Summary
RIG-I has a remarkable ability to specifically select viral 5’ppp dsRNAs for activation from a pool of cytosolic self-RNAs. The ATPase activity of RIG-I plays a role in RNA discrimination and activation, but the underlying mechanism was unclear. Using transient state kinetics, we have elucidated the ATPase-driven ‘kinetic proofreading’ mechanism of RIG-I activation and RNA discrimination, akin to DNA polymerases, ribosomes, and T-cell receptors. Even in the autoinhibited state of RIG-I, the C-terminal domain kinetically discriminates against self-RNAs by fast off-rates. ATP-binding facilitates dsRNA engagement, but interestingly makes RIG-I promiscuous, explaining the constitutive signaling by Singleton-Merton syndrome-linked mutants that bind ATP without hydrolysis. ATP hydrolysis dissociates self-RNAs faster than 5’ppp dsRNA, but more importantly, drives RIG-I oligomerization through translocation, that we show is regulated by helicase motif-IVa. RIG-I translocates directionally from dsRNA-end into stem-region and 5’ppp-end “throttles” translocation to provide a mechanism for threading and building a signaling-active oligomeric complex.
Introduction
The innate immune receptor RIG-I (Retinoic acid Inducible Gene I) recognizes viral RNAs in the cytoplasm and initiates a signaling pathway that creates an antiviral state with the production of interferons and inflammatory cytokines (Poeck et al., 2010; Sumpter et al., 2005; Yoneyama et al., 2004). A well-characterized RIG-I PAMP (Pathogen Associated Molecular Pattern) is blunt-ended dsRNA with 5’ triphosphate (5’ppp), a feature found in many viral RNA genomes and replication intermediates (Hornung et al., 2006; Rehwinkel et al., 2010; Schlee et al., 2009). RIG-I is also activated by dsRNAs with a 5’m7G cap (Cap-0); therefore, higher eukaryotes 2’-O-methylate the first nucleotide ribose near the cap (Cap-1) to evade RIG-I recognition (Devarkar et al., 2016; Schuberth-Wagner et al., 2015). Although RIG-I binds blunt-ended 5’ppp dsRNA with a greater affinity than dsRNAs with other end-modifications (Kohlway et al., 2013; Ramanathan et al., 2016), such as miRNAs carrying 3’ overhangs or dsRNA stems in tRNAs and rRNAs, binding affinities are not sufficiently different to explain the high selectivity in signaling for 5’ppp dsRNA. Therefore, mechanisms must exist to increase RNA discrimination. Indiscriminate RIG-I signaling from cellular RNAs creates a highly immunostimulatory and detrimental environment, as observed in atypical Singleton-Merten syndrome (SMS), where point mutations in RIG-I’s ATPase motifs render it constitutively active (Jang et al., 2015).
RIG-I has tandem N-terminal CARDs (Caspase Activation and Recruitment Domains), a central helicase domain (with Hel1, Hel2, and Hel2i subdomains), and a C-terminal RD (Repressor domain) (Figure 1A). Post-translational modifications, such as phosphorylation of CARDs (Nistal-Villan et al., 2010), regulate RIG-I activation. However, a key mechanism for limiting non-specific RNA binding is a structurally encoded autoinhibition mechanism (Jiang et al., 2011; Kowalinski et al., 2011; Lu et al., 2011; Luo et al., 2011; Wang et al., 2010). In the absence of RNA, the signaling CARDs are sequestered through extensive interactions between CARD2 and Hel2i (Kowalinski et al., 2011), which prevents CARDs from signaling and helicase domains from indiscriminately binding to dsRNA backbone. Accordingly, mutations in the CARD2:Hel2i interface decrease RNA selectivity (Ramanathan et al., 2016). 5’ppp dsRNA binding disrupts CARD2:Hel2i interactions, aiding in CARDs release, and ATP binding facilitates this process (Zheng et al., 2015). CARDs tetramerization is proposed to nucleate MAVS (Mitochondrial Antiviral Signaling protein) filament, which propagates the signal downstream (Jiang et al., 2012; Peisley et al., 2014; Peisley et al., 2013; Wu et al., 2014). However, the mechanism of RIG-I oligomerization on RNA leading to this proposed CARDs tetramerization is still unknown.
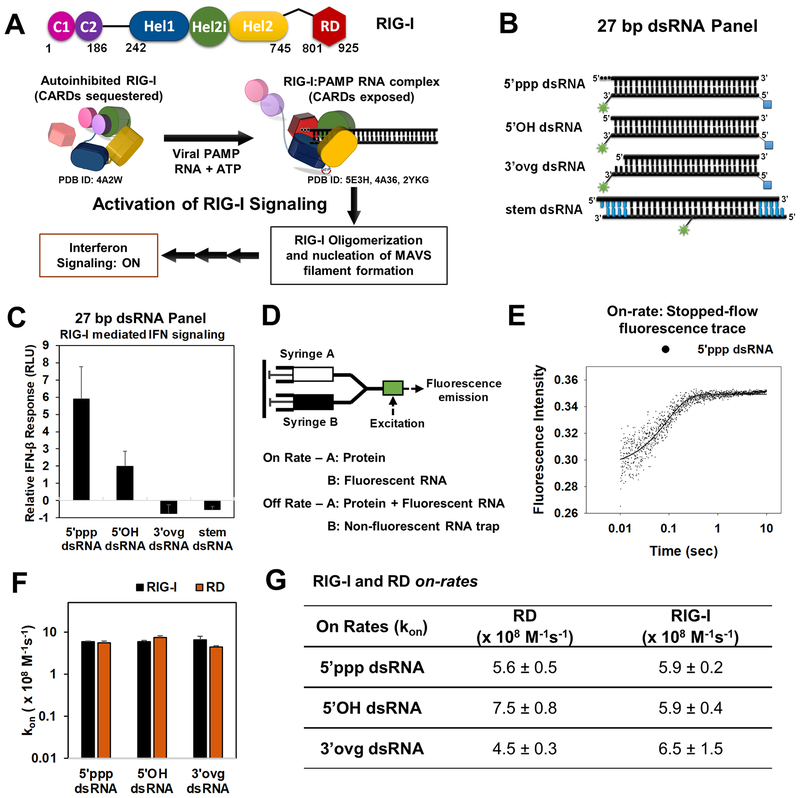
Figure 1. RIG-I activation pathway and RIG-I/RD on-rates on 5’ppp and non-PAMP dsRNAs.
(A) Schematic of a general activation mechanism of RIG-I. (B) Cartoon of the dsRNAs used in the study (green asterisk-DY547/Cy3; blue square-biotin; blue bars-DNA bases). (C) IFN-β reporter activation in HEK293T cells expressing RIG-I and transfected with the dsRNA panel from B. Error bars are SEM from quadruplicate sets. (D) Stopped-flow experimental set up for on-rate and off-rate measurements. (E) Representative stopped-flow time course shows an increase in fluorescence intensity upon binding of RIG-I (45 nM) to 5’ppp dsRNA (10 nM) at 25°C. (F, G) On-rates of RIG-I and RD. The associated errors from the linear fits are shown. See also Figure S1.
RIG-I belongs to the DEXD/H family of RNA helicases, and like all helicases, RIG-I has an ATPase activity. However, the role of ATPase activity in RIG-I activation remains controversial. Some ATPase-deficient mutants are inactive in RIG-I signaling whereas others are constitutively activated (Bamming and Horvath, 2009; Jang et al., 2015; Yoneyama and Fujita, 2007). Single molecule studies show ATPase-driven translocation of RIG-I on dsRNA without strand separation (Myong et al., 2009), but the role of such a translocation activity is not understood. Additionally, ATP binding and hydrolysis steps are proposed to proofread RNAs by dissociating RIG-I from non-PAMPs. One model suggested that ATP binding provides a proofreading function by preferentially dissociating RIG-I from non-PAMP RNAs (Rawling et al., 2015). Another model proposes that ATP hydrolysis offers this proofreading function (Lassig et al., 2015; Louber et al., 2015). The above models contradict each other and have not been quantitatively tested with parallel studies of PAMP and non-PAMP RNAs.
Here we employ a variety of quantitative biochemical tools, including transient state stopped-flow kinetics to measure the dynamic interactions of RIG-I with RNAs. Throughout these studies, we compare PAMP and non-PAMP dsRNAs and comprehensively investigate the roles of ATP binding and hydrolysis in RNA proofreading, RIG-I translocation, and oligomerization. Based on our combined results, we present a kinetic proofreading pathway used by RIG-I to selectively oligomerize and signal on PAMP RNAs and achieve discrimination beyond differences in binding energies of its ligands. The kinetic proofreading scheme presented here is akin to those governing DNA polymerases, ribosomes, and T-cell receptors, and powered by the intrinsic dynamics of RIG-I interactions with RNAs and an ATPase-driven translocation-throttling mechanism.
Results
To monitor the kinetics of RNA bound RIG-I, we used 27bp dsRNAs with various end-modifications mimicking PAMP or non-PAMP features, including a blunt-ended 5’ppp dsRNA, a blunt-ended 5’OH dsRNA, and a 3’ovg dsRNA carrying 5’OH and two nucleotide (nt) 3’ overhang (Figure 1B). RIG-I has a footprint of 8–10 bp (Jiang et al., 2011; Luo et al., 2011); thus, 27-bp dsRNAs are suitable for studying RIG-I translocation and oligomerization. The RNAs carried a fluorophore (DY547 or fluorescein) at the 3’-end of the RNA near the desired end-modification (5’ppp/5’OH/3’ovg). The presence of fluorophore has minimal effects on RNA binding (Figure S1A). A mimic of the RNA stem was prepared by flanking a 17 bp dsRNA with four base pairs of dsDNA and a 5’ overhang (Figure 1B). RIG-I has no affinity for dsDNA (Figure S1B); hence, this design prevents end-binding and only allows dsRNA stem-binding. A Cy3 fluorophore was placed in the middle of the 17 bp stem region to monitor RIG-I binding.
We tested these fluorophore-labeled 27-bp dsRNAs in cell-based RIG-I signaling assays. Transfection of 5’ppp dsRNA into human embryonic kidney 293T (HEK293T) cells expressing RIG-I showed robust activation of IFN-β promoter-driven luciferase reporter gene, confirming that 5’ppp dsRNA is a RIG-I PAMP (Figure 1C). The signaling response from 5’OH dsRNA was low, and signaling from 3’ovg and stem dsRNAs was negligible after subtracting background (without RNA transfection), indicating that the 5’OH dsRNA is a mild PAMP and the 3’ovg and stem dsRNAs are non-PAMPs. The Kd values of RNAs inversely correlated with their signaling activities (Figure S1C). Therefore, this set of dsRNAs are excellent mimics of RIG-I PAMP and non-PAMPs for detailed biophysical and biochemical analyses.
RIG-I binds RNA-ends at diffusion-limited on-rates
Stopped-flow kinetics were used to measure the dynamic interactions of RIG-I with RNAs. These experiments were conducted with wild-type (wt) RIG-I (1–925), RD (801–925), and Helicase-RD (232–925). To determine whether RIG-I selects 5’ppp dsRNA by fast association, slow dissociation, or both, we measured the on-rates and off-rates of PAMP and non-PAMP RNAs and determined the lifetimes of RIG-I on these RNAs. To learn how ATP binding and hydrolysis steps affect RNA selectivity, we measured lifetimes in the presence of a non-hydrolyzable ATP analog (ADP.BeF3) and ATP. Off-rates of RIG-I are sensitive to Mg2+ concentration, and slow in the absence of Mg2+ (Figure S1D–E); therefore, all measurements were carried out with a fixed concentration of Mg2+.
The on-rates were measured by mixing DY547-labeled dsRNA from one syringe of the stopped-flow apparatus with RD or RIG-I from another syringe (Figure 1D). A time-dependent increase in fluorescence intensity measured the RNA binding step in real time (Figure 1E). The on-rates were estimated from the slopes of the linear increase in RNA binding rates with protein concentrations (Figure S1F). Interestingly, the on-rates of RD and RIG-I were almost the same and close to the diffusion limit (5 − 7 × 108 M−1s−1) (Figure 1F–G) on all dsRNAs including 5’ppp, 5’OH, and 3’ovg. The almost identical on-rates of RD and RIG-I suggests that the RD module in RIG-I is free to sample dsRNA-ends, and the diffusion-limited on-rate for binding to all dsRNA-ends indicates that the initial binding event is rapid and indiscriminate.
RIG-I engages RNA in distinct RD-mode and Helicase-mode conformations
The off-rates were measured in a stopped-flow instrument by chasing DY547 dsRNA-protein complexes with an excess of trap RNA (non-fluorescent 12 bp 5’ppp hairpin RNA) (Figure 1D). The trap prevents rebinding of proteins to the fluorescent RNA and makes the reaction single-turnover. Control experiments showed that off-rates were independent of trap RNA concentration (Figure S2A) and type of fluorophore (Figure S2B). Unlike the on-rates that were the same for all dsRNAs, the off-rates of RD were highly sensitive to dsRNA-end modifications (Figure 2A, Table S1). The off-rate of 5’ppp dsRNA-end from RD was 0.15 s−1 whereas the off-rates of 5’OH and 3’ovg dsRNA-ends were much faster (21 s−1 and 120 s−1, respectively). Stem dsRNA did not show any interactions with RD (Figure S2C). Collectively, the on and off-rates demonstrate that RD kinetically discriminates against non-PAMP dsRNA-ends by faster dissociation.
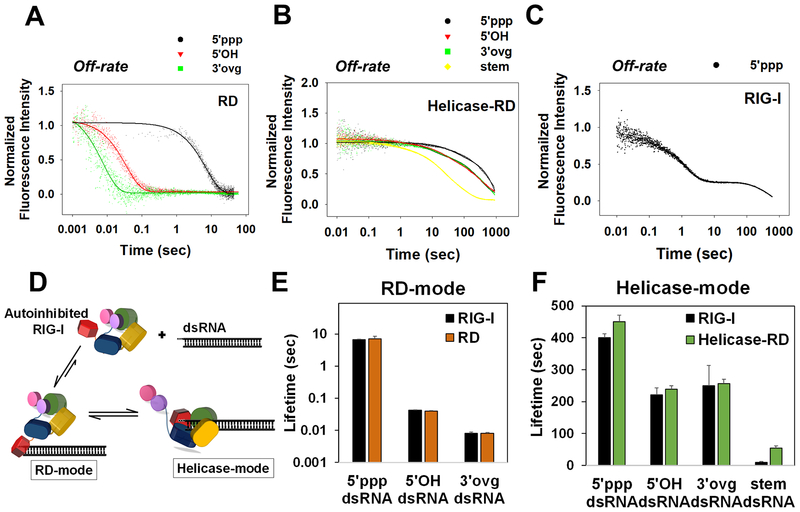
Figure 2. Off-rates of 5’ppp and non-PAMP RNAs from RD, Helicase-RD, and RIG-I.
(A-C) Stopped-flow kinetics of RNA dissociation. A preformed complex of RD (A), Helicase-RD (B), or RIG-I (C) with the specified fluorophore-tagged RNA was chased with a 10-fold excess of unlabeled 5’ppp ds12 hairpin RNA at 25°C. (D) RIG-I forms two distinct complexes with dsRNA: RD-mode and Helicase-mode conformations. (E-F) Lifetimes of RD-mode (E) and Helicase-mode (F) RIG-I complexes calculated from reciprocal of corresponding off-rates. Representative kinetic traces are an average of 4–6 traces. See also Figure S2 and Table S1.
Off-rates of RNAs from Helicase-RD (Figure 2B) were markedly slower than from RD. The off-rate of 5’ppp dsRNA from Helicase-RD was 0.002 s−1, which is 75-times slower than from RD. Interestingly, a significant proportion of 5’OH and 3’ovg dsRNAs formed long-lived complexes on Helicase-RD (Table S1). Furthermore, unlike RD, Helicase-RD bound to stem dsRNA (Figure 2B and S2C) and formed long-lived complexes. The non-discriminatory and slow off-rates of the CARD-less Helicase-RD is consistent with the model that the CARD2:Hel2i interactions prevent the helicase domains from non-specific interactions with the dsRNA backbone (Ramanathan et al., 2016).
Off-rates of dsRNAs from wt RIG-I were biphasic and showed RD-like (RD-mode) and Helicase-RD-like (Helicase-mode) kinetic phases (Figure 2C and S2D–F). RIG-I also bound to stem dsRNA and dissociated with multiple off-rates (Figure S2F, Table S1). On the 5’ppp dsRNA, the RD-mode represented 70% of the total population, and the 0.15 s−1 off-rate is the same as the off-rate of RD from 5’ppp dsRNA. The Helicase-mode represented 30% of the total population, and the 0.002 s−1 off-rate is identical to Helicase-RD off-rate from 5’ppp dsRNA. The 5’OH and 3’ovg dsRNAs showed the same pattern with RD-like and Helicase-RD-like off-rates (Figure S2D–E, Table S1).
From the above kinetic studies, we conclude that RIG-I binds to dsRNA-end in two distinct conformations - the RD-mode, where autoinhibited RIG-I engages dsRNA-end via RD, and the Helicase-mode, where helicase domains are also involved in binding the adjacent dsRNA backbone (Figure 2D). Although the RD-mode is established rapidly, it has a shorter lifetime than the Helicase-mode. The lifetime is calculated from the reciprocal of the off-rate, or if multiple phases were observed, from a weighted average of the off-rates. The lifetime of RD-mode is ~7 s on 5’ppp dsRNA and much shorter on 5’OH and 3’ovg dsRNAs (~40 and 8 ms, respectively) (Figure 2E). Lifetimes of Helicase-mode are much longer (250–400 s) on dsRNA-ends (5’ppp, 5’OH, and 3’ovg) than on stem dsRNA (~9 s) (Figure 2F). Thus, RD interactions with the dsRNA-end dramatically increase the kinetic stability of the stem region bound Helicase-mode complex.
ATP binding stabilizes, and ATP hydrolysis destabilizes RIG-I complexes
Next, we measured the off-rates of PAMP and non-PAMP RNAs in the presence of a non-hydrolyzable ATP analog (ADP.BeF3) and ATP. Stopped-flow experiments were carried out by preincubating RIG-I with fluorescent dsRNA in the presence of ADP.BeF3 or ATP and chasing the complex with an excess of non-fluorescent trap RNA. Experiments with ATP were carried out with an ATP regenerating system. RNA dissociation kinetics in the presence of nucleotides showed RD-mode and Helicase-mode kinetic phases (Figure 3A and S3A–C). The RD-mode lifetimes were unaffected by nucleotides (Figure 3B, Table S2), indicating that ATP binding to the helicase domains does not perturb RD interactions with the dsRNA-end.
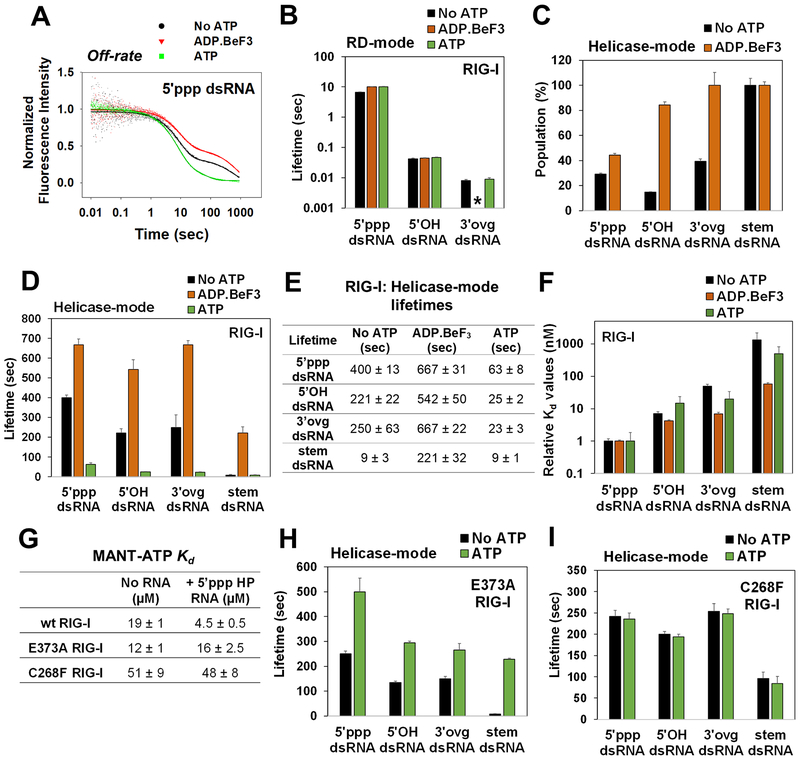
Figure 3. Off-rates of 5’ppp and non-PAMP RNAs in the presence of ADP.BeF3 and ATP.
(A) Representative off-rate kinetics of RIG-I from 5’ppp dsRNA. (B) Lifetimes of the RD-mode population. (*) indicates that an RD-mode population was not observed. Relative percentages (C) and lifetimes (D) of the Helicase-mode population of RIG-I. (E) Helicase-mode lifetimes of RIG-I:RNA complexes in the presence of the specified ATP analog (F) Kd values of RIG-I:RNA complexes normalized to the 5’ppp dsRNA:RIG-I complex. (G) MANT-ATP Kd values. (H-I) Lifetimes of the Helicase-mode population of E373A (H) and C268F RIG-I (I). Representative kinetic traces are an average of 4–6 traces. See also Figure S3 and Tables S2–S6.
Unlike RD-mode lifetimes, the Helicase-mode was sensitive to the nucleotides. ADP.BeF3 increased the relative ratio of the Helicase-mode over the RD-mode on all RNAs (Figure 3C, Table S2). The 5’ppp dsRNA showed a modest increase (15%), but 5’OH and 3’ovg dsRNA showed a pronounced increase (60–70%) in Helicase-mode population. ADP.BeF3 also increased the lifetimes of RIG-I on 5’ppp, 5’OH, and 3’ovg dsRNAs by 2–3 fold relative to minus nucleotides (Figure 3D orange bars and 3E). A more striking effect of ADP.BeF3 was observed on stem dsRNA, whose lifetime increased by 25-fold. Lifetimes were also measured in the presence of ADP and ATPYS, which confirmed that ADP.BeF3 represents the ATP-bound state (Figure S3D–E). Thus, ATP binding preferentially stabilizes the lifetimes of RIG-I on non-PAMPs, and this is inconsistent with the ATP-binding proofreading model (Rawling et al., 2015).
To obtain further evidence of a decrease in RNA selectivity upon ATP binding, we compared the Kd values of 5’ppp and non-PAMP dsRNA complexes with and without ADP.BeF3 (Table S3). Selectivity is the ratio of binding affinities of 5’ppp dsRNA and non-PAMP RNAs. Without nucleotides, RIG-I has a 7-fold, 50-fold, and 1300-fold higher selectivity for 5’ppp dsRNA over 5’OH, 3’ovg, and stem dsRNA, respectively (Figure 3F black bars). In the presence of ADP.BeF3, RNA selectivity for 5’ppp dsRNA over 5’OH, 3’ovg, and stem dsRNA dropped to 1.5-fold, 7-fold, and 50-fold, respectively, (Figure 3F orange bars). Thus, ATP binding does not proofread RNAs and instead makes RIG-I promiscuous.
Next, we measured the off-rates of RIG-I under ATP hydrolysis conditions. Interestingly, ATP hydrolysis decreased the lifetimes of all RIG-I-dsRNA complexes (Figure 3D green bar, and 3E). Under ATP hydrolysis conditions, the lifetime of 5’ppp dsRNA complex decreased by 10-fold compared to the lifetime in the ATP-bound state. Lifetimes of 5’OH, 3’ovg, and stem dsRNA complexes decreased by ~20–30 fold. Thus, ATP hydrolysis destabilizes non-PAMP complexes slightly more than the 5’ppp dsRNA complex. Compared to the lifetime on 5’ppp dsRNA (~63 s), RIG-I lifetimes are ~2.5 times shorter on 5’OH or 3’ovg dsRNA (20–25 s), and ~seven times shorter on stem dsRNA (~9 s) under ATP hydrolysis conditions (Figure 3E). Therefore, ATP hydrolysis contributes 3–7 fold to RNA proofreading by dissociation.
SMS linked RIG-I mutants are trapped on RNA in an ATP-bound Helicase-mode conformation
Two mutations (E373A and C268F) in the ATPase pocket of RIG-I are linked to the autoimmune disorder atypical SMS (Figure S3F) (Jang et al., 2015). Both mutants are ATPase-deficient and constitutively active in RIG-I signaling (Figure S3G–H). Both mutants bind ATP and similarly discriminate between dsRNA-ends to wt RIG-I (Figure 3G, Table S4).
Interestingly, the lifetimes of SMS mutants on PAMP and non-PAMP dsRNAs were much longer in the presence of ATP compared to wt RIG-I, and resembled the lifetimes of wt RIG-I with ADP.BeF3 (Tables S5 and S6, Figure 3H–I). E373A RIG-I formed strikingly long-lived complexes on stem dsRNA in the presence of ATP (Figure 3H), whereas C268F RIG-I formed a long-lived complex on stem dsRNA even in the absence of ATP (Figure 3I). Perhaps the aromatic phenylalanine substitution of C268 in the Walker A motif causes local changes that mimic the effects of ATP binding. Thus, the loss of ATPase activity without any effect on ATP binding traps the SMS mutants in long-lived ATP-bound complexes on stem RNAs, which explains their constitutively active signaling by cellular RNAs.
RIG-I is an ATPase driven dsRNA translocase closely tracking the 3’-strand
To understand the mechanism of RNA dissociation by ATP hydrolysis, we investigated the ATPase-driven translocation activity of RIG-I on PAMP and non-PAMP dsRNAs using biotin-streptavidin displacement assay and a fluorescence-based stopped-flow assay (Fischer et al., 2010; Morris et al., 2001; Tomko et al., 2010).
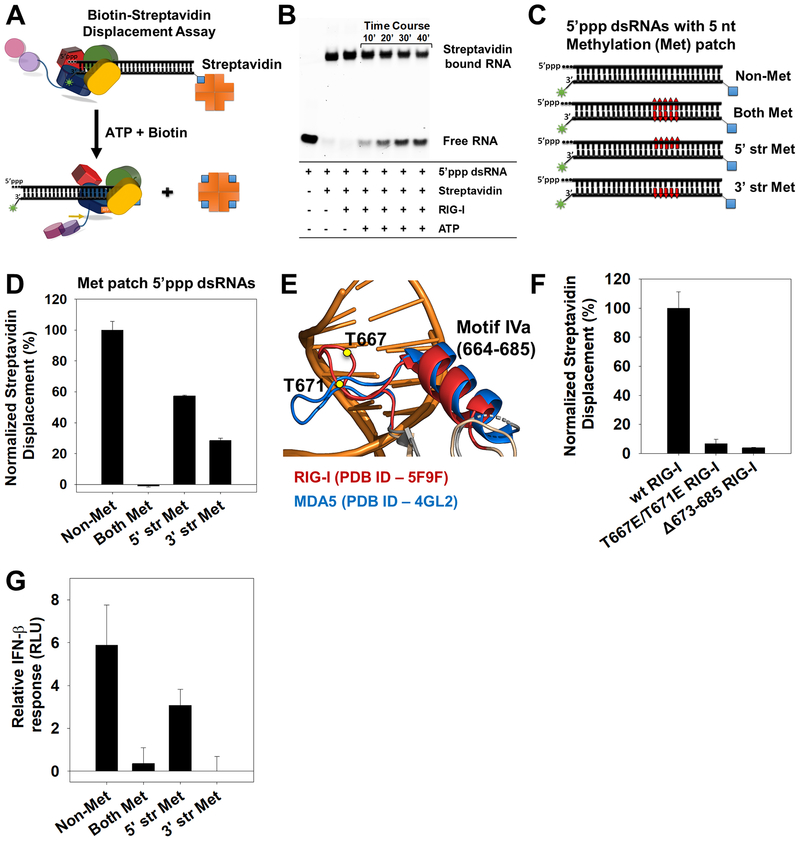
Biotin-streptavidin was conjugated to the 27bp dsRNA at the opposite end of the 5’-end modifications (Figure 4A). RIG-I displaced streptavidin from 5’ppp and 5’OH dsRNAs only under ATP hydrolysis conditions (Figure 4B and S4A). To determine the directionality of translocation, we introduced a 5-nt patch of 2’-O-methylated ribose nucleotides, 15 bp downstream from the 5’ppp end. Crystal structure of dsRNA bound RIG-I (PDB ID: 5E3H) shows multiple contacts with ribose 2’-OH groups of dsRNA; therefore, we expect 2’-O-methylation to hinder RIG-I binding. Three variants of 5’ppp dsRNA were made, one contained a methylation patch on both strands, and others contained a methylation patch either in the 5’ strand (containing 5’ppp) or in the 3’-strand (opposite to 5’ppp) (Figure 4C). RIG-I did not displace streptavidin from dsRNA that contained methylation patches on both strands (Figure 4D). When the methylation patch was on the 5’-strand, 60% streptavidin displacement was observed, and when the methylation patch was on the 3’ strand, 30% streptavidin displacement was observed, compared to the non-methylated variant (Figure 4D). These findings indicate that RIG-I is a dsRNA translocase that closely tracks along the 3’-strand as it moves away from the 5’ppp end into the stem region of dsRNA.
Figure 4. Directionality of RIG-I translocation and role of Helicase motif IVa.
(A) Biotin-streptavidin displacement assay schematic. (B) Time-course of biotin-streptavidin displacement from 5’ppp dsRNA (25 nM) by RIG-I (25 nM). (C) 27bp methylated patch 5’ppp dsRNAs to assess directionality of RIG-I translocation (Red bars- 5nt patch of 2’-O-methylated nucleotides placed 15bp downstream of the 5’ppp end). (D) Streptavidin displacement activity on methylated patch 5’ppp dsRNAs. Error bars are SEM from triplicates. (E) Helicase motif IVa of RIG-I (red) and MDA5 (blue). T667E and T671E in RIG-I are shown as yellow circles. (F) Streptavidin displacement by motif IVa mutants on the 5’ppp dsRNA. Error bars are SEM from triplicates. (G) IFN-β reporter activity of RIGI transfected HEK293T cells upon activation by the 2’-O-methylated 5’ppp dsRNAs. Error bars are SEM from quadruplicates. See also Figure S4.
RIG-I translocation is governed by Helicase motif IVa and is important for signaling
Helicase motif IVa (664–685) is a conserved region in the Hel2 domain of RIG-I, vital for RIG-I’s signaling activity (Devarkar et al., 2016). Moreover, DAPK1 kinase phosphorylates T667 and T671 in motif IVa and impairs RIG-I signaling (Willemsen et al., 2017). The motif forms an extended loop-helix structure in MDA5, where it interacts with the major groove of dsRNA (Figure 4E) and is critical for signaling (Wu et al., 2013). The phosphomimetic mutant T667E/T671E RIG-I is fully competent in 5’ppp dsRNA binding and exhibits ~50% of wt RIG-I’s ATPase activity (Figure S4B). Thus, loss of signaling activity is not due to defects in RNA binding or ATPase activity. Interestingly, biotinstreptavidin displacement assays showed that T667E/T671E RIG-I is defective in translocation and shows only 6% streptavidin displacement compared to wt RIG-I (Figure 4F). Deletion of the short alpha-helix region in motif IVa (Δ673–685) also reduced translocation significantly (Figure 4F). This indicates that translocation is vital for RIG-I’s signaling function.
To further test the hypothesis that RIG-I translocation is essential for signaling, we examined the signaling activity of dsRNAs showing varying degrees of translocation activity (Figure 4C–D). The 5’ppp dsRNA without any methylation supported translocation and showed robust stimulation of RIG-I signaling, whereas the 5’ppp dsRNA carrying a methylated patch on both strands was translocation deficient and signaling inactive (Figure 4G). The 5’ppp dsRNA with a methylation patch in the 5’-strand showed inefficient translocation and a mild signaling response, whereas the 3’-methylated strand variant showed a more significant defect in translocation and was signaling inactive. These data provide additional evidence that translocation plays an important role in the activation of RIG-I.
Translocation rate of RIG-I is dictated by 5’-end RNA modification
We used a fluorescence stopped-flow assay to monitor RIG-I translocation in real time and estimate the translocation rates on PAMP and non-PAMP RNAs. A Cy3 fluorophore was introduced in the 27-bp dsRNAs at 13th, 17th, 20th, or 24th bp position from the dsRNA-end containing either 5’ppp, 5’OH, or 3’ovg modification (Figure 5A). The transient state translocation kinetics were measured under single-round conditions, wherein a preformed complex of RIG-I with the specified Cy3-tagged dsRNA (1.5-fold excess over protein) was rapidly mixed with ATP and a 10-fold excess of the trap. The expected signal from the time courses of a translocating helicase is a peak of fluorescence intensity. As RIG-I translocates away from the 5’ppp end and arrives close to Cy3 in the dsRNA backbone, fluorescence intensity should increase, and when RIG-I moves away from Cy3 or dissociates, fluorescence should decrease.
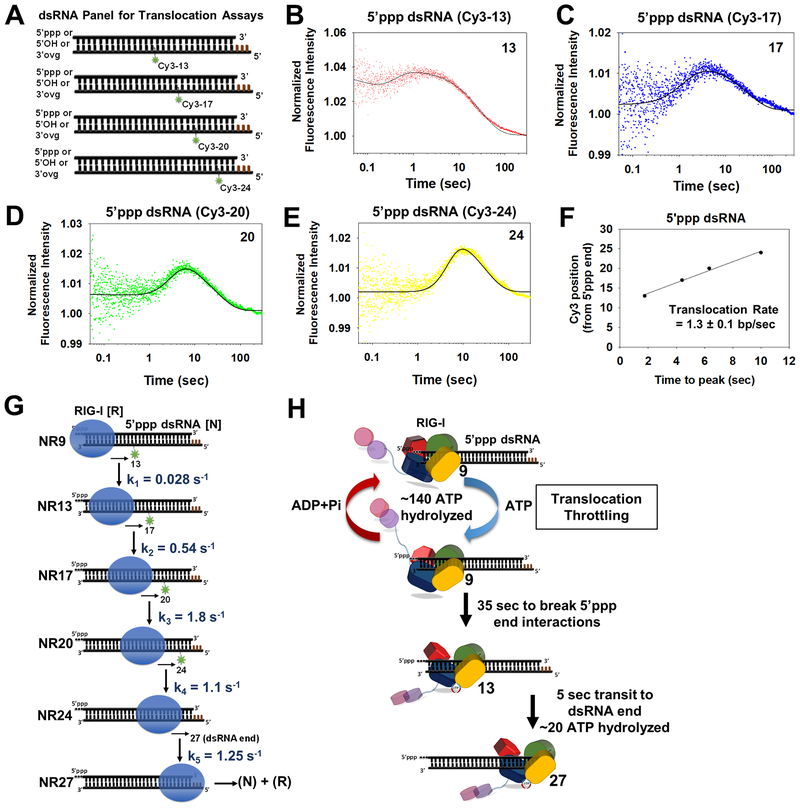
Figure 5. Stepping rates of RIG-I translocation.
(A) Schematic of a panel of Cy3 labeled 27bp dsRNAs for RIG-I translocation studies (Brown bars-DNA bases). (B-E) Transient state kinetics of RIG-I translocation on the 5’ppp dsRNA measured by Cy3 fluorescence intensity changes. The position of Cy3 from the RNA-end is denoted. Solid lines are fit to the model in G. (F) Time-to-peak increases linearly with Cy3 position and slope provides the average translocation rate. (G) The minimal translocation model fits the stopped-flow translocation data in B-E (solid lines) with the given stepping rates. (H) Translocation throttling mechanism of RIG-I at the 5’ppp end. See also Figure S5 and Table S7.
Such peaks of fluorescence intensity changes were observed in the time courses of 5’ppp dsRNAs labeled at various positions (Figure 5B–E). The 5’OH and 3’ovg dsRNAs also showed peaks (Figure S5A–B) but were less pronounced when Cy3 was present at a longer distance of 20 and 24bp from the RNA-end. The peak profile was not seen in the absence of ATP (Figure S5C). The time-to-peak increased linearly with an increasing distance of Cy3-position on the dsRNA, which is consistent with directional translocation of RIG-I on the dsRNAs (Figure 5F and S5D–E). Since RIG-I initiates translocation from one dsRNA-end, the slopes provided the average translocation rates (Dillingham et al., 2000; Tomko et al., 2010). RIG-I translocates with an average rate of 1.3 bp/s, 3.4 bp/s, and 3.9 bp/s on 5’ppp, 5’OH, and 3’ovg dsRNAs, respectively (Figure 5F and S5D–E). The y-intercepts were ~10 bp, which is consistent with the 8–10 bp footprint of RIG-I on dsRNA. Thus, RIG-I translocates ~3 times faster on 5’OH/3’ovg dsRNA than on 5’ppp dsRNA, which was unexpected, but explains the ~3 times faster dissociation rate of RIGI from 5’OH and 3’ovg dsRNAs under ATP hydrolysis conditions (Figure 3E).
5’ppp blunt-end throttles RIG-I translocation
To further understand the different translocation rates of RIG-I on 5’ppp and 5’OH/3’ovg dsRNAs, we globally fit the kinetic data to a 5-step translocation model (Figure 5G, 1 step/fluorophore position) using Kintek Explorer software. We assumed step-wise translocation (9→13→17→20→24→27) of RIG-I from the dsRNA-end into the stem region with rate constants k1 to k5 (Figure 5G). Each position corresponds to the location of the fluorophore on the dsRNA. We reasoned that at the start of the reaction, the leading edge of RIG-I is near the 9th bp from the dsRNA-end based on the ~9-bp footprint. For best fits, we considered that some amount of RIG-I was bound to each of the above internal positions and these populations dissociated upon addition of ATP with rate constants k6 to k9 (Table S7). The best fits estimated that roughly 90% of RIG-I was bound to RNA-end and 10% internally (Figure S5F).
The two most revealing findings from the global fitting were, a) the rate-limiting step of translocation is breaking the dsRNA-end interactions and moving into the stem-region (k1<<<k2-k5), and b) RIG-I moves away from the 5’ppp dsRNA-end with a ~3.5 times slower rate than from 5’OH/3’ovg dsRNA-end (Figure 5G, Table S7). This indicates that the 5’ppp blunt-end ‘throttles’ RIG-I translocation, forcing RIG-I to reside at the 5’ppp-end for most of its time spent on the dsRNA. RIG-I takes ~40 s to translocate from the 5’ppp end to the 27th bp at the other end, of which ~35 s are spent just reaching the 13th position (k1 = 0.028 s−1). On 5’OH and 3’ovg dsRNAs, RIG-I takes ~10 s and ~11 s to reach the 27th position, of which 9 s and 10 s are spent reaching the 13th position (k1 = 0.11 s−1 and 0.1 s−1), respectively. The composite rate of RIG-I translocation from the individual rate constants (k1 to k5) is ~0.7 bp/s on 5’ppp dsRNA, and ~2.7 bp/s and 2.4 bp/s on 5’OH and 3’ovg dsRNAs, respectively. These rates are close to the rates obtained from the time-to-peak analysis (Figure 5F and S5D–E).
During the 35 s of residence time at the 5’ppp RNA-end, we estimate that RIG-I hydrolyzes ~140 ATP molecules (Figure 5H). This is calculated from the steady-state ATPase rate of 4 ATP/s multiplied by the 35 s of residence time at the 5’ppp dsRNA-end (Figure S5G). Using similar calculations, we estimate that RIG-I hydrolyzes ~36 ATP molecules during its residence time at the 5’OH RNA-end and ~30 ATP molecules at 3’ovg RNA-end. Thus, RIG-I spends a significant amount of energy in the form of ATP hydrolysis to break its interactions with the 5’ppp dsRNA-end. Translocation-throttling at the 5’ppp-end with continued ATP binding and hydrolysis cycles provides a mechanism that can ensure complete release of CARDs until stabilized by downstream factors such as ubiquitin or by RIG-I oligomerization.
RIG-I forms dimers on the 27 bp dsRNA
We used the Electrophoretic Mobility Shift Assay (EMSA) to characterize RIG-I oligomerization on the 27-bp 5’ppp and non-PAMP dsRNAs. One end of the RNA was blocked by biotin-streptavidin to allow RIG-I oligomerization only from the end carrying the desired modification.
At equimolar concentrations of RIG-I and 5’ppp dsRNA and no ATP, we observed only RIG-I monomers on RNA (Figure 6A). However, when ATP was added, we observed RIG-I dimers, which were not present with ADP.BeF3, ADP.AlFx, or ADP. SEC-MALS (size exclusion chromatography-multiple angle light scattering) of 5’ppp dsRNA bound RIG-I verified that the observed EMSA species correspond to RIG-I monomer and dimer (Figure S6A–B). The translocation deficient mutant T667E/T671E failed to form dimers (Figure 6A), indicating that translocation is important for dimer formation. Similarly, no dimers were observed on 5’ppp dsRNA carrying methylation patch on both strands (Figure 6B). This result indicates that RIG-I dimers are formed by two RIG-I molecules bound to the same 27bp dsRNA and not via intermolecular association of two RNA-bound RIG-I monomers. We observed fewer dimers on 5’OH dsRNA and none on 3’ovg dsRNA or stem dsRNA in the presence of ATP, even at a 3-fold excess RIG-I concentration (Figure 6C–D). The ATPase-deficient SMS mutants formed dimers on stem dsRNA, and SEC-MALS analysis verified that the oligomeric species corresponds to a dimer (Figure S6C), but the ATPase-deficient K270A mutant did not form dimers on stem dsRNA (Figure 6D). Moreover, only 5’ppp and 5’OH dsRNAs that support RIG-I oligomerization are signaling active, and RNAs that bind RIG-I but do not support oligomerization, are signaling inactive. Thus, RIG-I oligomerization strongly correlates with signaling activity.
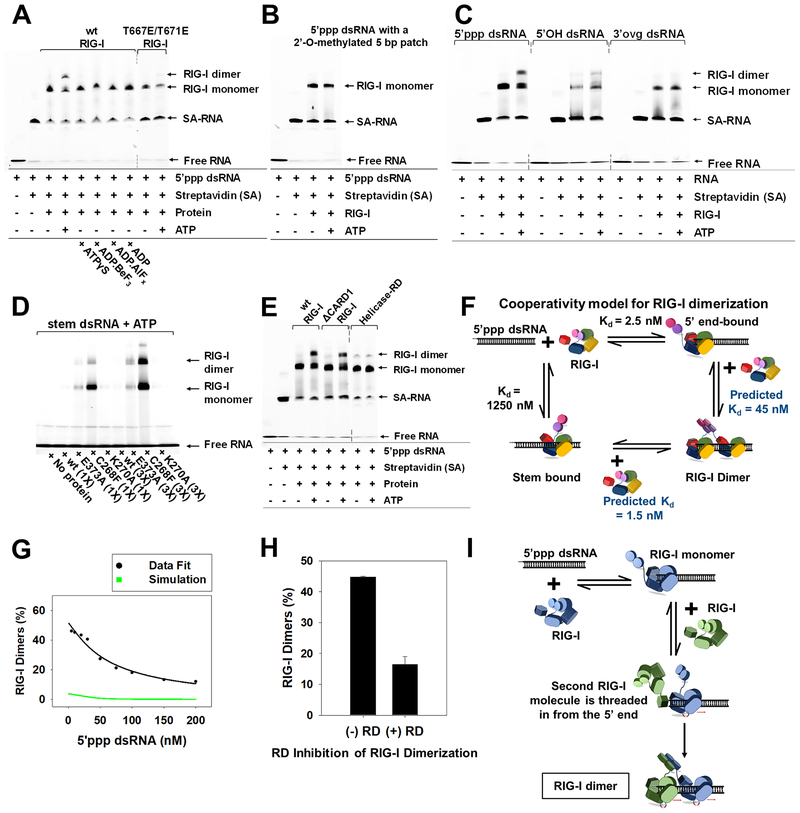
Figure 6. ATP facilitates cooperative RIG-I dimerization on 5’ppp dsRNA by 5’-end threading mechanism.
(A-B) EMSAs of RIG-I or specified mutant (25 nM) with DY547-tagged 5’ppp dsRNA (25 nM) (A) and methylation patch carrying 5’ppp dsRNA (25 nM) (B). (C) EMSAs of DY547 labeled 5’ppp dsRNA, 5’OH dsRNA and 3’ovg dsRNA (100 nM each) with RIG-I (100 nM). (D) EMSAs of Cy3 labeled stem dsRNA (100 nM) with RIG-I and specified mutants (100 and 300 nM each). (E) EMSA of DY547 labeled 5’ppp dsRNA (25 nM) with RIG-I or specified mutant (25 nM) in presence or absence of ATP. (F) Cooperativity model for RIG-I dimerization. (G) RIG-I (50 nM) was titrated with increasing concentrations of 5’ppp dsRNA (5–200 nM) with ATP (2mM) and EMSA was used to determine percent dimers. Green line represents the expected trend for non-cooperative RIG-I dimerization, and the black line shows the best fit to the cooperative dimerization model in E. (H) 5’ppp dsRNA (25 nM) and RIG-I (75 nM) were incubated with and without RD (75 nM) and ATP (2 mM), and percent RIG-I dimers from EMSA are plotted. Error bars are SEM from triplicates. (I) The 5’-end threading model is consistent with results in G. Dotted lines in EMSA gel blots represent deletion of lanes. See also Figure S6.
Mechanism of ATP facilitated RIG-I oligomerization
Dimerization on the 27bp 5’ppp dsRNA requires binding of two RIG-I molecules, one to the 5’ppp end and a second molecule to the adjacent stem region. Based on the weak affinity of RIG-I for dsRNA stem (1.25 μM with ATP, Table S3), it is unlikely that dimers are generated by independent binding of two RIG-I molecules to the 5’ppp dsRNA at 25 nM protein and RNA concentrations. Moreover, the presence of dimers at equimolar RIGI and RNA suggests cooperativity in RIG-I oligomerization. Interestingly, under equimolar conditions, Helicase-RD (232–925) showed very few dimers on 5’ppp dsRNA compared to wt RIG-I, whereas first-CARD deleted (97–925) RIG-I construct showed robust dimerization (Figure 6E). This indicates that the CARDs and/or the CARD2-Hel1 linker (~55 aa) might be involved in cooperative oligomerization.
To verify cooperativity in RIG-I dimerization, RIG-I was titrated with increasing concentrations of 5’ppp dsRNA in the presence of ATP. RIG-I dimers were quantified after EMSA and their RNA dependency was modeled using Kintek explorer (Figure 6F–G and S6D). For monomers, we used the experimentally measured 5’ppp-end Kd of 2.5 nM and stem-region Kd of 1.25 μM under ATP hydrolysis conditions (Table S3). For dimer formation by the non-cooperative mechanism, we used the same Kds for second RIG-I molecule binding to each of the two regions. As expected, the non-cooperative case predicted insignificant amount of dimer (Figure 6G, green) as compared to the experimentally measured dimer formation (Figure 6G, black). Therefore, we fit the dimerization data to the cooperative model (Douglass et al., 2013), which indicated that stem-binding affinity is ~28-fold greater (predicted Kd ~45 nM) when another molecule of RIG-I is bound to the 5’ppp-end. A similar titration estimated a lower cooperativity of ~18 for 5’OH dsRNA (Figure S6E). This suggests that there is positive cooperativity in RIG-I oligomerization.
RIG-I oligomerization by threading from the 5’ppp end
To further understand the mechanism of ATPase-driven RIG-I dimerization, we asked whether the second molecule of RIG-I is threaded from the 5’ppp-end or binds directly to the stem region adjacent to the first molecule (Figure S6F). We used the RD construct as a competitor to differentiate between these two models. RD binds avidly to 5’ppp end and shows no interactions with dsRNA stem (Figure S2C). Modeling predicted the following two scenarios (Figure S6F–G) - if RIG-I forms dimers by binding a second molecule directly to stem region adjacent to 5’ppp end-bound RIG-I, then the presence of RD will not affect RIG-I dimer:monomer ratio. If RIG-I dimerizes by threading from the 5’ppp end, then RD will compete and lower the dimer:monomer ratio. EMSAs of RIG-I and 5’ppp dsRNA in the presence of ATP showed a ~60% reduction in dimers upon addition of RD (equimolar to RIG-I) in comparison to dimers in the absence of RD (Figure 6H and S6H). These results support the threading mechanism of RIG-I dimerization (Figure 6I).
A kinetic proofreading pathway for RIG-I oligomerization and RNA discrimination
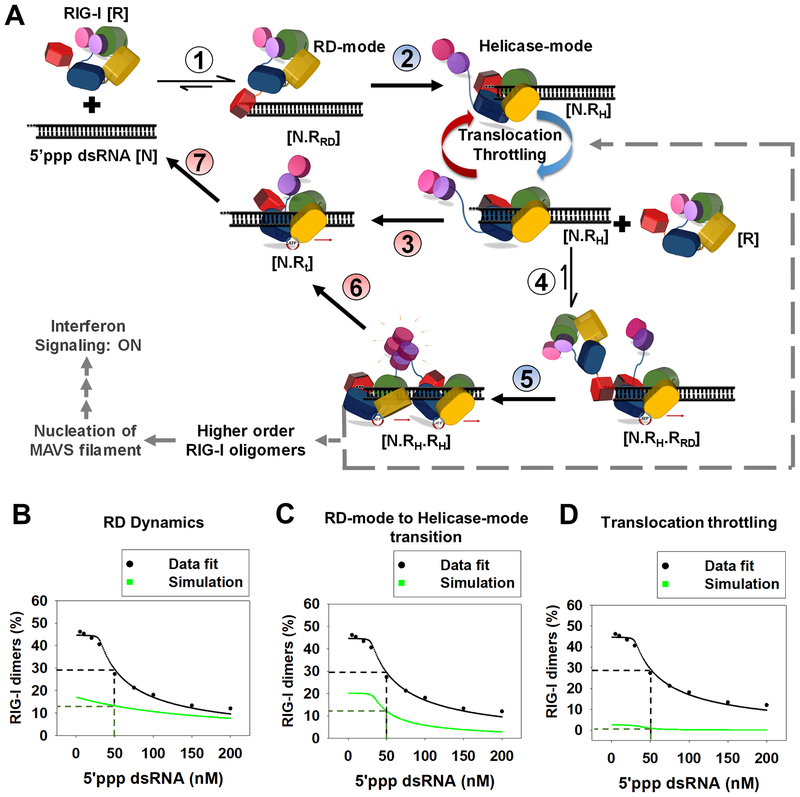
Our combined results suggest a multistep kinetic mechanism for RNA discrimination and selective RIG-I oligomerization, detailed in Figure 7A. We were able to test this 7-step kinetic model by fitting the experimentally measured decrease in RIG-I dimerization with increasing 5’ppp dsRNA concentration (Figure S6D, see STAR Methods). Data fitting identified three critical steps for efficient RIG-I dimerization: 1) RD on-rate and off-rate from the 5’-end, 2) the RD-mode (N.RRD) to Helicase-mode (N.RH) transition rate, and 3) translocation throttling at the 5’-end. We changed the rate constants of each of the above steps one at a time and monitored the outcome on RIG-I dimerization. Increasing the RD-mode off-rate from the one observed on 5’ppp end (0.15 s−1) to the one on 3’ovg end (125 s−1) reduced RIG-I dimerization by ~55% (at 50 nM RIG-I and RNA) (Figure 7B). Reducing the N.RRD to N.RH transition rate by 3-fold causes a ~60% decrease in dimerization of RIG-I (Figure 7C). Assuming no translocation throttling at the 5’-end; that is, assuming RIG-I translocates at a uniform rate (1.3 s−1 for steps 3, 6 and 7) reduced RIG-I dimerization by ~95% (Figure 7D). This kinetic proofreading pathway explains how minor dsRNA-end modifications can significantly modulate RIG-I oligomerization and signaling response.
Figure 7. Kinetic proofreading pathway for RIG-I oligomerization and RNA discrimination.
(A) The seven steps show the kinetic pathway of RIG-I oligomerization. (1) RIG-I forms the RD-mode complex. (2) The RD-mode complex transitions into an ATPase active Helicase-mode complex that undergoes multiple rounds of ATP hydrolysis at the 5’-end (Translocation-throttling). (3) Helicase-mode complex translocates away from the dsRNA-end, or (4) binds a second molecule of RIG-I in the RD-mode. (5) Second RIG-I molecule transitions to Helicase-mode resulting in RIG-I dimer on dsRNA. (6–7) RIG-I dimer can translocate and dissociate from the RNA. Steps shaded in blue (2,5) are facilitated by ATP binding and steps shaded in red are driven by ATP hydrolysis (3,6 and 7). (B-D) The decrease in RIG-I dimers with increasing 5’ppp dsRNA concentration (Figure S6D) fit well to the kinetic proofreading pathway from A (black). The experimentally measured values used for the fitting are: Step 1 and 4 (kon: 6 × 108 M−1s−1, koff: 0.15 s−1), Step 3 and 6 (0.028 s−1), step 7 (1.2 s−1); predicted rates from best fit for step 2 and 5 were 0.03 s−1 and 0.06 s−1, respectively. Simulations (green) were carried out by either substituting the RD-mode off-rates of 5’ppp dsRNA in the model with 3’ovg dsRNA off-rate (B), decreasing the N.RRD to N.RH transition rate by 3-fold (C), and removing all translocation throttling steps at the 5’ end for monomer and dimer (D).
Discussion
RIG-I recognizes viral PAMP RNAs in a cytoplasmic environment where it is surrounded by an abundance of cellular non-PAMP RNAs. Although RIG-I has a lower affinity for non-PAMPs, differences in the binding affinities for PAMP and non-PAMP RNAs (~20–500 fold) are not sufficient to explain the high selectivity of RIG-I activation (Devarkar et al., 2016; Kohlway et al., 2013; Ramanathan et al., 2016). Moreover, selectivity is most likely kinetically controlled because biological processes seldom reach equilibrium (Banerjee et al., 2017). Many biological systems, including DNA polymerases and ribosomes, use kinetic proofreading to increase fidelity beyond differences in equilibrium binding affinities of right and wrong substrates (Blanchard et al., 2004; Hopfield, 1974; Tsai and Johnson, 2006). T-cell receptors use kinetic proofreading to achieve high selectivity for signaling in response to foreign peptides (McKeithan, 1995). In a ‘kinetic proofreading’ mechanism, selectivity is increased through energy providing steps such as exonuclease, phosphorylation, or ATP hydrolysis. RIG-I has an ATPase activity, but the role of ATP binding and hydrolysis in RNA selection and RIG-I activation has been controversial. By carrying out comprehensive and quantitative studies of RNA binding kinetics, translocation, and RIG-I oligomerization on PAMP and non-PAMP RNAs, we show that ATP binding and hydrolysis have multiple roles in RIG-I activation and RNA selection.
In kinetic proofreading, a) the receptor discriminates between ligands by differential off-rates, b) intermediate steps create a time delay between ligand binding and activated complex formation, c) an energy source (e.g., ATP) drives at least one of the intermediate steps, and d) there are dissociation paths for ligands from the intermediate steps. Kinetic proofreading pathway for RIG-I presented here (Figure 7A) meets all the above characteristics.
RD discriminates non-PAMP RNAs by faster dissociation (step 1, Figure 7A). On-rates of RD are diffusion-limited and indiscriminate, and only off-rates are sensitive to dsRNA-end modifications (Figure 1G and 2A). This sets up a mechanism whereby autoinhibited RIG-I can quickly sample RNA-ends by a hopping mechanism to locate PAMP RNA-ends. ATP binding aids CARDs release (Zheng et al., 2015) by disrupting the autoinhibitory CARD2:Hel2i interface and engaging the Hel2i interface residues with the dsRNA backbone (step 2, Figure 7A). ATP binding, however, does not preferentially stabilize dsRNA interactions on PAMP, and in fact, stabilizes non-PAMPs more than PAMP (Figure 3D and 3E). Thus, contrary to the ATP-binding proofreading model (Rawling et al., 2015), ATP binding does not proofread RNAs by dissociation but makes RIG-I less selective. ATP hydrolysis, on the other hand, provides a proofreading function by dissociating non-PAMPs about 3–7 fold faster than 5’ppp dsRNA. However, this modest difference cannot fully account for the stringent selectivity of RIG-I mediated signaling response.
We show that ATP hydrolysis dissociates RIG-I from RNAs through translocation (step 3 and 7, Figure 7A). The directionality of translocation is away from the dsRNA-end into the stem region, which might seem counterintuitive, but this directionality provides a threading mechanism for RIG-I oligomerization on dsRNA. RIG-I is threaded from the 5’ppp-end via its RD (Figure 6H–I) and CARDs are involved in stabilizing the dimer (Figure 6E–F). Interestingly, RIG-I translocation is throttled at the 5’ppp dsRNA-end (Figure 5H), which increases the lifetime of RIG-I on dsRNA-end and facilitates loading of the second molecule of RIG-I. In the absence of throttling, RIG-I monomer would move away quickly from the 5’ppp-end into the stem region and dissociate, as it happens with non-PAMPs, decreasing the chances of stable dimer formation on dsRNA. We modeled the lack of translocation-throttling on RIG-I dimerization by assuming a uniform translocation rate for RIG-I on 5’ppp dsRNA, and the modeling predicted a 95% drop in RIG-I dimers (Figure 7D). The 5’ppp-end needs to be exposed, at least transiently, to enable RIG-I loading via the RD. During translocation-throttling, RIG-I hydrolyzes multiple ATP molecules; hence, ATPase-coupled conformational changes, such as one or two bp movements, could expose the 5’ppp-end and make it accessible for RIG-I loading. Thus, repeated cycles of threading via the translocation-throttling mechanism builds higher order RIG-I oligomers on longer PAMP dsRNA (step 4 and 5, Figure 7A). Since every added RIG-I molecule has to go through this kinetic pathway, the discrimination against non-PAMP RNAs increases exponentially with the length of RIG-I oligomer.
A critical function of the ATPase activity is to facilitate RIG-I dissociation from non-PAMPs and to build RIG-I oligomers selectively on PAMP RNAs. The kinetic proofreading mechanism for these steps is impaired in the SMS RIG-I mutants (E373A and C268F), which are capable of binding ATP but are defective in hydrolysis. The SMS mutants bind avidly to stem dsRNA, and moreover, are competent in forming oligomers on stem dsRNA, which explains constitutive activation by cellular RNAs (Lassig et al., 2015). K270A is also an ATPase-deficient RIG-I mutant, but unlike the SMS mutants, K270A RIG-I does not bind stem dsRNA, most likely due to a defect in ATP binding. Thus, unlike E373A and C268F, K270A RIG-I is signaling defective.
RIG-I follows the paradigm of DNA polymerases, ribosomes, and T-cell receptors for achieving a high fidelity for its correct ligand. DNA polymerase kinetically discriminates against incorrect nucleotides by faster off-rates (Tsai and Johnson, 2006), akin to the faster off-rates of the RD-mode complex of RIG-I from non-PAMPs vs PAMPs. The probability of incorrect nucleotide incorporation by DNA polymerase is low, analogous to the low probability of RIG-I engaging the non-PAMP RNAs in an ATPase-active and CARDs-exposed conformation. The DNA polymerase uses an energetically favorable exonuclease activity to remove the incorrect nucleotide, which can be related to RIG-I using the ATPase-driven translocation to dissociate from non-PAMP RNAs. And finally, analogous to DNA polymerases engaging a primer-template and synthesizing an exact complementary DNA strand, RIG-I forms oligomers only on PAMP RNAs to trigger a signaling response.
In summary, our findings address the controversial roles of ATP binding and hydrolysis in RIG-I activation. By characterizing RIG-I dynamics and translocation on PAMP and non-PAMP RNAs, we have delineated a kinetic proofreading pathway for RIGI ligand selection and oligomerization. Given the importance of RIG-I in viral infections, autoimmune diseases, and cancer immunotherapy, the findings presented here would be important in leveraging RIG-I in therapeutic approaches.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
For further information and requests for resources and reagents, please contact the Corresponding author, Smita S. Patel (patelss@rwjms.rutgers.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293T cells (Sex: Female) were a kind gift from Prof. Charles Rice, Rockefeller University, USA. This cell line was described previously (Ramanathan et al., 2016) to be devoid of RIG-I expression and did not respond to transfected double-stranded RNA or Poly I:C. Cells were grown at 37°C and 5% CO2 in DMEM with 10% FBS. RIG-I was transiently expressed in the cells by plasmid transfections and were then tested for IFN-β cell signaling response to various transfected RNA ligands.
METHOD DETAILS
Protein Expression and Purification.
All RIG-I protein constructs were expressed using pET28 SUMO vector in Escherichia coli strain Rosetta (DE3) (Novagen). The soluble fraction was purified from the cell lysate using a Ni2+-nitrilotriacetate (Qiagen) column, followed by Ulp1 protease digestion to remove the 6xHis-SUMO tag, and further purified by hydroxyapatite (CHT-II, Bio-Rad) and heparin sepharose column chromatography (GE Healthcare). Purified protein was dialyzed against 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, 10% glycerol overnight at 4°C, snap frozen in liquid nitrogen, and stored at −80 °C.
RNAs
All RNAs were chemically synthesized and HPLC purified by Bio-Synthesis Inc. or TriLink BioTechnologies. The synthetic RNAs were analyzed for purity by mass spectrometry and HPLC. The lyophilized RNA was resuspended in 20 mM potassium phosphate buffer pH 7.0. The concentration was determined from the absorbance in 7 M guanidinium HCl using the NanoDrop spectrophotometer at A260 and their extinction coefficients. Duplex RNA was prepared by mixing complementary ssRNAs in a 1:1.1 ratio, heating to 95°C for 1 min, and slow cooling to 4°C. The sequences of all the RNAs used in this study are tabulated in Supplementary Table S8.
RNA binding and ATPase Assay
The ATP hydrolysis activity was measured at constant RIG-I (10 nM or 25 nM) and increasing RNA concentration (1 nM - 1 μM) in the presence of 1 mM ATP (spiked with [γ−32P] ATP). A time course (0–60 min) of the ATPase reactions was performed in Buffer A (50 mM MOPS pH 7.4, 5 mM DTT, 5 mM MgCl2, 0.01% Tween20) at 25°C. The reactions were stopped at desired time points using 4 N formic acid and analyzed by PEI-Cellulose-F TLC (Merck) developed in 0.4 M potassium phosphate buffer (pH 3.4). The TLC plates were exposed to a phosphorimager plate, imaged on a Typhoon phosphor-imager, and quantified using the ImageQuant software. The ATPase rate was determined from the slopes of molar [Pi] produced versus time (s). The ATPase rates were then plotted as a function of RNA concentration and fitted to, ATPase rate = katpase × [PR]/[Pt]; where [Pt] is total protein concentration; [PR] is the amount of RIG-I/RNA complex formed and [R] is the RNA concentration being titrated. The hyperbolic equation (Equation 1) or quadratic equation (Equation 2) was used to determine [PR] and estimate the equilibrium dissociation constant (Kd) and maximal ATPase rate (katpase).
| (Equation 1) |
| (Equation 2) |
Fluorescence Intensity Titrations
Fluorescence intensity measurements were carried out using FluoroMax-4 spectrofluorimeter (Horiba Jobin Yvon) in Buffer A. Fluorophore-labeled blunt-ended dsRNAs (5 nM) or non-blunt ended dsRNAs (20 nM) were titrated with increasing protein, and the change in fluorescence intensity was measured at 25°C. Fluorescence emission was measured at 570 nm after excitation at 547 nm for DY547 or Cy3 labeled RNAs. The observed fluorescence intensity was plotted as a function of protein concentration (Pt) and fitted to equation 1 or 2 to obtain the equilibrium dissociation constant (Kd). The reported RNA Kd values were consistently observed in titrations repeated 2 to 3 times.
IFN-β Reporter Cell Signaling Assays
HEK293T cells were grown in 6-well plates to 60% confluence, and cotransfected with firefly luciferase reporter plasmid (pLuc125 / 2.5 μg), Renilla luciferase reporter plasmid (pRL-TK / 500 ng), and a plasmid carrying either the wt RIG-I gene or the desired mutant RIG-I gene or an empty plasmid under the constitutively active CMV promoter (pcDNA 3.1 / 2 μg). The firefly luciferase gene is under the interferon β promoter, and the Renilla luciferase plasmid is under the constitutively active TK promoter. The plasmid transfections were carried out with X-tremeGENE HP DNA Transfection Reagent (Roche). Cells were replated in 96-well plates the next day at 2 × 104 cells/well density and transfected with the specified RNA ligands (700 nM final concentration in 110 μl volume) using Lipofectamine transfection reagent (Life Technologies). After 20 hours the activities of firefly and Renilla luciferases were measured sequentially with the Dual-Luciferase reporter assay (Promega). Data was collected in quadruplicate sets, and the relative luciferase activities were calculated. The error bars represent the standard error of the mean (SEM).
On and Off Rate Measurements
The RNA off-rates were measured at 25°C using a stopped-flow instrument (Auto-SF 120, Kintek Corp, Austin, Tx). A mixture of fluorophore labelled dsRNA and protein in Buffer A (pre-incubated at 25°C for 10 min) from syringe A was mixed with a 10-fold excess of an RNA trap (unlabeled 5’ppp ds12 hairpin RNA) from syringe B. For 5’ppp, 5’OH and 3’ovg dsRNA, an equimolar concentration of protein and fluorescent dsRNA was used (50 nM each for 5’ppp and 5’OH dsRNA and 100 nM each for 3’ovg dsRNA). For the stem dsRNA, 200 nM of the Cy3 labeled dsRNA was mixed with 400 nM of protein. The fluorescence emission of DY547 or Cy3 was measured using a 570 nm band-pass filter after excitation at 547 nm, and the fluorescence emission of fluorescein was measured using a 515 long-pass filter after excitation at 495 nm. To study the effect of ATP or ATP analog on the off-rate kinetics, we added 2 mM ATP or 1 mM ADP.BeF3 (1 mM ADP + 0.5 mM BeCl2 and 5 mM NaF) to the RIG-I/RNA complex and to the RNA trap solution. ATP regeneration mix (0.1 mg/ml phosphocreatine kinase, 10 mM phosphocreatine and 1 mM MgCl2) was added in the off-rate experiments with ATP to limit the accumulation of ADP. The change in fluorescence intensity was plotted as a function of time, and the data were fit to a single exponential or sum of two to three exponentials to estimate the off-rates. The on-rates were measured at 25°C using the stopped-flow instrument. A fixed concentration (10 nM) of fluorophore-labeled RNA from syringe A was rapidly mixed with the protein from syringe B, and the experiments were carried out at increasing protein concentrations (45–200 nM). The fluorescence intensity was plotted as a function of time and fitted a single exponential equation to obtain the observed rates of binding. The observed binding rates were plotted against protein concentration and fitted a linear equation to estimate the on-rates.
Biotin-Streptavidin Displacement Assays
3’-DY547 labeled 27 bp dsRNAs (25 nM) carrying biotin at the 5’-end and desired 5’-modification at the opposite end were incubated with streptavidin (60 nM, New England Biolabs) in Buffer A, followed by RIG-I (25 nM) addition. The reactions were started by mixing this complex with ATP (2 mM) or ATP analog (1 mM) and free biotin (50 μM). The reactions were quenched with 10 mM EDTA and 0.5% SDS. The samples were run on a 4–20% TBE gel (Invitrogen) at 4°C and scanned using a Typhoon FLA 9500 laser-based scanner (GE Healthsciences). The reactions were run in triplicates, and the fraction of streptavidin displaced from the biotinylated RNAs was quantitated using ImageQuant TL software.
Electrophoretic Mobility Shift Assays (EMSA)
EMSA assays were carried out by incubating RIG-I (or RIG-I construct), fluorophore-tagged dsRNA (3’-DY547, 5’-Biotin) and a 4-fold excess of monovalent streptavidin at 4°C in Buffer A for 60 minutes. An equimolar concentration (25 nM) of RIG-I or the specified RIG-I construct and the fluorophore-tagged dsRNA was used for the EMSAs, unless stated otherwise. 2 mM ATP or 1 mM ATP analog was added to the reaction as specified in the figure legends, 15 minutes before loading the sample on the gel. Loading buffer (10× concentration is 1.5% Ficoll 400 in the Tris-borate buffer, pH 8.0) was added to the samples and run on a 4–16% gradient Native PAGE gel (Invitrogen) at 4°C. The gels were scanned at 532 nm using a Typhoon FLA 9500 laser-based scanner (GE).
SEC-MALS
E373A RIG-I (300 nM) was incubated with stem dsRNA (100 nM) for 30 min on ice. The complex was injected onto Superdex 200 Increase (GE Healthcare) Size Exclusion Chromatography (SEC) in the buffer of 50mM HEPES pH 7.5, 50mM NaCl, 1 mM MgCl2, 5mM DTT and 500μM ATP with a flow rate of 0.5 mL/min. The complex was characterized by SEC coupled with Multi-Angle Light Scattering (SEC-MALS). Sample eluted from the SEC passed through a miniDAWN TREOS detector (Wyatt Technology) and an Optilab T-rEX refractometer (Wyatt Technology). Light scattering intensity and eluent refractive index were recorded and analyzed by ASTRA v7.0.1.24 software to yield a weight-averaged molecular mass.
Measurement of RIG-I Translocation Rates
A panel of 27 bp dsRNAs carrying an internal Cy3 labeled nucleotide at 13th, 17th, 20th or 24th position from the 5’ppp, 5’OH or 3’ovg end was created to measure the RIG-I translocation rates on the dsRNA using a stopped-flow instrument (Auto-SF 120, Kintek Corp, Austin, Tx). A mixture of the internal Cy3 labelled dsRNA (30 nM for 5’ppp; 75 nM for 5’OH and 3’ovg dsRNA) and RIG-I (20 nM for 5’ppp; 50 nM for 5’OH and 3’ovg dsRNA) in Buffer A (pre-incubated at 25°C for 10 min) from syringe A was mixed with ATP (2 mM) and a 10-fold excess of RNA trap (unlabeled 5’ppp ds12 hairpin RNA) from syringe B. The fluorescence emission was measured using a 570 nm band-pass filter after excitation at 547 nm, as a function of time. Data were globally fitted using the Kintek Explorer software.
Kintek Explorer Global Data Fitting
The stopped-flow translocation kinetics were globally fit using the Kintek Explorer software to a minimal 5-step model. The dsRNA panel of 5’ppp, 5’OH and 3’ovg dsRNAs (Figure 5A) carried a Cy3 at the 13th, 17th, 20th and 24th position from the 5’ end. RIG-I prefers binding to dsRNA-end and has a footprint of ~9 bp on the dsRNA to form NR9. In our minimal model, the end-bound RIG-I (NR9) translocates to the opposite end of the 27 bp dsRNA in five steps: NR9→NR13→NR17→NR20→NR24→NR27 with rates k1-k5, respectively. For optimal fits, we assumed that a certain percentage of RIG-I was bound internally at the fluorophore position (NS13, NS17, NS20, and NS24) and these complexes dissociate at rates k6-k9. The best fits indicated that the NS complexes were ~10% of total population of RNA-bound complexes for all three dsRNAs. In 5’ppp dsRNA, a majority (5–10%) of the internally bound population was close to the 13th position, which could also be due to RNA-end bound RIG-I influencing Cy3 fluorescence at 13th position. In case of 3’ovg, the internally bound population (~10%) was distributed to all positions (13th-24th). The rates of each of the steps mentioned above for 5’ppp, 5’OH and 3’ovg dsRNA are provided in the Results section.
Kinetic Proofreading Model and Data Fitting
The 7-step kinetic proofreading model presented in Figure 7A is as follows - RIG-I (denoted as R) patrols the cytosol in an autoinhibited state and engages with RNA-ends (N) (5’ppp, 3’ovg, 5’ovg, etc.) via its C-terminal RD at diffusion-limited on-rates to form RD-mode complexes (N.RRD). If the RNA-end is non-PAMP, then RIG-I dissociates quickly, and if the RNA-end is a PAMP, then RIG-I forms a stable complex (step 1). The RD-mode (N.RRD) transitions into Helicase-mode (N.RH), and ATP binding facilitates this process (step 2). However, ATP hydrolysis drives RIG-I away from the RNA-end by translocation (N.Rt) (step 3). If RIG-I is bound to 5’ppp blunt-end, then translocation is throttled at the 5’ end, and the conformational changes associated with the numerous ATP binding and hydrolysis cycles transiently expose the 5’ppp end. This allows a second RIG-I molecule to engage with the 5’ppp end and make a dimer (N.RH.RRD) (step 4). The nascent RIG-I bound in RD-mode can fall off or transition to Helicase-mode (N.RH.RH) (step 5). This would require initially bound RIG-I to translocate into the interior stem region and provide room for the nascent molecule to fully engage with the dsRNA. This cycle of translocation-throttling and threading of RIG-I from the 5’ppp RNA-end can continue to assemble higher-order RIG-I oligomers on RNA. The N.RH.RH dimer is dynamic and translocates away from the RNA-end (step 6) to dissociate (step 7).
We tested this 7-step kinetic model (Figure 7A) by fitting the experimentally measured decrease in RIG-I dimerization with increasing 5’ppp dsRNA concentration (Figure S6D). We used the kinetic rate constants of RNA binding, translocation, and dissociation measured in this study. For steps 1 and 4, we used the on-rates and off-rates of the RD-mode complex (kon=6×108 M−1s−1, koff=0.15 s−1). We assumed that Helicase-mode monomer at the 5’ppp end translocates away from dsRNA-end (N.RH → N.Rt) with the experimentally measured RIG-I translocation rate k1 (steps 3 and 6, k1 = 0.028 s−1). We assumed that the dissociation of N.Rt to N+R occurs at the rate of translocation on stem RNA (average of k2-k5 for step 7, 1.2 s−1). Steps 2 and 5, where RD-mode transitions to the Helicase-mode are the only unknown rate constants (floating variables in modeling). Best fit indicated that the transition rate of the second RIG-I molecule (step 5) is faster than for the first RIG-I molecule (step 2), which is consistent with positive cooperativity in dimer formation. To minimize the number of steps in the model, we assumed that the transition is irreversible, which may not be the case.
QUANTIFICATION AND STATISTICAL ANALYSIS
The quantification of biotin-streptavidin displacement assays and EMSA gel blots was done using ImageQuant TL 8.1 software (GE Healthcare Life Sciences). The luciferase signals in cell-based signaling assays were measured using a luminometer (BioTek) and quantified in Microsoft Excel. The fluorescence intensity titrations were quantified and fitted using SigmaPlot v11.0 (Systat Software Inc.). Stopped-flow kinetic traces were fitted using the Kintek Stop-flow software and Sigmaplot v11.0. All the modelling was done using Kintek Explorer 6 software. The associated errors of the measurements and fitting and the number of sample sets (n) is noted in the corresponding figure legends.
DATA AND SOFTWARE AVAILABILITY
Raw gel scans can be accessed at - http://dx.doi.org/10.17632/7psjn7wrfp.1
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| ATP-γ-S, Lithium salt | Roche | Cat.# 11162306001 |
| ATP, [γ−32P] | Perkin Elmer | Cat.# NEG002A250U |
| Critical Commercial Assays | ||
| Dual-Luciferase Reporter Assay system | Promega | Cat.# E1980 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Agilent | Cat.# 200522 |
| Deposited Data | ||
| Raw Imaging Data | This Paper | http://dx.doi.org/10.17632/7psjn7wrfp.1 |
| Experimental Models: Cell Lines | ||
| HEK293T Cell Line | Dr. Marcotrigiano | Devarkar et al., PNAS 2016 |
| Oligonucleotides | ||
|
5′ppp ss27 RNA 5′-pppAUACGUCCUGAUAGUUAGUAUCCAUCG-3′ |
Biosynthesis | N/A |
|
5′OH ss27 RNA 5′-AUACGUCCUGAUAGUUAGUAUCCAUCG-3′ |
Dharmacon | N/A |
|
5′OH ss25 RNA (for 3′ovg) 5′-ACGUCCUGAUAGUUAGUAUCCAUCG-3′ |
Dharmacon | N/A |
|
complementary ss27 RNA 5′Bi-CGAUGGAUACUAACUAUCAGGACGUAU-3′ DY547 |
Dharmacon | N/A |
|
5′ppp ss27 methylated RNA 5′-pppAUACGUCCUGAUAGUmUmAmGmUmAUCCAUCG-3′ |
Biosynthesis | N/A |
|
complementary ss27 methylated RNA 5′Bi-CGAUGGAmUmAmCmUmAACUAUCAGGACGUAU-3′ DY547 |
Dharmacon | N/A |
|
ss26 RNA chi (for stem dsRNA) 5′-dAdTdAdCdGUCCUGAUAGUUAGUAUCdCdAdTdC-3′ |
Dharmacon | N/A |
|
complementary ss26 RNA chi intCy3 (for stem dsRNA) 5′-dCdGdAdTdGGAUACUAAC(Cy3)UAUCAGGAdCdGdTdA-3′ |
T riLink | N/A |
|
complementary ss30 RNA chi (intCy3 at 13/17/20/24 nt from 3′
end) (for stopped-flow translocation assays) 5′Bi-dGdCdTCGAUGGAUACUAACUAUCAGGACGUAU-3′ |
T riLink | N/A |
| Recombinant DNA | ||
| pET28b 6xHis-SUMO-RIG-I | Jiang, et al. 2011 | N/A |
| pET28b 6xHis-SUMO-HelRD | Jiang, et al. 2011 | N/A |
| pET28b 6xHis SUMO-T667/671E RIG-I | This paper | N/A |
| pET28b 6xHis-SUMO-Δ673–685 | Devarkar, et al. 2016 | N/A |
| pET28b 6xHis-SUMO-C268F | This paper | N/A |
| pET28b 6xH is-SUMO-E373A | This paper | N/A |
| pcDNA 3.1 wt RIG-I | Devarkar, et al. 2016 | N/A |
| pcDNA 3.1 empty vector | Devarkar, et al. 2016 | N/A |
| pcDNA 3.1 E373A RIG-I | This paper | N/A |
| pcDNA 3.1 C268F RIG-I | This paper | N/A |
| pRL-TK (Renilla luciferase plasmid) | Devarkar, et al. 2016 | N/A |
| pLuc125 (Firefly luciferase plasmid) | Devarkar, et al. 2016 | N/A |
| Software and Algorithms | ||
| Kintek Explorer Enzyme Kinetics Software | Kintek | https://www.kintekexplorer.com |
| Other | ||
| TLC PEI Cellulose F | Millipore | Cat.# 105579 |
Acknowledgments
We thank Dr. Mark Howarth (Department of Biochemistry, University of Oxford) for generously donating monovalent streptavidin. This work was supported by NIH grants GM111959 to S.S.P. and J.M., GM118086 to S.S.P., and the Intramural Research Program of NIAID and NIH to C.W. and J.M.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Bamming D, and Horvath CM (2009). Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem 284, 9700–9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Kolomeisky AB, and Igoshin OA (2017). Accuracy of Substrate Selection by Enzymes Is Controlled by Kinetic Discrimination. J Phys Chem Lett 8, 1552–1556. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, and Puglisi JD (2004). tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11, 1008–1014. [DOI] [PubMed] [Google Scholar]
- Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, Patel SS, and Marcotrigiano J (2016). Structural basis for m7G recognition and 2’-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A 113, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Wigley DB, and Webb MR (2000). Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry 39, 205–212. [DOI] [PubMed] [Google Scholar]
- Douglass EF Jr., Miller CJ, Sparer G, Shapiro H, and Spiegel DA (2013). A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc 135, 6092–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CJ, Wooten L, Tomko EJ, and Lohman TM (2010). Kinetics of motor protein translocation on single-stranded DNA. Methods Mol Biol 587, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ (1974). Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A 71, 4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. (2006). 5’-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Jang MA, Kim EK, Now H, Nguyen NT, Kim WJ, Yoo JY, Lee J, Jeong YM, Kim CH, Kim OH, et al. (2015). Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet 96, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr., Patel SS, and Marcotrigiano J (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, and Chen ZJ (2012). Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A, Luo D, Rawling DC, Ding SC, and Pyle AM (2013). Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep 14, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, and Cusack S (2011). Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435. [DOI] [PubMed] [Google Scholar]
- Lassig C, Matheisl S, Sparrer KM, de Oliveira Mann CC, Moldt M, Patel JR, Goldeck M, Hartmann G, Garcia-Sastre A, Hornung V, et al. (2015). ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louber J, Brunel J, Uchikawa E, Cusack S, and Gerlier D (2015). Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5. BMC Biol 13, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ranjith-Kumar CT, Hao L, Kao CC, and Li P (2011). Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5’ triphosphate. Nucleic Acids Res 39, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, and Pyle AM (2011). Structural insights into RNA recognition by RIG-I. Cell 147, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan TW (1995). Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A 92, 5042–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PD, Tackett AJ, and Raney KD (2001). Biotin-streptavidin-labeled oligonucleotides as probes of helicase mechanisms. Methods 23, 149–159. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, and Ha T (2009). Cytosolic viral sensor RIG-I is a 5’-triphosphate-dependent translocase on double-stranded RNA. Science 323, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistal-Villan E, Gack MU, Martinez-Delgado G, Maharaj NP, Inn KS, Yang H, Wang R, Aggarwal AK, Jung JU, and Garcia-Sastre A (2010). Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem 285, 20252–20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, and Hur S (2014). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, and Hur S (2013). RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell 51, 573–583. [DOI] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. (2010). Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol 11, 63–69. [DOI] [PubMed] [Google Scholar]
- Ramanathan A, Devarkar SC, Jiang F, Miller MT, Khan AG, Marcotrigiano J, and Patel SS (2016). The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection. Nucleic Acids Res 44, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawling DC, Fitzgerald ME, and Pyle AM (2015). Establishing the role of ATP for the function of the RIG-I innate immune sensor. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. (2010). RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140, 397–408. [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. (2009). Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, Goldeck M, Schmidt T, Schmid-Burgk JL, Kerber R, Wolter S, et al. (2015). A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N-2’O-Methylated Self RNA. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, and Gale M Jr. (2005). Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79, 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko EJ, Fischer CJ, and Lohman TM (2010). Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods 51, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, and Johnson KA (2006). A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. (2010). Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen J, Wicht O, Wolanski JC, Baur N, Bastian S, Haas DA, Matula P, Knapp B, Meyniel-Schicklin L, Wang C, et al. (2017). Phosphorylation-Dependent Feedback Inhibition of RIG-I by DAPK1 Identified by Kinome-wide siRNA Screening. Mol Cell 65, 403–415 e408. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, and Hur S (2013). Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, and Hur S (2014). Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell 55, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, and Fujita T (2007). Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem 282, 15315–15318. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, and Fujita T (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Zheng J, Yong HY, Panutdaporn N, Liu C, Tang K, and Luo D (2015). High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res 43, 1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw gel scans can be accessed at - http://dx.doi.org/10.17632/7psjn7wrfp.1