Abstract
Cell-derived microvesicles (MVs) are a recently discovered mechanism of cell-to-cell communication. Our previous data show that MVs secreted by equine amniotic mesenchymal-derived cells (AMCs) are involved in downregulation of proinflammatory genes in lipopolysaccharide-stressed equine tendon and endometrial cells. The aim of the present study was to evaluate whether AMC-MVs contain selected microRNAs (miRNAs) involved in inflammation. Two pools of cells, derived from 3 amniotic membranes each, and their respective MVs were collected. Small RNAs were extracted and deep sequenced, followed by miRNA in silico detection. The analysis identified 1,285 miRNAs, which were quantified both in AMCs and MVs. Among these miRNAs, 401 were classified as Equus caballus miRNAs, 257 were predicted by homology with other species (cow, sheep, and goat), and 627 were novel candidate miRNAs. Moreover, 146 miRNAs differentially expressed (DE) in AMCs and MVs were identified, 36 of which were known and the remaining were novel. Among the known DE miRNAs, 17 showed higher expression in MVs. Three of these were validated by real time polymerase chain reaction: eca-miR-26, eca-miR-146a, and eca-miR-223. Gene ontology analysis of validated targets showed that the DE miRNAs in cells and MVs could be involved both in immune system regulation by modulating interleukin signaling and in the inflammatory process. In conclusion, this study suggests a significant role of AMCs in modulating immune response through cell–cell communication via MV-shuttling miRNAs.
Keywords: amniotic-derived cells, equine, shedding vesicles, miRNA
Introduction
In cell therapy, for many years, mesenchymal stromal cells (MSCs) have been used for their action based on their ability to differentiate into reparative or replacement cell types, enhance the nutrient supply, and improve the survival and function of the endogenous cells via paracrine actions1–3. Considering the inhospitable environment of the lesion, due to the presence of inflammatory cells and molecules that lead to a premature death of stem cells used in cell therapy4, it is now suggested that paracrine signaling is mainly involved in repair. Indeed, some studies revealed that cell secretion alone, or conditioned medium (CM), without the stem cells themselves, led to tissue repair in various conditions that involve tissue/organ damage5 – 7. Each stem cell type, depending on its very typical nature, can secrete different factors including small soluble molecules (neurotransmitters, chemokines, cytokines, and hormones) that could behave in a paracrine or in an endocrine manner8. However, some factors, such as nucleic acid, cannot span the membranes freely and a vehicle should be involved to facilitate the crossing. Microvesicles (MVs), which are released by cells into the extracellular environment9,10, have been supposed as shuttles of the functional components for MSC paracrine action. The MVs, or extracellular vesicles, are membrane-bound vesicles that transfer molecules such as lipids, proteins, and nucleic acids from one cell to another, thereby influencing the recipient cell function11,12. The MVs are classified in shedding vesicles, if released by budding of cell membrane, and exosomes that form in multivesicular bodies are released by exocytosis. These 2 kinds of MVs are also different in size: 30 to 120 nm for exosomes and 100 to 1,000 nm for shedding vesicles10. The MVs have been studied in several biological processes and are now characterized as novel mediators of intercellular communication both in healthy systems and during disease pathogenesis13. Their proregenerative role is described in several models of tissue regeneration including regeneration of the kidney10,14, heart15, liver16,17, and nervous tissues18. Many other studies have been performed on the ability of MVs to carry information between cells, with the aim of ameliorating a pathological situation. Some of these studies demonstrated that MVs from different cell sources have immunological properties, since they are able to differentially modulate T, B, and natural killer (NK) cell functions19.
Our research group tested the CM, derived from equine amniotic mesenchymal cells (AMCs), in the treatment of spontaneous tendon lesion in sport horses, showing angiogenic and healing properties mediated by paracrine mechanisms6. These results overlap with those previously obtained treating the same pathology with only AMCs20.
It is important to emphasize that the CM obtained by amniotic membrane holds the same in vivo potential of cells. On the contrary, bone marrow–derived cells and their CM exert immunomodulatory potential only if the cells are cultured in the presence of activating conditions21. This increases the interest for the therapeutic potential of amniotic-derived cells based on their paracrine effects and on their released factors. Indeed, to understand the same therapeutic effects in the absence of cells, Lange-Consiglio et al.22 verified the presence and the type of MVs secreted by AMCs and classified these MVs as shedding vesicles. Then, they tested the action of AMC-MVs on tendon and endometrial cells stressed in vitro by lipopolysaccharide (LPS). They observed the incorporation of labeled MVs within both cell lines, the downregulation of tumor necrosis factor-α (TNF-α), interleukin 6 (IL-6), IL-1β, matrix metalloproteinase-1 (MMP-1), and MMP-13 genes and the restoration of transforming growth factor-β (TGF-β) expression22,23. Undoubtedly, MVs and their cargo play key roles in these effects, but the exact mechanisms regarding the interaction of MVs and the injured tissues need further investigation. The main question to explain all these data is “how do MVs act?” One of the most attractive characteristics of MVs is their ability to transfer RNAs (tRNAs) from one cell to another, thereby allowing the transferred RNAs to affect the target cells. In recent years, a remarkable finding demonstrated that MV-contained mRNAs could be transferred in recipient cells and translated into proteins9,24. Furthermore, MVs containing RNAs have been shown to be transferred from MSCs to injured cells and to contribute to tissue recovery25. Additionally, in 2010, MVs were shown to transfer microRNAs (miRNAs) between cells, and these miRNAs displayed RNA interference effects in the recipient cells26–28. This means that MVs may have a functional impact on the target cells through miRNA delivery. The miRNAs are a group of small (21–24 nt) noncoding RNAs that function as posttranscriptional regulators of gene expression by either triggering mRNA cleavage or repressing translation29–31. Cantaluppi et al.14 reported that, in MVs derived from endothelial progenitor cells, specific enrichment of mRNAs involved in cell proliferation, transcription, and immune regulation was detected, providing an example of selective uptake of RNA species into MVs. The mechanism of mRNA and miRNA compartmentalization within MVs has not been clarified; however, it is possible that ribonucleoproteins, that mediate the fate of RNAs within the cells, could be involved32. The transfer of miRNAs by MVs could be important in tissue regeneration; indeed, the positive effect of these MVs is inhibited by the knockdown of Dicer, essential for miRNA processing14. These data indicate an important and potential role of MVs-transferred miRNA in stimulating tissue regeneration.
In this context, in addition to previous characterization of AMC-derived MVs22, the aim of this study was to comparatively investigate the miRNA content in AMCs and their MVs to assess if also in this cell line a compartmentalization of miRNAs involved in anti-inflammatory processes exists.
Materials and Methods
Chemicals, cell culture media, and supplements were obtained from Sigma (Milano, Italy) unless otherwise specified, and tissue culture dishes were purchased from Euroclone (Milano, Italy).
Tissue Collection and Cell Isolation
All procedures to collect allanto-amniotic membrane were conducted following standard veterinary practice and in accordance with 2010/63 European Union directive on animal protection and Italian Law (D.L. No. 116/1992). Allanto-amniotic membranes were obtained at term from normal pregnancies of 6 mares and amniotic cells were obtained as described by Lange-Consiglio et al.33 Briefly, the amniotic membrane was stripped from the overlying allantois and cut into small pieces (about 9 cm2 each) before starting the enzymatic digestion. Then, amnion fragments were incubated for 9 min at 38.5 °C in phospate buffer saline (PBS) containing 2.4 U/mL dispase (Becton Dickinson, Milan, Italy). After a resting period (5–10 min) at room temperature in high-glucose Dulbecco’s modified Eagle’s medium (HG-DMEM; EuroClone), supplemented with 10% heat inactivated fetal bovine serum (FBS) and 2 mM l-glutamine, the fragments were digested with 0.93 mg/mL collagenase type I and 20 mg/mL DNase (Roche, Mannheim, Germany) for approximately 3 h at 38.5 °C. The amnion fragments were removed, and mobilized cells were passed through a 100 mm cell strainer before being collected by centrifugation at 200× g for 10 min. Cells cultures were established in HG-DMEM supplemented with 10% FBS, penicillin (100 UI/mL)-streptomycin (100 mg/mL), 0.25 mg/mL amphotericin B, 2 mM l-glutamine, and 10 ng/mL epidermal growth factor (EGF). Amniotic cells were cultured and expanded to passage (P) 3 to obtain MVs. At the same passage, 2 million of AMCs from each amniotic membrane were washed in PBS, treated with Trizol solution (Invitrogen, Carlsbad, CA, USA) and stored at −80 °C until use.
Isolation and Measurements of MVs
Microvesicles were obtained from the culture media of P3 AMCs cultured for a week with HG-DMEM supplemented with 10% MVs-deprived FBS, and overnight in HG-DMEM deprived of FBS and supplemented with 0.5% bovine serum albumin (BSA). The isolation and measurements of MVs were performed as previously described by Lange-Consiglio et al.22 Briefly, the overnight culture media were centrifuged at 2,000g for 20 min to remove debris, then at 100,000g (Beckman Coulter Optima L-100 K ultracentrifuge) for 1 h at 4 °C. The pellet was washed in serum-free medium 199 containing N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (HEPES) 25 mM and submitted to a second ultracentrifugation under the same conditions. The final pellet was immediately resuspended in HG-DMEM, and a sample of the resuspended pellet was taken for measurement of MV size and concentration. The remaining pellet was treated with RNase, with 800 μL of Trizol solution, stored at −80 °C, and used for RNA extraction. Size and concentration of MVs were evaluated by the Nanosight LM10 instrument (Nanoparticle Tracking Analysis [NTA], Nano-Sight Ltd., Amesbuty, United Kingdom), which permits discrimination of microparticles less than 1 µm in diameter. The software (NTA Version 2.0 analytic software) allows for the analysis of video images of particle movement under Brownian motion and the calculation of a diffusion coefficient, sphere equivalent, and hydrodynamic radius of particles by using the Strokes–Einstein equation.
RNA Isolation
Samples for RNA isolation were represented by 2 pools of amniotic cells (each obtained by pooling 3 amniotic membranes) and by the 2 corresponding pools of MVs. Total RNA was isolated from isolate MVs and from cells. Total RNA was purified by NucleoSpin® miRNA kit (Macherey-Nagel, Germany), following the protocol in combination with TRIzol® (Invitrogen, Carlsbad, CA, USA) lysis with small and large RNA in 1 fraction (total RNA). Concentration and quality of RNA were determined by Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). The isolated RNAs were stored at −80 °C until use.
Library Preparation and Sequencing
Four libraries were obtained either for MVs or cells, representing 2 different isolated pools and 2 technical replicates each. The libraries were prepared using TruSeq Small RNA Library Preparation kits, according to manufacturer’s instructions (Illumina). Libraries were then purified on a Pippin Prep system (Sage Science, Beverly, MA, USA) to recover from 125 to 167 nt fraction containing mature miRNAs. The quality and yield after sample preparation were measured with an Agilent 2200 Tape Station, High Sensitivity D1000. The obtained libraries were quantified by Real Time PCR with KAPA Library Quantification Kits (Kapa Biosystems, Inc., Wilmington, MA, USA). Libraries were sequenced on a single lane of Illumina Hiseq 2000 (San Diego, CA, USA).
miRNA Data Analysis
After quality check with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and trimming with Trimmomatic34. Illumina sequences were input to miRDeep235 for miRNA detection and discovery. Equus caballus miRNAs available at MirBase (http://www.mirbase.org/) were used to accomplish known miRNA detection. Known miRNAs from related species (sheep, cow, and goat) available at MirBase were also input into miRDeep2 to support the individuation of novel miRNAs. The miRDeep2 quantifier module was used to quantify expression and retrieve counts for the detected known and novel miRNAs. Differential expression analyses between samples were run with the Bioconductor edgeR package (version 2.4) (false discovery rate [FDR] < 0.000001)36. MicroRNA cluster analysis was performed with Genesis37. Box-plot graphic was generated with BoxPlotR38. MicroRNA target prediction and functional analysis were performed by Ingenuity Pathway Analysis (IPA, Ingenuity System, www.ingenuity.com). Human homologous miRNAs were analyzed with miRNA target filter (IPA) to attribute (experimentally observed) target genes. Finally, miRNA target mRNA and the corresponding experimental log ratios were used for pathway analysis.
miRNA validation by quantitative polymerase chain reaction (q-PCR)
Samples of RNA isolated from each organ were retrotranscribed with miScript II RT Kit following the manufacturer’s instructions (Qiagen, Inc., Valencia, CA, USA). Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) was carried out on cDNAs with 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Reactions were done in 10 μL volumes containing 0.5 μL of each forward and 0.5 μL of universal reverse primer (Qiagen, Inc.), 4 μL cDNA, and 5 μL 2× Power SYBR® Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocols. The primers used for eca-miR-146a, eca-miR-26, and eca-miR-223 were Hs_miR-146a_1, Hs_miR-26a_2, and Hs_miR-223_1 miScript Primer Assay (Qiagen, Inc.), respectively. For the normalization, the reference U6 small nuclear RNA Hs_RNU6-2_11 miScript Primer (Qiagen, Inc.) was used. Negative controls using water in place of sample were performed alongside each reaction. Reactions were run using the cycling parameters of 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Relative expression levels were calculated and significance for each treatment separately using the 2-Ct method39.
Statistical Analysis
A general linear model was used in the Bioconductor EdgeR package to generate lists of miRNAs with statistically significant different expression between sample groups. EdgeR uses negative binomial-based models to variation the quadratic mean–variance relationship that can be observed in sequencing reads data and to distinguish between biological and technical sources of variation40. Pathway analysis was performed with the Ingenuity Pathway 198 Analysis software (IPA, Ingenuity System, www.ingenuity.com).
Results
Cell Isolation
Cells were selected purely on their ability to adhere to plastic. The initial viability of AMCs was >90% as detected by Trypan blue exclusion. These cells show typical fibroblast-like morphology (Fig. 1). Previous molecular biological analyses on AMCs at P3 showed that these cells display a typical stem cell phenotype, with the expression of markers, such as CD29, CD44, CD106, CD105, and MHCI, but not CD34 and MHCII. Moreover, these cells have differentiation potential toward mesenchymal (adipogenic, chondrogenic, and osteogenic differentiation) and ectodermic lines (neurogenic differentiation) as reported by Lange-Consiglio et al.33
Figure 1.
Morphology of equine amniotic mesenchymal derived cells. Magnification 20×. Scale bar 20 nm.
Isolation and Measurements of MVs
The size of MVs ranged from 50 nm to 670 nm, with a mean size of 258 ± 55 nm for the 6 samples. The number of MVs ranged from 800 to 4,700 particles/cell, with a mean value of 2,550 ± 71 particles/cell (corresponding to 540 × 106 particles/mL of medium). Previously, a study using transmission electron microscopy22 revealed the presence of variably sized extracellular membranous vesicles budding from, or lying near, the source cell and characterized by an electron-lucent or moderately electron-dense content. The size of these MVs varied from about 100 nm to 1,000 nm, with a predominance of vesicles between 100 nm and 200 nm. The vesicles were roughly spherical. Multivesicular bodies in the early stages of maturation were occasionally detected. This suggests that the production of exosomes by these cells is less relevant than that of shedding vesicles. Because of size and morphological characteristics, the vesicles observed were mainly considered as shedding vesicles.
miRNA Analysis
Hiseq sequencing resulted in 184,147,508 reads with an average production of 23,018,439 reads per sample (range between 17,251,888 and 40,351,754). About 7.34% and 56.28% of the total reads were assigned to miRNA for MVs and cells, respectively (Table 1). In order to explore the miRNA content in MVs and cells, bioinformatic analyses of sequenced products were performed using miRDeep2 software (version 2.0.0.5). In all samples tested, 1,285 known and novel miRNA were identified and quantified. Among these miRNAs, 401 were classified as already known eca-miRNA, 257 were found by homology with miRNAs from other species and 627 were unknown predicted candidate miRNA (Online Supplementary Materials S1, S2, and S3).
Table 1.
Statistics of miRNA Sequencing for Microvesicles (MVs) and Cells.
| Total Sequences | Count Reads miRNA | % Identified as miRNA | |
|---|---|---|---|
| MVs 1a | 17582945 | 1041770 | 5.92 |
| MVs 1b | 17251888 | 1172231 | 6.79 |
| MVs 2a | 20883727 | 1687592 | 8.08 |
| MVs 2b | 18795945 | 1605210 | 8.54 |
| Cell 1a | 40351754 | 17474694 | 43.31 |
| Cell 1b | 23975838 | 12141959 | 50.64 |
| Cell 2a | 21985243 | 14780903 | 67.23 |
| Cell 2b | 23320168 | 16591980 | 71.15 |
| Total | 184147508 | 66496339 | 36.11 |
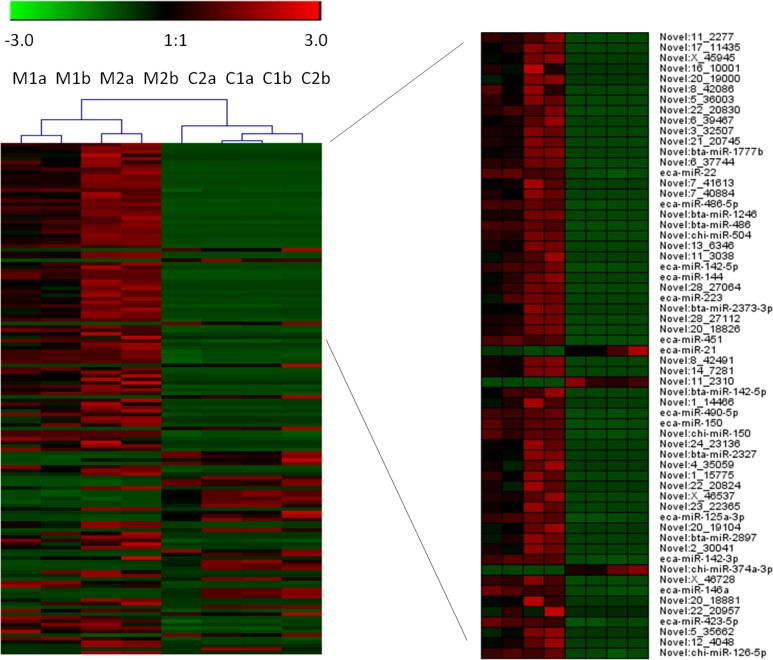
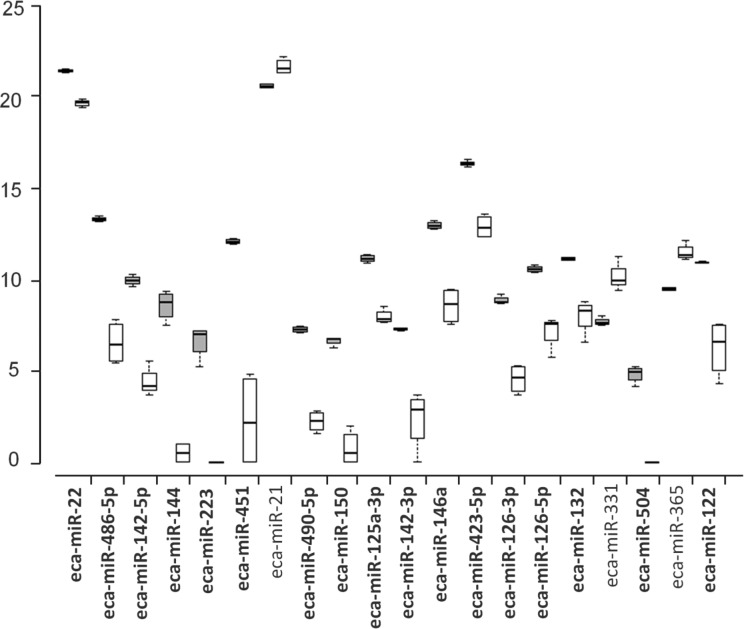
After applying a stringent filtering approach (FDR < 0.000001) to compare miRNA abundance in MVs and cells, 146 differentially expressed (DE) miRNAs were identified, 36 of which were known and the remaining were novel (Online Supplementary Material S4). A tree with a clear distinction between the MVs and cells was generated by cluster analysis (Fig. 2). Among the known DE miRNAs considered, 17 miRNAs showed greater expression in MVs. A view of the normalized expression of miRNAs in MVs and cells is reported in Fig. 3.
Figure 2.
Cluster analysis of the 146 differentially expressed (DE) miRNAs (FDR < 0.00001) microvesicles and cells. A subset of the first 60 DE microRNAs (ranked for FDR) were represented. M, microvescicle; C, cells; 1 and 2, biological replicates; a and b, technical replicates.
Figure 3.
Box plot showing the first 20 differentially expressed (DE) Equus caballus known microRNAs (ranked for FDR) in microvesicles (MVs; gray bar) and cells (white bar). Central lines inside the boxes indicate median values, box width indicates 25% and 75% quartile ranges around the median, “T” indicates the maximum and minimum values, and black dots represent outliers. N = 4 for group. miRNAs overexpressed in MVs are given in bold.
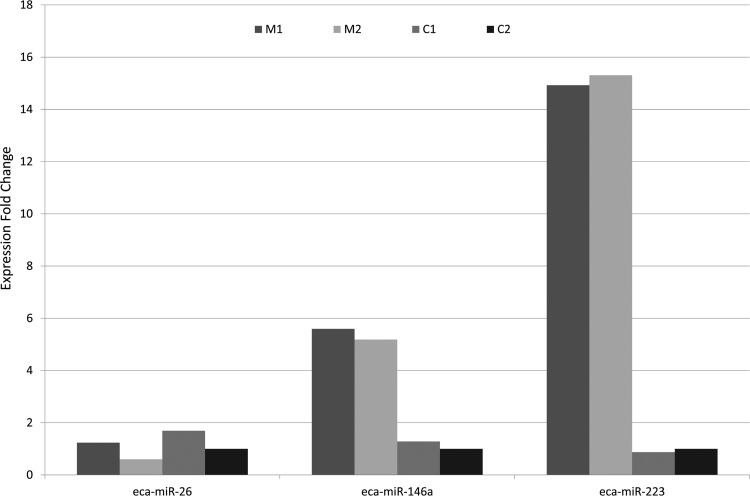
Three of these DE genes were validated by RT-PCR: eca-miR-26, eca-miR-146a, and eca-miR-223 (Fig. 4). For each test, qPCR results confirmed RNA-Seq data. Eca-miR-26, eca-miR-146a, and eca-miR-223 showed fold change ratio in qPCR between MVs versus cells of 0.68, 4.72, and 16.13, respectively. Considering RNA-Seq data, eca-miR-223 was observed only in MVs, whereas eca-miR-26 and eca-miR-146a showed a count ratio between MVs versus cells of 0.45 and 18.17, respectively.
Figure 4.
Validation of microvesicles (MVs) and cells microRNA (miRNA) expression. Three miRNAs (eca-miR-26, eca-miR-146a, and eca-miR-223) were selected for further qPCR assays. Consistent with previous sequencing results, the expression of miR-146 and miR-223 miRNAs is higher in MVs compared to cells. miR-26 is enriched in cells. M, microvesicles; C, cells.
Pathway Analysis of Predicted miRNA Targets
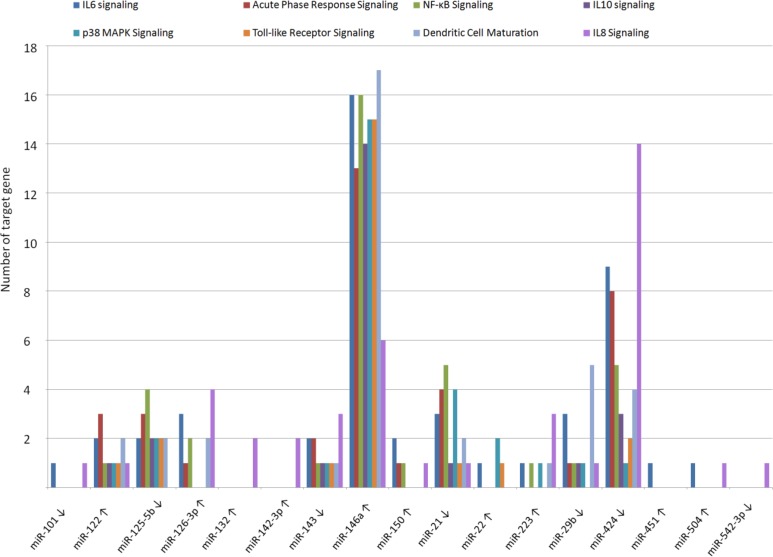
Pathways encompassing the target genes of the 36 known miRNAs were identified. Interestingly, DE-miRNAs modulate different pathways involved in the immune response (Table 2). The top canonical pathway, IL-6 signaling, and many others, for example, the role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis; the role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis; acute phase response signaling; NF-κB signaling; IL-10 signaling; p38 MAPK signaling; toll-like receptor signaling; dendritic cell maturation; IL-8 signaling; IL-12 signaling and production in macrophages; and HMGB1 signaling, are regulated by different DE-miRNAs. Among these, some miRNAs (122, 126-3p, 132, 142-3p, 146a, 150, 22, 223, 451, and 504) were observed to be overexpressed in MVs whereas others (101, 125b-5p, 143, 21, 29b, 424, and 542-3p) were predominantly expressed in cells (Fig. 5).
Table 2.
Canonical Pathway Analysis of mRNA Targets for the 36 Known Differentially Expressed miRNA (FDR < 0.000001).
| Canonical Pathways | −log (P Value) | z-Score |
|---|---|---|
| IL-6 signaling | 31.30 | 1.121 |
| Pancreatic adenocarcinoma signaling | 30.10 | −1.095 |
| Molecular mechanisms of cancer | 28.80 | NaN |
| Hepatic fibrosis/hepatic stellate cell activation | 26.60 | NaN |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 26.00 | NaN |
| p53 Signaling | 23.70 | −0.557 |
| Role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis | 21.40 | NaN |
| Chronic myeloid leukemia signaling | 21.10 | NaN |
| Acute phase response signaling | 20.70 | 0.87 |
| Glioma signaling | 20.30 | −1.89 |
| NF-κB signaling | 19.70 | 0.507 |
| Glioblastoma multiforme signaling | 19.60 | −1.732 |
| IL-10 signaling | 19.00 | NaN |
| p38 MAPK signaling | 18.40 | 1.732 |
| Toll-like receptor signaling | 18.00 | 1.886 |
| Dendritic cell maturation | 18.00 | 0.539 |
| PTEN signaling | 17.20 | 2.2 |
| Colorectal cancer metastasis signaling | 16.90 | 0.169 |
| PPAR signaling | 16.70 | −0.408 |
| Small cell lung cancer signaling | 16.70 | NaN |
| Prostate cancer signaling | 16.60 | NaN |
| IL-8 signaling | 16.60 | −0.174 |
| Melanoma signaling | 16.00 | NaN |
| Cell cycle: G1/S checkpoint regulation | 15.80 | 1.886 |
| Cholecystokinin/gastrin-mediated signaling | 15.80 | −0.816 |
| Hepatic cholestasis | 15.70 | NaN |
| Glucocorticoid receptor signaling | 15.50 | NaN |
| Aryl hydrocarbon receptor signaling | 15.20 | −0.447 |
| Non-small cell lung cancer signaling | 15.20 | −1.213 |
| Ovarian cancer signaling | 14.90 | NaN |
| Role of tissue factor in cancer | 14.80 | NaN |
| IL-12 signaling and production in macrophages | 14.80 | NaN |
| PI3K/AKT signaling | 14.60 | −1.279 |
| HGF signaling | 14.40 | −1.279 |
| PEDF signaling | 14.40 | −0.894 |
| Bladder cancer signaling | 14.10 | NaN |
| EGF signaling | 14.00 | −0.688 |
| HMGB1 signaling | 13.90 | −1.633 |
| Apoptosis signaling | 13.80 | −1.528 |
| Myc-mediated apoptosis signaling | 13.80 | NaN |
Note. Pathways related to inflammatory response are given in boldface. IL: interleukin; p53: protein 53; NF-kB: nuclear factor kappa-light-chain-enhancer of activated B cells; p38: protein 38; MAPK: mitogen-activated protein kinase; PTEN: phosphatase and tensin homolog; PPAR: peroxisome proliferator-activated receptor; PI3K/AKT: phosphoinositide-3-kinase/protein kinase B; HGF: epatocyte growth factor; PEDF: pigment epithelium-derived factor; EGF: epidermal growth factor; HMGB1: high-mobility group protein 1.
Figure 5.
Number of differentially expressed microRNA (miRNA) target genes belonging to different immune response pathways. ↑ indicates miRNA overexpressed in MVs, ↓indicates miRNA overexpressed in cells.
Discussion
In this work, we first performed miRNA sequencing of MVs and AMCs. Secreted MVs contain a low percentage of miRNAs (7.3%) compared to their cells of origin. Although RNA isolated from MVs is predominantly composed of small RNAs, Lin et al.41 reported that also MVs isolated from chondrocytes contain less miRNAs (6.1%) than cells, and the majority of small RNAs are unannotated in the databases. Moreover, library composition of small RNAs from deep sequencing of neuronal exosomes shows that more than 90% of sequencing reads are assigned to tRNA, whereas only about 2% are miRNAs42. The miRNA profiling of human MSCs of liver resident stem cells (HLSCs) and their related MVs show that, even though miRNA content is tissue-specific, MVs of different origins clearly cluster together, separately from cells, harboring a select pattern of miRNAs32. In addition, MVs contain ribonucleoproteins that are involved in the intracellular trafficking of RNA and this suggests a dynamic regulation of RNA compartmentalization in MVs. The Ago2 complexes, for example, selectively associate with miRNAs in the MVs, contributing to their stability43.
Accordingly, our results support the hypothesis that in AMCs, miRNAs are packaged within the MVs in a regulated manner, with an miRNA profile distinct from that of the parent cells. We found several miRNAs enriched hundreds or even thousands of times in MVs, whereas others had the tendency to remain to the same level of the parent cells. Among the more abundant miRNAs that we found in MVs, many have been previously reported to be secreted in MVs, as well. MiR-223, miR-142-3p, miR-451, miR-486, and miR-142-5p were enriched in MVs secreted from HLSCs31, whereas miR-451, miR-223, miR-144, miR-142-5p, miR-142-3p, miR-150, miR-126a-3p, and miR-132 were found to be prevalently present in MVs from chondrocytes41. Some of the miRNAs, that we found to be enriched in MVs from AMCs, were reported to be accumulated also in exosomes, for example, mir-150, miR-125a-3p, mir-451, mir-146a, mir-486, and miR14344,45. Previous studies indicated specific loading of miRNAs into exosomes45,46. Despite the discrepancies in their biogenesis process, it is likely that the system to sort miRNAs in exosome is well conserved and used to vehicle miRNAs also into MVs. It is noteworthy that our results are consistent with miRNA profiling of MVs and cells isolated from different species and analyzed using different miRNA detection platforms. Hence, it is unlikely that enrichment of specific miRNAs in MVs derived from AMCs is a technical artifact. In addition, real-time PCR validates eca-miR-26, eca-miR-146a, and eca-miR-223 expression in MVs and counterpart cells.
Microvesicles may act as mediators of cell-to-cell communication through miRNA delivery32, and Mehta and Baltimore47 have recently reviewed the role of miRNAs in regulating the immune system.
Our findings support the hypothesis that MVs secreted from AMCs contain miRNAs able to modulate the immune response. Target genes of miRNAs DE between MVs and cells are mainly associated with the immune response: for example, IL-6 signaling, acute-phase response signaling, NF-κB signaling, IL-10 signaling, p38 MAPK signaling, toll-like receptor signaling, IL-8 signaling, IL-12 signaling, and production in macrophages. In addition, many of the miRNAs enriched in AMC-MVs are observed to regulate the inflammatory response. For example, overexpression of miR-146 decreases the expression of the inflammatory cytokine IL-6 in LPS-stimulated macrophage cells. MiR-223 can negatively regulate the expression of many inflammatory genes in macrophage cells (i.e., IL-6, IL-1b, and TNF-α)48. Exosomal miR-146 and miR-223 are seen to contribute to cardioprotection in infarction and sepsis also in vivo49,50. Again, miR-150 decreases production of inflammatory cytokines, such as IL-2 and TNF-α, and elicits the induction of immune tolerance51; miRNA-122 inhibits the production of cytokines in human hepatic stellate cells52 and miR-126 promotes angiogenesis and inhibits vascular inflammation in endothelial cells53. Finally, other miRNAs involved in regulation of the immune response such as miR-125b-5p, miR-143, miR-21, miR-29b, and miR-424 are underrepresented in the MVs. Although these miRNAs show an anti-inflammatory effect if upregulated, the exact functional outcome is determined by multiple features including the cell type and the inducing signal, which ultimately affect the availability and ability to engage different target mRNAs and fine tune the response54,55.
Conclusions
Our work provides a deep characterization of miRNAs present in AMCs and their MVs. Compared to parent cells, a strong compartmentalization of specific miRNAs in MVs is observed. Our study suggests a potential role of MVs as regulatory elements in cell–cell communication, through the transfer of miRNAs, and their probable involvement in the anti-inflammatory process that we previously tested in vitro on stressed tendon and endometrial cells21,22. These results are promising for the use of MVs as possible future candidates in cell-free therapy but, obviously, the work is in progress. It will be necessary to monitor the ribonucleoproteins involved in the intracellular trafficking of RNAs and to compare the species of miRNAs contained in the MVs and in the cells of origin to obtain information on the mechanism of RNA accumulation within MVs. Moreover, it should be demonstrated that miRNAs contained inside MVs are transferred to target cells. This will require studying the miRNA cargo inside the target cells to confirm the presence of transferred miRNAs and to understand if these miRNAs are functional. Last but not least, it is important to underline that our study was performed with amniotic cells that represent an alternative and promising source of MSCs with the appeal of possibly producing off-the-shelf cells and cell-free products from a biological waste, which extrafetal membranes are considered.
Acknowledgments
The authors wish to thank Dott.ssa Maria Chiara Deregibus and Professor Giovanni Camussi (Department of Internal Medicine and Molecular Biotechnology Center, University of Torino, Torino, Italy) for their skilled assistance in Nanosight use.
Authors’ Note: Novel miRNA mature and precursor FASTA sequences, including miRNAs predicted by homology with miRNAs from other species and novel candidate miRNAs, are available in the Online Supplementary Materials S2 and S3. The online appendices are available at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/RRNEJC.
Ethical Approval: Animals were used.
Statement of Human and Animal Rights: Animals were used.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research was supported by grants from Università degli Studi di Milano (Milano, Italy), by MIUR GenHome project “Technological Resort for the Advancement of Animal Genomic Research,” and by the Italian Ministry of Education, Universities and Research for the project “Progetto Bandiera INTEROMICS—Sottoprogetto 1: Sviluppo di Infrastrutture Bioinformatiche per le applicazioni OMICS in Biomedicina.”
Supplemental Material: Supplementary material for this article is available online.
References
- 1. Dazzi F, Horwood NJ. Potential of mesenchymal stem cell therapy. Curr Opin Oncol. 2007;19(6):650–655. [DOI] [PubMed] [Google Scholar]
- 2. Hwang NS, Zhang C, Hwang YS, Varghese S. Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdiscip Rev Syst Biol Med. 2009;1(1):97–106. [DOI] [PubMed] [Google Scholar]
- 3. Satija NK, Singh VK, Verma YK, Gupta P, Sharma S, Afrin F, Sharma M, Sharma P, Tripathi RP, Gurudutta GU. Mesenchymal stem cell-based therapy: a new paradigm in regenerative medicine. J Cell Mol Med. 2009;13(11–12):4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leung VYL, Chan D, Cheung KMC. Regeneration of intervertebral disc by mesenchymal cells: potentials, limitations and future direction. Eur Spine J. 2006;15(Suppl 3):S406–S413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cargnoni A, Ressel L, Rossi D, Poli A, Arienti D, Lombardi G, Parolini O. Conditioned medium from amniotic mesenchymal tissue cells reduces progression of bleomycin-induced lung fibrosis. Cytotherapy. 2012;14(2):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lange-Consiglio A, Rossi D, Tassan S, Perego R, Cremonesi F, Parolini O. Conditioned medium from horse amniotic membrane-derived multipotent progenitor cells: immunomodulatory activity in vitro and first clinical application in tendon and ligament injuries in vivo. Stem Cell Dev. 2013;22(22):3015–3024. [DOI] [PubMed] [Google Scholar]
- 7. Yang D, Wang W, Li L, Peng Y, Chen P, Huang H, Guo Y, Xia X, Wang Y, Wang H, Wang WE, Zeng C. The relative contribution of paracrine effect versus direct differentiation on adipose-derived stem cell transplantation mediated cardiac repair. PLoS ONE. 2013;8(3):e59020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morhayim J, Baroncelli M, van Leeuwen JP. Extracellular vesicles: specialized bone messengers. Arch Biochem Biophys. 2014;561:38–45. [DOI] [PubMed] [Google Scholar]
- 9. Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20(5):847–856. [DOI] [PubMed] [Google Scholar]
- 10. Camussi G, Deregibus MC, Tetta C. Paracrine/endocrine mechanism of stem cells on kidney repair: role of microvesicle-mediated transfer of genetic information. Curr Opin Nephrol Hypertens. 2010;19(1):7–12. [DOI] [PubMed] [Google Scholar]
- 11. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;14(4):14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoon YJ, Kim OY, Gho YS. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014;47(10):531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantaluppi V, Gatti S, Medica D, Figliolini F, Bruno S, Deregibus MC, Sordi A, Biancone L, Tetta C, Camussi G. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82(4):412–427. [DOI] [PubMed] [Google Scholar]
- 15. Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, Pasterkamp G, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(4):214–222. [DOI] [PubMed] [Google Scholar]
- 16. Herrera MB, Fonsato V, Gatti S, Deregibus MC, Sordi A, Cantarella D, Calogero R, Bussolati B, Tetta C, Camussi G. Human liver stem cell derived microvesicles accelerate hepatic regeneration in hepatectomized rats. J Cell Mol Med. 2010;14(6b):1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Yan Y, Wang B, Qian H, Zhang X, Shen L, Wang M, Zhou Y, Zhu W, Li W, Xu W. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev 2013;22(6):845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Shang X, Zhang ZG, Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30(7):1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. di Trapani M, Bassi G, Midolo M, Gatti A, TakamKamga P, Cassaro A, Carusone R, Adamo A, Krampera M. Differential and transferable modulatory effects of mesenchymal stromal cell-derived extracellular vesicles on T, B and NK cell functions. Sci Rep. 2016;13(6):24120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lange-Consiglio A, Tassan S, Corradetti B, Meucci A, Bizzaro D, Cremonesi F. Investigating the potential of equine mesenchymal stem cells derived from amnion and bone marrow in equine tendon diseases treatment in vivo. Cytotherapy. 2013;15:1011–1020. [DOI] [PubMed] [Google Scholar]
- 21. Fierabracci A, Lazzari L, Muraca M, Parolini O. How far are we from the clinical use of placental-derived mesenchymal stem cells? Expert Opin Biol Ther. 2015;15(5):613–617. [DOI] [PubMed] [Google Scholar]
- 22. Lange-Consiglio A, Perrini C, Tasquier R, Deregibus MC, Camussi G, Pascucci L, Marini MG, Corradetti B, Bizzaro D, De Vita B, Romele P, et al. Equine amniotic microvesicles and their anti-inflammatory potential in a tenocyte model in vitro. Stem Cells Dev. 2016;25(8):610–621. [DOI] [PubMed] [Google Scholar]
- 23. Perrini C, Strillacci MG, Bagnato A, Esposti P, Marini MG, Corradetti B, Bizzaro D, Idda A, Ledda S, Capra E, Pizzi F, et al. Microvesicles secreted from equine amniotic-derived cells and their potential role in reducing inflammation in endometrial cells in an in vitro model. Stem Cell Res Ther. 2016;7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 25. Katsuda T, Ochiya T. Molecular signatures of mesenchymal stem cell-derived extracellular vesicle-mediated tissue repair. Stem Cell Res Ther. 2015;6:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. [DOI] [PubMed] [Google Scholar]
- 29. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. [DOI] [PubMed] [Google Scholar]
- 30. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. [DOI] [PubMed] [Google Scholar]
- 31. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 32. Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, Tetta C, Camussi G. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010;5(7):e11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lange-Consiglio A, Corradetti B, Bizzaro D, Magatti M, Ressel L, Tassan S, Parolini O, Cremonesi F. Characterization and potential applications of progenitor-like cells isolated from horse amniotic membrane. J Tissue Eng Regen Med. 2012;6(8):622–635. [DOI] [PubMed] [Google Scholar]
- 34. Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. [DOI] [PubMed] [Google Scholar]
- 36. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sturn A, Quackenbush J, Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. [DOI] [PubMed] [Google Scholar]
- 38. Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nat Methods. 2014;11(2):121–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 40. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin Z, Rodriguez NE, Zhao J, Ramey AN, Hyzy SL, Boyan BD, Schwartz Z. Selective enrichment of microRNAs in extracellular matrix vesicles produced by growth plate chondrocytes. Bone. 2016;88:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, Liu Y, Zhang CY, Zen K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7(10):e46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8(5):1432–1446. [DOI] [PubMed] [Google Scholar]
- 46. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279–294. [DOI] [PubMed] [Google Scholar]
- 48. Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125(23):2892–2903. [DOI] [PubMed] [Google Scholar]
- 49. Ibrahim AG, Cheng K, Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2014;2(5):606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, Gu H1, Qin D, Yang L, Huang W, Essandoh K, Wang Y, Caldwell CC, Peng T, Zingarelli B, Fan GC. Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis. Sci Rep. 2015;5:13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sang W, Wang Y, Zhang C, Zhang D, Sun C, Niu M, Zhang Z, Wei X, Pan B, Chen W, Yan D, Zeng L, et al. MiR-150 impairs inflammatory cytokine production by targeting ARRB-2 after blocking CD28/B7 costimulatory pathway. Immunol Lett. 2016;172:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakamura M, Kanda T, Sasaki R, Haga Y, Jiang X, Wu S, Nakamoto S, Yokosuka O. MicroRNA-122 inhibits the production of inflammatory cytokines by targeting the PKR activator PACT in human hepatic stellate cells. PLoS One. 2015;10(12):e0144295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu J, Zeng L, Huang J, Wang G, Lu H. miR-126 promotes angiogenesis and attenuates inflammation after contusion spinal cord injury in rats. Brain Res. 2015;1608:191–202. [DOI] [PubMed] [Google Scholar]
- 54. Hulsmans M, De Keyzer D, Holvoet P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J. 2011;25(8):2515–2527. [DOI] [PubMed] [Google Scholar]
- 55. Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016;35:103. [DOI] [PMC free article] [PubMed] [Google Scholar]