Abstract
Epidermal growth factor receptor variant III (EGFRvIII) is a tumor-specific cell surface antigen often expressed in glioblastoma and has drawn much attention as a possible therapeutic target. We performed immunohistochemistry on histology sections of surgical specimens taken from 67 cases with glioblastoma, isocitrate dehydrogenase-wild type, and evaluated the morphological characteristics and distribution of the EGFRvIII-positive tumor cells. We then evaluated the localization of EGFRvIII-expression within the tumor and peritumoral areas. EGFRvIII immunopositivity was detected in 15 specimens taken from 13 patients, including two recurrent specimens taken from the same patient at relapse. Immunofluorescence staining demonstrated that EGFRvIII-positive cells were present in cells positive for glial fibrillary acidic protein (GFAP), and some showed astrocytic differentiation with multiple fine processes and others did not shown. The EGFRvIII-positive cells were located in cellular areas of the tumor, but not in the invading zone. In the two recurrent cases, EGFRvIII-positive cells were markedly decreased in one case and retained in the other. With regard to overall survival, univariate analysis indicated that EGFRvIII-expression in patients with glioblastoma was not significantly associated with a favorable outcome. Double-labeling immunofluorescence staining of EGFRvIII and GFAP showed that processes of large, well differentiated, GFAP-positive glia extend to and surround less differentiated, EGFRvIII-positive glial cells in cellular areas of tumor. However, in the tumor periphery, EGFRvIII-positive tumor cells were not observed. This finding suggests that EGFRvIII is involved in tumor proliferation, but that invading glioma cells lose their EGFRvIII expression.
Keywords: glioblastoma, EGFRvIII, morphology, distribution, prognosis
Introduction
Glioblastoma is the most common histopathological type of primary brain tumor in adults. Despite recent application of multidisciplinary treatment involving a combination of surgical resection, chemotherapy, and radiation therapy, the prognosis of patients with glioblastoma is still poor and the median overall survival remains 14–15 months.1–5) At present, various potential therapeutics, including Rindopepimut – an anti-epidermal growth factor receptor variant III (EGFRvIII) peptide vaccine – are being assessed in clinical trials.6–9) EGFRvIII, the product of the EGFR gene with an in-frame deletion of exons 2–7 (del 2–7 EGFR, or ΔEGFR), is a tumor-specific cell surface antigen with a molecular mass of 145 kDa.6,10,11) EGFRvIII constitutively activates the STAT and PI3K-Akt pathways.12–17) and promotes angiogenesis and tumor growth.18–20) In glioblastoma, it has been shown that EGFRvIII is expressed in a subset of primary tumors,21) with molecular profiles of EGFR amplification and absence of isocitrate dehydrogenase 1 (IDH1) mutations.22–24)
In this study, we performed immunohistochemistry using a recently available antibody specific for EGFRvIII on histology sections of surgical specimens taken from patients with glioblastoma, IDH-wild-type, in order to evaluate the morphological characteristics and distribution of EGFRvIII-positive tumor cells, and also the significance of EGFRvIII expression.
Materials and Methods
Patients
We reviewed the medical records of 67 consecutive patients (34 males, 33 females; age at surgery, mean = 64.5 years) who were admitted to the Department of Neurosurgery, Niigata University Medical and Dental Hospital, Japan, between 2011 and 2017, and diagnosed pathologically as having glioblastoma, IDH-wild-type. In accordance with the methods stipulated in the WHO Classification of Tumors,25) immunohistochemistry for IDH1 and DNA sequencing for IDH1 and IDH2 were performed, as described previously.26) The clinical profiles of the patients are summarized in Tables 1 and 2.
Table 1.
Clinical profiles of patients in each group
| EGFRvIII | P | ||
|---|---|---|---|
| Positive | Negative | ||
| Total | 13 | 54 | |
| Age | 66.5 | 63.8 | 0.223 |
| Female sex | 4 (30.8%) | 28 (51.9%) | 0.540 |
| Laterality | |||
| Right | 8 (61.5%) | 27 (50.0%) | 0.736 |
| Left | 4 (30.8%) | 20 (37.0%) | 0.302 |
| Bilateral | 1 (7.7%) | 7 (13.0%) | 0.237 |
| Tumor location | |||
| Frontal | 5 (38.5%) | 26 (48.1%) | 0.758 |
| Temporal | 3 (23.1%) | 14 (25.9%) | 0.999 |
| Parietal | 1 (7.7%) | 3 (5.6%) | 0.999 |
| Occipital | 2 (15.4%) | 1 (1.9%) | 0.094 |
| Insular | 1 (7.6%) | 2 (3.7%) | 0.482 |
| Corpus callosum | 1 (7.6%) | 2 (3.7%) | 0.482 |
| Multiple | 0 | 6 (11.1%) | 0.588 |
| Surgery | |||
| GTR | 3 (23.1%) | 13 (24.0%) | 0.999 |
| STR | 8 (61.5%) | 16 (29.6%) | 0.051 |
| PR | 2 (15.4%) | 25 (46.3%) | 0.059 |
| Radiation therapy | 13 (100%) | 52 (96.3%) | 0.999 |
| Temozolomide | 12 (92.3%) | 51 (94.4%) | 0.999 |
| Bevacizumab | 5 (%) | 7 (13.0%) | 0.204 |
| Additional surgery | 2 (%) | 5 (9.3%) | 0.614 |
| MGMT positivity | 7 (53.8) | 24 (44.4%) | 0.556 |
GTR: gross total resection, PR: partial resection, STR: subtotal resection.
Table 2.
Clinical and surgical characteristics of 13 patients harboring EGFRvIII-positive glioblastomas
| Patient | Age | Sex | OP | MGMT | P1 (%) | P2 (%) | RT | TMZ | Rec | Treatment at recurrence | OS (m) | D/A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 59 | M | GTR | − | 36 | 0 | + | + | PD | Surgery, TMZ | 62 | A |
| 2 | 81 | M | PR | + | 19 | − | + | + | PD | TMZ | 14 | D |
| 3 | 75 | F | STR | − | 34 | − | + | + | PD | TMZ, Bev | 34 | D |
| 4 | 59 | M | GTR | + | 48 | 0 | + | + | PD | TMZ, RT | 13 | D |
| 5 | 67 | M | STR | − | 56 | 0 | + | + | PD | TMZ, Bev | 36 | D |
| 6 | 67 | F | PR | − | 31 | − | + | + | PD | TMZ | 21 | D |
| 7 | 81 | M | STR | + | 42 | − | + | − | PD | TMZ, RT, Bev | 13 | D |
| 8 | 72 | M | STR | − | 21 | − | + | + | PD | − | 8 | D |
| 9 | 76 | F | STR | − | 100 | 0 | + | + | PD | − | 16 | A |
| 10 | 48 | M | STR | + | 39 | − | + | + | PD | Bev | 12 | A |
| 11 | 69 | M | STR | + | 94 | 0 | + | + | PD | Surgery | 9 | D |
| 12 | 62 | M | STR | + | 49 | 0 | + | + | SD | − | 5 | A |
| 13 | 69 | M | GTR | + | 26 | − | + | + | PD | Bev, TMZ | 15 | A |
A: alive, Bev: bevacizumab, D: dead, GTR: gross total resection, OS: overall survival, P1: EGFRvIII-positive ratio in the cellular component, P2: EGFRvIII-positive ratio in the tumor periphery, PD: progressive disease, PR: partial resection, SD: stable disease, STR: subtotal resection, Rec: recurrence, RT: radiation therapy, TMZ: temozolomide.
In cases #11 and #13 (Table 2), tumor recurrence was detected by follow-up magnetic resonance imaging (MRI), and the additional resection of the tumor was performed 5 and 10 months after surgery, respectively.
Postoperative contrast enhancement MRI was performed within 48 h of surgery. Gross total resection, subtotal resection, and partial resection were defined as no residual tumor, 90–99% removal, and <90% removal of the tumor, respectively.
This study was approved by the Ethics Committee of Niigata University School of Medicine and written informed consent for use of the resected tissues for research purposes was obtained from all patients.
Histology and immunohistochemistry
Surgical specimens were fixed with 20% buffered formalin and embedded in paraffin. Histopathological examination was performed on serial 4-μm-thick sections stained with hematoxylin and eosin (HE) and the Klüver–Barrera (KB) method. Selected sections were processed for immunohistochemistry using the methods described elsewhere.26) As the primary antibodies, we used two mouse monoclonal antibodies against EGFRvIII (clone L8A4; Absolute Antibody, Oxford, UK; dilution 1:200, pretreated by heating and clone DH 8.3; Millipore, Temecula, CA, USA; 1:200), and mouse monoclonal antibodies against human IDH1 R132H (clone H09; Dianova, Hamburg, Germany; 1:100), and vimentin (clone V9, Dako, Glostrup, Denmark; 1:400), MGMT (clone MT3.1; Chemicon International, Temecula, CA, USA; 1:50), and a rabbit polyclonal antibody against glial fibrillary acidic protein (GFAP: Dako, Glostrup, Denmark; 1:1500). We evaluated the distribution and morphology of the vimentin-positive tumor cells. Diagnosis of glioblastoma was based on the criteria listed in the WHO Classification of Tumors of the Central Nervous System.25) MGMT immunoreactivity was evaluated in representative areas of the tumors showing the characteristic features defining their histological grades as described previously using a cutoff of 30%.26) Staining for EGFRvIII was considered positive when staining (usually punctate) was evident in the cytoplasm of tumor cells.
Multiplex ligation-dependent probe assay
To investigate the amplification of EGFR, we performed multiple ligation-dependent probe assay.27) We used five samples, “normal cortex”, “glioblastoma, IDH-wildtype showing EGFRvIII immunoreactivity (patient #10 in Table 2)”, other two “glioblastoma IDH-wildtype”, which showed no EGFRvIII immunoreactivity, and “anaplastic oligodendroglioma”. DNA was extracted from paraffin-embedded tumor tissue using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Detection of EGFR amplification was performed by a multiplex ligation-dependent probe assay (SALSA MLPA probemix P105-D2 Glioma-2, MRC Holland, Amsterdam, The Netherlands).
Western blotting
To confirm the reliability of the EGFRvIII antibody, we performed western blotting of tissue from one glioblastoma patient (#10 in Table 2), one recurrent glioblastoma patient and one patient diagnosed as having an anaplastic oligodendroglioma with IDH-mutation. Proteins extracted from fresh-frozen surgical specimens were separated by 10% SDS-PAGE, using the methods described previously.28) As the primary antibodies, we used a rabbit polyclonal antibody against EGFR (Millipore, Burlington, MA, USA, dilution 1:1000), a mouse monoclonal antibody against EGFRvIII (clone L8A4; Absolute Antibody, 1:200),6,29) a rabbit polyclonal antibody against phosphor-EGFR pTyr1086 (Invitrogen, Carlsbad, CA, USA, 1:1000) and antibody against β-actin (anti-β-Actin-HRP-DirecT, MBL, Nagoya, Japan, 1:2000).
Double-labeling immunofluorescence
A double-labeling immunofluorescence study was performed to assess the expression of EGFRvIII and GFAP in the tumor cells, using the paraffin sections described above. As the primary antibodies, we used the same mouse monoclonal antibody against EGFRvIII (1:50), and rabbit polyclonal antibody against GFAP (1:1000) that we had used for immunohistochemistry at different concentrations. The secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit IgG (Jackson ImunoResearch; 1:200). 4,6-Diamidino-2-phenylindole staining agent was used to visualize the nuclei by microscopy. The images were acquired using an Olympus FV1200 Laser Scanning Confocal Microscope (Olympus Life Science, Tokyo, Japan).
Statistical analysis
Data analysis was perfomed using GraphPad Prism 7.0 statistical software (http://graphpad-prism.software.informer.com/7.0). Overall survival (OS) was calculated from the time of surgery until death, or last follow-up according to the Kaplan–Meier method with long-rank test for comparison of survival between patients with EGFRvIII-positive and EGFRvIII-negative glioblastomas (Table 1). Unpaired Student’s t-test was used for continuous variables, and Fisher’s exact test for categorical variables. Differences at p ≤ 0.05 were considered as statistically significant.
Results
EGFR amplification and EGFRvIII detection
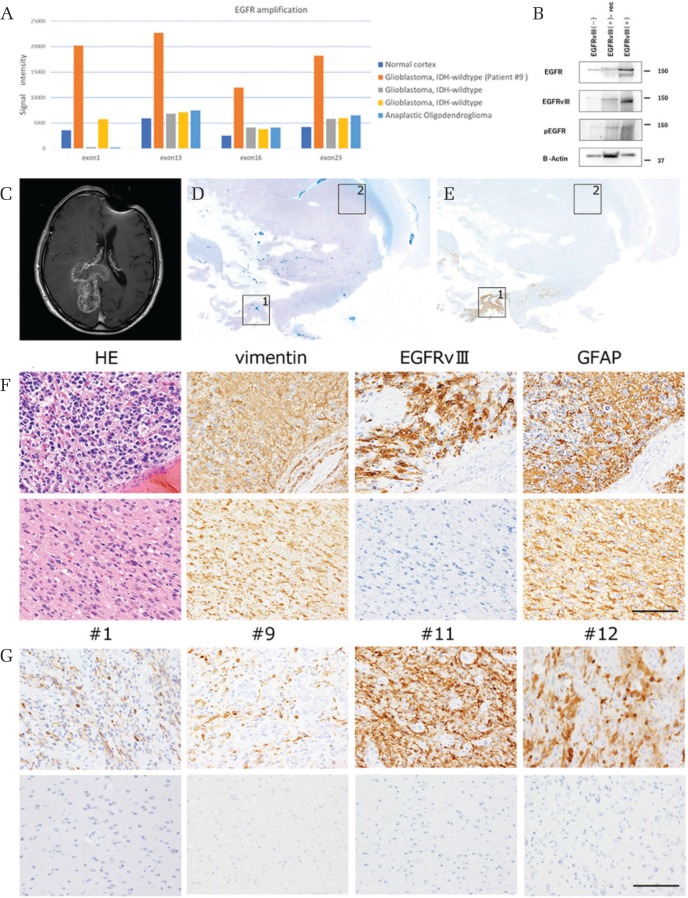
Epidermal growth factor receptor amplification was only detected in the specimen taken from patient #10, which showed EGFRvIII-positivity (Fig. 1A). In two cases of glioblastoma, IDH-wildtype without confirmed EGFRvIII-positivity and one case of anaplastic oligodendroglioma, IDH-mutant did not demonstrate EGFR amplification compared with normal cortex. Western blotting using the monoclonal antibody against EGFRvIII was performed, and we confirmed the presence of a single band at around 145 kDa (Fig. 1B) in protein taken from tumor in a patient with glioblastoma, IDH-wildtype patient (case #10), and recurrent glioblastoma, IDH-wildtype. Two bands were noted for EGFR, the lower band at 145 kDa corresponding with EGFRvIII consistent with previous reports.6,11) Only one band at 170 kDa, corresponding with wildtype EGFR, was found in tissue taken from a patient with anaplastic oligodendroglioma, IDH-mutant.
Fig. 1.
(A) Multiple ligation-dependent probe assay. Horizontal axis means EGFR exon number and vertical axis means signal intensity. One “glioblastoma, IDH-wildtype” patient (case #10 in Table 2) showed higher signal intensity compared with other specimens. (B) EGFR antibody recognized 170 and 145 kDa bands, which correspond to wtEGFR and EGFRvIII, respectively in case #10 and a recurrent glioblastoma, IDH-wildtype case, but not in an anaplastic oligodendroglioma, IDH-mutant case. EGFRvIII antibody recognized the only the 145 kDa band. (C–F) Representation of the features of case #5. (C) T1-weighted magnetic resonance image with contrast enhancement (MRI-T1CE) demonstrates a large tumor in the right occipital lobe. (D) A histology section of the resected brain. Klüver–Barrera stain. The central portion of the tumor indicated by square 1 shows a high nuclear concentration, whereas the peripheral portion indicated by square 2 exhibits relative myelin pallor. (E) A serial section immunostained with the EGFRvIII antibody. EGFRvIII immunoreactivity is seen in the cellular portion of tumor. (F) Higher-power magnification views of the cellular area (upper panels: corresponding to square 1 in D and E) and the peripheral portion (lower panels: corresponding to square 2 in D and E) of the tumor. In the cellular areas, highly cellular tumor cells show immunoreactivity for vimentin, EGFRvIII and GFAP, having multiple fine processes, whereas in the peripheral portion the infiltrating tumor cells show immunoreactivity for vimentin and GFAP, but not for EGFRvIII. Scale bars = 50 μm. (G) Representation of the features of cases #1, #9, #11, #12. The tumor cells expressing EGFRvIII existed in the central portion of the tumor (upper panels), but not in the peripheral portion (lower panels). The tumor cells heterogeneously expressed EGFRvIII in the tumor and the positive ratio at the site with the most positive cells was different between tumors (23–100%).
EGFRvIII expression
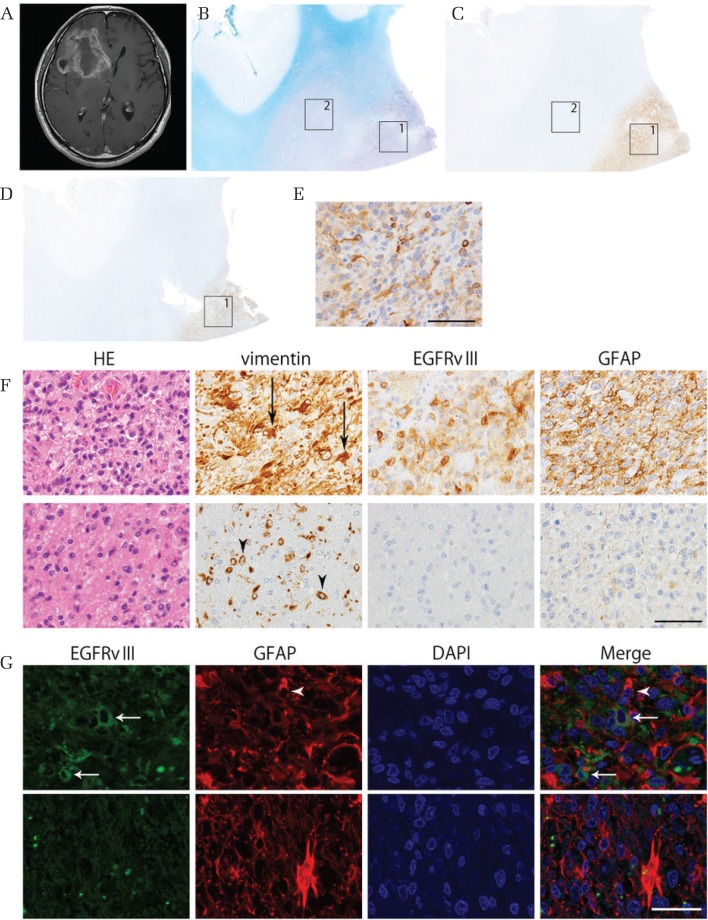
Epidermal growth factor receptor variant III immunoreactivity was detected in 15 specimens, including two specimens at the time of tumor recurrence, taken from 13 patients (Table 2: 19.4% of the 67 patients). The tumor cells heterogeneously expressed EGFRvIII in the tumor and the positive ratio at the site with the most positive cells was different between tumors (median 39%, range 19–100%, Fig. 1G and Table 2), whereas there was no staining (0%) in the periphery of six assessible EGFRvIII gliomas. EGFRvIII distribution was successfully assessed in six specimens (cases #1, 4, 5, 9, 11, 12) because the tumor resection range was wide and the storage condition was optimal. The tumor cells expressing EGFRvIII existed in cellular areas of the tumor, but not in cells of the invading zone (Figs. 1C–1G, 2A–2G). Cellular areas were defined as areas of the tumor with high cellularity, usually near the tumor core and away from the tumor periphery and perinecrotic areas (Figs. 1D–1F, 2B–2F). Two different antibodies against EGFRvIII were used, but similar staining was observed (Fig. 2C–2F). Positivity for both EGFRvIII and GFAP were seen in cellular areas with cells showing prominent features of astrocytic differentiation having multiple fine processes (Figs. 1C–1G, 2A–2G). EGFRvIII staining was not evident in the invading zone of the same specimens (Figs. 1E–1G, 2B–2G). Double-labeling immunofluorescence staining for EGFRvIII and GFAP showed that EGFRvIII and GFAP do not colocalize, but that processes of large, well differentiated, GFAP-positive glia extend to and surround less differentiated, EGFRvIII-positive glial cells in cellular areas of tumor (Fig. 2G).
Fig. 2.
Representation of the features of case #4. (A) T1-weighted magnetic resonance image with contrast enhancement (MRI-T1CE) demonstrates a large tumor in the right frontal lobe. (B) A histological section of the resected brain. Klüver–Barrera stain. The cellular portion of the tumor indicated by square 1 shows a high nuclear concentration, whereas the peripheral portion indicated by square 2 exhibits relative myelin pallor. (C) A serial section stained with the EGFRvIII antibody (Absolute Antibody). EGFRvIII immunoreactivity is seen in the cellular portion of tumor. (D) A section stained with the EGFRvIII antibody (Millipore). (E) Higher-power magnification immunostained with the EGFRvIII antibody (Millipore) (F) Higher-power magnification views of the cellular portion (upper panels: corresponding to square 1 in B and C) and the peripheral portion (lower panels: corresponding to square 2 in B and C) of the tumor. In the central portion, highly cellular tumor cells show immunoreactivity for vimentin, EGFRvIII and GFAP, having multiple fine processes (arrows), whereas in the peripheral portion the infiltrating tumor cells show immunoreactivity for vimentin, but not for EGFRvIII or GFAP, and have a small, rounded morphology (arrowheads). Scale bars = 25 μm. (G) Double-labeling immunofluorescence views of the central portion (upper panels: corresponding to square 1 in B and C) and the peripheral portion (lower panels: corresponding to square 2 in B and C) of the tumor. Processes of large, well differentiated, GFAP-positive glial processes (arrowhead) extend to and surround less differentiated, EGFRvIII-positive glial cells in cellular areas of tumor (arrow), whereas in the peripheral portion the infiltrating tumor cells show no immunoreactivity for EGFRvIII or GFAP. Scale bars = 25 μm.
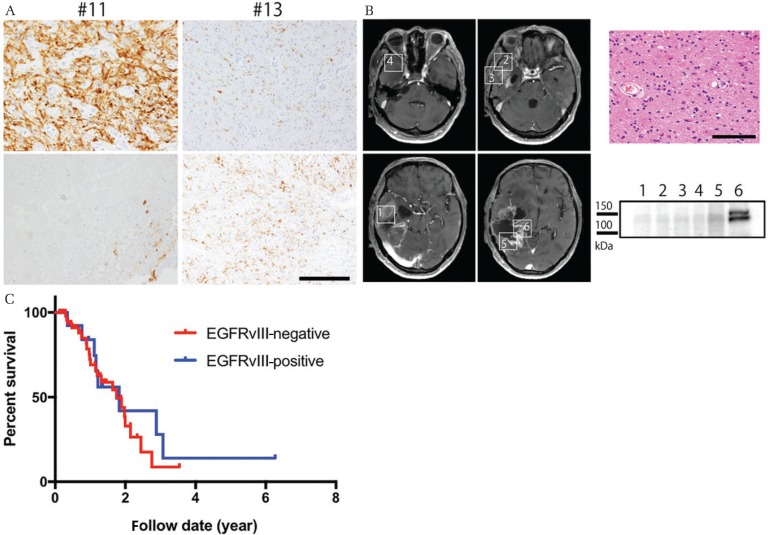
Two patients whose tumor initially showed EGFRvIII immunoreactivity relapsed and underwent second surgeries. EGFRvIII-positive cells were markedly decreased at relapse in one case (#11) and retained in the other (#13) (Fig. 3A), and EGFRvIII-positivity was confirmed by western blot only in an area where viable cells remained (Fig. 3B).
Fig. 3.
(A) Representation of the features of cases #11 and #13 at the time of initial surgery (upper panels) and second surgery (lower panels). In case #11, the tumor cells exhibit astrocytic characteristics and stain positive for EGFRvII, whereas only a few viable EGFRvIII-positive cells and necrosis are noted at relapse. In case #13, positive staining for EGFRvIII was retained at relapse. Scale bars = 50 μm. (B) Frozen tissues were taken from six different areas (areas 1–6) during surgery for relapse, with corresponding tissues for histopathological analysis. EGFRvIII was detected only from area 6, where cellular tumor cells were present (HE section) Scale bar = 50 μm. (C) Kaplan–Meier curves. Overall survival rate (OS) for cases with EGFRvIII-positive and EGFRvIII-negative glioblastoma. P-values were calculated by the long-rank test.
Prognosis
There was no difference in median OS between patients with EGFRvIII expression (1.83 years) and no EGFRvIII expression (1.75 years) (P = 0.547, Fig. 3C).
Discussion
Epidermal growth factor receptor variant III is known to promote angiogenesis through activation of c-myc18) and tumor growth through constitutive activation of the signal transducers and activators of transcription (STAT) and PI3K-Akt pathways.12–17) EGFR amplification is seen in about 40% of primary glioblastomas.30) EGFRvIII, a mutant EGF receptor, is overexpressed in 50–60% of EGFR-amplified glioblastomas, lacks the extracellular ligand-binding domain (exons 2–7 deletion) and is constitutively active.31) Some papers have reported the localization of EGFRvIII within gliomas to be more regional than EGFR.29–33) Physical interaction of EGFRvIII and EGFR, both paracrine and co-expressed within individual tumor cells, have also been elucidated.34,35) Recently, newer antibodies have been developed to detect EGFRvIII,36) and the analysis of localization of EGFRvIII within gliomas has become possible.
In this study, we found EGFRvIII expression in cellular areas of the tumor in a subset of primary glioblastomas, but not at the invading zone. Xenograft models or cultured cell models have shown that tumor cells expressing EGFRvIII are less invasive than EGFRvIII-negative cells.33,37,38) In some reports, tumor cells in the peritumoral brain have been shown to have higher invasive potential.39–41) Taken together, the evidence suggests that EGFRvIII-expressing proliferating tumor cells may lose their expression before they become invasive.
The grow-or-go phenomenon, whereby cell motility and proliferation are mutually exclusive, is well documented in cancer. Hypoxia has been implicated in the switch from proliferation to invasion.42–45) Our results suggest that EGFRvIII may also be an important factor determining whether cancer cells “go-or-grow”.
It is known that in some glioblastomas, sharply delineated, round, GFAP-negative foci arise in the background of a more differentiated lesion.46) This phenomenon is thought to be induced by altered genetic expression, but the exact mechanism is unknown. A similar phenomenon was observed in case #3 (Table 2), where small, round, GFAP-negative tumor cells were found in the infiltrating area of a glioblastoma with a more differentiated core expressing EGFRvIII. These small cells lacked EGFRvIII expression.
Although EGFRvIII expression in tumors is known to be heterogeneous,19,29,33,47) previous studies have focused mainly on the tumor mass, and not the invading zone. Interestingly, a study by Eskilsson et al.33) found EGFRvIII expression near the border of the tumor in two cases of glioblastoma. This study demonstrated the location of EGFRvIII expression at cellular areas of tumor, whereas invading glioma cells lack such expression. A recent review article by Eskilsson et al.48) highlights the heterogeneity of EGFR in glioblastoma and crosstalk between wtEGFR, which enhances tumor cell invasion, and EGFRvIII, which promotes angiogenic tumor cell invasion, and also reviews the potential therapeutic implications to targeted therapies.
In two recurrent cases, EGFRvIII-positive cells were markedly decreased in one case and retained in the other (Figs. 3A and 3B). This observation is not consistent with a detailed study of somatic mutations in glioblastomas at the time of initial diagnosis, which found that EGFRvIII was expressed in the initial tumors but disappeared at relapse.21) A recent report has indicated that EGFRvIII can be eliminated from extrachromosomal DNA of tumor cells when treated with EGFR tyrosine kinase inhibitors, but reappears after drug removal.49) The exact mechanism in which EGFRvIII is eliminated and reemerges needs to be elucidated. These data suggest the importance of evaluating EGFRvIII status not only in the initial tumor, but also at relapse when using EGFRvIII-targeted treatments in the recurrent setting.
There are conflicting reports about the prognostic relevance of EGFRvIII.32,50–53) Some studies have suggested that patients with EGFRvIII-expressing glioblastomas have a good prognosis, whereas others have found no difference in survival. We were unable to demonstrate any such survival difference in this study. The extent of tumor resection has also been reported to affect survival,54) but in this study there was no difference in the extent of resection between the two groups. Given the central location of EGFRvIII-expressing cells within the tumor and the lack of EGFRvIII-expressing cells at the time of relapse, we can speculate that infiltrative EGFRvIII-negative cells may have a prognostic effect.
In conclusion, in a subset of primary glioblastomas, EGFRvIII is expressed in cellular areas of tumor where the tumor cells show features of astrocytic differentiation and GFAP positivity, whereas invading cells show no EGFRvIII expression. This finding suggests that EGFRvIII is involved in tumor proliferation, whereas invading glioma cells lose their expression of EGFRvIII.
Footnotes
Conflicts of Interest Disclosure
The authors declare that they have no conflict of interest. The authors who are members of the Japan Neurosurgical Society (JNS) have registered online Self-reported COI Disclosure Statement Forms through the website for JNS members.
References
- 1).Stupp R, Mason WP, van den Bent MJ, et al. : Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996, 2005 [DOI] [PubMed] [Google Scholar]
- 2).Clarke J, Butowski N, Chang S: Recent advances in therapy for glioblastoma. Arch Neurol 67: 279–283, 2010 [DOI] [PubMed] [Google Scholar]
- 3).Preusser M, de Ribaupierre S, Wöhrer A, et al. : Current concepts and management of glioblastoma. Ann Neurol 70: 9–21, 2011 [DOI] [PubMed] [Google Scholar]
- 4).Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G, ESMO Guidelines Working Group : High-grade glioma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 25 Suppl 3: iii93–iii101, 2014. 24782454 [Google Scholar]
- 5).Reardon DA, Wen PY, Mellinghoff IK: Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro Oncol 16 Suppl 8: viii7–viii13, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Wikstrand CJ, Hale LP, Batra SK, et al. : Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res 55: 3140–3148, 1995 [PubMed] [Google Scholar]
- 7).Cheever MA, Allison JP, Ferris AS, et al. : The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 15: 5323–5337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Babu R, Adamson DC: Rindopepimut: an evidence-based review of its therapeutic potential in the treatment of EGFRvIII-positive glioblastoma. Core Evid 7: 93–103, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Gedeon PC, Choi BD, Sampson JH, Bigner DD: Rindopepimut: anti-EGFRvIII peptide vaccine, oncolytic. Drugs Future 38: 147–155, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Nishikawa R, Ji XD, Harmon RC, et al. : A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA 91: 7727–7731, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Gupta P, Han SY, Holgado-Madruga M, et al. : Development of an EGFRvIII specific recombinant antibody. BMC Biotechnol 10: 72, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Padfield E, Ellis HP, Kurian KM: Current therapeutic advances targeting EGFR and EGFRvIII in glioblastoma. Front Oncol 5: 5, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Gan HK, Cvrljevic AN, Johns TG: The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J 280: 5350–5370, 2013 [DOI] [PubMed] [Google Scholar]
- 14).Thorne AH, Zanca C, Furnari F: Epidermal growth factor receptor targeting and challenges in glioblastoma. Neuro-Oncol 18: 914–918, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Fan QW, Cheng CK, Gustafson WC, et al. : EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 24: 438–449, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Guo G, Gong K, Wohlfeld B, Hatanpaa KJ, Zhao D, Habib AA: Ligand-independent EGFR signaling. Cancer Res 75: 3436–3441, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Villa GR, Mischel PS: Old player, new partner: EGFRvIII and cytokine receptor signaling in glioblastoma. Nat Neurosci 19: 765–767, 2016 [DOI] [PubMed] [Google Scholar]
- 18).Katanasaka Y, Kodera Y, Kitamura Y, Morimoto T, Tamura T, Koizumi F: Epidermal growth factor receptor variant type III markedly accelerates angiogenesis and tumor growth via inducing c-myc mediated angiopoietin-like 4 expression in malignant glioma. Mol Cancer 12: 31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Inda MM, Bonavia R, Mukasa A, et al. : Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 24: 1731–1745, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Zanca C, Villa GR, Benitez JA, et al. : Glioblastoma cellular cross-talk converges on NF-κB to attenuate EGFR inhibitor sensitivity. Genes Dev 31: 1212–1227, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Wang J, Cazzato E, Ladewig E, et al. : Clonal evolution of glioblastoma under therapy. Nat Genet 48: 768–776, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Nobusawa S, Watanabe T, Kleihues P, Ohgaki H: IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 15: 6002–6007, 2009 [DOI] [PubMed] [Google Scholar]
- 23).Lo HW: EGFR-targeted therapy in malignant glioma: novel aspects and mechanisms of drug resistance. Curr Mol Pharmacol 3: 37–52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Maire CL, Ligon KL: Molecular pathologic diagnosis of epidermal growth factor receptor. Neuro Oncol 16 Suppl 8: viii1–viii6, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Louis DN, Ohgaki H, Wiestler OD, Cavenee WK: WHO Classification of Tumours of the Central Nervous System. 4th edn IARC, Lyon, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ogura R, Tsukamoto Y, Natsumeda M, et al. : Immunohistochemical profiles of IDH1, MGMT and P53: practical significance for prognostication of patients with diffuse gliomas. Neuropathology 35: 324–335, 2015 [DOI] [PubMed] [Google Scholar]
- 27).Natté R, van Eijk R, Eilers P, et al. : Multiplex ligation-dependent probe amplification for the detection of 1p and 19q chromosomal loss in oligodendroglial tumors. Brain Pathol 15: 192–197, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Miyahara H, Natsumeda M, Shiga A, et al. : Suppressed expression of autophagosomal protein LC3 in cortical tubers of tuberous sclerosis complex. Brain Pathol 23: 254–262, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H: Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol 14: 131–136, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Smith JS, Tachibana I, Passe SM, et al. : PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst 93: 1246–1256, 2001 [DOI] [PubMed] [Google Scholar]
- 31).Wong AJ, Ruppert JM, Bigner SH, et al. : Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA 89: 2965–2969, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Heimberger AB, Suki D, Yang D, Shi W, Aldape K: The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med 3: 38, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Eskilsson E, Rosland GV, Talasila KM, et al. : EGFRvIII mutations can emerge as late and heterogenous events in glioblastoma development and promote angiogenesis through Src activation. Neuro-oncology 18: 1644–1655, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Luwor RB, Zhu HJ, Walker F, et al. : The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene 23: 6095–6104, 2004 [DOI] [PubMed] [Google Scholar]
- 35).Fan Q, Cheng CK, Gustafson WC, et al. : EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 24: 438–449, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Li G, Mitra SS, Monje M, et al. : Expression of epidermal growth factor variant III (EGFRvIII) in pediatric diffuse intrinsic pontine gliomas. J Neurooncol 108: 395–402, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Lindberg OR, McKinney A, Engler JR, et al. : GBM heterogeneity as a function of variable epidermal growth factor receptor variant III activity. Oncotarget 7: 79101–79116, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Talasila KM, Soentgerath A, Euskirchen P, et al. : EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol 125: 683–698, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Glas M, Rath BH, Simon M, et al. : Residual tumor cells are unique cellular targets in glioblastoma. Ann Neurol 68: 264–269, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Ruiz-Ontañon P, Orgaz JL, Aldaz B, et al. : Cellular plasticity confers migratory and invasive advantages to a population of glioblastoma-initiating cells that infiltrate peritumoral tissue. Stem Cells 31: 1075–1085, 2013 [DOI] [PubMed] [Google Scholar]
- 41).Lemée JM, Clavreul A, Menei P: Intratumoral heterogeneity in glioblastoma: don’t forget the peritumoral brain zone. Neuro-oncology 17: 1322–1332, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM: Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 3: 347–361, 2003 [DOI] [PubMed] [Google Scholar]
- 43).Krishnamachary B, Berg-Dixon S, Kelly B, et al. : Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res 63: 1138–1143, 2003 [PubMed] [Google Scholar]
- 44).Pàez-Ribes M, Allen E, Hudock J, et al. : Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell 15: 220–231, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Kahlert UD, Suwala AK, Raabe EH, et al. : ZEB1 promotes invasion in human fetal neural stem cells and hypoxic glioma neurospheres. Brain Pathol 25: 724–732, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Schmitt HP: Rapid anaplastic transformation in gliomas of adulthood. “Selection” in neuro-oncogenesis. Pathol Res Pract 176: 313–323, 1983 [DOI] [PubMed] [Google Scholar]
- 47).Jeuken J, Sijben A, Alenda C, et al. : Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol 19: 661–667, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Eskilsson E, Røsland GV, Solecki G, et al. : EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro-oncology 20: 743–752, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Nathanson DA, Gini B, Mottahedeh J, et al. : Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343: 72–76, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Shinojima N, Tada K, Shiraishi S, et al. : Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res 63: 6962–6970, 2003 [PubMed] [Google Scholar]
- 51).Heimberger AB, Hlatky R, Suki D, et al. : Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 11: 1462–1466, 2005 [DOI] [PubMed] [Google Scholar]
- 52).Montano N, Cenci T, Martini M, et al. : Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia 13: 1113–1121, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Faulkner C, Palmer A, Williams H, et al. : EGFR and EGFRvIII analysis in glioblastoma as therapeutic biomarkers. Br J Neurosurg 29: 23–29, 2015 [DOI] [PubMed] [Google Scholar]
- 54).Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS: An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg 115: 3–8, 2011 [DOI] [PubMed] [Google Scholar]