Abstract
Human mesenchymal stem cells (hMSCs) are an effective tool in regenerative medicine notably for their intrinsic plentiful paracrine activity rather than differentiating properties. The hMSC secretome includes a wide spectrum of regulatory and trophic factors, encompassing several naked molecules as well as different kinds of extracellular vesicles (EVs). Among EVs, exosomes represent an intriguing population, able to shuttle proteins, transcription factors, and genetic materials, with a relevant role in cell-to-cell communication, modulating biological responses in recipient cells. In this context, the extracellular milieu can greatly impact the paracrine activity of stem cells, modifying their metabolism, and the dynamics of vesicle secretion. In the present study, we investigated the effects elicited on exosome patterning by tailored, ad hoc formulated lipid supplementation (Refeed®) in MSCs derived from human fetal membranes (hFM-MSCs). Wound healing experiments revealed that stem cell exposure to exosomes obtained from Refeed®-supplemented hFM-MSCs increased their migratory capability, although the amount of exosomes released after Refeed® supplementation was lower than that yielded from non-supplemented cells. We found that such a decrease was mainly due to a different rate of exosomal exocytosis rather than to an effect of the lipid supplement on the endocytic pathway. Endoplasmic reticulum homeostasis was modified by supplementation, through the upregulation of PKR-like ER kinase (PERK) and inositol-requiring enzyme 1α (IRE1α). Increased expression of these proteins did not lead to stress-induced, unfolded protein response (UPR)-mediated apoptosis, nor did it affect phosphorylation of p38 kinase, suggesting that PERK and IRE1α overexpression was due to augmented metabolic activities mediated by optimization of a cellular feeding network afforded through lipid supplementation. In summary, these results demonstrate how tailored lipid supplementation can successfully modify the paracrine features in hFM-MSCs, impacting both intracellular vesicle trafficking and secreted exosome number and function.
Keywords: mesenchymal stromal cells, exosomes, endoplasmic reticulum, lipid supplementation, membrane lipidomics
Introduction
Mesenchymal stem cells (MSCs) are a promising tool in regenerative medicine and different hypotheses about their possible use have been evaluated in the last decades. MSCs can be isolated from many organs and tissues, being virtually present in all parts of the body; MSCs are well-recognized cells with a great differentiation potential, being able to generate in vitro different mesenchymal lineage-derived cells, such as osteoblasts, chondrocytes, and adipocytes1, but also cardiac-like cells2, endothelial cells3,4, and even ectodermal lineage cells5.
Often, however, therapeutic benefits mediated by MSC transplantation appear to be mainly due to a secretome-based paracrine activity, rather than a substantial MSC differentiation6,7.
Secretome-mediated MSC beneficial effects are well documented in several clinical conditions8, such as cardiac diseases9–12, central nervous system disorders13–15, renal injury16, articular cartilage defects17–21, spontaneous tendon lesions22, and rheumatic diseases23. We have already demonstrated that transplantation of human MSCs (hMSCs) into infarcted rat hearts enhanced cardiac repair, increasing capillary density, normalizing left ventricular function, and decreasing scar tissue7. These pleiotropic effects were partially due to hMSC secretion of trophic mediators, such as vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF), acting in a paracrine way on different cellular elements of the heart.
It’s now clear that MSCs secrete a wide range of bioactive molecules, with various effects on tissue-resident cells, such as promoting angiogenesis24, enhancing proliferative capability, and inhibiting apoptosis25 and fibrosis26 and many others27. The secretome released from MSCs is not only formed by “naked” molecules (cytokines, chemokines, growth factors, and metabolites) but also by different kinds of extracellular membrane vesicles including exosomes, microvesicles, microparticles, nanovesicles, and others. Exosomes are a characterized population of extracellular vesicles (EVs), with a diameter ranging from 30 to 150 nm28,29, and their protein, RNA, and lipid compositions are catalogued in a dedicated database, ExoCarta30. Unlike microvesicles, that originate at the cellular surface and are released by direct budding of plasma membrane, exosomes are generated within multivesicular bodies (MVBs) through an endolysosomal pathway and released by membrane fusion of MVBs with plasma membrane. Due to its origin, exosome membrane presents endosomal proteins, such as CD9, CD63, and CD81, frequently used for immunoaffinity isolation31. The exact mechanism and regulation of exosome secretion is not yet clear32. There is some evidence that secretion is not completely constitutive but can be modulated by different endogenous and exogenous stimuli33. Furthermore, the exact mechanism of exosome internalization by neighboring cells has not been not fully elucidated. EVs released in the environment can be incorporated into recipient cells by different mechanisms including phagocytosis, endocytosis, pinocytosis, and fusion with plasma membrane34. Once englobed, exosomes could be led to different fates. In one way, exosomes merge into endosomes, undergo transcytosis, and are released into the extracellular space without any processing. In another way, fusion of endosomes with lysosomes compels exosomes to degradation35,36. Unfortunately, there is little evidence about regulatory mechanisms involved in exosome internalization even if exosome uptake appears to be cell type–specific37,38. In recent years, MSC-derived exosomes have received an increasing scientific interest due to their emerging regenerative potential. Furthermore, bypassing problems concerning cell transplantation, exosomes should be considered an appealing alternative to overcome current medical and legal obstacles in advanced therapies. A growing number of studies have investigated their role in regeneration of the cardiovascular system39,40, kidney, liver, and nervous system after acute injury41.
Placenta-derived tissues appear to be a promising source of mesenchymal stromal/stem cells (i.e., amniotic fluid, placenta, fetal membranes, and umbilical cord), due to their availability and easy recovery without any ethical concerns42, and exhibit characteristics comparable to MSCs isolated from other sources43–46.
Recently, we demonstrated that a tailored lipid supplementation (Refeed®) is able to improve in vitro functional properties of MSCs derived from human fetal membranes (hFM-MSCs) by recreating a cell membrane environment more similar to the in vivo physiological cell counterpart47. Refeed® supplementation, preserving membrane physiology, improved biological characteristics of cultured hFM-MSCs, such as proliferative potential, angiogenic ability, and immunomodulatory properties. Overall, Refeed® supplementation contributed to more efficient cell physiology and metabolism47.
The extracellular environment not only can significantly modify cell metabolism but, as a consequence, it can affect secretome patterning48,49. Moreover, biophysical characteristics of membranes can be highly influenced by lipid molecules, in terms of flexibility, robustness and membrane elasticity, and alteration of membrane lipid composition can impact EV release50.
In the present study, we focused on the effects of the previously mentioned tailored lipid supplementation on hFM-MSC-derived exosomes. To this end, we addressed different features and biological actions of exosomes secreted by hFM-MSCs that had been cultured in the absence or presence of Refeed®. Within this context, we also investigated the endocytosis and exocytosis pathways as well as the involvement of endoplasmic reticulum (ER) in exosome secretion.
Materials and Methods
Experimental Plan
We hypothesized that Refeed® supplementation could theoretically directly impact hFM-MSCs, modifying their biological responses as well as indirectly changing the behavior of hFM-MSC-secreted exosomes. To discriminate effects produced by Refeed® supplementation on exosome secretion versus those carried out on cellular metabolism, we designed an ad hoc experimental plan (Table 1). Therefore, for the experiments on hFM-MSCs performed during this study, we created 4 experimental groups: CC, control cells maintained in complete medium and treated with exosomes isolated from control cells; CR, control cells treated with exosomes isolated from Refeed®-supplemented cells; RC, Refeed®-supplemented cells treated with exosomes isolated from control cells; and RR, Refeed®-supplemented cells treated with exosomes isolated from Refeed®-supplemented cells.
Table 1.
Experimental Groups Used in the Present Study.
| Groups | Cell Cultures | Exosomes Source |
|---|---|---|
| CC | Control hFM-MSCs | Control hFM-MSCs |
| CR | Control hFM-MSCs | Refeed®-supplemented hFM-MSCs |
| RC | Refeed®-supplemented hFM-MSCs | Control hFM-MSCs |
| RR | Refeed®-supplemented hFM-MSCs | Refeed®-supplemented hFM-MSCs |
First, we evaluated the quantity and immunophenotype of the released exosomes. Then, we investigated the biological effects of the exosomes on hFM-MSCs, through tube formation as well as wound healing assays. Next, we explored exosome uptake and release kinetics, also with the aid of an exocytosis inhibitor, using membrane fluorescent probes and immunofluorescence. Lastly, we inspected the ER status quantifying the UPR cascade with Western blot and caspase activation in live imaging.
Cell Isolation and Culture
Human term placentas (n = 5) were obtained by scheduled caesarean sections, after written informed consent, according to the policy of St. Orsola-Malpighi University Hospital Ethical Committee (protocol number 1645/2014, ref. 35/2014/U/Tess).
Fetal membranes were processed, as previously described47, by enzymatic treatment with collagenase and trypsin. Isolated hFM-MSCs were seeded in standard medium, Dulbecco’s modified Eagle medium (DMEM; Lonza, Walkersville, MD, USA) containing 10% fetal bovine serum (FBS; Gibco, Life Technologies, Carlsbad, CA, USA), 2 mM Ultraglutamine and 1% penicillin-streptomycin solution (Lonza) with or without Refeed® supplementation, and they were grown at 37 °C with 5% CO2.
During passage 0, Refeed®-supplemented cells were treated, according to the manufacturer’s instructions, with one-third of the total dose in order to allow cells to adapt to the supplement. Starting from the first passage, treated cells were supplemented with a full dose of Refeed® until experiments concluded. Experiments were performed from cell passage four through ten.
Refeed® Supplements
Refeed® supplements (Remembrane Srl, Imola, Italy) are a completely defined combination of non-animal derived lipids and antioxidants solubilized in 1 mL of ethanol (Sigma-Aldrich, St. Louis, MO, USA). One milliliter of Refeed® was diluted in 500 mL of complete cell growth medium, the resulting ethanol concentration being <1% (vol/vol) in the final medium. Refeed® composition and use have been previously described47.
Exosome Purification and Quantification
For exosome isolation, hFM-MSCs were seeded onto T75 tissue culture flasks (Corning Inc., Corning, NY, USA) at a density of 8,000 cells/cm2. After 2 days, when cells reached ∼80% confluency, the culture medium was replaced with 12 mL DMEM supplemented with 10% exosome-depleted FBS (Gibco, Life Technologies), 2 mM Ultraglutamine and 1% penicillin-streptomycin solution, with or without Refeed® supplementation. After 3 additional days, conditioned media were collected to isolate exosomes, while cells were detached and counted. Supernatants were centrifuged at 2,000× g for 30 min to remove cells and debris. Cell-free supernatants and Total Exosome Isolation reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA) were combined in a 2:1 dilution, and exosomes were collected, following the manufacturer’s instructions. Briefly, supernatant/reagent mixtures were vortexed and incubated at 4 °C overnight. The next day, samples were centrifuged at 10,000× g for 1 h at 4 °C. Supernatants were aspirated and discarded, and, to normalize exosome concentration, pellets were resuspended in Dulbecco’s phosphate-buffered saline (DPBS; Gibco, Life Technologies) in a ratio of 100 µL DPBS : 1 × 106 cells and used for downstream analysis or stored at −20 °C.
Exosomes were stained with BODIPY® TR ceramide (Molecular Probes™, Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions. Exosome samples were incubated in a solution with a final dye concentration of 10 µM for 20 min at 37 °C (protected from light). Excess unincorporated dye was removed from labeled exosomes with Exosome Spin Columns (MW 3,000; Invitrogen, Life Technologies), following the standard protocol. Labeled exosomes were analyzed using the Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Based on the obtained quantification, for all following treatments, exosomes were added to cells taking into account the abovementioned proportion (100 µL exosome suspension : 1 × 106 cells) to respect autocrine patterning.
Exosome Cytofluorimetric Characterization
To perform flow cytometric detection, CD81-positive exosomes were isolated with Exosome-Human CD81 Flow Detection (from cell culture; Invitrogen, Life Technologies), following manufacturer’s instructions. Briefly, preenriched exosome solution, prepared using Total Exosome Isolation reagent, was added to a tube containing CD81 isolation beads previously washed and incubated at 4 °C overnight with end-over-end mixing. After incubation, the bead-bound exosomes were isolated with a magnetic separator (MagnaRack, Invitrogen) and washed once with Assay Buffer containing 0.1% bovine serum albumin (BSA, Sigma-Aldrich) in DPBS. Then, isolated CD81-positive exosomes were labeled with anti-human CD9-PerCP Cy5.5 or anti-human CD81-PE antibodies (Invitrogen, Life Technologies), and 150 µL of sample were detected with flow cytometer (Attune™ NxT Acoustic Focusing Cytometer, Life Technologies). Negative control was performed by repeating entirely the previously described staining procedure with DPBS (vehicle) instead of exosomes. Data collected from the experiments were analyzed using Attune NxT Software version 2.5 (Life Technologies). At least 3 independent experiments were performed for each point.
Tube Formation Assay
Analysis of capillary-like tube formation was performed using semisolid medium Cultrex® Basement Membrane Extract (BME, Trevigen, Gaithersburg, MD, USA). hFM-MSCs were detached, counted, and seeded at a density of 30,000 cells/cm2 in a BME-coated 96-well plate. Cells were maintained in basal medium (DMEM, 2 mM Ultraglutamine, 1% penicillin-streptomycin solution) supplemented with exosomes or vehicle (DPBS) as negative control. The plate was introduced into the incubation chamber (OKOlab, Naples, Italy) of the microscope station to maintain CO2, temperature, and humidity in controlled conditions, and time-lapse acquisitions were performed over 24 h with a Nikon Inverted Microscope Eclipse Ti-E (Nikon Instruments, Tokyo, Japan) equipped with a Digital Sight camera DS-03 linked to a computer driving the imaging software NIS-Elements version 4.30 (Nikon Instruments). Total well areas were captured, and tube length was measured using NIS-Elements software tools. At least 3 independent experiments were performed for each point.
Migration Assay
hFM-MSCs were seeded in 24-well plates at a density of 13,000 cells/cm2 and cultured until they reached confluence. Cell monolayers were scratched by a 100-µL sterile pipette tip. Fresh serum-free culture medium containing exosomes or vehicle (DPBS, negative control) was added. Images of the whole wound were taken immediately after the scratch and 16 h post-treatment with a Nikon Inverted Microscope Eclipse Ti-E equipped with a Digital Sight camera DS-03. Scratch areas were measured at 0 and 16 h, and scratch closure percentages were calculated using NIS-Elements software tools. At least 3 independent experiments were performed for each point.
Exosome Tracing Experiments
To trace trafficking, exosomes (or vehicle) were stained with BODIPY TR ceramide as previously described. Alternatively exosomes were labeled using PKH26 Red Fluorescent Cell Linker Kit (Sigma-Aldrich). Briefly, the purified exosome suspension was mixed with the stain solution (PKH26 diluted in Diluent C) at a final dye concentration of 2 µM and incubated for 5 min at room temperature (RT). BODIPY TR ceramide- or PKH26-stained exosomes were added to receiving cells in DMEM FluoroBrite (Gibco, Life Technologies) containing 1% penicillin-streptomycin solution and 2 mM Ultraglutamine. To better visualize exosome internalization, cells were counter-stained with vital dye PKH2 Green Cell Linker Kit (5 µM; Sigma-Aldrich), following the manufacturer’s instructions. Both wide-field fluorescence and z-stack confocal imaging were performed until 24 h.
In separate experiments, the staining solution (BODIPY TR ceramide, 1 µM) was directly added to the cells, omitting exosomes, to follow exocytosis. Unincorporated dye was washed away after 30 min and images were acquired in live imaging for 24 h. Just prior to acquisitions, cells were stained with ER-Tracker™ Blue-White DPX (1:1000 dilution; Molecular Probes™, Life Technologies) to visualize ER.
For inhibiting exocytosis, hFM-MSCs were pretreated for 4 h with Exo2 (50 µM; Sigma-Aldrich) in DMEM FluoroBrite (1% penicillin-streptomycin solution, 2 mM Ultraglutamine) before the addition of BODIPY TR ceramide. After 30 min-incubation with dye, cells were washed with DPBS and images acquired until 24 h. At least 3 independent experiments were performed for each point.
Immunofluorescence
After specific treatments, hFM-MSCs, grown on glass coverslips in 24-well plates, were washed with DPBS and fixed with 4% neutral buffered formaldehyde (Sigma-Aldrich) at RT for 10 min. After fixation, cells were washed with DPBS/0.25% Tween 20 (Sigma-Aldrich) and permeabilized with 1% Triton X-100 (Sigma-Aldrich) at RT for 10 min. Aspecific antibody binding sites were blocked by incubating with 4% BSA in DPBS for 1 h at RT. Cells were then labeled overnight at 4 °C in humid chamber in the presence of mouse anti-human CD81 (1:200 dilution; Invitrogen, Life Technologies). The next day, after 3 washes, cells were stained at RT for 30 min with goat antimouse Dye488 (1:1000 dilution; Bethyl). Nuclei were counter-stained with NucBlue® Fixed Cell ReadyProbes® Reagent (Molecular Probes™, Life Technologies). Fixed cell samples were mounted with antifade AF-400 (Immunological Sciences, Rome, Italy) and observed under a Nikon Inverted Microscope Eclipse Ti-E (Nikon Instruments). Images were acquired with a Digital Sight camera DS-03 through the imaging software NIS-Elements.
Western Blot
Exosomes were lysed in diluted 5× radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific), sonicated for 10 s, and incubated for 15 min at 4 °C. Lysed exosomes were then resuspended in 2× Laemmly Sample Buffer (Bio-Rad, Hercules, CA, USA) and boiled. Twenty microliter of lysed exosomes were separated to sodium dodecyl sulphate - polyacrylamide gel electrophoresis (SDS-PAGE) on a 4% to 20% Mini-PROTEAN® TGX Stain-Free™ Precast Protein Gel (Bio-Rad), and total protein content was detected in stain-free acquisition.
hFM-MSCs were lysed using Mammalian Protein Extraction Reagent (M-PER, Thermo Fisher Scientific) containing protease and phosphate inhibitors (Sigma-Aldrich). After lysis, protein concentrations were measured using Bradford Reagent (Sigma-Aldrich). Cellular lysates were resuspended in 4× Laemmly Sample Buffer and boiled. Fifteen microgram of cellular lysates were separated to SDS-PAGE on a 10% Mini-PROTEAN® TGX Stain-Free™ Precast Protein Gels (Bio-Rad) and transferred to a 0.2-µm nitrocellulose membrane (Bio-Rad) with Trans-Blot® Turbo™ Transfer System (Bio-Rad). After blocking, nitrocellulose membrane was incubated with a diluted primary antibody solution overnight at 4 °C. The next day, the membrane was washed and probed with horseradish peroxidase (HRP) conjugated secondary antibody (1:10000 dilution; AbCam, Cambridge, UK) for 1 h at RT. Bound antibodies were detected with the use of Clarity Western ECL Substrate (Bio-Rad) and quantified by densitometry with ChemiDoc Touch™ Imaging System (Bio-Rad). Detected signal for each sample was normalized to total protein detected in stain-free acquisition. The antibodies used for Western analysis were ER Stress Antibody Sampler Kit (PKR-like ER kinase [PERK], inositol-requiring enzyme 1α [IRE1α], Calnexin, Ero1-Lα, binding immunoglobulin protein [BiP], protein disulfide isomerase [PDI], and CHOP; 1:1000 dilution; Cell Signaling, Danvers, MA, USA), phospho-p38, and p38 (1:1000; Cell Signaling, Danvers, MA, USA).
Analysis of Caspase Activation
hFM-MSCs were grown in a 24-well plate and incubated in the presence of CellEvent™ Caspase-3/7 Green Detection Reagent (Life Technologies) for 60 min at 37 °C according to the manufacturer’s instructions. Detection reagent signal was observed in live cells using a standard FITC filter set in a Nikon Inverted Microscope Eclipse Ti-E.
Statistical Analysis
The statistical analysis was performed using GraphPad Prism version 6 (La Jolla, CA, USA). Data were evaluated by using a 2-tailed, unpaired Student t-test or analysis of variance as appropriate, with Tukey’s post hoc test, assuming a P value less than 0.05 as the limit of significance.
Results
Isolation, Characterization, and Quantification of hFM-MSC-Released Exosomes
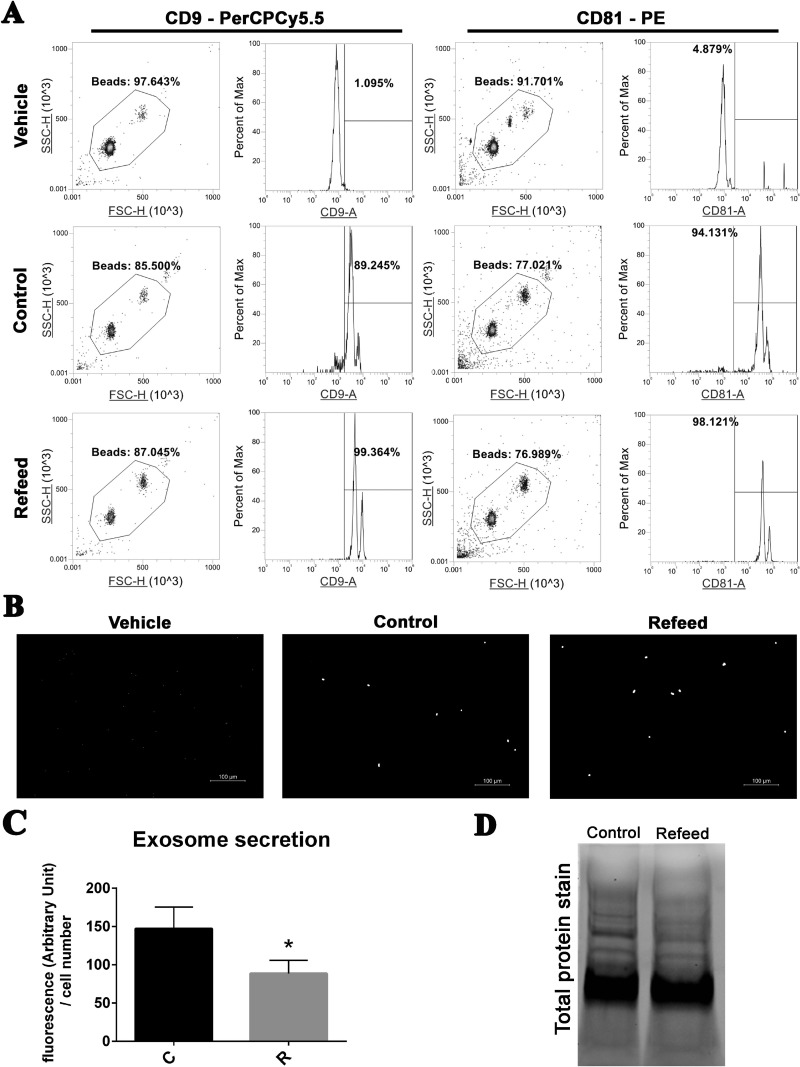
To perform experiments, hFM-MSCs were seeded in complete medium with (R) or without (C) Refeed®, replaced after 2 d by exosome depleted one, supplemented or not with Refeed®, as well. After 3 more days, supernatants were collected and EVs were isolated. Isolated EVs were characterized by the presence of exosome-specific tetraspanin membrane proteins, CD9 and CD81 (Fig. 1). Exosomes of a small size were not able to be visualized by laser in flow cytometric analysis. To circumvent this problem, exosomes were attached to antibody-coated beads before being analyzed. Exosomes were coupled with magnetic beads targeting human CD81 and analyzed for CD9 or CD81 immunoreactivity. The exosome-bead complexes could aggregate with each other, forming doublets, triplets,or even more. So, as suggested by the manufacturer (Life Technologies), we analyzed only a defined fraction of the bead population. As shown in Fig. 1A, both control and Refeed®-supplemented hFM-MSC exosomes were positive for CD9 and CD81, without substantial differences in fluorescence peaks. Exosome-bead complexes prepared for cytofluorimetric analyses were observed through a microscope at high magnification to visually confirm fluorescence collected data (Fig. 1B).
Fig. 1.
Exosome characterization and quantification in media of cells cultured with or without Refeed®. (A) Characterization of specific tetraspanin membrane proteins CD9 and CD81 by cytofluorimetric analysis in CD81-enriched exosomes derived from control (Control) or Refeed®-supplemented (Refeed®) cells; vehicle alone (Vehicle) analysis is also shown. Density plots showing gating strategy used to sort exosomes-anti-CD81 bead complexes (left panel). Histogram plots showing fluorescence intensity detected for CD9 or CD81 from the gated beads (right panel). The numbers above the gating bars on the histograms depict percentage of positive gated beads for the selected marker. Representative of 3 different analysis. (B) Representative wide-field fluorescence gray-scale images of exosomes-bead complexes stained with anti-CD81 PE antibody. (C) Quantification of exosomes stained with BODIPY® TR ceramide isolated from control (C) or Refeed®-supplemented (R) human fetal membranes MSCs. Fluorescence values were normalized on cell number of each sample. *Significantly different from control-derived exosomes (n = 6, statistical test: 2-tailed, unpaired Student’s t-test, *p < .05). (D) Stain-free acquisition of total protein content of control and Refeed® cell-derived exosomes. Representative of 3 different analyses.
Quantification of total exosomes obtained from cell culture supernatants of control and Refeed®-supplemented cells was also achieved. Exosomes were stained with BODYPY TR ceramide and assessed with fluorometer. Detected fluorescence was significantly higher in control exosome samples versus Refeed®-supplemented ones (Fig. 1C). In separate experiments, total proteins isolated from exosomes were visualized with stain-free gel technology, confirming an increase in exosome number in the supernatant of control cells compared to Refeed®-supplemented ones (Fig. 1D). Intriguingly, both observations underpinned a significant decrease in the number of exosomes in Refeed®-supplemented hFM-MSC supernatants. This surprising result prompted us to evaluate the impact of Refeed® treatment on biological activities.
Impact of Refeed® Supplementation on Biological Responses
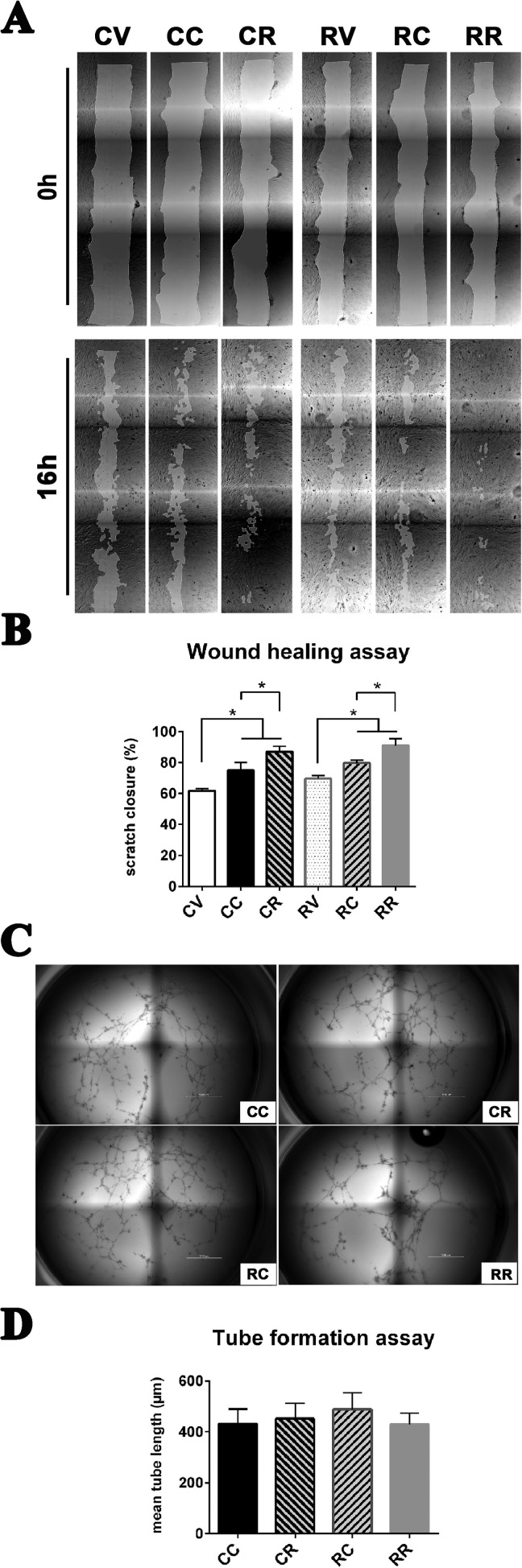
To evaluate biological effects of exosomes isolated from control or Refeed®-supplemented cells, we performed 2 different functional tests: wound healing assay with scratch method and tube formation assay on semisolid medium (Fig. 2).
Fig. 2.
Exosomes from Refeed®-supplemented hFM-MSCs enhance their migration ability without modifying vasculogenic properties. (A) Representative images (4× magnification) of the whole wound area in the scratch assay taken at 0 (upper panel) and 16 h (lower panel) after cell monolayer scratching and concurrent supplementation of vehicle or exosomes. Control cells treated Fig. 2 (continued). with vehicle (CV), with exosomes derived from control hFM-MSCs (CC), or with exosomes derived from Refeed®-supplemented hFM-MSCs (CR); Refeed®-supplemented cells treated with vehicle (RV), with exosomes derived from control hFM-MSCs (RC), or with exosomes derived from Refeed®-supplemented hFM-MSCs (RR). (B) The migration rate of control cells supplemented with exosomes derived from Refeed®-supplemented hFM-MSCs (CR and RR) was significantly higher compared with exosomes derived from control hFM-MSCs (CC and RC) or with vehicle (CV and RV). The migration rate is represented as percentage of scratch closure area. *Significant difference between two groups. Each bar represents the mean ± SD (n = 3, one-way analysis of variance with subsequent Tukey’s test, *p < .05). (C) In vitro capillarogenesis assessed in control (C) and Refeed®-supplemented (R) hFM-MSCs exposed to exosomes derived from control (CC or RC) or Refeed®-supplemented (CR or RR) cells. Representative images of total well area in tube formation assays after 12 h. (D) Bar graph showing quantification of tube formation at 12 h in CC, CR, RC, and RR cells. Each bar represents the mean ± SD (n = 6; one-way analysis of variance with subsequent Tukey’s test).
Wound healing assay
We investigated exosomes isolated from hFM-MSCs cultured with or without Refeed® for their ability to induce cellular migration and growth by using the scratch assay method (Fig. 2A). The migration rate of hFM-MSCs supplemented with Refeed® exosomes (CR and RR) was significantly higher compared to control exosomes (CC and RC) or vehicle (CV and RV), without a significant difference between control (CC vs. CR) and Refeed®-supplemented (RC vs. RR) cells (Fig. 2B). As the number of isolated exosomes was higher in control cells, this result showed that carried biological messages were different in quality and not proportional to the brute number of exosomes.
Tube formation assay
We examined the capability of exosomes isolated from control and Refeed®-supplemented hFM-MSCs to induce vasculogenesis in control and Refeed®-supplemented hFM-MSCs (Fig. 2C). Control or Refeed®-supplemented hFM-MSCs were seeded on BME, and exosomes obtained from control and Refeed®-supplemented cells were added to fresh basal medium. Exosome supplementation induced hFM-MSCs to organize into 3-dimensional vascular networks, but tube-forming capability was not influenced by Refeed® supplementation. Indeed, Refeed® and control cell-derived exosomes seemed to have same inductive effect on Refeed®-supplemented as well as on control hFM-MSCs, and no differences were detected between any of the groups (Fig. 2D).
Vesicle Trafficking Analyses
In order to explain the different amounts of exosomes in control versus Refeed®-supplemented cell supernatants, and to clarify their difference in biological activities, we investigated internalization and secretion mechanisms underpinning vesicle trafficking. In particular, we were interested in evaluating different kinetics of endocytosis as well as exocytosis, as both pathways are involved in exosome-mediated communication between cells.
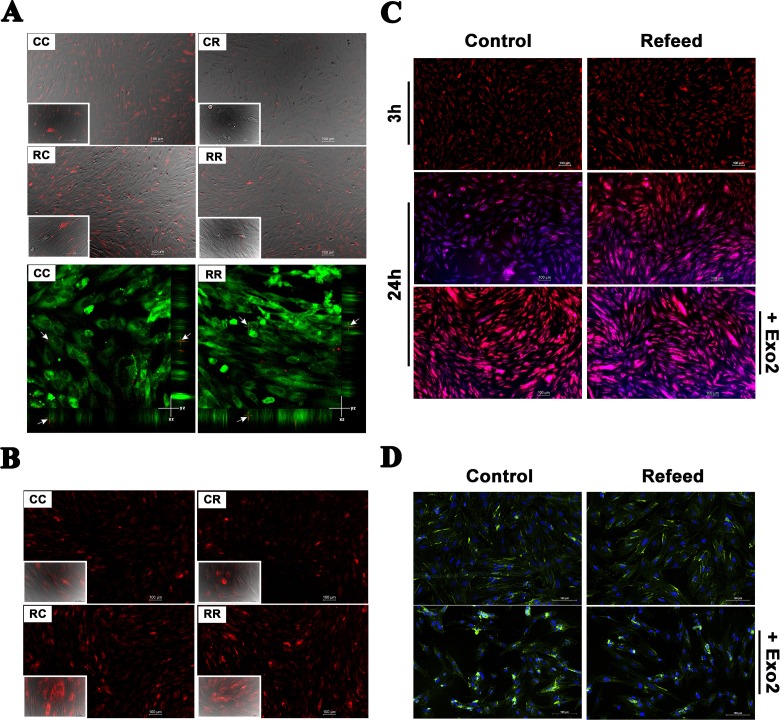
Exosomes isolated from Refeed®-supplemented and control hFM-MSCs were stained with BODIPY TR ceramide and added to control or Refeed®-supplemented hFM-MSCs. BODIPY TR ceramide is a fluorescent lipid probe commonly used for lipid trafficking studies in living cells, and, once internalized into recipient cells, it is able to produce selective staining of the Golgi complex. Labeled exosomes were added to the medium and continuously observed in time-lapse with a fluorescence microscope. After 3 h, most hFM-MSCs (both control and Refeed®-supplemented hFM-MSCs) exhibited intracellular fluorescence (not shown). No detectable differences in cellular uptake rate between control and Refeed®-supplemented cells (CC vs. RC), as well as between control and Refeed®-supplemented cell-derived exosomes (CC vs. CR, RC vs. RR), were detected. Unfortunately, weak fluorescence provided by BODIPY TR ceramide did not allow for an accurate microscopic visualization after exosome uptake inside cells, and it was not possible to take satisfying images with a confocal microscope. Therefore, we decided to stain exosomes with PKH26 and analyze cells after exosome supplementation (Fig. 3A). PKH26 provides fluorescent labeling of vesicles by incorporating aliphatic reporter molecules into the cell membrane lipid bilayer by selective partitioning. The analysis confirmed data collected previously with BODIPY TR ceramide: exosomes were localized into the cytoplasm and a clear intracellular localization was ascertained as validated by the images taken with a confocal microscope 20 min after exosome addition to cell medium (Fig. 3A). Furthermore, at 3 h, we were able to confirm that the overall quantity of exosomes was higher when they were isolated from control cells, but the uptake ability of seeded cells was almost the same, without any evident difference between CC versus RC or CR versus RR groups.
Fig. 3.
Vesicle trafficking is modified by Refeed® supplementation. Control and Refeed® cell-derived exosomes were added to culture medium of control and Refeed®-supplemented hFM-MSCs. (A) Three hours after exosome supplementation, acquisitions were performed. Live imaging merge of Differential Interference Contrast (DIC) and fluorescence channel of control or Refeed®-supplemented hFM-MSCs incubated with PKH26-labeled exosomes (red spots; derived from control [CC or RC] or Refeed®-supplemented cells [CR or RR]). Images confirm quantification data, as control exosomes appear more numerous in every captured field. Confocal live images (lower panel) at 20 min confirmed exosome internalization into cytoplasm of cells counterstained with PKH2 (green). The arrows highlight exosome x- and y-position projection on the z-stack. No differences in terms of uptake kinetic was noticeable. (B) Twenty-four hours after exosome adding, images show different fluorescent levels, which is cell dependent and not exosome origin dependent. Fluorescence live images of control or Refeed®-supplemented hFM-MSCs incubated with BODIPY® TR ceramide-labeled exosomes (red; derived from control [CC or RC] or Refeed®-supplemented cells [CR or RR]; inset: merge of bright field with fluorescence images at higher magnification [40×]; scale bar: 50 µm). (C) Fluorescence live images of control and Refeed®-supplemented cells at 3 and 24 h after staining with BODIPY TR ceramide alone (red). ER-TrackerTM Blue-White DPX (blue) was added just prior to acquisition. Cells stained with BODIPY TR ceramide treated with Exo2 (lower panels). Images show the different turnover of BODIPY TR ceramide dye at 24 h, with a higher clearance in control cells. This difference drops in presence of the inhibitor. (D) Fluorescence merged images of control and Refeed®-supplemented cells treated (lower panels) or not (upper panels) with Exo2. CD81 (green) expression in untreated control cells and Refeed®-supplemented ones is comparable. Exo2 treatment deeply affected control cells that accumulated CD81 in large intracellular compartments. CD81 accumulation is also visible in Refeed®-supplemented cells but to a lesser extent. Cell nuclei were counterstained with NucBlue® Fixed Cell ReadyProbes® Reagent (blue).
While the initial uptake kinetic was apparently unaffected, unexpected results showed up after 24 h of exosome supplementation when BODIPY TR ceramide was used to label vesicles (Fig. 3B). Control and Refeed®-supplemented cells were clearly different, with the latter ones brighter (RC and RR vs. CC and CR) no matter if added with control or Refeed® cell-derived exosomes. Furthermore, as expected, cells treated with labeled control cell-derived exosomes were brighter than those treated with Refeed®-supplemented cell-derived exosomes, but this was clearly due to the fact that control exosomes are more numerous and introduce more fluorescent dye into the cells. On the whole, these data showed us that, upon internalization, the dye was retained by Refeed®-supplemented cells and stored in the Golgi apparatus, while control cells possessed a different clearance kinetic of the internalized dye.
In the attempt to further dissect such different turnover kinetics, we decided to test uptake and release of BODIPY TR ceramide alone. We analyzed the effect of directly added BODIPY TR ceramide in the culture medium of control and Refeed®-supplemented cells in the absence of exosomes (Fig. 3C). As it was observed in the previous experiment, the uptake of BODIPY TR ceramide was similar in control and Refeed®-supplemented cells within the first 3 h after treatment.
Twenty-four hours after BODIPY TR ceramide addition, Refeed®-supplemented cells appeared more stained than control ones, showing a reduced turnover of vesicular and organelle components. To corroborate this idea, control and Refeed®-supplemented cells were added with the Exo2 molecule, a well-known exocytosis inhibitor, prior to supplementation with BODIPY TR Ceramide. As expected, Exo2 was not able to change fluorescence uptake kinetics at 3 h (not shown), but after 24 h, differences in fluorescence between control and Refeed®-supplemented cells were leveled and Exo2-treated control cells were as fluorescent as the Refeed® ones. As a result, the effect of the inhibitor on Refeed®-supplemented cells was not so evident, being Exo2-treated and not treated Refeed®-supplemented cells quite similar in fluorescence levels (Fig. 3C).
To confirm the basal reduction in exocytosis rate in Refeed®-supplemented cells, we also investigated the CD81 intracellular amount (Fig. 3D). Control and Refeed®-supplemented cells, treated or not with Exo2, were fixed and immunostained with anti-CD81 antibody. Accumulation of this tetraspanin in cytoplasmic compartments was comparable in control cells versus Refeed®-supplemented ones. On the contrary, treatment with Exo2 deeply affected control cells that largely accumulated CD81 in intracellular compartments, whereas it did not affect Refeed®-supplemented ones.
Lastly, all these data about trafficking support the idea of different exocytosis rates between control and Refeed®-supplemented cells, albeit the exocytosis process seemed to be regulated in both groups as a balanced turnover of CD81 in absence of Exo2 suggested.
ER Activation
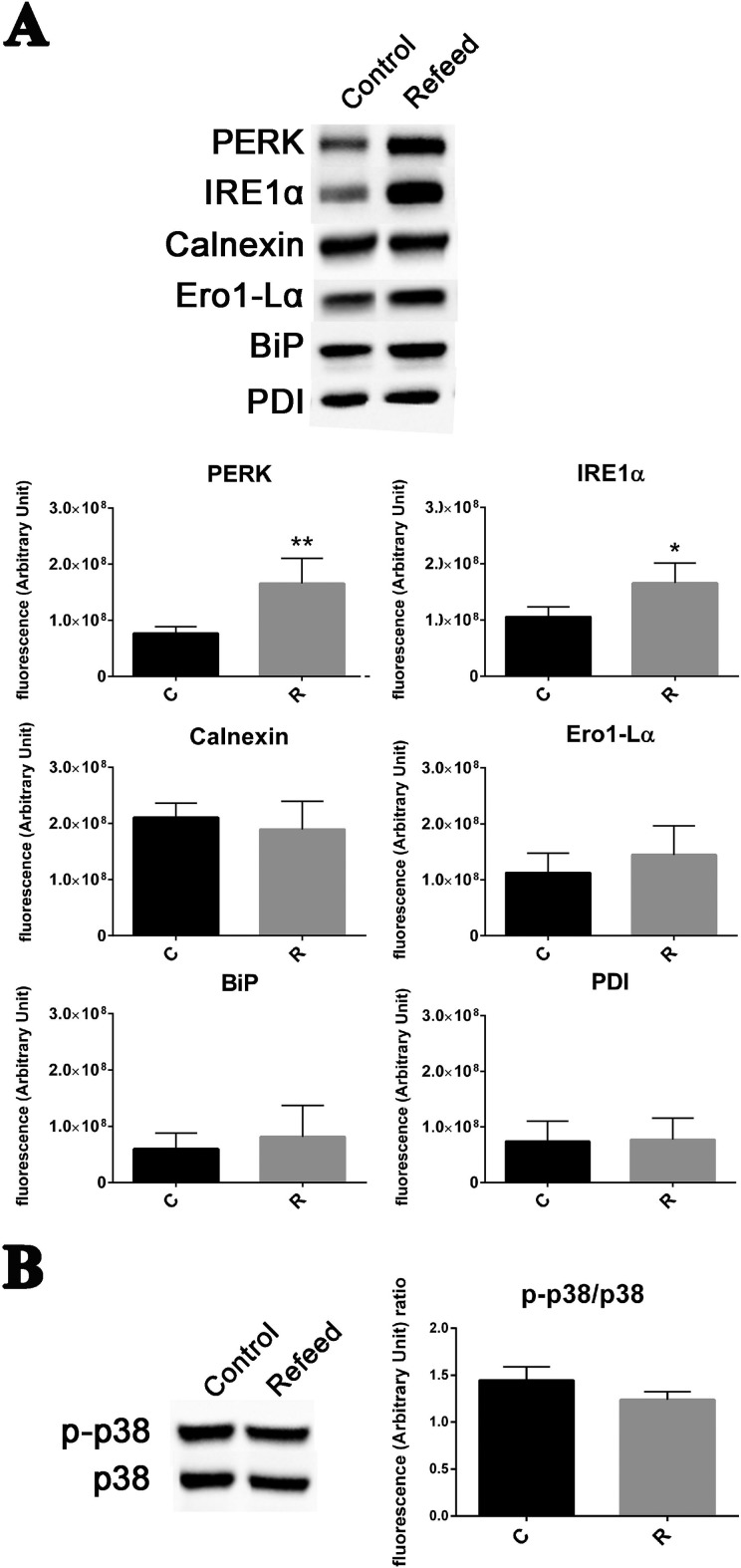
The ER plays a pivotal role in membrane lipid homeostasis and composition, and it is strongly involved in secretory pathways. So, we investigated how Refeed® supplementation may impact ER metabolism. ER responds to stress by evoking “unfolded protein response” (UPR), which is initiated by PERK and IRE1, two ER transmembrane proteins51. Many proteins are activated in the UPR pathway, in particular chaperones such as Calnexin and BiP, and several oxidoreductases of the PDI family51. Finally, ER stress can induce apoptosis, through the activation of the transcriptional factor CHOP (CCAAT-enhancer-binding protein)52.
As shown in Fig. 4A, PERK and IRE1α were upregulated in Refeed®-supplemented cells when compared with control ones, but no differences were detected for Calnexin, Ero1-Lα, BiP, or PDI. CHOP was not expressed both in Refeed®-supplemented and in control cells, suggesting that no apoptotic signals are invoked (data not shown). Phosphorylation of p38, another protein involved in ER stress response53, was not induced by Refeed® supplementation (Fig. 4B). Absence of apoptosis both in Refeed®-supplemented and in control cells was confirmed by verifying that caspase-3 and -7 were not activated (data not shown).
Fig. 4.
Refeed® supplementation increases endoplasmic reticulum activity in hFM-MSCs without activating proapoptotic pathways. (A) Representative Western blot analysis of PERK, IRE1α, Calnexin, Ero1-Lα, binding immunoglobulin protein (BiP), and protein disulfide isomerase (PDI) expression in control (Control) and Refeed®-supplemented (Refeed®) cells. The lower graphs show the comparison of relative densitometric values of the bands normalized to total protein detected in stain-free acquisition in control (C) and Refeed®-supplemented (R) cells (n = 4, statistical test: 2-tailed, unpaired Student’s t-test, *p < .05, **p < .01). (B) Western blot analysis of phospho-p38 and p38 expression in control and Refeed®-supplemented cells. The side graph show the comparison of relative densitometric values of p-p38 normalized on total p38 protein detected (n = 4, statistical test: 2-tailed, unpaired Student’s t test, *p < .05).
Discussion
MSCs live in a complex system of intercellular communication54, which arises through a specific secretome including a broad spectrum of cytokines, chemokines, and different types of vesicles, such as exosomes27,55, with bioactive effects on neighboring cells56–58. There is also emerging evidence that exosomes secreted under different cellular conditions may have different biological effects on the receiving cells33,59. We have previously shown that Refeed®, a tailored lipid supplement, increased cell proliferation, endothelial protein expression, and immunomodulatory properties in hFM-MSCs, with greater ability to express fully functional cell membrane molecules47.
In the present study, we show for the first time that a tailored lipid supplementation formulated ad hoc for hFM-MSCs modulates their EV release and trafficking. While Refeed®-mediated lipid supplementation did not affect exosomal expression of cell surface antigens, surprisingly it led to a significant decrease in the amount of exosomes released into the culture medium. The cellular secretome is influenced by intracellular and extracellular stimuli, both in physiological60 and diseased states61. Similarly, exosome release is modified under different stressful conditions, being enhanced during hypoxia62 and oxidative stress63 or deeply altered in quality and quantity in some pathologies, such as Alzheimer’s disease64 and cardiac disorders65. Accordingly, placenta-derived exosomes, which can be identified in maternal blood66, increased in the maternal circulation of patients with preeclampsia67 or gestational diabetes mellitus68. Nevertheless, a greater number of exosomes may not correspond to enhanced biological responses. In fact, the decrease in exosome release here observed in Refeed®-treated hFM-MSCs may ensue from the acquisition of a more physiological milieu owing to the lipid supplementation to the cell culture environment. The migration assay indicated that hFM-MSC-released exosomes enhanced cell migration in wound healing scratch assay. Moreover, exosomes from Refeed®-supplemented hFM-MSCs increased cell motility to a greater extent than the exosomes from control cells, both in Refeed®-supplemented and non supplemented hFM-MSCs. Thus, the biological activity of exosomes mainly correlates with the cell source rather than with their number, as they were even released to a lower extent in lipid-supplemented than non supplemented cells. It is possible that exosomes released by Refeed®-supplemented hFM-MSCs may be a subpopulation with enhanced biological properties, more useful in healing processes.
Exosomes from different cell types have been shown to be implicated in cell motility69,70 and migration71. Our data, while highlighting an innate ability of hFM-MSC-derived exosomes to induce migration, also provide evidence that human stem cell migration can be enhanced by exosomes released from Refeed®-supplemented hFM-MSCs.
Under our experimental conditions, hFM-MSCs did not exhibit major changes in their spontaneous angiogenic activity when exposed to exosomes obtained from either lipid supplemented or nonsupplemented cells. This observation is not in keeping with the angiogenic potential of MSC-derived exosomes reported elsewhere72. However, these studies have either used different target cells, including human umbilical vein endothelial cells (HUVECs) or microvascular placental derived endothelial cells, or cells grown under hypoxic/stressing conditions72. We cannot exclude that factors responsible for the modulation of the angiogenic properties in hFM-MSCs may be concentrated in the cell secretome outside the exosomal fraction and that adding hFM-MSC-derived exosomes to these cells may have not sorted additional angiogenic responses.
We tried to understand whether a lower number of exosomes in the medium of Refeed®-supplemented cells may be due to a decreased release or to a higher uptake by hFM-MSCs. The results obtained by adding BODIPY TR ceramide-stained control- or Refeed®-derived exosomes to control or Refeed®-supplemented hFM-MSCs suggest a comparable endocytosis rate at 3 h. Similar results were yielded in the presence of PKH26, another reporter molecule. On the contrary, after 24 h, Refeed®-supplemented cells were strongly stained compared to control cells. BODIPY TR ceramide, when it is not added directly to the culture medium, enters hFM-MSCs through stained exosome uptake. Once englobed, exosomes could be immediately redirected to the extracellular environment via transcytosis, or fused with lysosomes, digested and their component assigned to the proper cellular organelle34,36. BODIPY TR ceramide has a high affinity for Golgi apparatus73; in fact, ceramide is a membrane bound lipid synthesized in the ER and then stored in the Golgi apparatus where it can be primarily converted into more complex sphingomyelins and glycosphingolipids74. The decreased accumulation currently observed in control compared to Refeed®-supplemented cells can be due to different exosome trafficking through the cell cytoplasm, preferring a transcytosis metabolism and/or a faster clearance of ceramide from the Golgi apparatus. To discern between these two hypotheses, fluorescent staining was achieved by direct supplementation of BODIPY TR ceramide to the culture medium. We obtained the same results as the previous experiments, with Refeed®-supplemented hFM-MSCs stained more intensely than control cells. Both data suggest more pronounced exocytosis ability from control cells compared to Refeed®-supplemented ones. As further evidence, treatment of control and Refeed®-supplemented cells with Exo2, an exocytosis inhibitor, equalized staining, turning control cell fluorescence into a similar level as that detected in Refeed®-supplemented hFM-MSCs. Moreover, the intracellular amounts of CD81 did not differ between control and Refeed®-supplemented cells under standard conditions, while being increased in control cells after Exo2 treatment. The greater CD81 accumulation may have been caused by an elevated requirement for this tetraspanin in EV assembly due to an increased request in control cells. CD81 tetraspanins modulate exocytosis75, promoting the microvillus formation and extension and enhancing the exterior bending of the plasma membrane76. All our data strongly correlate with a faster exocytosis rate in control cells, which also probably required an accelerated CD81 turnover.
There is a strong link between exocytosis ability and intracellular organelle metabolism77. ER controls multiple functions, being the primary cell site for lipid synthesis and export78. Its perturbations in lipid storage/secretion ultimately contribute to many pathological conditions, such as obesity, diabetes, and atherosclerosis79. ER interacts with the other cellular organelles, also in relation to external stimuli80, in order to maintain cellular homeostasis. Over a long period, ER activation can correlate with adverse circumstances leading to the activation of the so-called UPR that can finally induce apoptosis81. To the contrary, a growing body of evidence indicates that UPR has a role in basal cellular homeostasis and that its activation does not necessarily lead to detrimental consequences82.
We found an increased expression of PERK and IRE1α in Refeed®-supplemented cells compared with control ones, whereas other proteins involved in UPR were equally expressed (Calnexin, BiP, etc.). Nonstress-mediated UPR activation is largely documented72 and can be advantageous under particular circumstances, for example, in supporting increased secretory pathways or in adjusting cellular physiology78. The UPR activation also occurs during differentiation processes83. Indeed, PERK is highly expressed in secretory cells, and it is essential for the viability of exocrine and endocrine pancreatic cells84. Intriguingly, IRE1α is crucial in developmental processes85, and its genetic deletion affects proliferation, development, and ER physiology in the placenta86. We did not find any other indication of stress responses in Refeed®-supplemented hFM-MSCs. Stress protein phospho-p38 was not overexpressed in Refeed®-supplemented cells and no caspase activity was detected. Also, ER stress-activated proapoptotic protein CHOP87 was not found in control and Refeed®-supplemented cells. It is likely that this greater ER activity was correlated with a more active cellular metabolism, as it has been documented in our previous work47.
The presence of new lipid material as in Refeed®-supplemented stem cells may deeply impact on ER and Golgi dynamics also thought to be affected by IRE1α pathway88. The overexpression of PERK and IRE1α observed in Refeed®-supplemented hFM-MSCs may be therefore associated with increased membrane synthesis, reorganization, and trafficking. Membrane biogenesis and reorganization following new lipid uptake may have relevant biomedical implication as shown in macrophages where remodeling of Golgi apparatus promotes transient cytokine synthesis and secretion89. We have already shown that Refeed® supplementation increases membrane fluidity and plasticity in hFM-MSCs by partially restoring the omega6 fatty acid membrane reservoire47. It is now evident that the entire cell membrane network is not homogeneous in its lipid content90,91. In fact, membranes of different organelles show a typical lipid composition tailored to suit their specialized tasks. One of the main lipid subcellular gradients present in mammalian cells, which parallels the main membrane traffic routes, is the gradual enrichment in saturated lipid species at the expense of unsaturated ones along the organelles of the secretory pathway90,91. Hence, it is possible to speculate that Refeed®-supplemented omega6 polyunsaturated fatty acids are mostly incorporated in ER and Golgi membrane networks of in vitro cultured hFM-MSCs. In fact, data reported in this article demonstrate that these fundamental organelles show improved efficiency in most biological processes tested, a possible consequence of the increased fluidity and plasticity of their membrane network, restored by Refeed® supplementation.
Conclusion
Tailored lipid supplementation is able to reinstate a physiological membrane phenotype in hFM-MSCs and to influence a multitude of cell pathways and properties, highlighting the central role of the membrane system in cell physiology. Among the influenced properties, the impact on vesicle trafficking should be carefully considered when therapeutic agents such as cells or exosomes are developed as tools for regenerative medicine applications. Prospective precision/personalized medicine therapies could be afforded by using therapeutic approaches based on modified EVs, obtained by the use of ad hoc lipid formulations tailored to specific clinical applications.
Footnotes
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: Human term placentas (n = 5) were obtained by scheduled caesarean sections, after written informed consent, according to the policy of St. Orsola-Malpighi University Hospital Ethical Committee (protocol number 1645/2014, ref. 35/2014/U/Tess).
Statement of Informed Consent: Human term placentas were obtained by scheduled caesarean sections, after written informed consent, according to the policy of St. Orsola-Malpighi University Hospital Ethical Committee (protocol number 1645/2014, ref. 35/2014/U/Tess).
Authors’ Contribution: The first two authors and last two authors listed contributed equally to this work. CC and CZ conceived, designed the study, performed the experiments and prepared the manuscript; EO, RT, VT, MR, PP performed the experiments and analyzed the data; AC, GS, FA interpreted the data and prepared the manuscript; LB and CV conceived the study, interpreted the data and prepared the manuscript. All authors have read and approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Alexandros Chatgilialoglu and Paola Poggi are cofounders and shareholders of Remembrane Srl, a manufacturer of lipid supplements for use in in vitro culturing. The remaining authors declare that they have no competing interests.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: University of Bologna grants (RFO2015-2016) and Ettore Sansavini Health Science Foundation ONLUS.
References
- 1. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. [DOI] [PubMed] [Google Scholar]
- 2. Li Q, Qi LJ, Guo ZK, Li H, Zuo HB, Li NN. CD73+ adipose-derived mesenchymal stem cells possess higher potential to differentiate into cardiomyocytes in vitro. J Mol Histol. 2013;44(4):411–422. [DOI] [PubMed] [Google Scholar]
- 3. Alviano F, Fossati V, Marchionni C, Arpinati M, Bonsi L, Franchina M, Lanzoni G, Cantoni S, Cavallini C, Bianchi F, et al. Term Amniotic membrane is a high throughput source for multipotent Mesenchymal Stem Cells with the ability to differentiate into endothelial cells in vitro. BMC Dev Biol. 2007;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oswald J, Boxberger S, Jorgensen B, Feldmann S, Ehninger G, Bornhauser M, Werner C. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells. 2004;22(3):377–384. [DOI] [PubMed] [Google Scholar]
- 5. Donega V, Nijboer CH, Braccioli L, Slaper-Cortenbach I, Kavelaars A, van Bel F, Heijnen CJ. Intranasal administration of human MSC for ischemic brain injury in the mouse: in vitro and in vivo neuroregenerative functions. PLoS One. 2014;9(11): e112339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linero I, Chaparro O. Paracrine effect of mesenchymal stem cells derived from human adipose tissue in bone regeneration. PLoS One. 2014;9(9): e107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ventura C, Cantoni S, Bianchi F, Lionetti V, Cavallini C, Scarlata I, Foroni L, Maioli M, Bonsi L, Alviano F, et al. Hyaluronan mixed esters of butyric and retinoic acid drive cardiac and endothelial fate in term placenta human mesenchymal stem cells and enhance cardiac repair in infarcted rat hearts. J Biol Chem. 2007;282(19):14243–14252. [DOI] [PubMed] [Google Scholar]
- 8. Liang X, Ding Y, Zhang Y, Tse HF, Lian Q. Paracrine mechanisms of mesenchymal stem cell-based therapy: current status and perspectives. Cell Transplant. 2014;23(9):1045–1059. [DOI] [PubMed] [Google Scholar]
- 9. Lazzarini E, Balbi C, Altieri P, Pfeffer U, Gambini E, Canepa M, Varesio L, Bosco MC, Coviello D, Pompilio G, et al. The human amniotic fluid stem cell secretome effectively counteracts doxorubicin-induced cardiotoxicity. Sci Rep. 2016;6:29994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang J, Wang J, Guo L, Kong X, Yang J, Zheng F, Zhang L, Huang Y. Mesenchymal stem cells modified with stromal cell-derived factor 1 alpha improve cardiac remodeling via paracrine activation of hepatocyte growth factor in a rat model of myocardial infarction. Mol Cells. 2010;29(1):9–19. [DOI] [PubMed] [Google Scholar]
- 11. Gnecchi M, He H, Noiseux N, Liang OD, Zhang L, Morello F, Mu H, Melo LG, Pratt RE, Ingwall JS, et al. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20:661–669. [DOI] [PubMed] [Google Scholar]
- 12. Liu YH, Peng KY, Chiu YW, Ho YL, Wang YH, Shun CT, Huang SY, Lin YS, de Vries AA, Pijnappels DA, et al. Human placenta-derived multipotent cells (hPDMCs) Modulate cardiac injury: from bench to small and large animal myocardial ischemia studies. Cell Transplant. 2015;24(12):2463–2478. [DOI] [PubMed] [Google Scholar]
- 13. Teixeira FG, Carvalho MM, Panchalingam KM, Rodrigues AJ, Mendes-Pinheiro B, Anjo S, Manadas B, Behie LA, Sousa N, Salgado AJ. Impact of the secretome of human mesenchymal stem cells on brain structure and animal behavior in a rat model of Parkinson’s disease. Stem Cells Transl Med. 2017;6(2):634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drago D, Cossetti C, Iraci N, Gaude E, Musco G, Bachi A, Pluchino S. The stem cell secretome and its role in brain repair. Biochimie. 2013;95:2271–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu KJ, Yu SJ, Chiang CW, Cho KH, Lee YW, Yen BL, Kuo LW, Wang Y. Transplantation of human placenta-derived multipotent stem cells reduces ischemic brain injury in adult rats. Cell Transplant. 2015;24(3):459–470. [DOI] [PubMed] [Google Scholar]
- 16. Bi B, Schmitt R, Israilova M, Nishio H, Cantley LG. Stromal cells protect against acute tubular injury via an endocrine effect. J Am Soc Nephrol. 2007;18:2486–2496. [DOI] [PubMed] [Google Scholar]
- 17. Skoloudik L, Chrobok V, Kalfert D, Koci Z, Sykova E, Chumak T, Popelar J, Syka J, Laco J, Dedková J, et al. Human multipotent mesenchymal stromal cells in the treatment of postoperative temporal bone defect: an animal model. Cell Transplant. 2016;25(7):1405–1414. [DOI] [PubMed] [Google Scholar]
- 18. Chang YH, Liu HW, Wu KC, Ding DC. Mesenchymal stem cells and their clinical applications in osteoarthritis. Cell Transplant. 2016;25(5):937–950. [DOI] [PubMed] [Google Scholar]
- 19. Stoddart MJ, Bara J, Alini M. Cells and secretome—towards endogenous cell re-activation for cartilage repair. Adv Drug Deliv Rev. 2015;84:135–145. [DOI] [PubMed] [Google Scholar]
- 20. van Buul GM, Villafuertes E, Bos PK, Waarsing JH, Kops N, Narcisi R, Weinans H, Verhaar JA, Bernsen MR, van Osch GJ. Mesenchymal stem cells secrete factors that inhibit inflammatory processes in short-term osteoarthritic synovium and cartilage explant culture. Osteoarthritis Cartilage. 2012;20(10):1186–1196. [DOI] [PubMed] [Google Scholar]
- 21. Wu L, Leijten JC, Georgi N, Post JN, van Blitterswijk CA, Karperien M. Trophic effects of mesenchymal stem cells increase chondrocyte proliferation and matrix formation. Tissue Eng Part A. 2011;17(9-10):1425–1436. [DOI] [PubMed] [Google Scholar]
- 22. Lange-Consiglio A, Rossi D, Tassan S, Perego R, Cremonesi F, Parolini O. Conditioned medium from horse amniotic membrane-derived multipotent progenitor cells: immunomodulatory activity in vitro and first clinical application in tendon and ligament injuries in vivo. Stem Cells Dev. 2013;22(22):3015–3024. [DOI] [PubMed] [Google Scholar]
- 23. Maumus M, Jorgensen C, Noel D. Mesenchymal stem cells in regenerative medicine applied to rheumatic diseases: role of secretome and exosomes. Biochimie. 2013;95(12):2229–2234. [DOI] [PubMed] [Google Scholar]
- 24. Chen L, Tredget EE, Wu PY, Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. 2008;3(4): e1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khubutiya MS, Vagabov AV, Temnov AA, Sklifas AN. Paracrine mechanisms of proliferative, anti-apoptotic and anti-inflammatory effects of mesenchymal stromal cells in models of acute organ injury. Cytotherapy. 2014;16(5):579–585. [DOI] [PubMed] [Google Scholar]
- 26. Li T, Yan Y, Wang B, Qian H, Zhang X, Shen L, Wang M, Zhou Y, Zhu W, Li W, et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev. 2013;22(6):845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Satija NK, Singh VK, Verma YK, Gupta P, Sharma S, Afrin F, Sharma M, Sharma P, Tripathi RP, Gurudutta GU. Mesenchymal stem cell-based therapy: a new paradigm in regenerative medicine. J Cell Mol Med. 2009;13(11–12):4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtiö J, El Andaloussi S, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, et al. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8): E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. [DOI] [PubMed] [Google Scholar]
- 33. Feng Y, Huang W, Wani M, Yu X, Ashraf M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PLoS One. 2014;9(2): e88685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3 doi:10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider A, Simons M. Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013;352(1):33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124(pt 3):447–458. [DOI] [PubMed] [Google Scholar]
- 38. Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288(24):17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Hu YW, Zheng L, Wang Q. Characteristics and roles of exosomes in cardiovascular disease. DNA Cell Biol. 2017;36(3):202–211. [DOI] [PubMed] [Google Scholar]
- 40. Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, Torre T, Siclari F, Moccetti T, Vassalli G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103(4):530–541. [DOI] [PubMed] [Google Scholar]
- 41. Marote A, Teixeira FG, Mendes-Pinheiro B, Salgado AJ. MSCs-derived exosomes: cell-secreted nanovesicles with regenerative potential. Front Pharmacol. 2016;7:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Evangelista M, Soncini M, Parolini O. Placenta-derived stem cells: new hope for cell therapy? Cytotechnology. 2008;58(1):33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parolini O, Alviano F, Bagnara GP, Bilic G, Bühring HJ, Evangelista M, Hennerbichler S, Liu B, Magatti M, Mao N, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells. 2008;26(2):300–311. [DOI] [PubMed] [Google Scholar]
- 44. Chen G, Yue A, Ruan Z, Yin Y, Wang R, Ren Y, Zhu L. Comparison of biological characteristics of mesenchymal stem cells derived from maternal origin placenta and Wharton’s jelly. Stem Cell Res Ther. 2015;6:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu W, Morschauser A, Zhang X, Lu X, Gleason J, He S, Chen HJ, Jankovic V, Ye Q, Labazzo K, et al. Human placenta-derived adherent cells induce tolerogenic immune responses. Clin Transl Immunology. 2014;3(5): e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fierabracci A, Del Fattore A, Luciano R, Muraca M, Teti A, Muraca M. Recent advances in mesenchymal stem cell immunomodulation: the role of microvesicles. Cell Transplant. 2015;24(2):133–149. [DOI] [PubMed] [Google Scholar]
- 47. Chatgilialoglu A, Rossi M, Alviano F, Poggi P, Zannini C, Marchionni C, Ricci F, Tazzari PL, Taglioli V, Calder PC, et al. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res Ther. 2017;8(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakopoulou A, Kritis A, Andreadis D, Papachristou E, Leyhausen G, Koidis P, Geurtsen W, Tsiftsoglou A. Angiogenic potential and secretome of human apical papilla mesenchymal stem cells in various stress microenvironments. Stem Cells Dev. 2015;24(21):2496–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xing J, Hou T, Jin H, Luo F, Change Z, Li Z, Xie Z, Xu J. Inflammatory microenvironment changes the secretory profile of mesenchymal stem cells to recruit mesenchymal stem cells. Cell Physiol Biochem. 2014;33(4):905–919. [DOI] [PubMed] [Google Scholar]
- 50. Najafinobar N, Mellander LJ, Kurczy ME, Dunevall J, Angerer TB, Fletcher JS, Cans AS. Cholesterol alters the dynamics of release in protein independent cell models for exocytosis. Sci Rep. 2016;6:33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim DS, Kim JH, Lee GH, Kim HT, Lim JM, Chae SW, Chae HJ, Kim HR. p38 Mitogen-activated protein kinase is involved in endoplasmic reticulum stress-induced cell death and autophagy in human gingival fibroblasts. Biol Pharm Bull. 2010;33(4):545–549. [DOI] [PubMed] [Google Scholar]
- 54. Krinner A, Roeder I. Quantification and modeling of stem cell-niche interaction. Adv Exp Med Biol. 2014;844:11–36. [DOI] [PubMed] [Google Scholar]
- 55. Meirelles Lda S, Fontes AM, Covas DT, Caplan AI. Mechanisms involved in the therapeutic properties of mesenchymal stem cells. Cytokine Growth Factor Rev. 2009;20(5–6):419–427. [DOI] [PubMed] [Google Scholar]
- 56. Koniusz S, Andrzejewska A, Muraca M, Srivastava AK, Janowski M, Lukomska B. Extracellular vesicles in physiology, pathology, and therapy of the immune and central nervous system, with focus on extracellular vesicles derived from mesenchymal stem cells as therapeutic tools. Front Cell Neurosci. 2016;10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bruno S, Deregibus MC, Camussi G. The secretome of mesenchymal stromal cells: role of extracellular vesicles in immunomodulation. Immunol Lett. 2015;168(2):154–158. [DOI] [PubMed] [Google Scholar]
- 58. Heldring N, Mäger I, Wood MJ, Le Blanc K, Andaloussi SE. Therapeutic potential of multipotent mesenchymal stromal cells and their extracellular vesicles. Hum Gene Ther. 2015;26(8):506–517. [DOI] [PubMed] [Google Scholar]
- 59. Li L, Jin S, Zhang Y. Ischemic preconditioning potentiates the protective effect of mesenchymal stem cells on endotoxin-induced acute lung injury in mice through secretion of exosome. Int J Clin Exp Med. 2015;8(3):3825–3832. [PMC free article] [PubMed] [Google Scholar]
- 60. Gardner OF, Fahy N, Alini M, Stoddart MJ. Differences in human mesenchymal stem cell secretomes during chondrogenic induction. Eur Cell Mater. 2016;31:221–235. [DOI] [PubMed] [Google Scholar]
- 61. Noman MZ, Hasmim M, Messai Y, Terry S, Kieda C, Janji B, Chouaib S. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol. 2015;309(9): C569–C579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neven KY, Nawrot TS, Bollati V. Extracellular vesicles: how the external and internal environment can shape cell-to-cell communication. Curr Environ Health Rep. 2017;4(1):30–37. [DOI] [PubMed] [Google Scholar]
- 63. Medvedev R, Hildt E, Ploen D. Look who’s talking-the crosstalk between oxidative stress and autophagy supports exosomal-dependent release of HCV particles. Cell Biol Toxicol. 2017;33(3):211–231. [DOI] [PubMed] [Google Scholar]
- 64. Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR. Plasma exosomal miRNAs in persons with and without Alzheimer disease: alteredexpression and prospects for biomarkers. PLoS One. 2015;10(10): e0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Amabile N, Rautou PE, Tedgui A, Boulanger CM. Microparticles: key protagonists in cardiovascular disorders. Semin Thromb Hemost. 2010;36(8):907–916. [DOI] [PubMed] [Google Scholar]
- 66. Sabapatha A, Gercel-Taylor C, Taylor DD. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol. 2006;56(5-6):345–355. [DOI] [PubMed] [Google Scholar]
- 67. Orozco AF, Jorgez CJ, Ramos-Perez WD, Popek EJ, Yu X, Kozinetz CA, Bischoff FZ, Lewis DE. Placental release of distinct DNA-associated micro-particles into maternal circulation: reflective of gestation time and preeclampsia. Placenta. 2009;30(10):891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Salomon C, Scholz-Romero K, Sarker S, Sweeney E, Kobayashi M, Correa P, Longo S, Duncombe G, Mitchell MD, Rice GE, et al. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65(3):598–609. [DOI] [PubMed] [Google Scholar]
- 69. Hu L, Wang J, Zhou X, Xiong Z, Zhao J, Yu R, Huang F, Zhang H, Chen L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci Rep. 2016;6:32993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rani S, Ritter T. The exosome—a naturally secreted nanoparticle and its application to wound healing. Adv Mater. 2016;28(27):5542–5552. [DOI] [PubMed] [Google Scholar]
- 71. Salomon C, Yee S, Scholz-Romero K, Kobayashi M, Vaswani K, Kvaskoff D, Illanes SE, Mitchell MD, Rice GE. Extravillous trophoblast cells-derived exosomes promote vascular smooth muscle cell migration. Front Pharmacol. 2014;5:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Salomon C, Ryan J, Sobrevia L, Kobayashi M, Ashman K, Mitchell M, Rice GE. Exosomal signaling during hypoxia mediates microvascular endothelial cell migration and vasculogenesis. PLoS One. 2013;8(7): e68451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Meisslitzer-Ruppitsch C, Röhrl C, Ranftler C, Neumüller J, Vetterlein M, Ellinger A, Pavelka M. The ceramide-enriched trans-Golgi compartments reorganize together with other parts of the Golgi apparatus in response to ATP-depletion. Histochem Cell Biol. 2011;135(2):159–171. [DOI] [PubMed] [Google Scholar]
- 74. Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol. 2010;688:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8(2):89–96. [DOI] [PubMed] [Google Scholar]
- 76. Fritzsching B, Schwer B, Kartenbeck J, Pedal A, Horejsi V, Ott M. Release and intercellular transfer of cell surface CD81 via microparticles. J Immunol. 2002;169(10):5531–5537. [DOI] [PubMed] [Google Scholar]
- 77. Derby MC, Gleeson PA. New insights into membrane trafficking and protein sorting. Int Rev Cytol. 2007;261:47–116. [DOI] [PubMed] [Google Scholar]
- 78. Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189(5):783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lagace TA, Ridgway ND. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833(11):2499–2510. [DOI] [PubMed] [Google Scholar]
- 80. Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol. 2009;41(10):1805–1816. [DOI] [PubMed] [Google Scholar]
- 81. Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167(1):35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. [DOI] [PubMed] [Google Scholar]
- 84. Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. [DOI] [PubMed] [Google Scholar]
- 85. Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115(2):268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106(39):16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833(12):3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Volmer R, Ron D. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol. 2015;33:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tian Y, Pate C, Andreolotti A, Wang L, Tuomanen E, Boyd K, Claro E, Jackowski S. Cytokine secretion requires phosphatidylcholine synthesis. J Cell Biol. 2008;181(6):945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Antonny B, Vanni S, Shindou H, Ferreira T. From zero to six double bonds: phospholipid unsaturation and organelle function. Trends Cell Biol. 2015;25(7):427–436. [DOI] [PubMed] [Google Scholar]
- 91. Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510(7503):48–57. [DOI] [PubMed] [Google Scholar]