Abstract
Enzymes are commonly used as a biochemical means to liberate cells from a host of tissues for use in in vitro studies and/or in vivo transplantations. However, very little understanding exists of the biological and functional effects that enzymes have on cells during the process of releasing the native cells from a given tissue. One specific reason for this is that no technology has existed as a nonenzymatic control to compare baseline biology and function for a given processed tissue. We have developed a sterile, onetime use, disposable system (referred to as the AuxoCell Processing System or AC:Px®) that allows for processing of solid tissue in a closed, standardized system using mechanical means to liberate cells without the need and/or use of any biochemical, enzymatic digestion. In this report, for the first time, we directly compare the cellular outputs derived from processing the same umbilical cord tissue (UCT) in the presence and absence of collagenase. In the presence of collagenase, we observed on average, approximately a 2.7-fold reduction in native mesenchymal stem/stromal cell (MSC) yields and a reduction in MSC-specific markers CD90, CD29, CD105, CD73, CD44, CD36, CD49b, CD49a, CD146, CD295, and CD166 and in endothelial marker CD31. These data directly exhibit that the use of collagenase to process UCT to release cells impacts cell recovery with respect to number and cell surface marker expression and, hence, could affect the in vivo function of the recovered native cellular population.
Keywords: umbilical cord tissue, mesenchymal stem/stromal cells, collagenase, AC:Px® System, enzymatic digestion
Introduction
Stem cells hold great clinical promise, especially those from perinatal sources—including umbilical cord blood (UCB), umbilical cord tissue (UCT), placenta, amnion, and chorion1–7—since these tissues possess young, fetal-derived (and in some cases maternal-derived) stem cells that can be collected postgestation. In order to harness the great potential of these cells and their products in treating ailing patients clinically, regulatory-friendly collection, processing, cryopreservation/thaw, and quality controls need to be developed and implemented today. Although UCB processing is relatively more mature and standardized, this is not the case with solid perinatal tissues. The most variable step in the aforementioned list for solid tissue is processing.
Currently, there are multiple ways that processing laboratories, including public and private tissue banks, as well as cell therapy manufacturers, process solid perinatal and nonperinatal tissues. For instance, for UCT, certain groups collect a small 2- to 5-cm segment of the UCT, manually mince the tissue with scissors in a petri dish and place the minced tissue fragments into cryovials with appropriate cryoprotectant and protein in order to cryopreserve the minced tissue for later processing and manipulation. When the unit is needed for therapeutic use, the manually minced tissue is thawed, washed, and placed into culture to derive an ex vivo expanded, manipulated mesenchymal stem/stromal cell (MSC) product. This process will take weeks to derive the needed cell numbers for transplantation and cells are sometimes cultured in the presence of fetal bovine serum (FBS), which could be a bottleneck for autologous transplantation and clinical utility, with more stringent regulatory approval1,2,8. Although often overlooked by the scientific community, there is also evidence that ex vivo expansion accelerates stem cell exhaustion and/or senescence, resulting in significantly reduced in vivo stem cell functional activity (i.e., potency)1,2,8,9. This could explain why such high numbers of ex vivo expanded MSCs are needed for transplantation since the potency of the population is reduced with each ex vivo cell division. Since the postthaw native cell recovery of cryopreserved minced tissue is approximately 15%1,2,10 likely due to the lack of sufficient diffusion of cryoprotectant into a large, complex tissue, one viable clinical product from minced tissue is an ex vivo expanded cellular product. Due to the nature of the minced tissue product, cell-based quality determinations based on individual units are difficult to attain. The primary advantage of this method, however, is that it is an inexpensive precryopreservation UCT processing method.
Another method employed by tissue banks to process UCT is to process the entire UCT to recover the heterogeneous native, primary cells that comprise the tissue1. The UCT can be used in its entirety or with the vessels dissected. Briefly, the UCT can be mechanically minced (using UC scissors) and subsequently, enzymatically digested (typically using collagenase) to collect the native cell population including the native MSCs. Mechanically mincing the tissue increases the surface area for tissue digestion and, therefore, reduces the incubation time with enzyme. However, longer incubation times can obviate the need to initially, mechanically mince the tissue, given that enough enzyme is added. The native cells can then be collected; quality control tested for viability, cell number, and cell surface protein markers of interest; and then cryopreserved in a 25-mL cryobag for cryogenic storage until thawed for use. Based on current regulation, this product could be used clinically, given that appropriate safety and efficacy investigations are conducted under appropriate regulatory oversight and applications for a specific indication of use. The advantage of this processing method is that it results in a native cellular product that can be verified for quality, with the qualified product ready for use immediately, if granted approval from regulatory agencies. Furthermore, the native stem cells that exist within this product represent the stem cells with the greatest potency1,2,7–9. The disadvantage of this method is that it costs relatively more to process the entire tissue to recover the native cells, compared to simply mincing and cryopreserving the tissue segment.
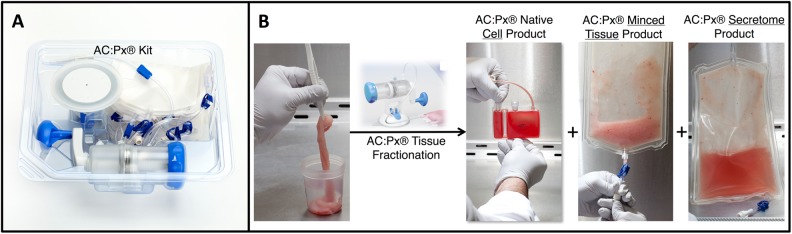
A more recent method uses mechanical means to sufficiently mince the UCT to release the native MSCs without the need for any enzymatic digestion. The technology is a novel, proprietary system, referred to as the AuxoCell Processing (AC:Px) System (Fig. 1A). The AC:Px System is a sterile, closed, single-use, completely disposable system to process solid tissue to release cells in a laboratory, outside of a patient setting. The AC:Px System consists of a closed mincer (i.e., semiautomated scissors) and a series of closed bags. The AC:Px System allows—for the first time—standardized processing of solid tissues without the use of enzymes in a closed system—validated by our group and others. Due to the design of the AC:Px System, processing takes approximately 60 min, resulting in 3 products from the same UCT unit (Fig. 1B). The 3 products include the native cells, minced tissue, and decellularized Wharton’s jelly. The native cells, minced tissue, and decellularized Wharton’s jelly can all be cryopreserved for future use in various applications. For instance, the native cells can be used directly, while the minced tissue can produce ex vivo expanded cell products by further manipulation and culturing to derive ex vivo expanded MSCs, and the decellularized Wharton’s jelly product can be used similarly to allografts. However, as with any other cellular therapy product, these products will have to go through the appropriate regulatory approvals. The invention of the AC:Px System allows for the first time the processing of solid tissues, including UCT, without the need for any biochemical, enzymatic digestion.
It has been hypothesized by our group and others that the use of enzymes in the processing of solid tissues can harm the cells that are recovered for use in subsequent in vitro and in vivo cellular therapy studies. Several recent publications have provided preliminary evidence of the adverse effects of various enzymes on recovered cells. Autengruber et al. observed that collagenase and dispase affect both cell surface markers and immune cell function to varying degrees, when they compared the cell surface receptor and immune function differences of splenocytes derived from either collagenase or dispase at low and high concentrations11. However, in order to fully understand the effects of enzymes on cells, ideally, it would be best to compare against nonenzymatic-treated controls in lieu of enzymes at various concentrations. For the first time, we have the technology to directly determine the effect of enzyme digestion on native cell liberation from the same UCT unit by comparing the enzyme-treated (collagenase) sample with a non-enzyme-treated sample using the AC:Px System, since the AC:Px uses only mechanical means and does not depend on enzymes to release the cells from the tissue.
Materials and Methods
Tissue Collection and Preparation
UCT units were obtained from healthy mothers who donated their de-identified UCT after giving written informed consent. UCT was collected after term delivery and UCB collection. Once collected, the UCT was placed in a sterile specimen jar and delivered to the laboratory for processing. Upon arrival at the processing laboratory, 10 mL of antibiotics (25-μg/mL gentamicin [Invitrogen, Grand Island, NY, USA], 100-IU/mL penicillin [Invitrogen], 100-μg/mL streptomycin [Invitrogen], 0.25-μg/mL amphotericin B [Invitrogen], 10-μg/mL ciprofloxacin [Mediatech, Manassas, VA, USA]) was added to the unit and placed at 4 °C until processed—which never exceeded 72 h postdelivery. Immediately prior to processing, the specimen jar containing the unit was taken out of 4°C, sterilized using 70% ethanol and placed in the biosafety cabinet (BSC). The unit was carefully taken out of the specimen jar and placed on a sterile petri dish using sterile stainless steel forceps (B. Braun Aesculap, Tuttlingen, Germany). Using a sterile pair of umbilical cord scissors (B. Braun Aesculap), the unit was cut into equal quarter segments. Every other quarter was placed into one of the 2 tared tubes—in order to control intratissue variability. The tared weight of each unit was recorded in a 50-mL conical tube (BD Falcon, Bedford, MA, USA) in order to enable normalization and quantification for the number of native MSCs per gram of tissue processed. Tubes were randomly designated for either enzymatic digestion or processing using the AC:Px System.
Enzymatic Digestion
Umbilical cords designated for enzymatic digestion were minced using umbilical cord scissors in their entirety (B. Braun Aesculap) until the tissue was finely minced. The minced tissue was placed into a tared 50-mL conical tube (BD Falcon) for weight determination postmincing. A 10-mL solution of 2.5-mg/mL collagenase NB6 (Serva, Heidelberg, Germany) in 2-mM calcium chloride (Amresco, Solon, OH, USA) in Dulbecco’s phosphate-buffered saline (DPBS; Invitrogen) was added to the 50-mL conical tube containing the minced UCT. The minced tissue was placed in a 37°C incubator with a rocking platform (VWR, Radnor, PA, USA) for 3 h. In postincubation, the unit was diluted with 30-mL DPBS and filtered through a 100-μm Steriflip (Millipore, Bedford, MA, USA). After the initial pass through the filter, 50-mL DPBS was added back to the tube containing the digested minced tissue for subsequent washes and refiltered through the same Steriflip filter for a total of 3 washes. The filtered cell product was placed in a 225-mL conical tube (BD Falcon) and centrifuged for 20 min at 750g in an Allegra X15R (Beckman Coulter, Danvers, MA, USA) centrifuge. In postcentrifugation, the supernatant (i.e., decellularized Wharton’s jelly) was decanted and collected into several 50-mL conical tubes. The cell pellet was resuspended in 22-mL CryoStor Base (CSB; BioLife Solutions, Bothell, WA, USA) medium. The resuspended cell solution was filtered through a 40-μm tube top filter (BD Falcon). The final volume was measured and, if needed, brought up to 22-mL with CSB medium. From the 22-mL final native cell unit, a 2-mL aliquot was taken for ex vivo MSC expansion and quality control determinations using flow cytometry. The remaining 20-mL was cryopreserved for postthaw ex vivo MSC expansion and flow cytometric analysis. The remaining undigested minced tissue was collected from the Steriflip filter for ex vivo MSC expansion (using an explant method) and cryopreservation. The decanted supernatant, postcentrifuge represents the decellularized Wharton’s jelly and was stored at −80°C in 50-mL conical tubes.
Mechanical Digestion Using the AC:Px System
UCTs designated for nonenzymatic processing were placed in the AC:Px (AuxoCell, Cambridge, MA, USA) System. Briefly, the entire tissue was placed in the input chamber of the AC:Px Mincer with the output chamber filled with 0.9% sodium chloride (B. Braun, Irvine, CA, USA) saline. After subsequent mincing and washes with saline, the postminced UCT was transferred into the supplied series of AC:Px bag sets in order to filter and centrifuge the native cellular product. Filtration took place in the AC:Px filter bag that filters using a 100-μm mesh, and subsequent centrifugation took place in the AC:Px centrifuge bag, clipped on a 97-mm blood bag centrifuge adaptor (Beckman Coulter) suspended, using the AC:Px centrifuge clip (AuxoCell). The cells were centrifuged for 20 min at 750g in an Allegra X15R (Beckman Coulter) benchtop centrifuge. In postcentrifugation, the supernatant (i.e., decellularized Wharton’s jelly) was decanted into the AC:Px filter bag with the cell pellet resuspended in 22-mL CSB (BioLife Solutions) medium. The resuspended cell solution was filtered through the remainder of the AC:Px bag set that includes a 40-μm filter bag. The final volume was measured and brought up to 22 mL, if needed. From the 22-mL sample volume, a 2-mL aliquot was taken for ex vivo MSC expansion and quality control determinations using flow cytometry. The remaining 20 mL was cryopreserved for postthaw ex vivo MSC expansion and flow cytometric analysis. The minced tissue was collected from the AC:Px for ex vivo MSC expansion (using an explant method) and cryopreservation. The decanted supernatant, postcentrifuge represents the decellularized Wharton’s jelly and was stored at −80°C in 50-mL conical tubes.
Ex vivo MSC Expansion Cultures from Native Cells
Native cells recovered from UCT processed with the AC:Px System or in the presence of collagenase were seeded into 12-well plates, 60-mm dishes, or T25 flasks (BD Falcon) in CTS™ StemPro MSC SFM (Invitrogen), per the manufacturer’s instructions. The working medium contained CTS StemPro MSC SFM basal medium, 25-μg/mL gentamicin, 100-IU/mL penicillin, 100-μg/mL streptomycin, 0.25-μg/mL amphotericin B (Invitrogen), 10-μg/mL ciprofloxacin (Mediatech), CTS StemPro™ MSC SFM supplement, and 1% GlutaMAX (Invitrogen). CTS CELLstart™ attachment substrate (Invitrogen) was coated onto culture surfaces per the manufacturer’s instructions and incubated at 37°C for 2 h. In postincubation, the substrate was carefully aspirated without disturbing the coated monolayer. For culture expansion, AB human serum (Mediatech) was carefully added to coat the CELLstart monolayer surface and placed into the incubator for 10 min. In postincubation, the fully prepared CTS StemPro MSC SFM was added to the culture vessel with the subsequent addition of the native/primary cells at a concentration of 2,500 cells/cm2. The culture vessels were placed back into a 37 °C, 5% CO2 humidified incubator for a period of 10 to 14 d with no medium changes or additions. After cells reached 70% to 90% confluency, the cells were washed once with DPBS and recovered using TrypLE™ (Invitrogen).
Explant Ex vivo MSC Expansion Cultures from Minced Tissue
Minced tissue recovered from the not fully enzymatically digested UCT or from the AC:Px System was removed directly from the Steriflip filter or from the AC:Px System filter bag, respectively. Minced tissue pieces were seeded into 100-mm culture dishes (BD Falcon) using an adapted explant method8,12. Briefly, dishes were coated with 1-mL of decellularized Wharton’s jelly collected from the specific UCT unit, along with 1-mL of RPMI-1640 (Invitrogen) medium (containing 20% MSC qualified FBS; Invitrogen), 100-IU/mL penicillin, 100-μg/mL streptomycin, 0.25-μg/mL amphotericin B (Invitrogen), and 10-μg/mL ciprofloxacin (Mediatech), and placed in a 37°C, 5% CO2 humidified incubator for 10 min. In postincubation, a 1- to 2-mL aliquot of the minced tissue was placed on the culture surface and spread by carefully swirling to disperse the minced tissue within the dish. The culture dishes were placed in a 37 °C, 5% CO2 humidified incubator with the addition of 5-mL of medium dropwise (in order to prevent disruption of the seeded minced tissue pieces) to each dish every 4 d and incubated for a period of 10 to 14 d. After cells reached 70% to 90% confluency, the cells were washed once with DPBS and recovered using TrypLE (Invitrogen).
Cryopreservation
Native cells were cryopreserved using CSB (BioLife Solutions) medium per the manufacturer’s recommendations. Briefly, native cells resuspended in a 20-mL final cryopreservation volume in CSB were combined with 5-mL BloodStor (55% dimethyl sulfoxide supplemented with 5% dextran 40; BioLife Solutions) and cryopreserved the 25-mL total volume in a 25-mL cryobag (Pall, Covina, CA, USA). The bag was sealed using a Sebra (Haemonetics, Braintree, MA, USA) sealer and placed at 4 °C for 30 min. After the 30 min at 4 °C, the unit was transferred to −20°C for 60 min and then transferred to −80°C for an additional 60 min until the unit was ready for long-term liquid nitrogen cryostorage (Thermolyne, locator 4, Thermo Fisher, Waltham, MA, USA).
Minced tissue was cryopreserved in a solution of FBS and BloodStor. Briefly, a mixture of 20-mL FBS and 5-mL BloodStor was prepared and added to approximately 15 mL of the minced tissue sample in a 50-mL conical tube. The mixture was then transferred into multiple 2-mL or 5-mL cryovials and placed at 4°C for 30 min. After the 30 min at 4°C, the unit was transferred to −20°C for 60 min and then transferred to −80°C for an additional 60 min until the unit was ready for long-term liquid nitrogen cryostorage.
Postcryopreservation Thaw
Cryopreserved native/primary cells or minced tissue that was taken out of liquid nitrogen storage for postcryopreservation processing was quickly thawed (approximately 30 s) while simultaneously shaken in a 37°C water bath (Shel Lab, Cornelius, OR, USA). In postthaw, the outside of the cryobag or cryotube was sterilized using 70% ethanol and placed in the BSC. The unit was opened and transferred into a 50-mL conical tube. An equal volume of BioLife’s cell thawing media (10% Dextran-40 in 5% Dextrose; BioLife Solution) was added dropwise to the unit. The unit was then placed in a centrifuge (Allegra X15R, Beckman Coulter) and spun for 10 min at 500g. In postcentrifugation, the supernatant was decanted with the cells resuspended in the respective culture medium.
Flow Cytometry
Native and culture expanded cells were stained for various human MSC-associated markers CD45, CD90, CD29, CD73, CD49b, CD49a, CD49d, CD44, CD295, CD106, CD166, CD36, CD146 (BD Biosciences, Franklin Lakes, NJ, USA), and CD105 (R&D Systems, Minneapolis, MN, USA); endothelial marker CD31 (BD Biosciences); and embryonic stem cell (ESC) marker stage-specific antigen-4 (SSEA-4, R&D Systems). All samples were analyzed on an Attune Acoustic (Applied Biosystems, Waltham, MA, USA) flow cytometer with mouse IgG antibodies (BD Biosciences) as isotype controls for determining nonspecific binding. For each flow cytometry evaluation, a minimum of 500,000 cells were stained, and at least 50,000 events were collected and analyzed. Cells were collected and blocked using fluorescent-activated cell sorting (FACS) buffer (1× phosphate-buffered saline; DPBS, 3% FBS, and 0.1% sodium azide). After centrifugation and decanting of the buffer, 5 μg of each antibody was added to cells in 200-μL FACS buffer along with 10-μL Viaprobe (7 amino-actinomycin-D [7AAD]; BD Biosciences) followed by incubation for 30 min at 4°C in the dark. In postincubation, FACS buffer was again added to bind free, unbound antibody in the solution. After another centrifugation and decanting, the cells were resuspended in 500-μL FACS buffer. Flow cytometry analysis was performed using an Attune flow cytometer and configured with a Blue Excitation Laser (488 nm) and Red Excitation Laser (638 nm). Appropriate isotype and unstained cells were used as controls. Cell number quantification was performed by adding a known volume of CountBright™ fluorescent counting beads (Invitrogen) to each tube. Compensation beads (BD Biosciences) were prepared per manufacturer’s instructions for each fluorochrome used and run on the Attune once all voltages for each channel were set. Compensation was not set on the Attune during cell collection but rather during final data analysis on FlowJo software (version 10.4.0) (Tree Star, Inc., San Carlos, CA, USA) in order to minimize risk of errors during acquisition.
Microscopy
A Trinocular Inverted Microscope (VWR) was used for routine cell viewing, and an EVOS Cell Imaging System (Thermo Fisher) was used for digital imaging.
Statistical Analysis
The Student’s t-test was used to determine the statistical confidence of observed differences. Statistical evaluations of differences between enzyme-treated cell yields and nonenzyme (AC:Px) control cell yields were performed using 1-group Student’s t-test (StatView, SAS Institute, Cary, NC, USA) or otherwise noted. Differences were considered statistically significant at P < 0.05. Standard error of the mean is reported for all averages.
Results
UCT can be processed using the AC:Px System (Fig. 1A) and fractionated into 3 products. These 3 products are the native cells, minced tissue, and decellularized Wharton’s jelly, which are derived from the same unit (Fig. 1B) without the use of any enzymes. The native cells represent the heterogeneous population of primary cells that make up and are found within the in utero UCT. The minced tissue contains native cells and the Wharton’s jelly within the 3-dimensional tissue. The Wharton’s jelly contains growth factors, extracellular matrix proteins (i.e., collagen), cytokines, and glycosaminoglycans (i.e., hyaluronic acid) that are all present within the in utero UCT13–19. In order to investigate the biological effects of collagenase during UCT processing, UCT were split into 2 mass equivalent units. One half of the unit was processed in the presence of collagenase (+collagenase), while the other was processed in the absence of collagenase (−collagenase; AC:Px). Processing with collagenase yielded on average 2.65 × 105 ± 0.45 × 105 live native MSCs (7AAD−, CD45−, CD29+, and CD90+) per gram of tissue processed. In comparison, the other half unit of UCT, when processed using the AC:Px System without collagenase, on average resulted in 7.20 × 105 ± 0.53 × 105 live native MSCs per gram of tissue processed (Table 1). Processing the same unit without collagenase (with the AC:Px System) yielded, on average, a 2.7-fold higher native live MSC recovery, compared to the same unit processed using collagenase (P < 0.01, n = 31). The total nucleated cell number and percentage viability of cells collected manually using collagenase were 2.96 × 106 ± 4.16 × 105 and 88.6% ± 2.0%, respectively. For cells recovered from the AC:Px System (no collagenase), the total nucleated cell number and the percentage of viability were 4.27 × 106 ± 4.00 × 105 and 84.4% ± 1.5%, respectively (Table 2). In order to better understand the effect of collagenase on cell yield, we placed a sample of the unit processed with the AC:Px in the presence of collagenase. Interestingly, the live native cell yield from collagenase-treated UCT processed using the AC:Px System resulted in a similar reduction in the recovery of live native MSC yield per gram of tissue processed as that of the collagenase-treated condition without the use of the AC:Px System. Collagenase-treated samples taken from a processed unit from the AC:Px System resulted in 3.79 × 105 ± 0.58 × 105 live native MSCs per gram of tissue processed (n = 6; Table 1), compared to 2.65 × 105 ± 0.45 × 105 live native MSCs per gram recovered using manual mincing and collagenase digestion. These yields from both collagenase treatments were not statistically different. Seemingly, any treatment of UCT with collagenase—independent of processing method—results in a reduced live native MSC yield per gram of tissue processed, when compared to processing with the AC:Px System in the absence of collagenase (7.20 × 105 ± 0.53 × 105; Table 1).
Fig. 1.
The AuxoCell Processing System (AC:Px®) comes in a sterile, onetime use, disposable sterile kit (A) that allows for solid tissue processing in a closed, standardized method using mechanical means to liberate cells without the need and use of any biochemical enzymatic digestion. The AC:Px System fractionates any particular solid tissue, including umbilical cord tissue, into 3 specific products from the same tissue (B): native/primary cellular, minced tissue, and secretome (i.e., decellularized Wharton’s jelly) products.
Table 1.
Native MSC Yields from Umbilical Cord Tissue (UCT) Processed with the AuxoCell Processing (AC:Px) System (Both in the Absence and Presence of Collagenase) and Manually Processing in the Presence of Collagenase Were Determined.
| Live Native MSCs per gram | ||||
|---|---|---|---|---|
| AC:Px® | Collagenase | Average | SEM | N |
| − | + | 2.65 × 105 | 0.45 × 105 | 31 |
| + | − | 7.20 × 105 | 0.53 × 105 | 31 |
| + | + | 3.79 × 105 | 0.58 × 105 | 6 |
Note: Umbilical cord tissue (UCT) was cut into equivalent halves and manually minced with umbilical cord scissors for collagenase treatment. Meanwhile, the other UCT half was placed in its entirety into the AC:Px Mincer for mechanical mincing, without the need or use of collagenase. In order to further understand collagenase effects on recovered UCT native cells, the native cells and minced tissue products from the AC:Px System from several units were further processed in the presence of collagenase. The average cell yields shown are the number of live native mesenchymal stem/stromal cells (MSCs) recovered per gram of processed UCT ± the standard error of the mean (SEM). Live native MSCs represent 7AAD−, CD45−, CD29+, and CD90+ cells in each of the recovered populations.
Live native MSCs = 7AAD−/CD45−/CD29+/CD90+.
Table 2.
Native Total Nucleated Cell Yields and Percent Viability from Umbilical Cord Tissue (UCT) Processed with the AuxoCell Processing System (in the Absence of Collagenase) and Manual Processing in the Presence of Collagenase Were Determined.
| Total Nucleated Cell Number per gram | ||||
|---|---|---|---|---|
| AC:Px® | Collagenase | Average | SEM | N |
| − | + | 2.97E + 06 | 4.16E + 05 | 31 |
| + | − | 4.27E + 06 | 4.00E + 05 | 31 |
Note: The average total nucleated cell yields per gram of processed UCT and percent viability are shown plus or minus the standard error of the mean (SEM).
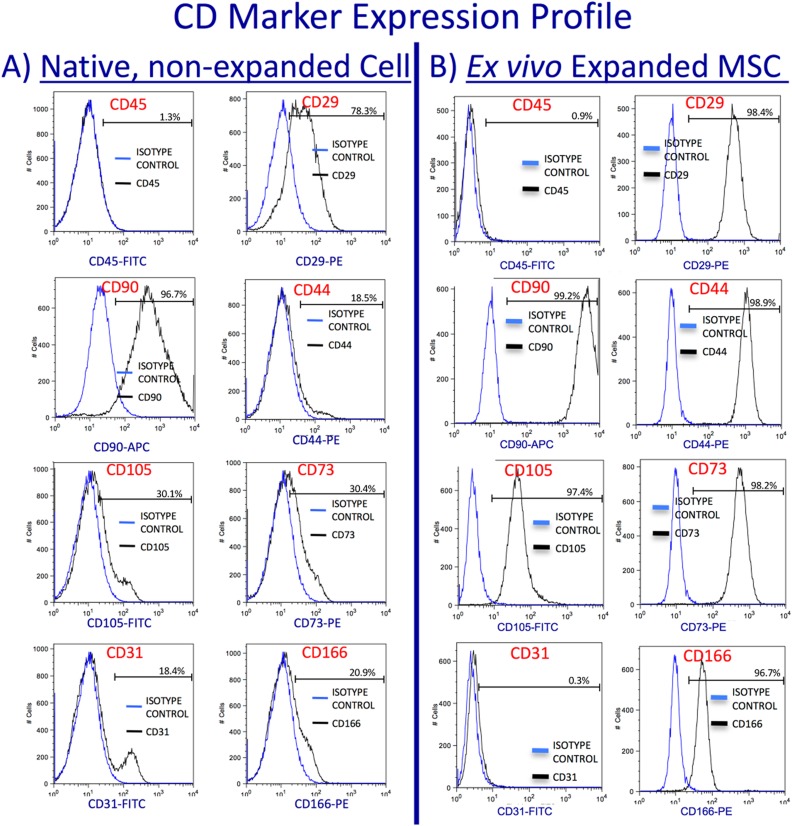
The heterogeneous population of native cells derived from the AC:Px System without the use of collagenase expresses MSC markers CD90 (Thy-1), CD29 (integrin beta1), and vascular cells adhesion molecule (VCAM) at high levels; CD105 (endoglin), CD44 (hyaluronic acid receptor), CD73 (ecto-5′-nucleotidase), and CD166 (activated leukocyte cell adhesion molecule) at relatively lower levels; and lack expression of CD45 (human leukocyte antigen [HLA]). The native cells also contain cells that express CD31 (platelet endothelial cell adhesion molecule; von Willebrand factor receptor; Fig. 2A). Once these cells are placed into culture for selective ex vivo expansion of MSCs, however, MSC markers (CD90+, CD29+, CD105+, CD44+, CD73+, CD166+, and CD45−) are expressed at levels expected on in vitro expanded MSCs (Fig. 2B)1,20. The native cells can be cryopreserved and thawed with on average 90.5% ± 7.8% postthaw recovery. Ex vivo expanded MSCs can be derived from either freshly derived (Fig. 3A) or cryopreserved (Fig. 3B) native cells processed using the AC:Px System.
Fig. 2.
Native, nonexpanded umbilical cord tissue (UCT) cells recovered from the AuxoCell Processing System were stained for mesenchymal stem/stromal cell surface receptors CD45, CD29, CD90, CD44, CD105, CD73, and CD166 and the endothelial marker CD31. The native UCT cells (A) express high levels of CD29 and CD90; low to moderate levels of CD44, CD105, CD73, CD166, and CD31; and no to very low expression of CD45. Once these native cells are placed into culture to ex vivo expand mesenchymal stem/stromal cell (MSCs), the expanded MSCs (B) express the expected levels of CD29, CD90, CD44, CD105, CD73, and CD166, with no expression of CD45 and CD31. The blue and black histograms represent the isotype control and receptor expression profiles for each antibody investigated, respectively. The percent positive for each marker is represented on each histogram.
Fig. 3.
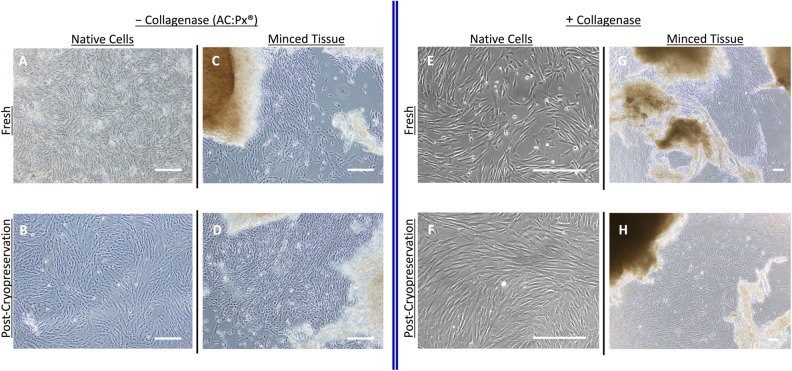
Freshly isolated native cells recovered from both the AuxoCell Processing System (AC:Px) (A) and collagenase digestion (E) can be placed into mesenchymal stem/stromal cell (MSC) culture medium and expanded ex vivo. In addition, native cells recovered from both the AC:Px System (B) and collagenase digestion (F) were cryopreserved, thawed, and placed into culture to derive ex vivo expanded MSCs. Freshly isolated minced tissue recovered from both the AC:Px System (C) and collagenase digestion (G) was used as explants to ex vivo expand MSCs, as well as from cryopreserved and thawed UCT minced tissue units from the AC:Px System (D) or collagenase digestion (H). Each image was taken with a phase contrast microscope. The white bar in each image represents 100 μm.
Minced tissue derived from the AC:Px System can also be placed into culture to derive ex vivo expanded MSCs using an explant method from both fresh (Fig. 3C) and postcryopreserved (Fig. 3D) explants. MSCs migrate out of the minced explant tissue in vitro, adhere to the plastic culture surface and expand resulting in cultured MSCs that express markers expected from in vitro expanded MSCs derived from native cells. These markers include greater than 95% expression of CD90, CD29, CD105, CD44, CD73, and CD166, while lacking expression of CD45. Accordingly, ex vivo expanded MSCs can be derived from both fresh (Fig. 3E) and cryopreserved (Fig. 3F) native cells recovered from UCT processed in the presence of collagenase. As described earlier, MSCs can migrate out from minced tissue pieces when placed into culture and adhere to the culture vessel. Cultured MSCs can also be derived from fresh (Fig. 3G) and cryopreserved (Fig. 3H) minced tissue recovered from collagenase-treated UCT.
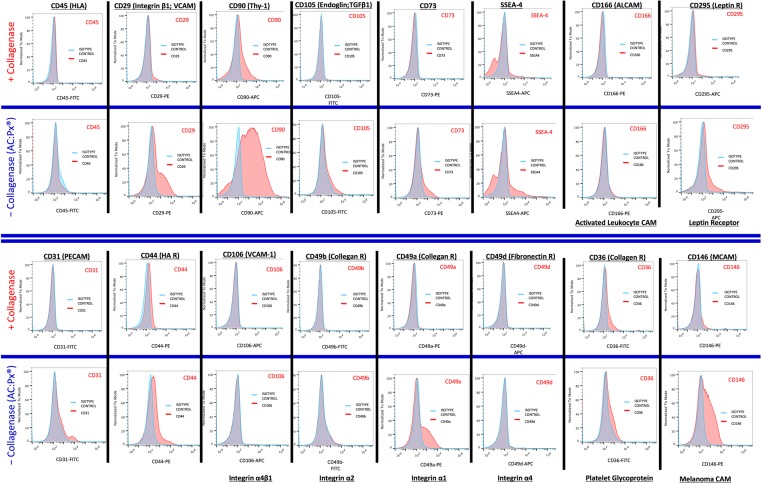
In order to further understand the effect of collagenase on the recovered native UCT cells, the native cells recovered from both processing in the presence and absence of collagenase were analyzed for various cell surface protein markers. Figure 4 illustrates the cell surface markers that are expressed on a typical UCT processed side-by-side, either by collagenase digestion or with the AC:Px System (without collagenase treatment). Quantitatively, specific markers show remarkable differences in expression (Table 3). Markers such as CD105, CD73, CD49b (collagen receptor), CD49a (collagen receptor), CD31, and CD166 are expressed on cells that are processed with the AC:Px System (without collagenase), while lower on cells processed using collagenase. Other markers such as CD90, CD29 (collagen receptor), SSEA-4, CD44, CD295 (leptin)21, CD36 (collagen receptor), and CD146 (melanoma cell adhesion molecule) show 2.0, 3.2, 1.3, 2.4, 3.6, 2.9, and 3.7 times greater expression in the cells recovered from the AC:Px System relative to cells recovered in the presence of collagenase (Table 3). Markers CD106 (VCAM-1) and CD49d (fibronectin receptor) show slight quantitative differences in expression in the absence of collagenase (Fig. 4, Table 3).
Fig. 4.
Native cells recovered from umbilical cord tissue (UCT) in the absence (using the AuxoCell Processing System) and presence of collagenase were stained with various antibodies against specific cell surface proteins and analyzed on an Attune™ Acoustic Flow Cytometer. Qualitative analysis of the cell surface proteins CD45, CD29, CD90, CD105, CD73, stage specific embryonic antigen-4, CD166, CD295, CD31, CD44, CD106, CD49b, CD49a, CD49d, CD36, and CD146 expression levels is exhibited after liberation from the same UCT with (first and third rows) or without (second and fourth rows) collagenase. The blue and red histograms represent the isotype control and receptor expression profiles for each antibody investigated, respectively.
Table 3.
Quantitative Analysis of the Cell Surface Protein Expression Levels Are Exhibited after Liberation from the Same Umbilical Cord Tissue in the Presence or Absence (AC:Px) of Collagenase.
| AVERAGE | SEM | N | |
|---|---|---|---|
| CD105+ | 6.3 | 3.1 | 10 |
| CD31+ | 12.8 | 6.8 | 10 |
| CD49b+ | 7.0 | 5.2 | 10 |
| CD36+ | 2.9 | 0.7 | 10 |
| CD29+ | 3.2 | 0.9 | 10 |
| CD73+ | 1.2 | 3.2 | 10 |
| CD44+ | 2.4 | 0.7 | 10 |
| CD49a+ | 9.6 | 4.8 | 10 |
| CD146+ | 3.7 | 1.0 | 10 |
| CD166+ | 8.8 | 6.0 | 10 |
| CD90+ | 2.0 | 0.4 | 10 |
| SSEA4+ | 1.3 | 0.3 | 10 |
| CD106+ | 1.2 | 1.1 | 10 |
| CD49d+ | 1.7 | 1.1 | 10 |
| CD235a+ | 2.1 | 1.1 | 10 |
| CD295+ | 3.6 | 1.3 | 10 |
Note: The average values shown represent the relative ratio expression of each respective marker ± the standard error of the mean (SEM). The relative ratio represents the ratio of the positive expression of each specific marker from the cells recovered from the AuxoCell Processing (AC:Px) System (absence of collagenase) to that of the manual processing method (presence of collagenase).
Discussion
The hypothesis has existed for many years now among scientists and regulators that enzyme treatment of tissues may result in biological changes of the recovered cell populations intended for cell-based therapies. In particular, certain cells from the heterogeneous native cell population may not be able to survive enzymatic treatment (e.g., erythrocytes, MSCs, endothelial cells). However, there has been no technology to directly provide evidence to investigate this hypothesize until now. The invention of the AC:Px System allows for standardized processing of solid tissues without the use of enzymes and acts as a control in investigating the impact of enzymes on cell recovery. No standard protocol exists to enzymatically process a given tissue, due to a multitude of variables, including tissue type, enzyme choice, incubation time, and enzyme concentration. It is for this reason that it is critical to compare any enzyme treatment to a non-enzyme-treated control. The AC:Px System is a sterile, single-use, disposable technology that allows for standardized solid tissue processing without the need for biochemical, enzymatic breakdown. Instead, the AC:Px System utilizes mechanical means to efficiently break down the tissue into small minced tissue pieces and, in the process, releases the heterogeneous native cell populations that comprise the tissue. Using this novel technology, we have the opportunity to directly determine the biological effects that collagenase treatment has on cells recovered from UCT, by comparing the results from the same tissue sample processed without enzymatic digestion using the AC:Px System. We report here for the first time that collagenase treatment of UCT results in lower cell yields and differences in cell receptor expression—when compared to noncollagenase processed UCT.
UCT can be processed using the AC:Px System (Fig. 1A) and fractionated into 3 products (native cells, minced tissue, and decellularized Wharton’s jelly)—all from the same unit—without the need or use of any enzymatic-dependent tissue breakdown (Fig. 1B). The native cells recovered from the AC:Px System represent the heterogeneous population of cells that make up and are found within the in utero UCT and contain cells that express MSC markers CD90+, CD29+, CD105+/−, CD73+/−, CD44+/−, and CD166+/− while also lacking CD45 expression (Fig. 2A). These cells, once placed into MSC growth medium in an ex vivo cell expansion vessel, result in ex vivo expanded MSCs that express CD90+, CD29+, CD105+, CD73+, CD44+, CD166+, and CD45− at expected levels20 (Fig. 2B, 3). One possible reason why cells increase their MSC marker expression when cultured, as compared to their initial expression levels, could be due to the selective pressure that cells are put under by the culturing conditions (e.g., 2-D flasks, medium). This selective pressure allows for selective adherence and expansion of CD90+, CD29+, CD105+, CD73+, CD44+, CD166+, and CD45− from the native cell population. The process of ex vivo expanding MSCs from the heterogeneous native UCT cell population, along with this selective pressure, yields an artificial cell product (relative to the starting in utero native, primary cell population) in that the MSCs recovered from ex vivo expansion are created under artificial conditions (e.g., incubator, medium, animal-derived serum). The combination of this selective and relatively artificial expansion could enable greater receptor turnover and expression for these particular MSC markers when placed into culture.
There is no significant difference in cell morphology or cell receptor protein expression, once the cells are placed into in vitro culture—independent of whether the UCT unit is processed in the presence or absence (AC:Px) of collagenase or whether the unit is expanded pre- or postcryopreservation (Fig. 3). One notable qualitative difference observed with the native cells collected from the AC:Px System was that the population possesses significantly greater number of erythrocytes (based on visual observations under a microscope) since the AC:Px indiscriminately collects all the native cells that make up the UCT, which includes erythrocytes from remnant coagulated UCB within the UCT vessels. In extremely erythrocyte-rich samples, the presence of erythrocytes hinders MSC adherence and expansion in vitro, precryopreservation, since the erythrocytes settle quicker and, at times, completely cover the surface of the culture vessel. However, MSC expansion from these erythrocyte-rich samples postcryopreservation were relatively more efficient (data not shown), presumably due to the significant reduction in erythrocytes during the cryopreservation freeze/thaw cycle (Fig. 3), consistent with observations from other groups that cryopreservation compromises the membranes on erythrocytes22. Furthermore, native cells recovered from UCT using enzymatic digestion result in a cell population that contains relatively lower number of erythrocytes by compromising erythrocyte membranes23,24, which could be one rationale as to why erythrocyte contamination does not impact ex vivo expansion from collagenase-treated UCTs.
We observed, under enzymatic digestion conditions using collagenase, an approximately 2.7-fold reduction in live native MSCs—when compared to no enzyme controls processed using the AC:Px System (Table 1). This reduction in recovered cells is not due to the differences in the process, but rather due to the collagenase digestion since we observed comparable cell recovery reductions with manual processing method using enzymatic digestion and AC:Px (with enzymatic digestion; Table 1). This difference is statistically significant (P < 0.01), suggesting that collagenase does indeed effect the number of native MSCs recovered. This significant reduction in observed cell yields with collagenase digestion—compared to noncollagenase processed UCT—could be due to the enzyme digesting not only the tissue but also the native cells. Although in our studies, enzymatic digestion was not longer than 3 h, presumably with these observations, with longer collagenase incubation or incubation of isolated cells in collagenase, the native cell yields could significantly decrease due to the digestion of native cells, themselves. Enzymatic digestion of the native cells that comprise a tissue could be the reason why certain tissues/organs—such as heart25 and cartilage26—are sensitive to enzymatic digestion since very little or no native cells are recovered postdigestion when processed in the presence of enzymes.
In order to understand the qualitative and quantitative differences in the makeup of the cell population between enzymatic treated and untreated (processed using the AC:Px System), we stained the recovered cells from each respective processings for various cell receptor proteins. Qualitatively (Fig. 4) and quantitavely (Table 3) specific markers show remarkable reduction in expression in the presence of collagenase. Markers such as CD105, CD29, CD73, CD90, SSEA-4, CD44, CD295, CD36, CD49b, CD49a, CD31, CD146, and CD166 exhibit higher expression on native cells recovered from the AC:Px System (without collagenase), compared to the native cells recovered using collagenase digestion (Table 3). Not surprising, collagen receptors CD49b/CD29 (VLA-4) and CD49a/CD29 are particularly sensitive to, and could be presumably, cleaved by collagenase. The cleaving of these receptors may affect the biological function of the recovered native cells since the collagen family of receptors functions to bind collagen, the most abundant extracellular matrix protein in humans27. The collagen receptor regulates cell adhesion, proliferation, migration, and coagulation cascades. They also regulate matrix metalloproteinases to control the extracellular matrix (ECM)28. Cleavage of type I collagen receptors has been shown to effect communication between cells and ligands in the ECM, creating a differential interaction to promote responses that drive pathological processes including fibrosis, inflammation, and tumor progression27–29. Cleaving of collagen receptors certainly can affect the biological function and/or malfunction of these cells and lead to misleading results in either in vitro or in vivo studies.
Other groups have observed similar enzymatic effects and have exhibited biological, functional changes of cells derived using enzymatic digestion from various tissues. However, unlike our study, where we compare our results to non-enzyme-treated controls, their observations are based on studies that arise from differing concentrations of various enzymes. In particular, Autengruber et al. concluded that enzymatic tissue digestion had profound effects on the expression of a variety of cell-surface receptors, directly effecting phenotypic analysis, targeted cell isolation using FACS or MACS, and immune cell function in in vitro cell culture experiments11. Specifically, they looked at the effect of collagenase or dispase on the isolation of primary mouse splenocytes at both low and high concentrations. They found that collagenase treatment resulted in relatively minor effects in splenocyte cell surface receptor differences, compared to dispase treatment. However, splenocyte isolation using dispase considerably affected a majority of the cell surface proteins, relative to collagenase treatment. Interestingly, the effects were dose dependent based on either low or high dispase treatment. This effect lingered even 24 h after in vitro culture as evident in the reduced expression of markers and also the impaired antigen-specific proliferation of CD4+ and CD8+ T cells. They also suggested that reduced cell surface protein expression due to enzyme-mediated digestion effects key immunological functions such as cell adhesion, signaling, and pathogen recognition and negatively impacts immune cell function giving rise to misleading in vitro data. Furthermore, enzyme-mediated cleavage of cell surface receptors may be the possible reason there is often a discrepancy between data obtained by immunohistochemical staining of 3-dimensional tissue, gene expression data, and data obtained from flow cytometry following staining cells derived from tissue that has been processed using enzymatic digestion11. However, they did not have the means to process splenocytes without the use of enzymes, as a control for baseline expression and function of the native cells. Similar marker effects have been observed in the enzymatic isolation of lung CD4+ T cells and pulmonary CD8+ T cells30.
Additional groups have observed similar enzyme-dependent effects, albeit at various levels, on cell surface receptor proteins on different cell isolations. Ford et al. observed almost complete loss in CD4 and CD25 cell surface protein levels in isolating murine T cells due to enzymatic digestion. Specifically, they used dispase to enzymatically digest central nervous tissue from Lewis strain rats to isolate central nervous system (CNS) microglia and other leukocytes. Compared to postactivated CD4+ T cells before enzymatic digestion, postdigestion isolations revealed almost complete loss of expression of CD4, CD25, and a number of other surface antigens31. White et al. found lost CD4 and CD8 expression after isolating human female reproductive tract cells using a cocktail of enzymes (comprised of pancreatin, hyaluronidase, and collagenase) while maintaining CD45, CD3, CD14, and CD19 expression32. Yoon et al. hypothesized that enzymatic digestion impacts functional activity by exhibiting the effect of enzyme on growth factor production. In order to investigate enzyme-dependent effects, they compared fibroblast growth factors (bFGFs) production levels of culture and expanded UCT MSCs derived from both enzymatic isolation and explant methods33. When comparing cellular characteristics and bFGF levels, they concluded that MSCs expanded from explanted UCT, released significantly higher bFGF levels during the first week of passaging, compared to UCT MSCs derived from enzymatic digestion. Since bFGF stimulates self-renewal, adhesion, and cell survival of ESCs34 and is essential for MSC differentiation35, it is no surprise that these levels significantly decreased with prolonged passaging, possibly due to culture-dependent senescence1,9. Interestingly, they observed lower expression levels of mitotic-related genes with the MSCs derived from enzyme digested UCTs, along with lower cellular yields at day 033. Tabatabaei et al. observed that even different vendors of trypsin used for enzymatic digestion of human amnion affected the expression of CD105 and HLA-I in derived amniotic epithelial cells36. The authors concluded that discrepancies with levels of HLA-I expression from differing investigators3,37–40 could be due to the type and specific activity of the trypsin used during cell liberation and isolation36. Other groups have also investigated nonenzymatic methods to avoid any change to the cells biology in other perinatal tissues. At the expense of cell yields, Canazza et al. developed a nonenzymatic method to derive ex vivo cultured MSCs from periumbilical adipose tissue in order to allow for translation into preclinical studies without affecting the cells biological features and basic functions41. Taken altogether, these studies exhibit the various biological and functional impacts of enzymatic digestion in liberating cells from tissue.
Biochemical means have been employed for several decades to break down the tissue to recover primary cells. One specific reason for this is that the cell yields are traditionally higher when tissue is processed with enzyme compared to solely mechanical means42. For instance, in the recovery of the stromal vascular fraction from adipose tissue, yields of up to 13 × 105 nucleated cells/milliliter of lipoaspirate processed have been observed for collagenase-based processings43, compared to 2.4 × 105 nucleated cells/milliliter of lipoaspirate processed using mechanical, nonenzymatic means to recover cells. This observation could be due to lack of sufficient mechanical means to break down the tissue to release and recover the native cells. At least in the case for UCT processing, the AC:Px System gives rise to a significantly higher MSC yield when processed using solely mechanical (and not biochemical) means compared, side-by-side, with collagenase breakdown of UCT. Investigating adipose tissue (along with other tissues) processed with the AC:Px System side-by-side with enzymatic digestion will give more insight on the comprehensiveness of this observation.
For the first time, we have shown the qualitative and quantitative effects that collagenase treatment has in liberating cells from UCT by comparing against a non-enzyme-treated control in the form of the novel technology, the AC:Px System. The observed adverse effects of collagenase bring into question the biological and functional enzyme–mediated effects on cells that are currently being processed in the presence of enzymes for current and/or future clinical use. Specifically, tissue banks are currently processing UCT and other tissues using enzyme(s) to liberate cells that will be cryopreserved and used directly in the clinical setting. This study suggests that enzymatic digestion affects the quality and quantity of the cell-based therapies destined for the clinics by effecting the cell surface receptor profile, which will certainly affect cellular function in vitro and in vivo. This study should beg the question as to whether cellular and noncellular products derived in the presence of enzymes should be further regulated by the worldwide regulatory agencies. Moreover, although additional studies are needed and will be conducted, it can be hypothesized that enzymes may be adversely affecting the function of noncellular proteins in the Wharton’s jelly, rich in various ECM proteins (i.e., collagen), growth factors, cytokines, and glycosaminoglycans (i.e., hyaluronic acid)13 that could be sensitive to enzymes. The decellularized Wharton’s jelly product from the AC:Px System could be clinically advantageous since the decellularized Wharton’s jelly components are not collected in the presence of enzymes—and, therefore, could be of higher quality. Future studies will further investigate the biological and functional effects of enzymatic digestion on liberating cells from umbilical cord tissue and other tissues as well as their respective Wharton’s jelly proteins.
Footnotes
Authors’ Note: R.R.T. designed, conducted, analyzed, and wrote the findings in this report. K.J.C. and C.L.C. reviewed the manuscript. R.R.T., K.J.C., and C.L.C. are employees and stakeholders in AuxoCell Laboratories, Inc., the inventor and manufacturer of the AC:Px System.
Ethical Approval: Ethical Approval is not appicable for this article.
Statement of Human and Animal Rights: Statement of Human and Animal Rights is not appicable for this article.
Statement of Informed Consent: Umbilical Cord Tissue units for studies were obtained from healthy mothers who donated their de-identified UCT after giving written informed consent.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Taghizadeh RR. Perinatal mesenchymal stem cell banking for umbilical cord blood transplantation and regenerative medicine In: Cetrulo KJ, Cetrulo CL, Jr., Taghizadeh RR, editors. Perinatal stem cells. Vol. 2 Hoboken, New Jersey, USA: John Wiley & Sons, Inc.; 2013. pp. 53–69. [Google Scholar]
- 2. Taghizadeh RR, Cetrulo KJ, Cetrulo CL. Wharton’s jelly stem cells: future clinical applications. Placenta. 2011;32(Suppl 4):S311–S315. [DOI] [PubMed] [Google Scholar]
- 3. Murphy S, Rosli S, Acharya R, Mathias L, Lim R, Wallace E, Jenkin G. Amnion epithelial cell isolation and characterization for clinical use: Current protocols in stem cell biology. Bhatia M, Elefanty A, Fisher SJ, editors. Hoboken, New Jersey, USA: John Wiley & Sons, Inc.; 2007: Unit 1E6. [DOI] [PubMed] [Google Scholar]
- 4. Bailo M, Soncini M, Vertua E, Signoroni PB, Sanzone S, Lombardi G, Arienti D, Calamani F, Zatti D, Paul P, et al. Engraftment potential of human amnion and chorion cells derived from term placenta. Transplantation. 2004;78(10):1439–1448. [DOI] [PubMed] [Google Scholar]
- 5. Parolini O, Alviano F, Bagnara GP, Bilic G, Bühring H-J, Evangelista M, Hennerbichler S, Liu B, Magatti M, Mao N, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international workshop on placenta derived stem cells. Stem Cells. 2008;26(2):300–311. [DOI] [PubMed] [Google Scholar]
- 6. Soncini M, Vertua E, Gibelli L, Zorzi F, Denegri M, Albertini A, Wengler GS, Parolini O. Isolation and characterization of mesenchymal cells from human fetal membranes. J Tissue Eng Regen Med. 2007;1(4):296–305. [DOI] [PubMed] [Google Scholar]
- 7. Taghizadeh RR, Sherley JL. Expanding the therapeutic potential of umbilical cord blood hematopoietic stem cells In: Cetrulo CL, Cetrulo K, Cetrulo CL, Jr, editors. Perinatal stem cells. Hoboken, NJ: Wiley-Blackwell; 2009. pp. 21–27. [Google Scholar]
- 8. Friedman R, Betancur M, Boissel L, Tuncer H, Cetrulo C, Klingemann H. Umbilical cord mesenchymal stem cells: adjuvants for human cell transplantation. Biol Blood Marrow Transplant. 2007;13(12):1477–1486. [DOI] [PubMed] [Google Scholar]
- 9. Taghizadeh RR, Pollok KE, Betancur M, Boissel L, Cetrulo KJ, Marino T, Wolfberg A, Klingemann HG, Cetrulo CL. Wharton’s jelly derived mesenchymal stem cells: regenerative medicine beyond umbilical cord blood. Placenta 2011;32(Supp 4): S339. [Google Scholar]
- 10. Briddell R, Litkenhaus F, Foertsch G, Fuhrmann A, Foster K, Falcon Girard K, Fiscus B, Boehm A, Brown M, Pettit M, et al. Recovery of viable MSCs isolated from fresh umbilical cord tissue, measured after cryopreservation, is on average 8-fold higher when compared to recovery of viable MSCs isolated from previously cryopreserved umbilical cord tissue. Blood. 2015;118(21):4398. [Google Scholar]
- 11. Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol. 2012;2(2):11–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boissel L, Betancur M, Klingemann H, Marchand J. Umbilical Cord Mesenchymal Stem Cells In: Cetrulo CL, Cetrulo K, Cetrulo CL, Jr, editors. Perinatal Stem Cells. Hoboken, NJ: Wiley-Blackwell; 2009. pp. 69–75. [Google Scholar]
- 13. Jadalannagari S, Converse G, McFall C, Buse E, Filla M, Villar MT, Artigues A, Mellot AJ, Wang J, Detamore MS, et al. Decellularized Wharton’s jelly from human umbilical cord as a novel 3D scaffolding material for tissue engineering applications. PLoS One. 2017;12(2):e0172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sobolewski K, Bankowski E, Chyczewski L, Jaworski S. Collagen and glycosaminoglycans of Wharton’s jelly. Biol Neonate. 1997;71(1):11–21. [DOI] [PubMed] [Google Scholar]
- 15. Karahuseyinoglu S, Cinar O, Kilic E, Kara F, Akay GG, Demiralp DÖ, Tukun A, Uckan D, Can A. Biology of stem cells in human umbilical cord stroma: In situ and in vitro surveys. Stem Cells. 2007;25(2):319–331. [DOI] [PubMed] [Google Scholar]
- 16. Wharton T. Adenographia: sive glandularum totius corporis descriptio [Adenographia: the arrangement of the glands of the whole of the body, description]. London, UK: Publisher Sumptibus Joannis Ravesteinii; 1656. [Google Scholar]
- 17. Bankowski E, Sobolewski K, Romanowicz L, Chyczewski L, Jaworski S. Collagen and glycosaminoglycans of Wharton’s jelly and their alterations in EPH-gestosis. Eur J Obstet Gynecol Reprod Biol. 1996;66(2):109–117. [DOI] [PubMed] [Google Scholar]
- 18. Davies JE, Walker JT, Keating A. Concise review: Wharton’s jelly: the rich, but enigmatic, source of mesenchymal stromal cells. Stem Cells Transl Med. 2017;6(7):1620–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sobolewski K, Bańkowski E, Chyczewski L, Jaworski S. Collagen and Glycosaminoglycans of Wharton’s jelly. Neonatology. 1997;71(1):11–21. [DOI] [PubMed] [Google Scholar]
- 20. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. [DOI] [PubMed] [Google Scholar]
- 21. Zhou Bo O, Yue R, Murphy Malea M, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henkelman S, Lagerberg JWM, Graaff R, Rakhorst G, Van Oeveren W. The effects of cryopreservation on red blood cell rheologic properties. Transfusion. 2010;50(11):2393–2401. [DOI] [PubMed] [Google Scholar]
- 23. Yamaguchi T, Matsumoto M, Kimoto E. Hemolytic properties under hydrostatic pressure of neuraminidaseor protease-treated human erythrocytes. J Biochem. 1993;114(4):576–581. [DOI] [PubMed] [Google Scholar]
- 24. Sharma S, Gokhale SM. Enzymatic cleavage of cell surface proteins of pig and cow erythrocytes and its effect on concanavalin-mediated agglutinability. Indian J Biochem Biophys. 2014;51(5):378–387. [PubMed] [Google Scholar]
- 25. Chen L, Pan Y, Zhang L, Wang Y, Weintraub N, Tang Y. Two-step protocol for isolation and culture of cardiospheres. Methods Mol Biol. 2013;1036:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakob M, Démarteau O, Schäfer D, Stumm M, Heberer M, Martin I. Enzymatic digestion of adult human articular cartilage yields a small fraction of the total available cells. Connect Tissue Res. 2003;44(3-4):173–80. [DOI] [PubMed] [Google Scholar]
- 27. Madsen DH, Engelholm LH, Ingvarsen S, Hillig T, Wagenaar-Miller RA, Kjøller L, Gårdsvoll H, Høyer-Hansen G, Holmbeck K, Bugge TH, et al. Extracellular collagenases and the endocytic receptor, urokinase plasminogen activator receptor-associated protein/endo180, cooperate in fibroblast-mediated collagen degradation. J Biol Chem. 2007;282(37):27037–27045. [DOI] [PubMed] [Google Scholar]
- 28. Leitinger B, Hohenester E. Mammalian collagen receptors. Matrix Biol. 2007;26(3):146–155. [DOI] [PubMed] [Google Scholar]
- 29. Mazur A, Holthoff E, Vadali S, Kelly T, Post SR. Cleavage of type I collagen by fibroblast activation protein-α enhances class A scavenger receptor mediated macrophage adhesion. PLoS One. 2016;11(3):e0150287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bruder D, Westendorf AM, Geffers R, Gruber AD, Gereke M, Enelow RI, Buer J. CD4 T lymphocyte–mediated lung disease. Am J Respir Crit Care Med. 2004;170(11):1145–1152. [DOI] [PubMed] [Google Scholar]
- 31. Ford AL, Foulcher E, Goodsall AL, Sedgwick JD. Tissue digestion with dispase substantially reduces lymphocyte and macrophage cell-surface antigen expression. J Immunol Methods. 1996;194(1):71–75. [DOI] [PubMed] [Google Scholar]
- 32. White HD, Prabhala RH, Humphrey SL, Crassi KM, Richardson JM, Wira CR. A method for the dispersal and characterization of leukocytes from the human female reproductive tract. Am J Reprod Immunol. 2000;44(2):96–103. [DOI] [PubMed] [Google Scholar]
- 33. Yoon JH, Roh EY, Shin S, Jung NH, Song EY, Chang JY, Kim BJ, Jeon HW. Comparison of explant-derived and enzymatic digestion-derived MSCs and the growth factors from Wharton’s jelly. Biomed Res Int. 2013;2013:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eiselleova L, Matulka K, Kriz V, Kunova M, Schmidtova Z, Neradil J, Tichy B, Dvorakova D, Pospisilova S, Hampl A, et al. A complex role for FGF-2 in self-renewal, survival, and adhesion of human embryonic stem cells. Stem Cells. 2009;27(8):1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ng F, Boucher S, Koh S, Sastry KSR, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, et al. PDGF, TGF-β, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112(2):295. [DOI] [PubMed] [Google Scholar]
- 36. Tabatabaei M, Mosaffa N, Nikoo S, Bozorgmehr M, Ghods R, Kazemnejad S, Rezania S, Keshavarzi B, Arefi S, Ramezani-Tehrani F, et al. Isolation and partial characterization of human amniotic epithelial cells: the effect of trypsin. Avicenna J Med Biotechnol. 2014;6(1):10–20. [PMC free article] [PubMed] [Google Scholar]
- 37. Ilancheran S, Michalska A, Peh G, Wallace EM, Pera M, Manuelpillai U. Stem cells derived from human fetal membranes display multilineage differentiation potential. Biol Reprod. 2007;77(3):577–588. [DOI] [PubMed] [Google Scholar]
- 38. Liu YH, Vaghjiani V, Tee JY, To K, Cui P, Oh DY, Manuelpillai U, Toh BH, Chan J. Amniotic epithelial cells from the human placenta potently suppress a mouse model of multiple sclerosis. PLoS One. 2012;7(4):e35758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikoo S, Ebtekar M, Jeddi-Tehrani M, Shervin A, Bozorgmehr M, Kazemnejad S, Zarnani AH. Effect of menstrual blood-derived stromal stem cells on proliferative capacity of peripheral blood mononuclear cells in allogeneic mixed lymphocyte reaction. J Obstet Gynaecol Res. 2012;38(5):804–809. [DOI] [PubMed] [Google Scholar]
- 40. Perin L, Giuliani S, Jin D, Sedrakyan S, Carraro G, Habibian R, Warburton D, Atala A, De Filippo RE. Renal differentiation of amniotic fluid stem cells. Cell Prolif. 2007;40(6):936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Canazza A, Bedini G, Caremoli F, Nava S, Latorre E, Tosetti V, Taiana M, Dossena M, Bersano A, Pareyson D, et al. A novel efficient method to isolate human adipose-derived stromal cells from periumbilical biopsies without enzymatic digestion. CellR4. 2015;3(1):e1397. [Google Scholar]
- 42. Aronowitz JA, Lockhart RA, Hakakian CS. Mechanical versus enzymatic isolation of stromal vascular fraction cells from adipose tissue. Springerplus. 2015;4:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshimura K, Shigeura T, Matsumoto D, Sato T, Takaki Y, Aiba-Kojima E, Sato K, Inoue K, Nagase T, Koshima I, et al. Characterization of freshly isolated and cultured cells derived from the fatty and fluid portions of liposuction aspirates. J Cell Physiol. 2006;208(1):64–76. [DOI] [PubMed] [Google Scholar]