Abstract
The microtubule-associated protein Tau, generated by the MAPT gene is involved in dozens of neurodegenerative conditions (“tauopathies”), including Alzheimer’s disease (AD) and frontotemporal lobar degeneration/frontotemporal dementia (FTLD/FTD). The pre-mRNA of MAPT is well studied and its aberrant pre-mRNA splicing is associated with frontotemporal dementia. Using a PCR screen of RNA from human brain tissues, we found that the MAPT locus generates circular RNAs through a backsplicing mechanism from exon 12 to either exon 10 or 7. MAPT circular RNAs are localized in the cytosol and contain open reading frames encoding Tau protein fragments. Transfected into cultured HEK 293 cells, the MAPT exon 10 is alternatively spliced and proteins involved in its regulation, such as clk2, SRSF7/9G8, PP1 (protein phosphatase 1) and NIPP1 (nuclear inhibitor of PP1) reduce the abundance of the circular MAPT exon 12→10 backsplice RNA. In summary, we report the identification of new bona fide human brain RNAs produced from the MAPT locus. These may be a component of normal human brain Tau regulation and, since the circular RNAs could generate high molecular weight proteins with multiple microtubule binding sites, they could contribute to taupathies.
Keywords: circular RNAs, tau, Alzheimer’s disease, alternative pre-mRNA splicing, gene expression
1. Introduction
The human microtubule-associated protein Tau is highly expressed in brain and promotes the assembly and stabilization of microtubules [1]. Tau protein can mis-fold into paired helical filaments and neurofibrillary tangles, which characterize a group of neurodegenerative diseases known as tauopathies, that include Alzheimer’s disease (AD), frontotemporal lobar degeneration (FTLD-TAU), progressive supranuclear palsy (PSP), chronic traumatic encephalopathy (CTE), and primary age-related tauopathy (PART) [2]. The clearest connection between the MAPT gene and neurodegeneration is found in FTLD-MAPT, as the disease is caused by many different known mutations in the MAPT locus on chromosome 17 [3]. Studies in mice indicate that tau protein is necessary for amyloid-beta induced neuronal cell death [4], and thus plays a central role in AD. Tau protein aggregates can apparently spread through neuronal networks, suggesting that “prion-like” transcellular propagation is an important mechanism of Tau pathology throughout the brain [5].
The human MAPT gene contains 16 exons, with exons 2, 3, 4a, 6, 8 and 10 being alternatively spliced cassette exons (Figure 1A). Alternative splicing of these exons in the normal adult human brain generates six major protein isoforms. These isoforms differ at the N-terminus due to alternative exons 2 and 3 in the tau projection domain and in the number of microtubule binding repeats due to alternative splicing of exon 10. Exon 10 encodes one of the four microtubule binding sites, and its alternative usage generates tau isoforms with either 3 or 4 binding sites (3R, 4R) that differ in their affinity to microtubules [6] and could thus ‘fine tune’ the interaction between the Tau protein and microtubules.
Figure 1: The human MAPT gene generates circular RNAs through exon 12 backsplicing.
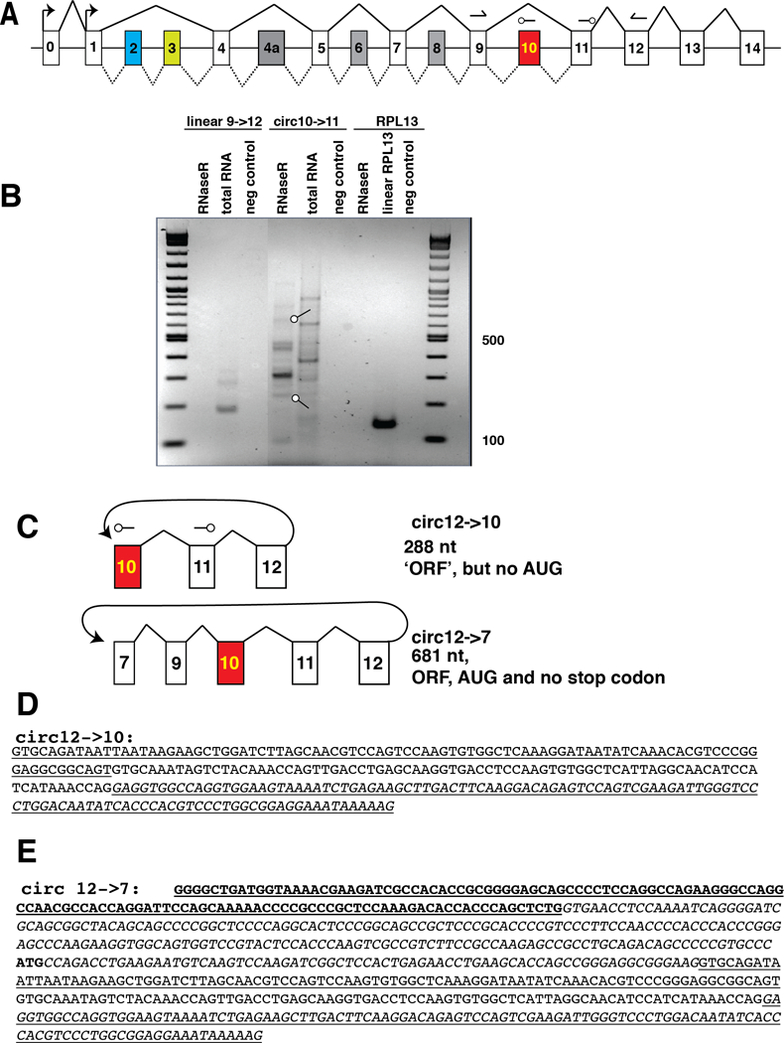
A. Schematic structure of the human MAPT gene, Constitutive exons are in white, alternative exons are colored. Splicing patterns are indicated by lines. The two transcriptional start sites in exons 0 and 1 are indicated by arrows. The numbering of exons is from [27]. The arrows in exons 9 and 11 indicate the position of primers to amplify linear MAPT mRNA, arrows in exons 10 and 11 indicate primers to detect circular RNAs. The arrowheads indicate the 3’ ends.B. Circular RNAs generated through backsplicing of exon 12, 10 μg of total human hippocampus RNA was amplified with primers pointing outwards in exon 10 and 11. Prior to RT-PCR, an equal part of the RNA was digested with RNase R. The negative control was performed without reverse transcription. The PCR-products were gel purified, subcloned and sequenced. Two bands indicated by arrows were circular RNAs generated through backsplicing of exon 12. The other bands were PCR artifacts, i.e. amplicons from different gene regions.C. Structure of the RNAs generated through exon 12 backsplicing.D. Sequence and predicted ORFs for circ12→10 The RNA part corresponding to exon 10 is underlined, exon 11 is in regular letters and exon 12 is underlined and italic.E. Sequence and predicted ORFs for circ12→7. Exon 7 is bold, exon 9 italic, exon 10 is underlined, exon 11 is in regular letters and exon 12 is underlined and italic. The in frame AUG is shown in bold.
At least 19 mutations causing FTLD-MAPT have been identified in exon 10 and its 5’ splice site [3]. Pathological changes in exon 10 usage without mutations in exon 10 result in tauopathies, for example PSP and corticobasal degeneration (CBD) are characterized by 4R Tau, whereas Pick’s disease is characterized by 3R Tau. Post mortem studies of AD brains indicated a slight increase of exon 10 usage [7–9], as well as deregulation of protein factors that regulate exon 10 splicing, namely the SR-like protein tra2-beta1 that promotes exon 10 usage and its kinase, clk2, that inhibits exon 10 usage [7, 10].
In addition to the well known linear RNAs, pre-mRNAs generate circular RNAs through a backsplicing mechanism, where a downstream 5’ splice site is joined with an upstream 3’ splice site [11]. In most cases, circRNAs are generated when the pre-mRNA forms a loop containing the exons undergoing backsplicing. This loop can be formed by either a large lariat or more commonly through intramolecular RNA base pairing, leading to double stranded RNA regions as short as 30–40 nt in length. Frequently, repeat elements that have regions of base complementarity provide the basis of loop formation. In humans, these elements are often Alu elements [12, 13], comprising around 11% of the human genome [14]. Due to their self complementarity Alu elements form extensive double stranded RNA structures in pre-mRNA, which can influence alternative splicing [15] and promote the formation of circRNAs [12]. With about 50 highly expressed exceptions, circRNAs comprise only 1–5% of their linear counterparts. Since they lack a 3’ or 5’ end, they escape the exonnucleic degradation of linear RNAs and are thus more stable. The analysis of a few circRNAs showed that they mainly reside in the cytosol, where they can function as microRNA sponges [13], and can undergo translation in the presence of an internal ribosomal entry site [16], possibly aided through methylation of adenine bases at the N6 position. In addition, intron-containing circRNAs have been implicated in transcriptional control in the nucleus [17].
Here, we used a PCR approach to identify circular RNAs from the human tau locus that contain the alternatively spliced exon 10. These circular RNAs are low abundant, comprising less than 1% of the linear tau RNA. The data shows that not all RNAs generated from the human tau locus have been identified.
2. Material and Methods
2.1. Primers
9 -> 11 Forward: TGT CAA GTC CAA GAT CGG CT
9 -> 11 Reverse: CTG GCC ACC TCC TGG TTT
11 -> 10 Forward: GAC CTC CAA GTG TGG CTC AT
11 -> 10 Reverse: TGG ACT GGA CGT TGC TAA GA
RPL13 Forward: GCC ATC GTG GCT AAA CAG GTA
RPL13 Reverse: GTT GGT GTT CAT CCG CTT GC
Tau 12->10 reverse: cag ctt ctt att aat tat ctg cac ctt tt
Tau 10->11 forward: gag gcg gca gtg tgc aa
HIPK3f: tcg gcc agt cat gta tca aa
HIPK3r: tgc ttg gct cta ctt tga gtt tc
Tau exon12 Rev: ccc aat ctt cga ctg gac tc
Tau exon 9 Forw: tgt caa gtc caa gat cgg ct
2.2. Minigene generation
The tau exon 9–12 minigene was generated using Gibson cloning by assembling exons 9, 10, 11 and 12 in pcDNA3.1 using these primers:
Vector1 AAGCTTAAGTTTAAACGCTAGCCAGCTTG
Vector 2 CTCGAGTCTAGAGGGCCCGTTTAAACC
Exon 9f ctggctagcgtttaaacttaagcttACGCTGGCCGCAGGGATT
Exon 9r cctgtggatttttGGCCCACGAGTGGAGATGC
Exon 10f ccactcgtgggccAAAAATCCACAGGTGATTCTGATGCC
Exon 10r gagtggggtatctGCGGCAGCCCAGTCTCAG
Exon 11f actgggctgccgcAGATACCCCACTCCTGCCTTTCCA
Exon 11r aacatcctgtaaaccatgaccccacAGTAGCTGGGACTACAGGCG
Exon 12f GTGGGGTCATGGTTTACAG
Exon 12r tttaaacgggccctctagactcgagAAACTGCAGTGACTTAGGCC
2.3. RNA isolation
Samples were derived from short-postmortem interval (PMI) autopsies. All methods conformed with a University of Kentucky IRB protocol. Premortem clinical evaluations and pathological assessments were as described previously [18]. The inclusion criteria that were applied: PMI <4hrs; no evidence of frontotemporal dementia; no cancer in the brain parenchyma; and no large infarctions in the brain, or microinfarcts found within 3cm of the brain tissue samples. We also obtained information on agonal events for each subject, and additional criteria for exclusion from the study were an extended interval of premortem hypoxia, any medical ventilator use, brain edema, or large infarct. The characterization of the samples is shown in Supplemental Figure 1. The RNA isolation was performed using Trizol and the PureLink RNA mini kit from Ambion by Life Technologies.
2.4. Cell fractionation
Was performed using PARIS kit (Protein and RNA isolation system) from Thermo Fisher Scientific/Invitrogen according to the manufacturer’s instructions.
2.5. Transfection assays
were performed as described [19].
2.6. RT-PCR
was performed as described [20].
2.7. RNAse protection
was performed as described [21].
3. Results
3.1. Tau generates circular RNAs through exon 12 backsplicing
No human circular MAPT RNAs have been reported in databases. Given the extensive alternative pre-mRNA processing of the human tau gene (Figure 1A), we used a PCR approach to search for possible human circular MAPT RNAs. We concentrated on exon 10, since this exon is alternatively spliced and deregulated in both Alzheimer’s disease and FTLD-MAPT [7, 8]. To amplify circular RNAs, we used a reverse primer upstream of the forward primer, i.e. a reverse primer in exon 10 and a forward primer in exon 11, which amplifies circular, but not linear RNAs (Figure 1A, C). To further enrich for circular RNAs, we digested the RNA with RNase R, a bacterial RNA exonuclease that removes linear RNAs [9].
As expected, RNAse R treatment removed the signal for linear MAPT, detected by primers in exons 9 and 11 and RPL3 mRNA, but enriched the signal for circular RNAs (Figure 1B). The bands from RNAse R treated RNA generated by the circular RNA primers were subcloned and sequenced. Two of the bands corresponded to circular RNAs made from exon12 back splicing, either to exon 10 or 7 (Figure 1C). The exon sequences present in the linear MAPT mRNA were completely present in the circular RNAs, indicating usage of the linear splice sites. Both circRNAs were divisible by 3 (288, and 681 nt, respectively) and circ12→7 contained an in frame AUG start codon. No AUG start codon was present in circ12→10 (Figure 1D, E). The other bands contained non-canonical splice sites and thus their mechanism of generation is unclear and they could be PCR artifacts.
3.2. A minigene spanning exons 9 to 12 generates circ12→10
We concentrated on circ12→10, as it was the most abundant RNA. To determine the sequences necessary for its formation, we created a minigene consisting of exons 9, 10, 11 and 12, each flanked by 1–2 kb of intronic sequence (Figure 2A). This minigene was transfected into HEK293 cells and circular RNAs were detected using PCR exon junction primers selective for the 12→10 backsplice, consisting of 10→12 reverse primer and a forward primer in exon 10 (Figure 2B). In addition, we amplified linear RNA, using primers in exons 9 and 11 (Figure 1A). The minigene contained the repetitive elements surrounding exon 12 and an intronic repetitive element upstream of exon 9. After minigene transfection, we could detect both the exon 9 to 11 linear RNA containing the alternative exon 10, as well as the circ12→10 backsplice RNA (Figure 2C). Next we tested the expression of circRNAs using a different method. We choose RNase protection analysis that has a linear readout, but is less sensitive than RT-PCR. Using a uniformly radioactively labeled probe exhibiting sequence complementarity towards the exon12→10 backsplice junction (Figure 2B, D), we could detect a faint signal in RNA from transfected HEK293 cells, which was enriched by RNAse R treatment (Figure 2E). In addition, the probe detected parts of exon 10 and 12, which derived from linear RNA indicating that RNAse R treatment does not remove all linear RNAs. A signal of the same length could also be detected from 50 μg of total human cortex RNA. The circular RNA signal was less than 1% of the linear RNA signal in transfected cells, indicating that despite our ability to detect circ12→10 using RT-PCR, this RNA is very weakly expressed (Figure 2E). Since our minigene can produce circular RNA, we conclude that all cis-acting sequence elements to generate the tau circ12→10 RNA are present in exons 9 to 12 and their immediate intronic vicinity.
Figure 2: A minigene containing exons 9–12 generates circ12→10.
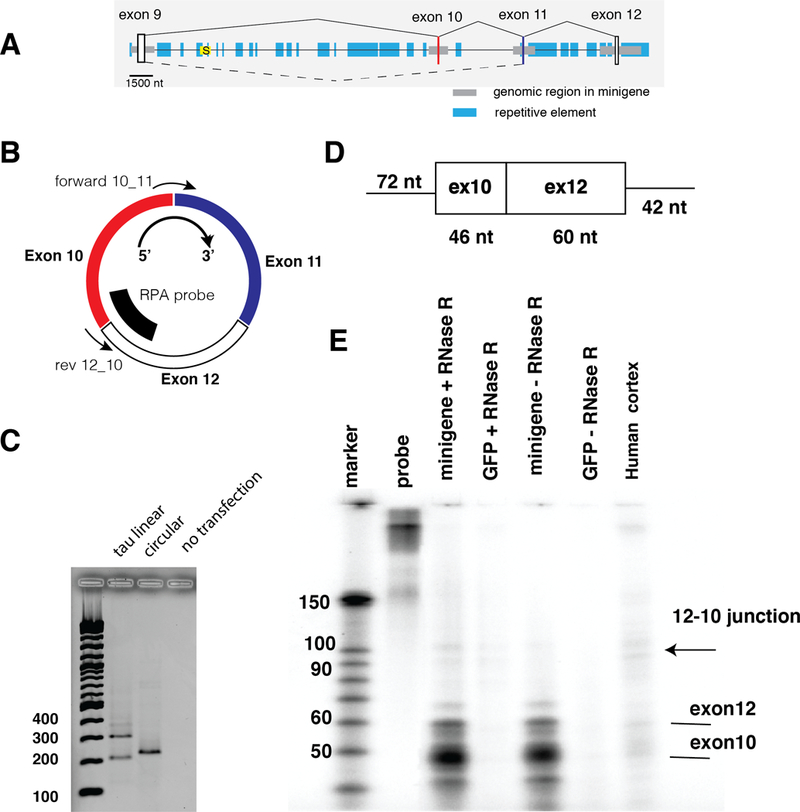
A. Structure of the minigene. Exons are indicated as vertical boxes, horizontal gray boxes indicate the genomic regions used for cloning the minigene. Blue boxes indicate repetitive elements defined by the UCSC genome browser repeat masker. The yellow box with an ‘S’ depicts the intronless Saitohin reading frame. The drawing is to scale, as indicated.B. Detection of circ12→10. The orientation of the circular RNA is clockwise 5’→3’. The location of the detection primer for10_11 and rev12_10 is indicated. The probe used for RPA is shown as a bold line.C. Detection of RNAs made from the exon 9–12 minigene. 1 μg of the minigene was transfected into HEK293 cells and after 24 hrs, RNAs were detected by RT-PCR. The negative control are untransfected HEK293 cells, using circRNA primers.D. RNase protection probe to detect circ12→10 RNA. The T7 antisense RNA is shown with the nucleotide lengths indicated., E. RNAse protection using RNA from transfected cells as well as human cortex. 50 μg total RNA was used, which was digested with RNaseR, as indicated.
3.3. Tau circ12→10 is regulated by clk2 and sensitive to mutations in exon 10
Tau circ12→10 contains exon 10, which is alternatively spliced in linear MAPT. Several exonic mutations causing FTLD/FTD have been identified that change exon 10 usage [3]. In addition, exon 10 is regulated by several trans-acting factors, most notable tra2-beta1/TRA2B[10], promoting its inclusion as well as the cdc2-like kinase CLK2 [7, 10], the SR-protein SRSF7/9G8, protein phosphatase 1 (PP1) and its nuclear inhibiter NIPP1 promoting exon 10 skipping [22–24].
To test a possible regulation, we transfected the tau 9–12 minigene with plasmids expressing EGFP, CLK2, the inactive CLK2 variant CLK2KR, tra2-beta, DYRK, 9G8, SRPK1, NIPP1 and PP1. The SR-protein kinase SRPK1 [ref], was tested as it phosphorylates tra2-beta1. To investigate the effect of an FTLD/FTD mutation on the formation of the circular RNA, we introduced the N279K mutation promoting exon 10 inclusion into the minigene [3].
First, we tested the effect of the trans-acting factors in MAPT linear RNA using PCR primers in exon 9 and 12. CLK2, 9G8, NIPP1 and PP1 caused exon 10 skipping, as expected from previous data using exon 9–11 reporter genes (Figure 3A). This experiment was repeated with a minigene harboring the N279K mutation. This exon 10 mutation (ATAA to AGAA) creates a stronger tra2-beta1 binding site, as tra2-beta1 binds to NGAA sequences [25]. The generation of this tra2-beta1 binding site promotes inclusion of exon 10, by promoting the formation of tra2-beta1 dependent splicing enhancer complexes. Importantly, the N279K mutation causes FTLD/FTD [3]. Testing the trans-acting factors on this minigene revealed that the effect on exon 10 skipping was strongly reduced for CLK2, 9G8, NIPP1 and PP1.
Figure 3: The abundance of exon 10 containing circ RNA is regulated by the kinase clk2.
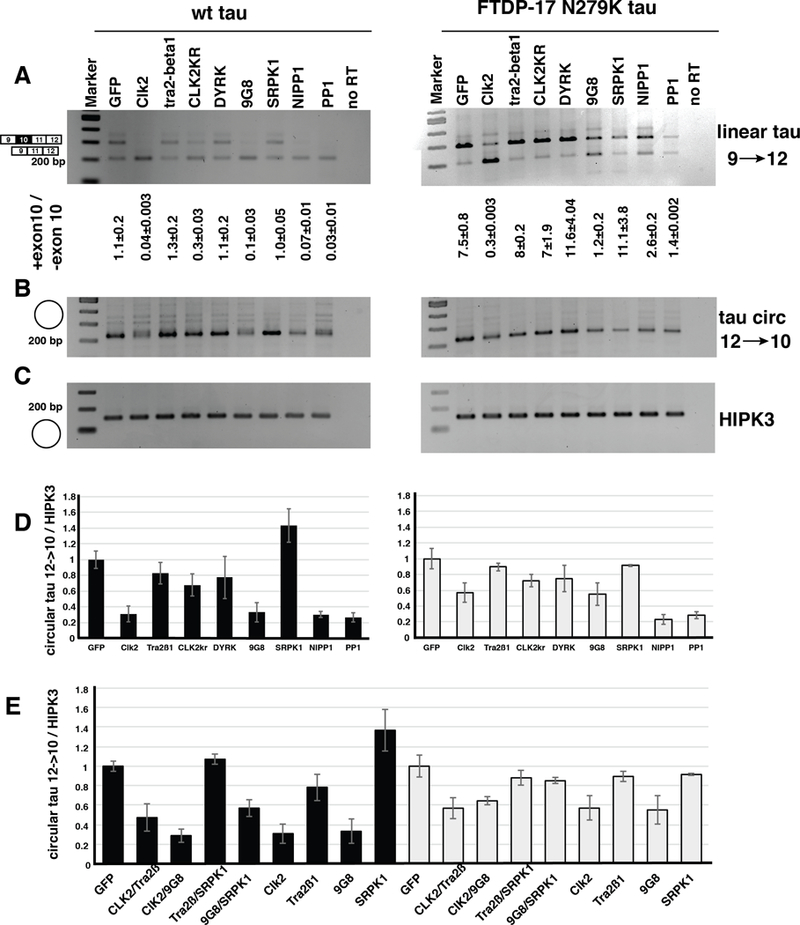
One μg of the tau exon 9–12 minigene was transfected into HEK293 cells together with 1 μg of the plasmids expressing the trans-acting factors indicated.A. Linear RNA was amplified using primers in exon 9 and 11, B. Circular RNA was amplified using primers rev12–10 and forward_exon 10. C. The circular HIPK3 RNA was amplified as a loading control, D. Quantification of the circtau to circHIPK3 ratio, E. Quantification of a cotransfection of the SR-proteins tra2-beta1 and 9G8 in the presence of their kinases CLK2 and SRPK1. 500 ng of each expressing vectors were cotransfected and three independent experiments were analyzed.
RNA from these experiments was next tested for the expression of the tau12→10 circRNA. CLK2 and 9G8, NIPP1 and PP1 strongly reduced the expression of this circRNA. A similar effect was observed when we used the N279K minigene. The trans-acting factors had no detectable effect on the HIPK3 circular RNA (Figure 3C), and we thus quantified our data by calculating the ratio between circtau12→10 and circHIPK3. The analysis of three independent experiments showed a statistically significant reduction of circtau12→10 caused by 9G8, NIPP1 and PP1 in both minigenes and by CLK2 in the wild-type minigene context (Figure 3D).
Finally, we investigated possible synergistic effects between the SR-proteins and their kinases and cotransfected CLK2 and SRPK1 together with tra2-beta1 and 9G8 expression clones. SRPK1 slightly increased the effect of tra2-beta1 and 9G8 on tau12→10 circRNA formation in the wild-type exon 10 context and had no detectable effect on the N279K mutations. In contrast, clk2 reduced the effect of tra2-beta1 in both wild-type and N279K background, whereas it had no effect on 9G8 (Figure 3E).
The data indicate that the abundance of circtau12→10 RNA can be regulated by the cell through trans-acting factor expression, whose actions is in turn regulated through their kinases. In addition, an FTLD/FTD mutation that promotes exon 10 inclusion also influences tau circ12→10 RNA formation.
3.4. Tau circ12→10 RNA is predominantly localized in the cytosol
We detected tau12→10 circRNA in the human neuroblastoma cells SH-SY5Y allowing to analyze the cellular localization of the endogenous RNA. We employed cell fractionation using different lysis of plasma and nuclear membranes followed by RT-PCR. The tau circ12→10 RNA was exclusively localized in the cytosol (Figure 4), similar to other circRNAs analyzed [12]. circHIPK3 RNA showed a similar cytosolic localization, whereas the C/D box snoRNA SNORD2 was exclusively nuclear.
Figure 4: circtau12→10 is cytosolic.
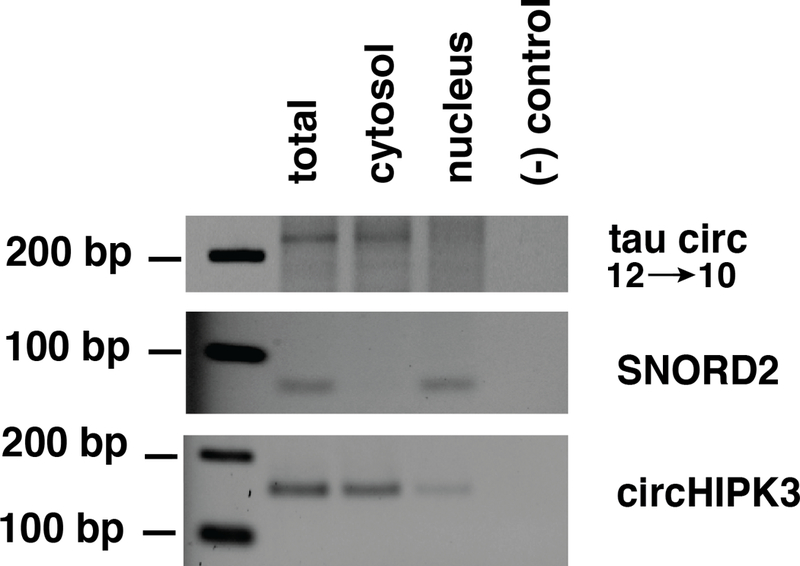
SH-SY5Y cells were separated into cytosol and nucleus and RNA isolated from each fraction. A. Detection of circ12→10, B. Detection of SNORD2, a C/D box snoRNA with nuclear localization, C. Detection of circHIPK3, a circular RNA known to be cytosolic.
3.5. No significant correlation between circ12→10 expression and Braak stages
Tau exon 10 and Clk2 splicing is deregulated in sporadic Alzheimer’s disease [7, 10]. We therefore tested brain samples from 15 subjects for a correlation between tau circ10→12 circRNA and Braak stages (Supplemental Data 1, Figure 5A-C). RNA from the gray matter of superior and middle temporal gyri was used. The amount of tau circ12→10 was normalized to circular HIPK3 or linear RPL3 mRNA. Although we found expression of tau circ 12→10 in all subjects, there was no statistical significant correlation with Braak stages, when the circ12→10 RNA was normalized to RPL3 or circHIPK3 RNA (Figure 5D). For the statistical analysis, only RNAs from samples with RIN numbers larger than seven were used. In addition, we tested these samples for expression of the kinase clk2 using primers spanning its alternative exon 4. Skipping of this exon generates a inactive kinase isoform. Again, there was no correlation between the amount of full-length clk2 and tau12→10 circRNA (Supplemental Figure 2).
Figure 5: The ratio between circtau 12→10 and circHIPK3 differs between individuals.
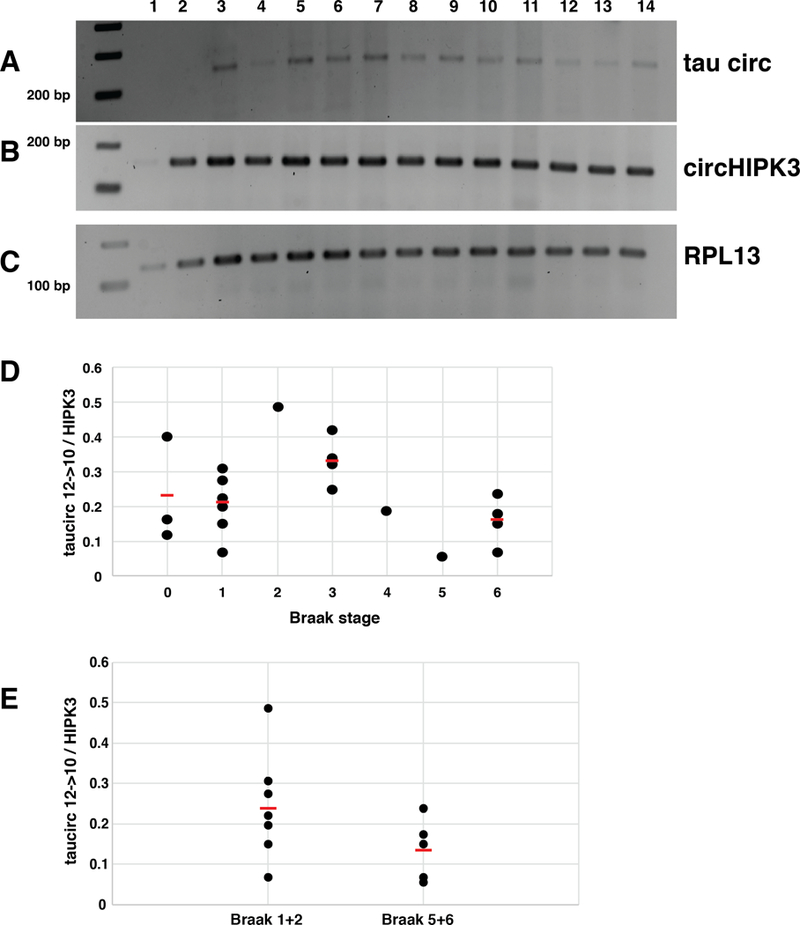
Temporal cortex from 15 patients with Braak stages 0 to 6 were analyzed by RT-PCR amplifying (A) cricTau12→10 and (B) the abundant circHIPK3 RNA, as well as (C) linear RPL3 mRNA. D shows the quantification of circtau 12→10 normalized to RPL3. There are no statistical significant changes between the Braak stages.
4. Discussion
4.1. MAPT as a contributor to human disease
The MAPT gene and its polypeptide products are acutely relevant to human diseases. A deregulation of MAPT is involved in Alzheimer’s Disease, Chronic Traumatic Encephalopathy, Frontotemporal Lobar Degeneration-tau, and many other ‘tauopathies’ that are not only common and clinically devastating but also are slated to rise appreciably in coming decades as western populations age [26].
4.2. Circular RNAs from the MAPT locus could be species specific
Here we present evidence that the human MAPT locus generates circular RNAs through backsplicing from exon 12 to either exon 10 or 7. Our data show that the regulation of MAPT gene expression and its molecular biology are still incompletely understood, despite numerous studies focusing on alternative splicing of MAPT [27].
We concentrated on circ12→10, as it is the most predominant circular RNA. Currently, human circular tau RNAs have not been reported in database. For mouse, a backsplice from exon six to four has been identified [28, 29]. We could not amplify the human orthologue for this mouse circ6→4 RNA, nor could we identify mouse orthologues for our described 12→10 and 12→7 circRNAs. It is thus possible that MAPT generates species-specific circRNAs, similar to exon 10 usage that is alternatively spliced in adult humans, whereas it is constitutively used in adult mouse [27].
CircRNA formation through backsplicing is facilitated by repetitive elements that form regions of complementarity in the pre-mRNA, allowing to position exons for backsplicing [12]. Human MAPT contains at least 83 Alu elements, 56 on the sense strand and 27 on the antisense strand [30]. Since Alu elements are primate-specific, they could cause a difference in circular RNA formation between mouse and humans.
We generated a minigene consisting of exons 9 to 12 with about 2 kb flanking intron each. We found that this construct forms circtau12→10 RNA when transfected into HEK293 cells, indicating that these regions are sufficient to generate the circular RNA. Importantly, in this construct, exon 10 is not surrounded by repetitive elements, suggesting that far-distance interaction, for example between repeats near exon 12 and 9 present in our minigene could form the basis for circRNA formation.
4.3. Possible functions of tau circRNAs
To gain insight into the possible function of tau circ12→10, we first determined its cellular localization in human SH-SY5Y neuroblastoma cells. Similar to other circular RNAs [12], tau circ 12→10 is almost exclusively cytosolic. Inspection of the sequence of tau circ 12→10 and circtau12→7 showed that both RNAs contain a number of nucleotides divisible by three. Circtau12→10 contains a reading frame without a stop or start codon. This reading frame is identical to a portion of the tau protein containing the microtubule binding site encoded by exon 10. Despite the absence of an AUG codon, it is conceptually possible that translation could occur, for example one of the two in frame AUA triplets could be edited by ADAR1 or 2 to AUI, where the inosine (I) is read as a G by the initiator tRNA, leading to protein synthesis. Further, repeat-associated non-ATG (RAN) translation [31] from CAGs that are also present in tau circ 12→10 are possible and finally a translation of the RNA could theoretically occur in mitochondria, that recognize AUA start codons [32]. Initiation at any of these non-AUG start codons could generate a tau protein fragment containing a microtubule binding site. Since the circtau12→10 RNA is divisible by three, it could be translated through a rolling circle mechanism that has been shown for model circular RNAs [33, 34]. A similar translational mechanism is possible for Circtau12→7 that contains an in frame start codon, but no stop codon. However, both circular Tau transcripts are very weakly expressed. Thus, their translation will be a rare event. If translation occurs, the expected product would be a high molecular weight tau multimer containing several microtubule binding sites.
4.4. Relation to disease
The alternative splicing regulation of tau exon 10 has been extensively studied, due to exon 10’s involvement in human disease. Mutations that interfere with the inclusion ratio of tau exon 10 cause frontotemporal dementia [3] and exon 10 usage as well as CLK2 splicing isoforms are changed in Alzheimer’s disease [7]. We thus tested the expression of circtau12→10 in AD brains of various Braak stages, using gray matter of the superior and middle temporal gyrus (SMTG), but did not find a statistically significant correlation between circtau12→10 expression, normalized to circHIPK3 or RPL3. In a minigene, we re-created one FTDP-17 mutation that strengthens exon 10 usage and found that it generates an amount of taucirc12→10 RNA similar to the wild-type in transfection assays, indication that mutations causing frontotemporal dementia can also form circular tau RNAs.
In summary, our data show that the MAPT locus can generate circRNAs that could play a role in neurodegenerative diseases, which warrants further investigation.
Supplementary Material
Acknowledgments
This work was supported by the NIH, 1R21NS098186–01A1
References
- [1].Wang Y, Mandelkow E, Tau in physiology and pathology, Nat Rev Neurosci, 17 (2016) 5–21. [DOI] [PubMed] [Google Scholar]
- [2].Lee VM, Goedert M, Trojanowski JQ, Neurodegenerative tauopathies, Annu Rev Neurosci, 24 (2001) 1121–1159. [DOI] [PubMed] [Google Scholar]
- [3].van Swieten J, Spillantini MG, Hereditary frontotemporal dementia caused by Tau gene mutations, Brain Pathol, 17 (2007) 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L, Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model, Science, 316 (2007) 750–754. [DOI] [PubMed] [Google Scholar]
- [5].Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, Diamond MI, Proteopathic tau seeding predicts tauopathy in vivo, Proc Natl Acad Sci U S A, 111 (2014) E4376–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goedert M, Jakes R, Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization, EMBO J, 9 (1990) 4225–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Moller HJ, Andreadis A, Henkel K, Stamm S, The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer’s disease, J. Neurochem, 96 (2006) 635–644. [DOI] [PubMed] [Google Scholar]
- [8].Conrad C, Zhu J, Schoenfeld D, Fang Z, Ingelsson M, Stamm S, Church G, Hyman BT, Single molecule profiling of tau gene expression in Alzheimer’s disease, J Neurochem, 103 (2007) 1228–1236. [DOI] [PubMed] [Google Scholar]
- [9].Suzuki H, Tsukahara T, A view of pre-mRNA splicing from RNase R resistant RNAs, Int J Mol Sci, 15 (2014) 9331–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hartmann AM, Rujescu D, Giannakouros T, Nikolakaki E, Goedert M, Mandelkow EM, Gao QS, Andreadis A, Stamm S, Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors, Mol Cell Neurosci, 18 (2001) 80–90. [DOI] [PubMed] [Google Scholar]
- [11].Chen LL, The biogenesis and emerging roles of circular RNAs, Nat Rev Mol Cell Biol, 17 (2016) 205–211. [DOI] [PubMed] [Google Scholar]
- [12].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, Circular RNAs are abundant, conserved, and associated with ALU repeats, Rna, 19 (2013) 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, Natural RNA circles function as efficient microRNA sponges, Nature, 495 (2013) 384–388. [DOI] [PubMed] [Google Scholar]
- [14].Deininger P, Alu elements: know the SINEs, Genome Biol, 12 (2011) 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G, Intronic Alus influence alternative splicing, PLoS Genet, 4 (2008) e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y, Wang Z, Efficient backsplicing produces translatable circular mRNAs, RNA, 21 (2015) 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G, Exon-intron circular RNAs regulate transcription in the nucleus, Nat Struct Mol Biol, 22 (2015) 256–264. [DOI] [PubMed] [Google Scholar]
- [18].Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS, Abner EL, Markesbery WR, Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity, J Neuropathol Exp Neurol, 66 (2007) 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stoss O, Stoilov P, Hartmann AM, Nayler O, Stamm S, The in vivo minigene approach to analyze tissue-specific splicing, Brain Research Protocols, 4 (1999) 383–394. [DOI] [PubMed] [Google Scholar]
- [20].Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S, Dual function of C/D box snoRNAs in rRNA modification and alternative pre-mRNA splicing Proc Natl Acad Sci U S A, 113 (2016) E1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen M, Eyras E, Wu J, Khanna A, Josiah S, Rederstorff M, Zhang MQ, Stamm S, Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression, Nucleic acids research, 39 (2011) 9720–9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ding S, Shi J, Qian W, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F, Regulation of alternative splicing of tau exon 10 by 9G8 and Dyrk1A, Neurobiol Aging, 33 (2012) 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao L, Wang J, Wang Y, Andreadis A, SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations, Mol Cell Neurosci, 34 (2007) 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S, Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing, Hum Mol Genet, (2008) 52–70. [DOI] [PubMed] [Google Scholar]
- [25].Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH, Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1, Nature structural & molecular biology, 18 (2011) 443–450. [DOI] [PubMed] [Google Scholar]
- [26].Iqbal K, Liu F, Gong CX, Tau and neurodegenerative disease: the story so far, Nat Rev Neurol, 12 (2016) 15–27. [DOI] [PubMed] [Google Scholar]
- [27].Andreadis A, Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases, Biochem. Biophys. Acta, 1739 (2005) 91–103. [DOI] [PubMed] [Google Scholar]
- [28].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N, Circular RNAs are a large class of animal RNAs with regulatory potency, Nature, 495 (2013) 333–338. [DOI] [PubMed] [Google Scholar]
- [29].Glazar P, Papavasileiou P, Rajewsky N, circBase: a database for circular RNAs, RNA, 20 (2014) 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Caillet-Boudin ML, Buee L, Sergeant N, Lefebvre B, Regulation of human MAPT gene expression, Mol Neurodegener, 10 (2015) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cleary JD, Ranum LP, Repeat-associated non-ATG (RAN) translation in neurological disease, Hum Mol Genet, 22 (2013) R45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Suzuki T, Nagao A, Suzuki T, Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases, Annu Rev Genet, 45 (2011) 299–329. [DOI] [PubMed] [Google Scholar]
- [33].Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, Sako Y, Ito Y, Abe H, Rolling circle amplification in a prokaryotic translation system using small circular RNA, Angew Chem Int Ed Engl, 52 (2013) 7004–7008. [DOI] [PubMed] [Google Scholar]
- [34].Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H, Rolling Circle Translation of Circular RNA in Living Human Cells, Sci Rep, 5 (2015) 16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


