Abstract
In the mammalian taste system, the taste receptor type 2 (T2R) family mediates bitter taste, and the taste receptor type 1 (T1R) family mediates sweet and umami tastes (the heterodimer of T1R2/T1R3 forms the sweet taste receptor, and the heterodimer of T1R1/T1R3 forms the umami taste receptor). In the chicken genome, bitter (T2R1, T2R2, and T2R7) and umami (T1R1 and T1R3) taste receptor genes have been found. However, the localization of these taste receptors in the taste buds of chickens has not been elucidated. In the present study, we demonstrated that the bitter taste receptor T2R7 and the umami taste receptor subunit T1R1 were expressed specifically in the taste buds of chickens labeled by Vimentin, a molecular marker for chicken taste buds. We analyzed the distributions of T2R7 and T1R1 on the oral epithelial sheets of chickens and among 3 different oral tissues of chickens: the palate, the base of the oral cavity, and the posterior tongue. We found that the distribution patterns and numbers were similar between taste bud clusters expressing these receptors and those expressing Vimentin. These results indicated broad distributions of T2R7 and T1R1 in the gustatory tissues of the chicken oral cavity. In addition, 3D-reconstructed images clearly revealed that high levels of T2R7 and T1R1 were expressed in Vimentin-negative taste bud cells. Taken together, the present results indicated the presence of bitter and umami sensing systems in the taste buds of chickens, and broad distribution of T2R7 and T1R1 in the chicken oral cavity.
Keywords: taste receptor, bitter, umami, chickens, taste buds
1. Introduction
Among the 5 basic tastes, bitter, sweet, and umami are sensed by the G-protein-coupled receptors (GPCRs) [1]. Bitter taste is sensed by the taste receptor type 2 (T2R) family, and sweet and umami tastes are sensed by the heterodimer of the taste receptor type 1 (T1R) family [2,3]. T1Rs (T1R1, T1R2, and T1R3) form the heterodimer of T1R2/T1R3 for sensing sweet taste and the heterodimer of T1R1/T1R3 for sensing umami taste [3]. In the chicken genome, only 3 T2R genes (T2R1, T2R2, and T2R7) have been found, in contrast to the many T2R genes in humans (25) and mice (35) [4]. The T1R genes for the umami taste receptor (T1R1 and T1R3) have also been detected in the chicken genome, but the T1R2 gene for the sweet taste receptor is missing [4].
Recently, the mRNAs of bitter (T2R1, T2R2, and T2R7) and umami (T1R1 and T1R3) taste receptors were detected in the oral tissues of chickens [5,6]. Cell-based assays have revealed that the chicken T2Rs and T1R1/T1R3 respond to various tastants, and chicken T2Rs are blocked by an antagonist, 6-methoxyflavanone, using HEK293T cells transiently expressing these receptors [7–9]. In addition, taste bud cells from the oral tissues of chickens respond to bitter and umami stimuli [10]. Consistently, behavioral analyses have shown that the agonists of chicken T2Rs and T1R1/T1R3, and 6-methoxyflavanone, affect the behavioral responses of chickens [9,11–13]. These findings suggest that chickens have functional bitter and umami sensing systems via T2Rs and T1Rs. However, the distributions of these receptor proteins in the taste buds of chickens have not been elucidated.
Previously, scanning electron microscopy (SEM) analysis and taste bud labeling with molecular markers in oral epithelial sheets of chickens revealed that the taste buds were distributed in 3 regions of the oral tissues: the palate, the base of the oral cavity, and the posterior tongue [14,15]. In this study, we demonstrated the localization of T2R7 and T1R1 in the taste buds of chickens by using Vimentin, the molecular marker for chicken taste buds, which labels all chicken taste buds, although not all taste bud cells [15,16]. We also found that the distribution patterns and numbers were similar between taste bud clusters expressing these receptors and those expressing Vimentin on the oral epithelial sheets of chickens, suggesting that T2R7 and T1R1 were broadly expressed in most of the taste bud clusters in chickens. T2R7 and T1R1 were also detected in taste buds labeled with Vimentin in all 3 regions of the chicken oral cavity. Interestingly, Vimentin and these receptors were hardly merged with each other at the taste bud cell level, and 3D image analyses clearly revealed that these receptors were expressed mostly in Vimentin-negative (Vimentin−) taste bud cells. Taken together, the present results suggested that chicken taste buds possess fundamental bitter and umami sensing mechanisms, and the mechanisms were broadly present in the chicken oral cavity.
2. Materials and methods
2.1. Animals and tissue collection
This use of animals has been approved by The University of Georgia Institutional Animal Care and Use Committee, and was in compliance with the National Institutes of Health Guidelines for the care and use of animals in research. This study also followed the Guide for Animal Experiments issued by Kyushu University, Japan’s Law Concerning the Human Care and Control of Animals (Law No. 105; October 1, 1973), and the Japanese Government Notification on the Feeding and Safekeeping of Animals (Notification No. 6; March 27, 1980).
Rhode Island Red strain 4-day-old chicks were used for immunohistochemistry (IHC) on oral epithelial sheets. The chicks were euthanized by decapitation, and the oral tissues in the palate and base of the oral cavity were dissected and processed for the peeling of the oral epithelial sheet as described below. Rhode Island Red strain, 3- to 4-week-old chicks were used for the IHC on frozen sections. The chicks were sacrificed with an overdose of pentobarbital sodium solution, and tissues were collected from the palate, the base of the oral cavity, and the posterior tongue. These tissues were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek Japan, Tokyo) and freshly frozen at −80°C. Frozen sections were cut at 12- to 14-μm thickness on a Leica CM1850 cryostat (Leica Instruments, Nussloch, Germany) and mounted onto MAS-coated glass slides (Matsunami Glass, Osaka, Japan). Sections were fixed with acetone for 10 min at 4°C and dried for 60 min at room temperature. After a brief rehydration by 0.1 M phosphate-buffered saline (PBS), the sections were processed for the IHC analyses as described below.
2.2. Oral epithelial sheet peeling
The oral epithelial sheets of chickens were prepared from the palate and base of the oral cavity using our previously reported methods [15] with some modifications. Briefly, an enzyme mixture of 1 mg/ml of collagenase A (Roche Diagnostics, Rotkreuz, Switzerland) and 2.5 mg/ml of dispase II (Roche Diagnostics) was injected into the sub-epithelial space of the palate and the base of the oral cavity, followed by incubation for 90 min for the palate and 60–90 min for the base of the oral cavity at 37°C. Following enzymatic tissue digestion, the tissues were immediately fixed with methanol for 20 min at −20°C, followed by a brief rinse in PBS. The soft tissue regions containing taste buds were dissected from the beaks, and the epithelial sheets of the palate and base of the oral cavity were peeled off from the underlying connective tissue. After thorough rinsing in PBS, the oral epithelial sheets were processed for the IHC.
2.3. Immunohistochemistry
Double fluorescence IHC on the oral epithelial sheets was performed to examine whether the taste receptors were co-localized with Vimentin in the taste bud clusters on the sheets. The primary antibodies were custom-made rabbit anti-chicken T2R7 (residues 171–184, GIFWKTNEEIRKHF) (1:1000) and rabbit anti-chicken T1R1 (residues 18–28, RPSPAEPRDGA) (1:500) antisera (Scrum, Tokyo), as well as mouse anti-Vimentin (1:500) (V9; Thermo Fischer Scientific). The secondary antibodies were Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG) (1:500) and Alexa Fluor 647 donkey anti-mouse IgG (1:500) (Jackson Immuno Research Laboratories, West Grove, PA). Nonspecific staining was blocked by 10% normal donkey serum (NDS) in PBS containing 0.3% Triton-X 100 (PBS-X) overnight at 4°C. The sheets were incubated with primary antibodies in 1% NDS in PBS-X for 72–120 hr at 4°C. The sheets were rinsed with PBS (30 min) 3 times. The sheets were then incubated with the secondary antibodies in 1% NDS in PBS-X 24–48 hr at 4°C. After rinsing with PBS (30 min) 3 times, the sheets were photomicrographed using a SZX2-ILLT Olympus stereomicroscope with CellSens software (Olympus, Life Sciences, Tokyo), or 2-photon microscopy, as described below.
Double fluorescence IHC on frozen sections was performed to validate the expression of the taste receptors with Vimentin in the chicken taste bud cells. The primary antibodies were custom-made rabbit anti-chicken T2R7 (1:100–500) and rabbit anti-chicken T1R1 (1:100) antisera (Scrum), as described above, and mouse anti-Vimentin (1:100) (Abcam 28028, Vim3B4; Abcam, Cambridge, MA). The secondary antibodies were Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) (1:500) and Alexa Fluor 594 goat anti-mouse IgG (1:500) (Thermo Fischer Scientific). In brief, nonspecific staining was blocked by 1.5% normal goat serum (NGS) (Vector Laboratories, Burlingame, CA) in PBS-X for 30 min at room temperature. The sections were incubated with the primary antibodies in 1.5% NGS in PBS-X overnight at 4°C. The sections were rinsed with PBS (5 min) 3 times. The sections were then incubated with the secondary antibodies in PBS for 40 min at room temperature. Following rinses with PBS (5 min × 3), the sections were mounted using VECTASHIELD with DAPI (Vector Laboratories). A confocal laser-scanning microscope system (A1R; Nikon Instech, Tokyo) with NIS-Elements AR 3.2 imaging software was used for the observation.
2.4. Antibody specificity
The specificities of the primary antibodies for T2R7 and T1R1, newly generated in this study, were checked by replacing the primary antibodies with 1.5% NGS in PBS-X. When verifying the specificities of T2R7 and T1R1 antibodies in the taste buds, we always used Vimentin antibody to identify the taste buds. We also performed the pre-adsorption tests for the T2R7 and T1R1 antibodies. These diluted antibodies (1:100) were pre-incubated with 1.2 mg/ml of excess peptides (T2R7: GIFWKTNEEIRKHF; T1R1: RPSPAEPRDGA) (Scrum) in PBS overnight at room temperature, and we performed IHC on frozen sections using these pre-adsorbed antibodies with added 1.5% NGS and 0.3% Triton X-100. As a result, the immunoreactivities of T2R7 and T1R1 were largely diminished by the replacement or pre-adsorption (data not shown).
2.5. Quantification and statistical analysis
The taste bud clusters in the palatine papilla region (ppr) of the palate were quantified. The quantification was carried out manually using photomicrographs obtained from an Olympus stereomicroscope; the same investigator performed each quantification to ensure consistency among groups. The quantification data were represented as means ± standard error (SE) (n = 3–5). A paired Student t-test was performed to test significant differences between the number of bud clusters expressing Vimentin and those expressing T2R7 or T1R1. A P value < 0.05 was considered significant.
2.6. 2-Photon microscopy and 3D image reconstruction of chicken taste buds
Oral epithelial sheets from the base of the oral cavity, stained by Vimentin, and T2R7 or T1R1 antibodies were used for 3D imaging. The 3D images were taken with a home-built 2-photon microscope, based on our previous reports [15]. Briefly, a ti-sapphire laser (Chameleon II; Coherent, Santa Clara, CA) was used at 850 nm to generate 130 fs pulses at 80 MHz. The beam power was modulated using a Pockels cell (Conoptics, Danbury, CT) and the beam was scanned over the sample by a resonant-galvanometer (fast axis – slow axis) scanner (RESSCAN-GEN; Sutter Instrument, Novato, CA). A 60x Olympus (LUMFLN60x) water immersion objective with NA of 1.1 was used for imaging. Z-scanning was performed using an X-Y-Z stage from Sutter Instrument (MPC-200). The signal emitted from the sample was separated into 2 channels: 697/58 nm and 509/22 nm filters were used to collect signals from Alexa Fluor 647 and Alexa Fluor 488, respectively. Photon multiplier tubes (PMT) from Hamamatsu were used to collect the signal. A transimpedence amplifier (Model 59–178; Edmund Optics, Barrington, NJ) was used for each channel to convert the current output of PMTs into an amplified voltage. National Instruments DAQ cards and FPGA modules were used to control and synchronize the system and to digitize the detected signal. The MATLAB-based open-source software ScanImage was employed to control the microscope [17,18].
3. Results
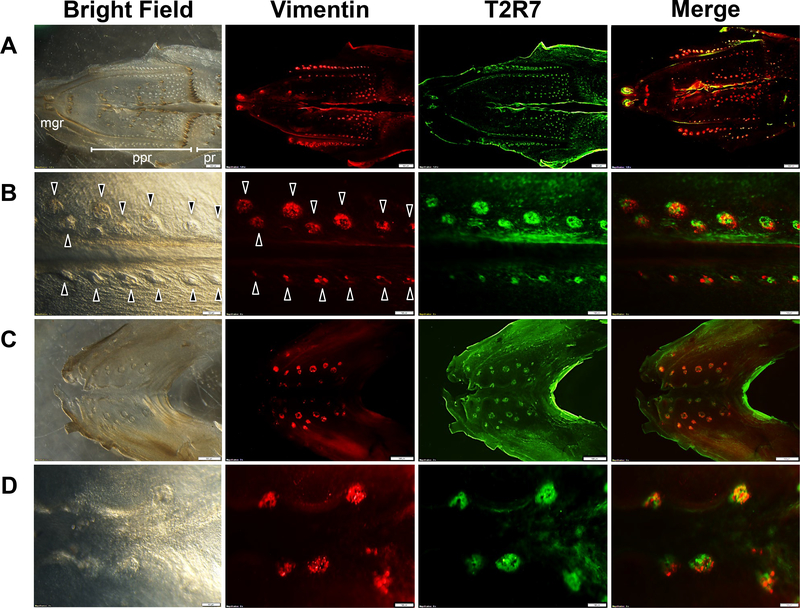
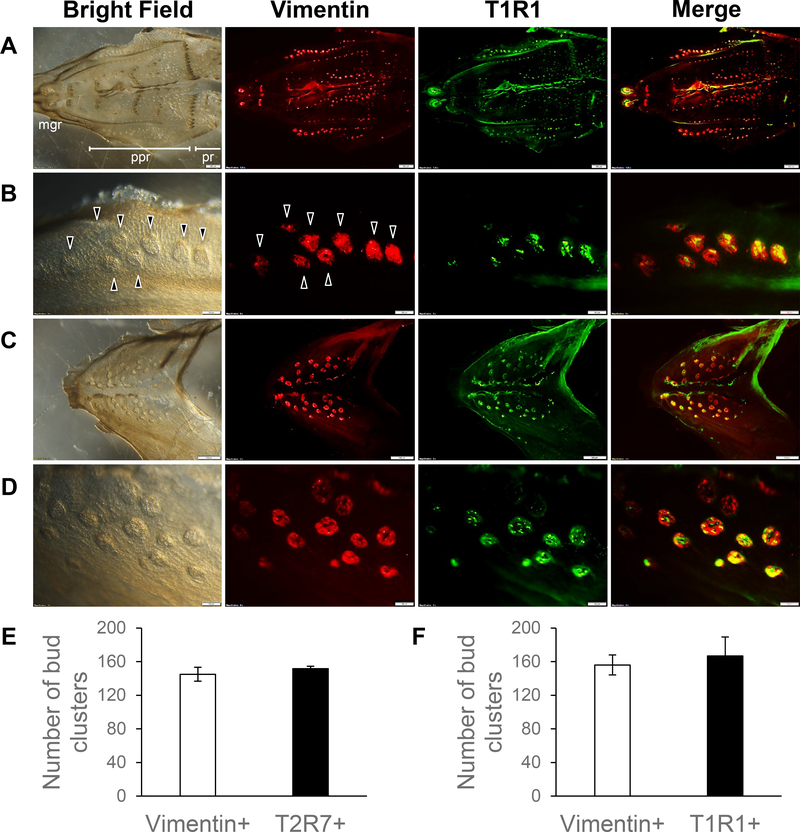
The specificities of the T2R7 and T1R1 antibodies, newly generated in this study, were validated by omitting the primary antibodies and the pre-adsorption tests (data not shown). To investigate whether T2R7 and T1R1 were expressed specifically in the taste buds of chickens, we used Vimentin, a molecular marker for the taste buds of chickens, according to a previous study showing that Vimentin labels all the taste buds of chickens [15,16]. Here, we characterized the distribution of T2R7 and T1R1 in the taste bud clusters on the oral epithelial sheets of chickens, and found that they were co-localized with Vimentin-expressing taste bud clusters (Fig. 1,2). T2R7 and T1R1 were found in most of the regions of oral epithelial sheets, including the maxillary gland region (mgr), palatine papilla region (ppr), and posterior region (pr) of the palate (Fig. 1A,2A), and at the base of the oral cavity (Fig. 1C,2C), where the taste bud clusters expressing Vimentin were located in a previous report [16]. The taste bud clusters expressing these receptors or Vimentin on the oral epithelial sheets were counted (representative taste bud clusters, identified by Vimentin expression or by tissue morphology under bright-field optics, are shown by arrowheads in Fig. 1B,2B). We found that the number of Vimentin-expressing taste bud clusters in ppr was not different from the number of those expressing T2R7 or the number of those expressing T1R1 (Fig. 2E,F).
Fig. 1.
An oral epithelial sheet of the palate shown at low magnification to illustrate the 3 different regions, i.e., the maxillary gland region (mgr), palatine papilla region (ppr), and posterior region (pr) (A), and at high magnification (B). An oral epithelial sheet from the base of the oral cavity is also shown at low (C) and (D) high magnification. The bright-field image, distribution of Vimentin (red) and T2R7 (green) immunosignals, and merged images of oral epithelial sheets of chickens are shown. Merged images show the overlay of Vimentin (red) and T2R7 (green). Arrowheads shown in B indicate representative taste bud clusters characterized by the Vimentin signals or by the morphology from the bright-field image. Scale bars: 500 μm in (A,C) and 100 μm in (B,D).
Fig. 2.
Oral epithelial sheets from the palate at low (A) and high (B) magnification, and from the base of the oral cavity at low (C) and high magnification (D). The bright-field image, distribution of Vimentin (red) and T1R1 (green) immunosignals, and merged images of oral epithelial sheets of chickens are shown. The merged image shows the overlay of Vimentin (red) and T1R1 (green). Arrowheads shown in B indicate representative taste bud clusters characterized by the Vimentin signal or by the morphology from the bright-field image. Scale bars: 500 μm in (A,C) and 100 μm in (B,D). Quantitative analyses of the numbers of Vimentin-expressing (Vimentin+) bud clusters and T2R7-expressing (T2R7+) bud clusters in the ppr (E) or the numbers of Vimentin+ bud clusters and T1R1-expressing (T1R1+) bud clusters in the ppr (F) are represented as means ± standard error (SE) (n = 3–5). There were no significant differences between the numbers of Vimentin+ bud clusters and T2R7+ bud clusters (P > 0.05) (E), or between the numbers of Vimentin+ bud clusters and T1R1+ bud clusters (P > 0.05) by paired Student t-test (F).
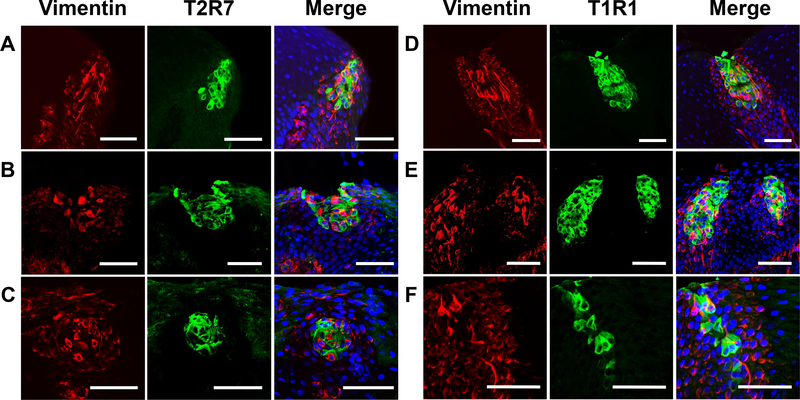
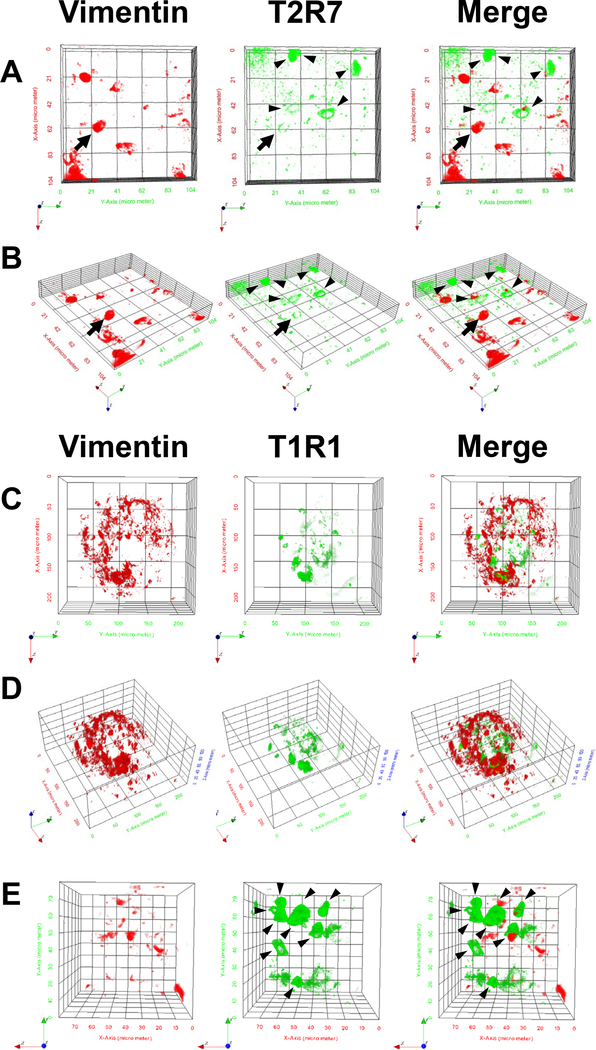
Next, we determined that T2R7 and T1R1 were distributed among all 3 regions of the oral tissues, the palate (Fig. 3A,D), the base of the oral cavity (Fig. 3B,E), and the posterior tongue (Fig. 3C,F), where the taste buds were distributed in a previous report [14]. T2R7 and T1R1 were expressed in the taste bud region labeled by Vimentin (Fig. 3A-F), suggesting that these receptors were expressed specifically in the taste buds of chickens. However, because Vimentin labels a subset of taste bud cells [16], the signals of these receptors and Vimentin were hardly merged with each other at the taste bud cell level (Fig. 3A-F). In addition, 3D-reconstructed images taken under a 2-photon microscope clearly showed that T2R7 and T1R1 were expressed largely in Vimentin− taste bud cells (Fig. 4A,B,E, arrowheads), although we also observed a single taste receptor cell expressing Vimentin at the taste bud cell level (Fig. 4A,B, arrow).
Fig. 3.
Distributions of Vimentin (red) and T2R7 (green) (A-C) or T1R1 (green) (D-F) immunosignals in the oral tissues of chickens in the palate (A,D), at the base of the oral cavity (B,E), and in the posterior tongue (C,F). Merged images show the overlay of Vimentin (red), T2R7 (green), or T1R1 (green), and DAPI (blue) to stain nuclei. Scale bars: 50 μm in all images (A-F).
Fig. 4.
3D-reconstructed images of chicken taste buds stained by Vimentin (red) and T2R7 (green) (A,B) or T1R1 (green) (C-E) taken under a 2-photon microscope are shown at high magnification to illustrate individual taste bud cells (A,B,E) and at low magnification (C,D). Panel (B) shows an inclined picture of (A) and panel (D) shows an inclined image of (C). Arrowheads show representative single taste receptor cells (A,B,E), as distinct from Vimentin-expressing cells, and arrows show a cell expressing both Vimentin and T2R7 (A,B). Panel (E) shows zoomed in version of taste bud cells in adjacent locations to the taste bud shown in (C,D).
4. Discussion
In the previous genome analyses, bitter taste receptor genes T2R1, T2R2, and T2R7 and umami taste receptor genes T1R1 and T1R3 were found in chickens [4]. However, although the mRNA expressions of these genes were detected in the oral tissues of chickens [5,6], the proteins and their localization in the taste buds of chickens have not been identified. The present study demonstrated that the bitter taste receptor T2R7 and the umami taste receptor subunit T1R1 were expressed in the taste buds of chickens, as characterized by Vimentin, a molecular marker for the taste buds of chickens [15,16]. Combined with the previous behavioral assays demonstrating that chickens respond to various tastants that activate chicken T2Rs and T1R1/T1R3 in cell-based assays [7–9,11–13], the present results strongly suggest that chicken taste buds have functional bitter and umami sensing systems.
A previous study revealed that the taste buds of chickens are located on the palate, the base of the oral cavity, and the posterior tongue, and this topographic distribution corresponds to the regions that food comes into contact with during its transport in the oral cavity [19]. In the present study, we found that T2R7 and T1R1 were expressed in all 3 regions of the oral tissues of chickens, i.e., the palate, the base of the oral cavity, and the posterior tongue. This result suggests that T2R7 and T1R1 can frequently interact with food throughout the regions of the oral cavity while the chicken is eating. In addition, in the present study we revealed that T2R7 and T1R1 were expressed in the taste bud clusters on the oral epithelial sheets immunoreacted with Vimentin. T2R7 and T1R1 were expressed in Vimentin-labeled taste buds in all the regions of the palate—i.e., mgr, ppr, and pr—and at the base of the oral cavity [15]. Further, the numbers of bud clusters expressing these receptors and the number of bud clusters expressing Vimentin were similar. Considering the previous reports that showed that Vimentin labeled all the taste buds [15,16], we can infer that T2R7 and T1R1 were expressed broadly in taste buds in the chicken oral cavity.
Previous reports demonstrated that Vimentin is expressed in all the taste buds but not in all taste bud cells [16]. In the present study, we observed that T2R7 and T1R1 were localized in Vimentin-labeled taste buds. However, there was little coincidence between the distribution of these receptors and that of Vimentin in the taste bud cells. In addition, 3D imaging analyses clearly revealed that the taste receptors were expressed mostly in Vimentin− taste bud cells. A previous study using RNA-Seq analyses of murine-cultured taste organoids revealed that the mRNA levels of T2Rs and T1Rs were upregulated at the late stage of growth [20], suggesting that T2Rs and T1Rs are mature taste cell markers. On the other hand, whereas Vimentin labels a population of chicken taste bud cells and human taste bud primordia and marginal cells of the bud [16,21], the function of Vimentin-expressing cells has not been clearly elucidated. Considering the present results, the taste bud cell populations, labeled by different markers, e.g., T2R7, T1R1, and Vimentin, may serve distinct functions in taste perception.
In conclusion, we found that the bitter taste receptor T2R7 and umami taste receptor subunit T1R1 were expressed in the taste buds of chickens, labeled by Vimentin, a molecular marker for the taste buds of chickens [15,16]. These receptors were observed in the taste bud clusters on the oral epithelial sheets of chickens and among all 3 regions of the oral tissues of chickens. We also found that the distribution patterns and numbers were similar between taste bud clusters expressing these receptors and those expressing Vimentin, suggesting that these receptors were broadly distributed in the taste buds in the oral cavity. In addition, 3D imaging analyses clearly revealed that taste receptors were expressed mostly in Vimentin− taste bud cells. Together with the findings from the previous cell-based assays [7–9,11] and behavioral assays [9,11–13], the present results suggest that the functional bitter and umami sensing mechanisms mediated by T2R7 and T1R1 are present in the taste buds of chickens, and these receptors could be a target for controlling the feeding behavior of chickens in poultry farming.
Highlights.
T2R7 and T1R1 were expressed specifically in chicken taste buds expressing Vimentin.
T2R7 and T1R1 were distributed in the taste bud clusters expressing Vimentin
3D images showed T2R7 and T1R1 were expressed in Vimentin-negative taste bud cells.
Acknowledgments
This study was supported by grants to F. Kawabata from JSPS KAKENHI (#26850207), the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (#26703), and the Sugiyama Sangyo-Kagaku General Incorporated Foundation; by a grant to H.X. Liu from the National Institutes of Health (NIDCD R01 DC012308); and by grants to Y. Yoshida from the JSPS Research Fellowship (#17J03180) and the JSPS Overseas Challenge Program for Young Researchers (#201780018).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Roper SD, Chaudhari N, Taste buds: cells, signals and synapses, Nat. Rev. Neurosci 18 (2017) 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Muller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJP, The receptors and coding logic for bitter taste, Nature 434 (2005) 225–229. [DOI] [PubMed] [Google Scholar]
- [3].Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJP, Zuker CS, The receptors for mammalian sweet and umami taste, Cell 115 (2003) 255–266. [DOI] [PubMed] [Google Scholar]
- [4].Shi P, Zhang J, Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes, Mol. Biol. Evol 23 (2006) 292–300. [DOI] [PubMed] [Google Scholar]
- [5].Cheled-Shoval SL, Druyan S, Uni Z, Bitter, sweet and umami taste receptors and downstream signaling effectors: Expression in embryonic and growing chicken gastrointestinal tract, Poult. Sci 94 (2015) 1928–1941. [DOI] [PubMed] [Google Scholar]
- [6].Yoshida Y, Kawabata Y, Kawabata F, Nishimura S, Tabata S, Expression of multiple umami taste receptors in oral and gastrointestinal tissues, and umami taste synergism in chickens, Biochem. Biophys. Res. Commun 466 (2015) 346–349. [DOI] [PubMed] [Google Scholar]
- [7].Behrens M, Korsching SI, Meyerhof W, Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes, Mol. Biol. Evol 31 (2014) 3216–3227. [DOI] [PubMed] [Google Scholar]
- [8].Baldwin MW, Toda Y, Nakagita T, O’Connell MJ, Klasing KC, Misaka T, Edwards SV, Liberles SD, Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor, Science 345 (2014) 929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dey B, Kawabata F, Kawabata Y, Yoshida Y, Nishimura S, Tabata S, Identification of functional bitter taste receptors and their antagonist in chickens, Biochem. Biophys. Res. Commun 482 (2017) 693–699. [DOI] [PubMed] [Google Scholar]
- [10].Kudo K, Kawabata F, Nomura T, Aridome A, Nishimura S, Tabata S, Isolation of chicken taste buds for real-time Ca2+ imaging, Anim. Sci. J 85 (2014) 904–909. [DOI] [PubMed] [Google Scholar]
- [11].Hirose N, Kawabata Y, Kawabata F, Nishimura S, Tabata S, Bitter taste receptor T2R1 activities were compatible with behavioral sensitivity to bitterness in chickens, Biochem. Biophys. Res. Commun 460 (2015) 464–468. [DOI] [PubMed] [Google Scholar]
- [12].Yoshida Y, Kawabata F, Kawabata Y, Nishimura S, Tabata S, Expression levels of taste-related genes in palate and tongue tip, and involvement of transient receptor potential subfamily M member 5 (TRPM5) in taste sense in chickens, Anim. Sci. J 89 (2018) 441–447. [DOI] [PubMed] [Google Scholar]
- [13].Yoshida Y, Kawabata F, Kawabata Y, Nishimura S, Tabata S, Short-term perception of and conditioned taste aversion to umami taste, and oral expression patterns of umami taste receptors in chickens, Physiol. Behav 191 (2018) 29–36. [DOI] [PubMed] [Google Scholar]
- [14].Kudo K, Nishimura S, Tabata S, Distribution of taste buds in layer-type chickens: Scanning electron microscopic observations, Anim. Sci. J 79 (2008) 680–685. [Google Scholar]
- [15].Rajapaksha P, Wang Z, Venkatesan N, Tehrani KF, Payne J, Swetenburg RL, Kawabata F, Tabata S, Mortensen LJ, Stice SL, Beckstead R, Liu HX, Labeling and analysis of chicken taste buds using molecular markers in oral epithelial sheets, Sci. Rep 6 (2016) 37247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Venkatesan N, Rajapaksha P, Payne J, Goodfellow F, Wang Z, Kawabata F, Tabata S, Stice S, Beckstead R, Liu HX, Distribution of alpha-Gustducin and Vimentin in premature and mature taste buds in chickens, Biochem. Biophys. Res. Commun 479 (2016) 305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pologruto T, Sabatani B, Svoboda K, ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2 (2003) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mortensen L, Alt C, Turcotte R, Masek M, Liu TM, Cote D, Xu C, Intini G, Lin C, Femtosecond laser bone ablation with a high repetition rate fiber laser source. Biomed. Opt. Express 6 (2015) 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ganchrow D, Ganchrow JR, Number and distribution of taste buds in the oral cavity of hatchling chicks, Physiol. Behav 34(1985) 889–894. [DOI] [PubMed] [Google Scholar]
- [20].Ren W, Aihara E, Lei W, Gheewala N, Uchiyama H, Margolskee RF, Iwatsuki K, Jiang P, Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation, Sci. Rep 7 (2017) 4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Witt M, Kasper M, Distribution of cytokeratin filaments and vimentin in developing human taste buds, Anat. Embryol 199 (1999) 291–299. [DOI] [PubMed] [Google Scholar]