 8.8% of genes within these regions are significantly down-regulated, with 4.4% of genes significantly up-regulated (adjusted P < .05). Genes within regions that change from B in HCEC to A in HCEC+7 are up-regulated to a larger extent. 22.3% of genes within these regions are significantly up-regulated, with 1% of genes significantly down-regulated (adjusted P < .05). HCEC revealed 133 TADs on chromosome 4, which were reduced to 109 TADs as a consequence of chromosome 7 aneuploidy. Change was also observed in TAD structure: the segregation of the domains was less pronounced in the HCEC+7 cells. This effect was not observed on other chromosomes. Genome wide comparison of gene expression and chromatin structure in the HCEC and HCEC+7 cells is shown in Supplementary Figure 3, A–K.
Trisomy 7 Affects Chromosomal Organization in the Interphase Nucleus as Measured by 3D-FISH
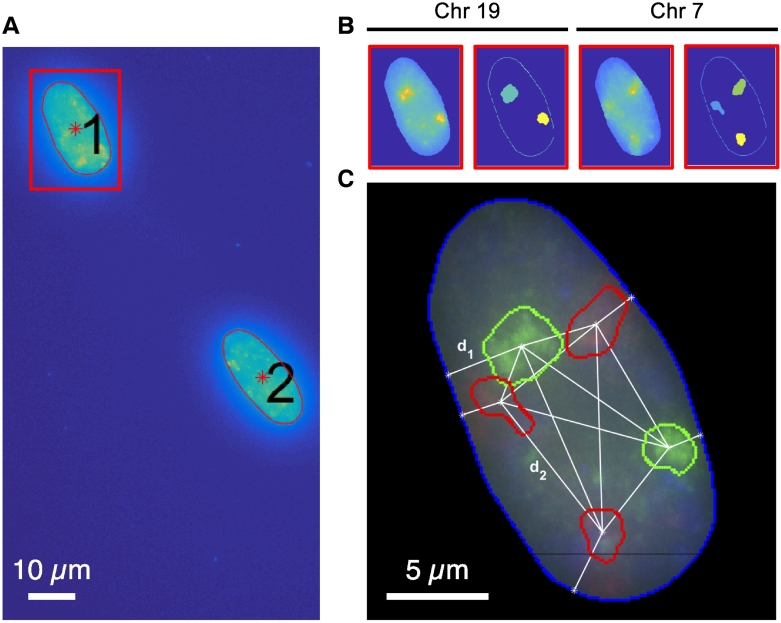
We next determined whether an extra copy of chromosome 7 affects the positioning of that chromosome in the interphase nucleus using an independent method. Therefore, we performed dual-color 3D-FISH on morphologically preserved HCEC and HCEC+7 nuclei. In addition to chromosome 7, we also determined the positioning of chromosome 19 as a control. Chromosome 19 has the highest gene density of all human chromosomes and is positioned closer towards the center of the interphase nucleus [36]. To objectively evaluate chromosome territory (CT) positioning and to enable statistical comparison between HCEC and HCEC+7 cells, 3D images were processed via a MATLAB script (see Material and Methods). The shape of the nuclei was detected and analyzed individually, the number of CTs was counted, and the size and distance between CTs and the nuclear periphery was calculated. An example of the image reconstruction and the position measurements is shown in Figure 4. The size of the CTs of chromosome 7 were slightly smaller in the HCEC+7 cells (HCEC: 5.96 ± 1.62 μm3; HCEC+7: 4.92 ± 1.54 μm3) although this trend did not reach statistical significance. In turn, the size of the chromosome territories of chromosome 19 tended to be slightly bigger in the HCEC+7 cells compared to the diploid cells (HCEC: 6.26 ± 1.70 μm3; HCEC+7: 7.54 ± 2.56 μm3) without any statistical significance. As expected the nuclear position of chromosome 19 was more internal than chromosome 7, which is gene poorer, regardless of an extra copy of chromosome 7. However, the positioning of the CTs of chromosome 7 was more variable in the HCEC+7 cells as measured by the distance to the edge of the nucleus to the centroid of the CT (HCEC: 2.10 ± 0.96 μm; HCEC+7: 2.37 ± 1.34 μm). The nuclei of the HCEC+7 cells were significantly bigger compared to the nuclei of the diploid cells (1CT: 139.06 ± 35.24 μm3; 1CT+ 7: 173.23 ± 48.82 μm3, P < .00085).
Figure 4.
3D-FISH image analysis of chromosome territories (CT). (A) Cell nuclei were automatically selected from DAPI images. (B) Fluorescent channels corresponding to chromosome 7 and 19 were analyzed, and CTs were automatically selected. (C) CT size, distance from CT to nucleus edge (d_1), and distance between CTs (d_2) was extracted.
Trisomy 7 Results in Global Gene Expression Changes
To determine the consequences of nuclear structural changes as a result of trisomy 7, we performed transcriptome profiling by RNA sequencing. The acquisition of an extra copy of chromosome 7 resulted in global gene expression changes. Chromosome 7 had the highest proportion of genes differentially expressed, as would be expected with additional gene copies, though not all genes were upregulated (Supplementary Figure 4).
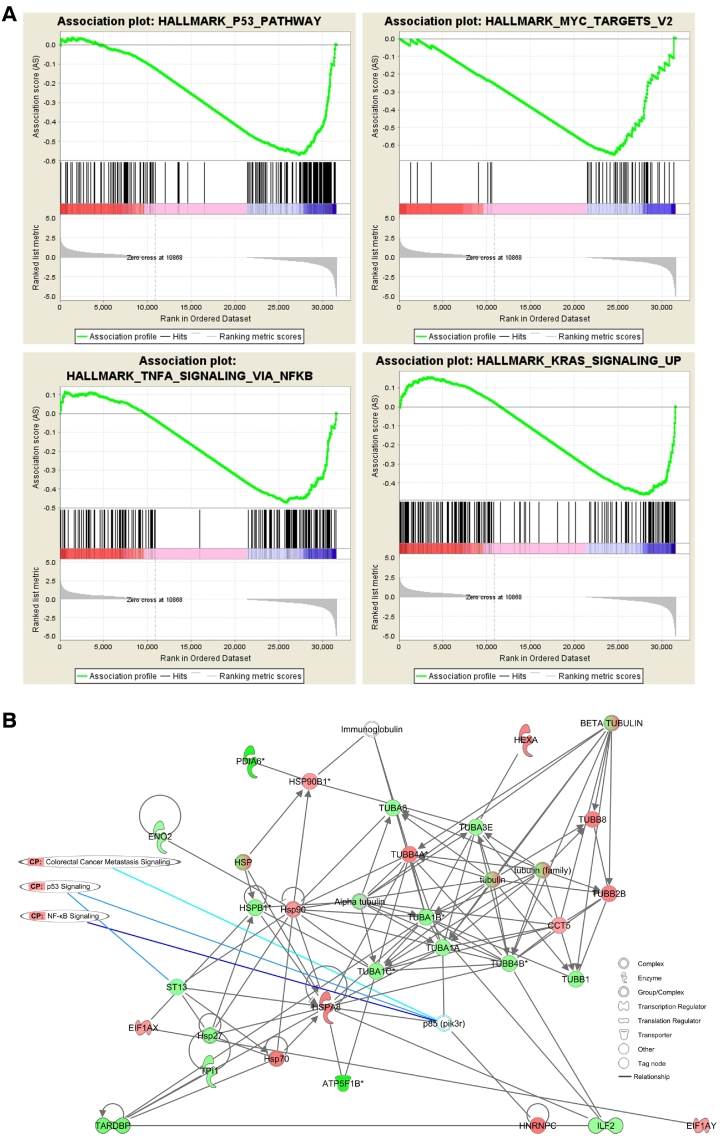
Next, we wanted to interrogate the functional space of gene expression changes. This was performed using gene set association analysis of RNA-seq data (GSAASeqSP) (Figure 5A). From this analysis, we identified 2 signaling networks that were significantly enriched in the HCEC+7 cells with an FDR q-value <0.005, and others that are strongly associated with the phenotype. The top 4 gene sets are crucial networks in colorectal carcinogenesis, namely, P53-pathway (0.003), MYC-targets (0.003), TNF-α via NF-κB (0.126), and KRAS signaling up (0.160).
Figure 5.
Identification of differentially regulated pathways. (A) GSAA analysis. The top four signaling networks that were enriched in the HCEC+7 cells are shown; P53-pathway (0.003), MYC-targets (0.003), TNF-α via NF-κB (0.126), KRAS signaling up (0.160). (B) IPA network analysis of differentially expressed proteins identified by 2-DIGE and MS. Red indicates over-expression, green indicates under-expression.
More detailed analysis of genes coded on chromosome 7, which are linked to colorectal carcinogenesis, revealed that HGF was significantly down-regulated in HCEC+7 cells, whereas MET was significantly up-regulated. In line, GSAA analysis showed an up-regulation of KRAS signaling, which is downstream of the HGF/MET axis. Though neither HGF nor MET showed a change in A/B compartment, MET is located within the region that shows a clear loss of chromatin patterning (Supplementary Figure 5).
Trisomy 7 Induces Global Changes of the Proteome in HCEC+7 cells
Finally, we analyzed the consequences of trisomy 7 on the proteome using 2D-gelelectrophoresis and subsequent mass spectrometry (MS) analysis. We found 96 protein spots to be significantly differentially expressed between the HCEC and HCEC+7 cells. We could identify a total of 148 proteins by subsequent MS analysis. Two or more proteins were identified in 20 of the protein spots. Twenty-eight protein spots could not be identified by either MALDI or ESI mass spectrometry.
The expression was significantly up-regulated for 48 and significantly down-regulated for 100 of the identified differentially expressed proteins in the HCEC+7 cells. Subsequent IPA analysis resulted in three cancer-associated IPA networks of which the top network (score 63) was Cancer, Gastrointestinal Disease and Neurological Disease (Figure 5B). The IPA-Network was closely associated with canonical pathways of TP53-, NF-κB- and colorectal cancer-signaling, which is consistent with the gene expression changes. A colon specific IPA analysis revealed the network Cell Death and Survival and Cancer (score 20) with TP53 as a central node (Supplementary Figure 6).
Discussion
Tissue-specific chromosomal aneuploidies emerge at early stages of tumorigenesis [9], [10], [37]. During the transformation of normal colon to polyps and eventually invasive carcinoma, cells frequently acquire extra copies of chromosome 7 at early stages of malignant transformation [9], [10], [11], [12]. We therefore attempted to understand the global consequences of trisomy 7 in colon cells on both nuclear organization and function.
We used Hi-C to study global changes to the nucleome; targeted 3D-FISH with chromosome painting probes and reconstruction of chromosome territories. Finally, the consequences of chromosome 7 trisomy on cellular function were measured using RNA sequencing and multiplex fluorescent two-dimensional gel-electrophoresis (2-D DIGE). These analyses were applied to an isogenic model system of immortalized, karyotypically normal colorectal epithelial cells (HCEC) that acquire an extra copy of chromosome 7 after cell culture under serum-starvation conditions [15]. We used this model system because it allows studying the consequences of trisomy 7 without potentially confounding additional cytogenetic changes commonly present in established cancer cells. In line, we confirmed that the presence of trisomy 7 did not result in additional chromosomal aberrations by SKY, array CGH and multiplex interphase FISH (miFISH).
Hi-C identified changes specific to chromosome 7, however, additional chromosomes were affected by distinct chromatin organization modifications. The increase in direct contacts are a result of an additional copy of chromosome 7. An increase in chromosome 7 Hi-C counts was observed genome-wide, inferring that the extra copy of chromosome 7 had no detectable preference in its position relative to other chromosomes. This is in line with our CT reconstruction analysis using 3D-FISH which did not show changes in chromosomal positioning. These results are consistent with previous results from our laboratories reporting comprehensive Hi-C maps of the colorectal cancer cell line HT-29 [18]. In addition, we showed that the Hi-C maps faithfully recapitulate 2D changes determined by high resolution molecular cytogenetic analyses, i.e., SKY, aCGH and FISH to chromosome number and structure.
Changes of the nucleome of the HCEC+7 cells were not restricted to chromosome 7. Genome-wide analysis revealed many regions with differences in structure and function, with the strongest changes observed on chromosome 4 and 14. On both of these chromosomes, A/B compartment changes were observed. On chromosome 4 we not only observed switches in the A/B compartments, but also changes in TAD boundaries. The number of TADs decreased from 133 in the diploid cells to 109 in the HCEC+7 cells. In addition, we observed structural changes. The mechanisms for these changes are presently not known. However, from our results it appears obvious that understanding the consequences of chromosomal trisomies requires genome wide analyses.
Changes in the nuclear structure resulted in a change in function defined by gene expression levels. Genes that switched from A to B regions were generally down-regulated, whereas genes that switch from B to A regions were generally upregulated. One example is the switch of a region on chromosome 14 (chr14:62.4Mb–63.8Mb) that changed from the A to B which is possibly reflective of hypermethylation. Hypermethylation is associated with heterochromatin and B compartmentalization. Furthermore, KCNH5 has been shown to hypermethylated previously in cancers [35], [38]. How this region is targeted after the trisomy 7 event is unknown. The loss of compartment structure observed on chromosome 4 extended from 91 Mb to 158.9 Mb and was generally associated with A to B transition. Again, the compartment switch resulted in a change in gene expression in a directionality that would be intuitive. The mechanism for this targeted loss of chromatin structure is unknown, however, we surmise that global gene expression changes have a structural correlate.
We found a significant down-regulation of HGF-mRNA expression in HCEC+7 cells, while MET was significantly up-regulated. HGF and MET are both located in a region on chromosome 7 that showed a loss of chromatin patterning, however, we did not observe a switch in A/B compartment. The hepatocyte growth factor (HGF) specifically binds to the receptor tyrosine kinase “mesenchymal to epithelial transition” (MET), which is a proto-oncogene [39], [40]. MET-mRNA quantification in primary CRC suggested that MET overexpression plays an important role in the development of loco-regional invasiveness in early stage of CRC development [41]. Consistent to changes in HGF and MET expression, we found an up-regulation of KRAS signaling by GSAA gene enrichment analysis, although it did not reach statistical significance.
Besides KRAS signaling, trisomy 7 in normal colon cells results in dysregulation of further signaling pathways crucial for CRC genesis such as the P53-pathway and TNF-α via NF-κB.
In line, protein profiling identified a total of 148 differentially regulated proteins that correspond to functional IPA networks of Cancer and Gastrointestinal Diseases as well as the canonical P53- and NF-κB signaling pathways. Although protein profiling is not able to cover the complete proteome of a given cell, the phenotypic results reflect a close association between the additional copy of chromosome 7 and impending malignant transformation.
Our data demonstrate that structural nuclear changes caused by an extra chromosome 7 entails the dysregulation of several pathways associated with colorectal carcinogenesis. Interestingly, trisomy 7 by itself is not carcinogenic as demonstrated here by us using murine xenograft models in nu/nu mice and others [15]. As trisomy 7 is commonly found in early stages of carcinogenesis in the colon such as adenomas, we conclude from our results that this chromosomal aberration is a perquisite facilitating subsequent malignant transformation. We show that this specific chromosomal aneuploidy has a profound impact on nuclear structure and function, both, locally and genome wide.
Acknowledgments
R.B. was supported by a Mildred Scheel postdoctoral scholarship of the German Cancer Aid (Deutsche Krebshilfe). We are grateful to Dr. Jerry Shay (UT Southwestern) for sharing the HCEC cell lines. We thank Casey Dagnall (Cancer Genomics Research Laboratory, Gaithersburg, MD) for STR-profiling and the CCR sequencing core facility for RNA sequencing. We are grateful to Sonja Hartwig and Stefan Lehr (Institute of Clinical Biochemistry and Pathobiochemistry, German Diabetes Center at the Heinrich-Heine-University Düsseldorf, Germany) for MS analyses. This study was supported by the intramural research program, National Institutes of Health, National Cancer Institute. We are grateful to Buddy Chen for editorial assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.02.003.
Contributor Information
Indika Rajapakse, Email: indikar@umich.edu.
Thomas Ried, Email: riedt@mail.nih.gov.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ried T. Homage to Theodor Boveri (1862-1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas. Environ Mol Mutagen. 2009;50:593–601. doi: 10.1002/em.20526. [DOI] [PubMed] [Google Scholar]
- 3.Balaban GB, Herlyn M, Clark WH, Jr., Nowell PC. Karyotypic evolution in human malignant melanoma. Cancer Genet Cytogenet. 1986;19:113–122. doi: 10.1016/0165-4608(86)90378-x. [DOI] [PubMed] [Google Scholar]
- 4.Magnani I, Guerneri S, Pollo B, Cirenei N, Colombo BM, Broggi G, Galli C, Bugiani O, DiDonato S, Finocchiaro G. Increasing complexity of the karyotype in 50 human gliomas. Progressive evolution and de novo occurrence of cytogenetic alterations. Cancer Genet Cytogenet. 1994;75:77–89. doi: 10.1016/0165-4608(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 5.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosom Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 6.El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res. 2000;60:3899–3903. [PubMed] [Google Scholar]
- 7.Lai LA, Paulson TG, Li X, Sanchez CA, Maley C, Odze RD, Reid BJ, Rabinovitch PS. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosom Cancer. 2007;46:532–542. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 8.Di Capua Sacoto C, Budia Alba A, Alapont Alacreu JM, Ruiz Cerda JL, Jimenez Cruz JF. In vivo aneuploidization during the expansion of renal adenocarcinoma. Urol Int. 2011;86:466–469. doi: 10.1159/000324101. [DOI] [PubMed] [Google Scholar]
- 9.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosom Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Bomme L, Lothe RA, Bardi G, Fenger C, Kronborg O, Heim S. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int J Cancer J Int du Cancer. 2001;92:816–823. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- 11.Grade M, Becker H, Liersch T, Ried T, Ghadimi BM. Molecular cytogenetics: Genomic imbalances in colorectal cancer and their clinical impact Cellular oncology. J Int Soc Cell Oncol. 2006;28:71–84. doi: 10.1155/2006/173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosom Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler D, Heselmeyer-Haddad K, Hirsch D, Hernandez LS, Torres I, Wangsa D, Hu Y, Zapata L, Rueschoff J, Belle S. Single-cell genetic analysis of clonal dynamics in colorectal adenomas indicates CDX2 gain as a predictor of recurrence. Int J Cancer J Int du Cancer. 2019;144:1561–1573. doi: 10.1002/ijc.31869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roig AI, Eskiocak U, Hight SK, Kim SB, Delgado O, Souza RF, Spechler SJ, Wright WE, Shay JW. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138(1012-1021):e1011–e1015. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 15.Ly P, Eskiocak U, Kim SB, Roig AI, Hight SK, Lulla DR, Zou YS, Batten K, Wright WE, Shay JW. Characterization of aneuploid populations with trisomy 7 and 20 derived from diploid human colonic epithelial cells. Neoplasia. 2011;13:348–357. doi: 10.1593/neo.101580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Kim S, Jia G, Buhmeida A, Dallol A, Wright WE, Fornace AJ, Al-Qahtani M, Shay JW. Exome Sequencing of Normal and Isogenic Transformed Human Colonic Epithelial Cells (HCECs) Reveals Novel Genes Potentially Involved in the Early Stages of Colorectal Tumorigenesis. BMC Genomics. 2015;16(Suppl 1):S8. doi: 10.1186/1471-2164-16-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ried T, Rajapakse I. The 4D Nucleome. Methods. 2017;123(1-2) doi: 10.1016/j.ymeth.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 18.Seaman L, Chen H, Brown M, Wangsa D, Patterson G, Camps J, Omenn GS, Ried T, Rajapakse I. Nucleome Analysis Reveals Structure-Function Relationships for Colon Cancer. Mol Cancer Res : MCR. 2017;15:821–830. doi: 10.1158/1541-7786.MCR-16-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc. 2006;1:3129–3142. doi: 10.1038/nprot.2006.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, Prindiville SA, Calzone KA, Soballe PW, Hu Y. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol. 2012;181:1807–1822. doi: 10.1016/j.ajpath.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Q, Mukherjee S, Furey TS. GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Sci Rep. 2014;4:6347. doi: 10.1038/srep06347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Chen J, Muir LA, Ronquist S, Meixner W, Ljungman M, Ried T, Smale S, Rajapakse I. Functional organization of the human 4D Nucleome. Proc Natl Acad Sci U S A. 2015;112:8002–8007. doi: 10.1073/pnas.1505822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Hero AO, III, Rajapakse I. Spectral identification of topological domains. Bioinformatics. 2016;32:2151–2158. doi: 10.1093/bioinformatics/btw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grade M, Ghadimi BM, Varma S, Simon R, Wangsa D, Barenboim-Stapleton L, Liersch T, Becker H, Ried T, Difilippantonio MJ. Aneuploidy-dependent massive deregulation of the cellular transcriptome and apparent divergence of the Wnt/beta-catenin signaling pathway in human rectal carcinomas. Cancer Res. 2006;66:267–282. doi: 10.1158/0008-5472.CAN-05-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 34.Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J. The consequences of chromosomal aneuploidy on the transcriptome of cancer cells. Biochim Biophys Acta. 2012;1819:784–793. doi: 10.1016/j.bbagrm.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawes SE, Stern JE, Feng Q, Wiens LW, Rasey JS, Lu H, Kiviat NB, Vesselle H. DNA hypermethylation of tumors from non-small cell lung cancer (NSCLC) patients is associated with gender and histologic type. Lung Cancer. 2010;69:172–179. doi: 10.1016/j.lungcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouadid-Ahidouch H, Rodat-Despoix L, Matifat F, Morin G, Ahidouch A. DNA methylation of channel-related genes in cancers. Biochim Biophys Acta. 2015;1848:2621–2628. doi: 10.1016/j.bbamem.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci U S A. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi H, Bilchik A, Saha S, Turner R, Wiese D, Tanaka M, Kuo C, Wang HJ, Hoon DS. c-MET expression level in primary colon cancer: a predictor of tumor invasion and lymph node metastases. Clin Cancer Res J. Am Assoc Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures