Abstract
To cluster anal microbiota and define microbial patterns associated with biological, clinical, and behavioral correlates among Nigerian men who have sex with men (MSM) living with or at risk for HIV. In this cross-sectional pilot study, the 15 most abundant 16S taxa in the anal microbiota of 113 MSM underwent unsupervised K-means clustering and z-score comparisons to define similarities and dissimilarities among 4 microbiota taxonomic profiles. Distributions of oncogenic HPV (high-risk human papillomavirus [HR-HPV]), concurrent HIV, antiretroviral therapy (ART), and other clinical and behavioral data were evaluated using Fisher's exact and Kruskal–Wallis tests to determine biological signatures of cluster membership. Prevotella was consistently represented in each cluster, but the average composition ranged from 14% to 44%. Cluster 2 was enriched with a member of the Fusobacteria phylum, Sneathia (29%). More participants of cluster 2 were HIV infected and taking ART (83%, 5/6), were virally suppressed (80%, 4/5), had HPV-16 (66.7%, 4/6), and reported no vaginal sex partners (83%, 5/6). HPV-35, a highly prevalent oncogenic HPV in Nigeria, was observed in all clusters except cluster 2 (0%, 0/6). Other covariates were similar across clusters (all p > .05). K-means unsupervised clustering, a canonical pattern recognition method, generalized the microbial community composition and structure while accounting for among sample variability. Further studies are needed to evaluate whether an anal microbial community enriched with members of the Fusobacteria phylum is associated with HIV-infected MSM who are virally suppressed and have a concurrent HPV-16.
Keywords: K-means clustering, MSM, Sneathia, Fusobacterium, HPV-16
Introduction
Commensal bacteria and their metabolites are important symbiotic partners in the host's immune response to local pathogenic viruses.1 Shifts in the microbial composition have been described for men who have sex with men (MSM) living with HIV as compared with HIV-uninfected MSM.2–4 During HIV infection as compared with healthy controls, there is a reduction in commensal genera, such as Faecalibacterium, Roseburia, Blautia, and Ruminococcus and an enrichment in other taxa, such as Prevotella, Fusobacteria, Anaerococcus, Porphyromonas, and Campylobacter in the lower gastrointestinal tract and rectum.2–7 Additional studies suggest that these compositional changes are influenced by anal sex, antiretroviral therapy (ART), and dietary patterns.2, 8–11 Whether these compositional changes are an indication of a less “healthy” microbiome and/or a loss of local immunity has not been well established.
One way to determine the downstream effect of an altered microbial composition is to evaluate its impact on the natural history of a local viral infection. Human papillomavirus (HPV) is a pervasive infection of the anogenital tract that undergoes states of clearance, regression, and/or recurrence depending on local immunity.12,13 It remains unclear whether an altered microbiota favors persistence or clearance of oncogenic HPV (high-risk human papillomavirus [HR-HPV]), and/or contributes to the higher burden of anal cancer, most attributed to HPV-16,14 among HIV-infected MSM on ART.
There has been limited study of the relationship between HIV-altered anal mucosa and the pathogenesis of HR-HPV. One small study among 42 HIV-infected MSM evaluated the association of bacterial taxa from biopsied samples with normal low-grade squamous intraepithelial lesions and high-grade squamous intraepithelial lesions (HSIL), finding that minority species including Peptostreptococcus, Campylobacter, and Gardnerella were associated with anal precancer. However, there was no consistency in the taxa across stages of precancer.15 Like most rectal microbial studies, Serrano-Villar et al. compared the enrichment or depletion of individual taxa.2,7–9,15,16
Individual taxa do not necessarily represent the ecologic niche that embodies cooperative and competitive interactions among multiple bacteria.17–19 Data from vaginal microbiome studies suggest that community state types, derived from unsupervised hierarchical clustering, characterize the microbial patterns and provide insight on local immunity.20,21 For HPV pathogenesis, a characteristic cluster or a type of microbial community would be expected to occur at all stages of anal dysplasia if part of the causal pathway.
The objective of this study was to define anal microbial patterns for MSM and their correlations with HR-HPV, HIV, and other biological and behavioral characteristics. These correlates were explored as indicators of the anal microbial communities much like Nugent score and pH are used as indicators of the vaginal microbiota.
Materials and Methods
Study design and population
Sampling and analytic approaches for this cross-sectional study of the anal mucosa microbiota among Nigerian MSM have been previously described.4,22 In brief, the prevalence of HR-HPV and the composition of the mucosal microbiota were characterized from a subset of archived baseline anal swab samples from the Abuja site of the parent cohort, TRUST/RV368 (n = 130). Selection criteria included completion of HIV testing at baseline, an available rectal swab sample, and among the first 165 enrolled participants. Data on demographics, sexual behaviors, prevalent and incident HIV, and sexually transmitted infections (STIs) were available from the parent cohort.23,24
All participants provided written informed consent. The study was approved by the Federal Capital Territory Health Research Ethics Committee in Nigeria and the University of Maryland Baltimore Institutional Review Board.
Laboratory procedures
In the parent cohort, whole blood was tested for HIV using rapid test kits (Abbott Determine HIV-1/2, Chembio HIV-1/2 Stat Pak, and Trinity biotech Uni-Gold HIV test for discordant results) as outlined by the parallel testing algorithm for high-risk individuals in Nigeria.25 HIV-uninfected individuals were tested for HIV at every quarterly visit. If a participant was HIV positive, HIV RNA was quantified using the COBAS TaqMan HIV-1 Test (Roche Molecular Diagnostics, Pleasanton, CA) and CD4 count was estimated using the Partec CyFlow Counter. Rectal Neisseria gonorrhoeae (NG) and Chlamydia trachomatis (CT) were detected using the Aptima Combo 2 CT/NG Assay (Hologic, San Diego, CA) from anal swabs that were inserted by a doctor ∼2″ into the anorectum and rotated for sample collection.
For our cross-sectional study, 37 high- and low-risk HPV genotypes were detected using the Linear Array HPV Genotyping Test (Roche Molecular Diagnostics, Indianapolis, IN). High-risk HPV included 13 type-specific infections: 16, 18, 31, 35, 39, 45, 51, 52, 56, 58, 59, and 68.22 HPV-52 was considered positive if there was no cross-reaction with HPV-33, HPV-35, or HPV-58. HPV-52 status in men with HPV-33, HPV-35, or HPV-58 is, therefore, unknown. The remaining 24 genotypes detected were considered low risk. Quality control samples were included during DNA extraction, polymerase chain reaction (PCR) amplification, and HPV genotype detection steps. Any sample missing human β-globin, an internal control target, was excluded from the analysis because of insufficient cellular material to detect HPV.
Anal microbiota data were generated by sequencing the V3 and V4 hypervariable regions of the 16S ribosomal RNA (rRNA) on the Illumina MiSeq platform. Negative and positive controls were included in both extraction and PCR steps. In addition, negative controls without a template were included for each barcoded primer pair. To maintain consistency, 50 ng of DNA and equimolar amounts of PCR amplicons were used for each sample. QIIME, USEARCH, and UCHIME were used for further sequence processing steps.26–28 Quality control of the sequence reads included review of the sequence tag (barcode) and 16S rRNA gene primer, a minimal length of 200 bp, and a 60% or more match to previously determined 16S rRNA. Sequences were clustered into operational taxonomic units (OTUs) with ≥97% similarity and classified to the taxonomic level of Genera using the RDP classifier in QIIME and the Greengenes database (gg13.8).29
Microbial community analyses
First, OTUs with at least 10 or more reads in >5% (n = 7) of samples were retained for clustering analysis, reducing the total OTUs from 213 to 67. Second, samples with at least 500 reads were included in the analysis, reducing the sample size from 130 to 113. Third, the relative abundance of the 15 most abundant genera4 was clustered into 4 characteristic signatures using K-means clustering. These genera included Prevotella, Faecalibacterium, Peptoniphilus, Roseburia, WAL_1855D, Finegoldia, Sneathia, Bacteroides, Anaerococcus, Porphyromonas, Ruminococcaceae-unclassified, Fusobacterium, Campylobacter, Blautia, and Ruminococcus.
K-means clustering randomly assigns K number of centroid means and then calculates the Euclidian distance between the individual data point and the closest centroid. Clusters begin to form, comprising the centroid and the closest data points. After initial clustering, a new centroid mean is calculated and the Euclidian distances for the individuals are recalculated to see whether they remain closest to the centroid mean or belong to another centroid. Individual data points may shift into a new cluster. This process of recalculating the centroid and the distances is repeated until all data points are clustered with the smallest distances to the K number of centroid means. K-means clustering analysis was performed using SPSS software (version 23; SPSS, Inc., Chicago, IL).
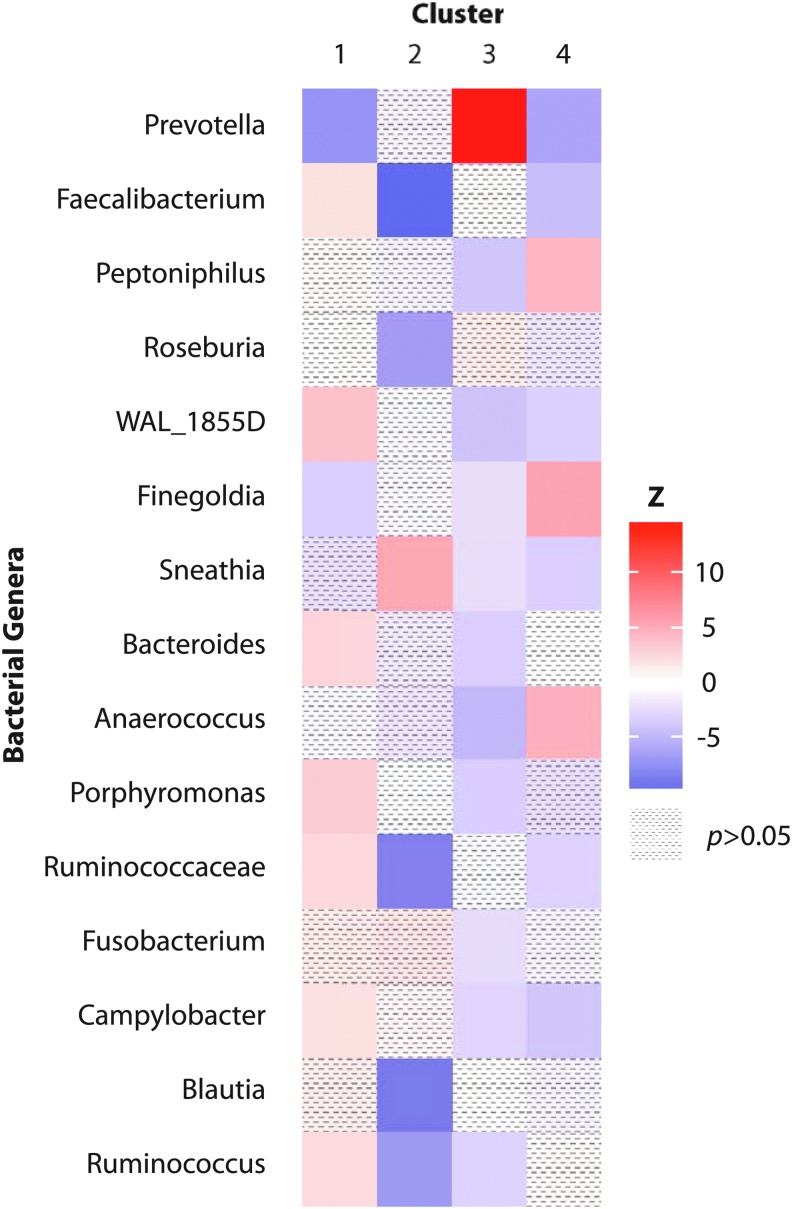
To evaluate the compositional differences across clusters, the centroid means of each genera were converted into z-scores. Z-scores were estimated as the difference between the centroid mean of the genera in the cluster as compared with the average of the centroids outside of that cluster, standardized by the joint standard error (meani − meant)/√(semi2 + semt2). A correlation matrix heat map was generated using RStudio (v. 1.0.153) with the ggplot2 package (v.2.1.0). Centroid means that were similar to one another were shaded in white. Centroid means in the clusters that were above the population average were shaded in red and those below the population average were shaded in blue. Z-scores more than two standard errors from the mean of zero were considered statistically significant (p < .05).
The primary exposure variable was microbial cluster. Concurrency was defined as having multiple sexual partnerships at the same time, further categorized as none, men only, or both men and women. Viral suppression was defined as those with <1,000 copies/mL of HIV RNA. Fisher's exact and Kruskal–Wallis tests were used to explore distributions of baseline demographics, clinical characteristics, sexual risk behavior, oncogenic HPV, prevalent and incident STIs, and antibiotic use by microbial cluster. Associations with p < .05 were considered statistically significant and those with p < .10 were suggestive of a trend. A bivariate analytic approach was used, as opposed to multivariable analysis, because of a limited sample size. Analyses were performed using Stata Statistical Software: Release 13 (StataCorp LP, College Station, TX).
Results
Cluster composition
Patterns of bacterial frequency from a total of 113 participants were distributed into 4 clusters based on the 15 most abundant genera in the population (Table 1). The composition of the cluster included the centroid mean of each taxon for all participants categorized within that cluster (Table 1). Prevotella was consistently represented in each cluster, ranging from 14% to 44%. Cluster 1 (n = 58) had 20% Prevotella, 7% Faecalibacterium, and 6% WAL_1855D. Cluster 2 (n = 6) had 24% Prevotella and 29% Sneathia. Cluster 3 (n = 38) had 44% Prevotella, 6% Faecalibacterium, and 5% Roseburia. Cluster 4 (n = 11) had 14% Prevotella, 9% Peptoniphilus, 22% Finegoldia, and 11% Anaerococcus. All remaining taxa within the clusters had centroid means <5%.
Table 1.
Centroid Means of 15 Most Abundant Genera After K-Means Clustering (n = 113)
| Phylum | Genera | Cluster 1 N = 58 | Cluster 2 N = 6 | Cluster 3 N = 38 | Cluster 4 N = 11 |
|---|---|---|---|---|---|
| Bacteroidetes | Prevotella | 20.0 | 24.0 | 44.3 | 13.8 |
| Firmicutes | Faecalibacterium | 7.1 | 0.9 | 6.4 | 3.4 |
| Firmicutes | Peptoniphilus | 4.2 | 3.0 | 2.3 | 8.5 |
| Firmicutes | Roseburia | 4.4 | 0.9 | 5.1 | 2.9 |
| Firmicutes | WAL_1855D | 5.5 | 2.7 | 1.3 | 1.4 |
| Firmicutes | Finegoldia | 3.0 | 4.0 | 3.5 | 21.6 |
| Fusobacteria | Sneathia | 1.2 | 28.9 | 0.8 | 0.2 |
| Bacteroidetes | Bacteroides | 4.0 | 1.4 | 1.4 | 2.8 |
| Firmicutes | Anaerococcus | 3.0 | 2.0 | 1.5 | 10.7 |
| Bacteroidetes | Porphyromonas | 3.0 | 1.8 | 0.9 | 0.9 |
| Firmicutes | Ruminococcaceae-unclassified | 2.5 | 0.6 | 2.0 | 1.2 |
| Fusobacteria | Fusobacterium | 2.7 | 4.3 | 1.1 | 1.5 |
| Proteobacteria | Campylobacter | 2.6 | 4.5 | 1.1 | 0.7 |
| Firmicutes | Blautia | 2.7 | 0.5 | 2.5 | 2.0 |
| Firmicutes | Ruminococcus | 3.0 | 0.4 | 1.5 | 3.1 |
When comparing the z-scores for each taxon within the clusters, cluster 1 had a significantly higher abundance of multiple genera than the other clusters (Faecalibacterium, WAL-1855D, Bacteroides, Porphyromonas, Ruminococcaceae-unclassified, Campylobacter, and Ruminococcus) and a lower abundance of Prevotella (Fig. 1). Cluster 2 had a high abundance of a single genus, Sneathia. Cluster 3 had one of the strongest differences in abundance, indicated by a z-score >10, of a single genus, Prevotella. Cluster 4 had a high abundance of a few genera (Peptoniphilus, Finegoldia, and Anaerococcus) and lower abundance of Prevotella.
FIG. 1.
Z-score distributions of 15 centroid means by cluster. Note: Z-scores represent the deviation in the centroid mean for each cluster-specific genera relative to the population centroid mean, standardized by the joint standard error. Genera with values between −2 and +2 were no different from the population average (p > .05). Centroid means in the clusters that were above the population average are shaded in red and those below the population average are shaded in blue.
Participant characteristics by microbiota cluster
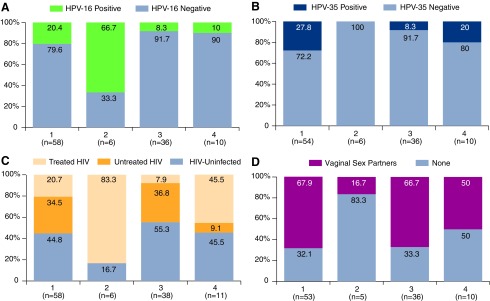
The majority of participants were in their 20s (65%), not married (84%), self-identified as bisexual (70%), and had their anal sexual debut within the past 9 years (57%) (Supplementary Table S1). Age, marital status, sexual orientation, years since first anal sex, number of receptive partnerships, condomless sex, and having concurrent partnerships were not significantly associated with clustering of anal microbiota (all p > .10) (Table 2 and Supplementary Table S2). Those with a cluster 2 microbiota trended toward self-reporting not having any vaginal sex partners (83% compared with 32%–50% in the other clusters) (Fig. 2).
Table 2.
Baseline Demographic, Behavioral, and Clinical Characteristics by Cluster Membership
| Cluster 1 N = 58 | Cluster 2 N = 6 | Cluster 3 N = 38 | Cluster 4 N = 11 | ||
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | p* | |
| Age (years) | .85 | ||||
| 16–19 | 9 (15.5) | 0 (0.0) | 8 (21.1) | 1 (9.1) | |
| 20–29 | 37 (63.8) | 5 (83.3) | 22 (57.9) | 9 (81.8) | |
| 30+ | 12 (20.7) | 1 (16.7) | 8 (21.1) | 1 (9.1) | |
| No. of receptive partnershipsa | .80 | ||||
| None | 12 (20.7) | 3 (50.0) | 8 (22.9) | 3 (27.3) | |
| 1–3 | 20 (34.5) | 1 (16.7) | 14 (40.0) | 4 (36.4) | |
| 4+ | 26 (44.8) | 2 (33.3) | 13 (37.1) | 4 (36.4) | |
| Condomless receptive sex | .77 | ||||
| No | 21 (45.7) | 2 (66.7) | 14 (48.3) | 3 (33.3) | |
| Yes | 25 (54.4) | 1 (33.3) | 15 (51.7) | 6 (66.7) | |
| Any vaginal sex partners | .07 | ||||
| No | 17 (32.1) | 5 (83.3) | 12 (33.3) | 5 (50.0) | |
| Yes | 36 (67.9) | 1 (16.7) | 24 (66.7) | 5 (50.0) | |
| HIV and ART status | <.01 | ||||
| HIV negative | 26 (44.8) | 1 (16.7) | 21 (55.3) | 5 (45.5) | |
| Untreated HIV positive | 20 (34.5) | 0 (0.0) | 14 (36.8) | 1 (9.1) | |
| Treated HIV positive | 12 (20.7) | 5 (83.3) | 3 (7.9) | 5 (45.5) | |
| HIV RNA viral load (copies/mL)b | <.01 | ||||
| <1,000 | 9 (28.1) | 4 (80.0) | 4 (23.5) | 5 (83.3) | |
| ≥1,000 | 23 (71.9) | 1 (20.0) | 13 (76.5) | 1 (16.7) | |
| High-risk HPV | |||||
| 16 | 11 (20.4) | 4 (66.7) | 3 (8.3) | 1 (10.0) | .01 |
| 18 | 11 (20.4) | 0 (0.0) | 7 (19.4) | 1 (10.0) | .79 |
| 31 | 4 (7.4) | 1 (16.7) | 2 (5.6) | 1 (10.0) | .58 |
| 33 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | — |
| 35 | 15 (27.8) | 0 (0.0) | 3 (8.3) | 2 (20.0) | .07 |
| 39 | 11 (20.4) | 1 (16.7) | 4 (11.1) | 1 (10.0) | .69 |
| 45 | 12 (22.2) | 1 (16.7) | 7 (19.4) | 1 (10.0) | .90 |
| 51 | 10 (18.5) | 0 (0.0) | 5 (13.9) | 2 (20.0) | .79 |
| 52 | 2 (3.7) | 0 (0.0) | 3 (8.3) | 2 (20.0) | .19 |
| 56 | 6 (11.1) | 0 (0.0) | 1 (2.8) | 0 (0.0) | .43 |
| 58 | 14 (25.9) | 2 (33.3) | 6 (16.7) | 1 (10.0) | .52 |
| 59 | 7 (13.0) | 0 (0.0) | 7 (19.4) | 0 (0.0) | .44 |
| 68 | 11 (20.4) | 0 (0.0) | 3 (8.3) | 2 (20.0) | .30 |
| Any | 36 (66.7) | 4 (66.7) | 20 (55.6) | 6 (60.0) | .75 |
| Prevalent rectal STIs | |||||
| NG | 14 (24.6) | 1 (16.7) | 15 (39.5) | 2 (18.2) | .35 |
| CT | 10 (17.2) | 0 (0.0) | 6 (15.8) | 1 (9.1) | .84 |
| Incident STIs | |||||
| HIVc | 4 (15.4) | 0 (0.0) | 3 (14.3) | 0 (0.0) | 1.00 |
| Rectal NGd | 14 (31.8) | 2 (50.0) | 8 (29.6) | 4 (50.0) | .64 |
| Rectal CTd | 10 (24.4) | 2 (50.0) | 5 (19.2) | 2 (28.6) | .56 |
| Antibiotics | |||||
| Prior treatmente | 10 (17.2) | 1 (16.7) | 7 (18.4) | 2 (18.2) | 1.00 |
| Current cotrimoxazole | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | — |
In past year.
Among HIV-infected only.
Among HIV-uninfected at baseline only.
New STI laboratory diagnosis after a negative diagnosis.
By healthcare provider in past year for symptomatic STIs.
Fisher's exact test, bolded are significant (p < .05).
ART, antiretroviral therapy; CT, Chlamydia trachomatis; NG, Neisseria gonorrheoae; HPV, human papillomavirus; STI, sexually transmitted infection.
FIG. 2.
Prevalence of HPV, ART, and HIV strata, and vaginal sex partners. Note: (A) Proportion with prevalent HPV-16. (B) Proportion with prevalent HPV-35. (C) Proportion with treated, untreated, and HIV uninfected. (D) Proportion self-reporting vaginal sex partners. ART, antiretroviral therapy; HPV, human papillomavirus.
Clinical characteristics by microbiota cluster
Approximately half of the participants were HIV uninfected (47%), a third had untreated HIV (31%), and one-fifth were taking ART for their HIV infections (22%) (Supplementary Table S1). HIV and ART status differed by clusters with cluster 2 having the highest proportion of MSM taking ART (83%) with viral suppression (80%). Cluster 4 had the second highest proportion of MSM taking ART (46%) with viral suppression (83%). The remaining clusters had higher proportions of MSM with untreated HIV (35%–37%) and less viral suppression (24%–28%) (Table 2 and Fig. 2).
Nearly two-thirds of the participants had at least one HR-HPV (62%) (Supplementary Table S1). Cluster 2 had the highest proportion of MSM with prevalent HPV-16 (67% as compared with 8%–20% in the other clusters) (Fig. 2). HPV-35 did not co-occur with HPV-16 but was prevalent in all the remaining clusters (0% as compared with 8%–28%) (Fig. 2). Having any type of HPV infection, multiple HPV infections, other high-risk HPVs or HPV genotypes associated with warts did not significantly differ by cluster membership (Supplementary Table S2 and Table 2).
Self-reported years since HIV diagnosis did not differ by cluster membership (Supplementary Table S2). Although only a portion of the HIV-infected participants entering the cohort study knew their status and could report how many years it had been since their first HIV-positive test [59% (19/32) in cluster 1, 100% (5/5) in cluster 2, 35% (6/17) in cluster 3, and 83% (5/6) in cluster 4]. For all those who knew their status from a healthcare provider before study entry (n = 35), 88% self-reported <5 years and 11% reported 6–8 years. CD4 T cell counts, prevalent rectal gonorrhea or chlamydia, antibiotics, incident HIV, incident rectal gonorrhea, or chlamydia did not differ by anal microbial cluster (Table 2 and Supplementary Table S2).
Discussion
This study describes the anal microbiota as clustered communities based on their taxonomic profile similarities and explores STIs and specific sexual risk practices as potential correlates among Nigerian MSM living with and without HIV. Microbial communities among MSM were enriched with Prevotella, consistent with prior studies,2,4,8,9 but interestingly had varying levels of abundance. Much of the prior work reported on differences in individual taxa. Our study evaluated whether these same taxa were shifted in our microbial communities to infer compositional similarities.
Cluster 3 was observed for a number of HIV-uninfected MSM and consistent with prior studies it comprised many commensal species (Faecalibacterium, Roseburia, and Blautia) and fewer minority taxa (Fusobacterium, Anaerococcus, Porphyromonas, and Campylobacter).5–7 Men who do not engage in receptive sex have a predominance of Bacteroides, rather than Prevotella, as was observed in cluster 1 where the highest proportion of MSM reported vaginal sex partners.2,8,9 Cluster 2 comprised HIV-infected MSM on ART who were virally suppressed and less likely to have vaginal sex partners. Interestingly, this cluster had a reduction in commensal taxa (Faecalibacterium, Roseburia, Blautia, and Ruminococcus) and a high abundance of minority taxa (a member of the Fusobacteria phyla, Sneathia).
Our exploratory analysis with oncogenic HPV suggested that a higher proportion of MSM with prevalent HPV-16 were associated with a cluster enriched with Sneathia. Sneathia is from the family of Fusobacteriaceae and was formerly grouped with Leptotrichia.30 Sneathia is not a highly abundant bacteria, but is becoming recognized as part of the genitourinary tract in both men and women and has been associated with a variety of clinical conditions such as bacterial vaginosis,31,32 urethritis,33 preterm labor,34 HIV acquisition,35 and cervical cancer.36,37 Leptotrichia amnionii, Sneathia amnii, and Sneathia sanguinegens are among the most studied Sneathia bacterial group members. Sneathia and members of the phylum, Fusobacteria, have virulence factors, such as adhesion molecule FadA, that enable them to colonize the mucosa, invade epithelial cells, and alter local immunity.30,38–40 Further species-level or even strain-level taxonomic resolution will further assist our understanding of its pathogenic attributes and potentials.
Data from studies of the vaginal microbiome suggest that Sneathia and other anaerobic bacteria may interact with the local epithelium in a way that allows oncogenic HPV to persist. In a study of HPV-discordant monozygotic twins, Fusobacteria and Sneathia species were more abundant in the HPV-positive twins than the HPV-negative twins.41 An anaerobic vaginal microbial community has been associated with persistent HPV infection,42 a critical risk factor for progression to HSIL, leading to cancer. In cross-sectional analyses among women, Sneathia has been associated with precancerous lesions,37,43 and Sneathia and Fusobacteria spp. have been associated with cervical cancer.36,37 Although all HR-HPVs are similar genetically, HPV-16 confers the highest carcinogenic risk and is the cause of >90% of anal cancers.14,44,45 To see HPV-16 emerge with a Sneathia-enriched cluster independent of the other HR-HPVs parallels earlier studies that there may be an interesting immunologic interaction. However, this observation arose from a very small sample size and should be interpreted with caution. Furthermore, the vaginal microbial community has a different function and influence of hormones, and thus may not be analogous to the anal microbial community.
Suppressive ART appeared to be associated with microbial clusters that had a higher expression of anaerobic microbiota not seen in clusters 1 and 3 where there was more untreated HIV or no HIV. Although our findings are limited by sample size, suppressive ART could potentially impact the composition of the microbial community. Prior studies have suggested that ART impacts the anal microbiota4,10,46–48 and is unable to restore the microbial community to a level comparable with HIV uninfected.6,7,16,49 Oral zidovudine was observed to have antibacterial effects but this study did not look at alterations in the microbial community.50 When tenofovir was applied locally as a rectal gel in a phase 1 randomized clinical trial, the Combination HIV Antiretroviral Rectal Microbicide (CHARM)-01 study, prevalence of five rectal bacteria declined.51 Although only a few species were impacted, the authors were restricted in the bacteria they could detect with culture assays as compared with 16S rRNA sequencing. Recent evidence suggests that bacteria from a community indicative of bacterial vaginosis degraded tenofovir from a microbicide gel applied in the vaginal tract.52 However, ART was delivered topically, and oral ART (pre-exposure prophylaxis) has not been shown to affect prevention of HIV for women with bacterial vaginosis.53 Further studies are needed to evaluate the role of oral ART on the microbial community and its impact on local immunity.
There were a few limitations to this study. The sample sizes for cluster 2 and cluster 4 were small and the associations may be spurious. However, the objective of this study was to assess the feasibility of reducing the dimensionality of the microbial data in a way that represented it most similarly to its structure in the data space. This approach uses all of the microbial data as compared with prior studies in men that tested differences in individual bacterial species. Furthermore, this study has access to an information-rich data set that enabled exploration of correlates of the anal microbial clusters. The anal mucosa is not dominated by lactic acid-producing Lactobacillus, limiting the applicability of Nugent score or pH to independently confirm the data structure of the clustering algorithms. The cross-sectional design of this study only provides a “snapshot” in time and does not allow analysis of any temporal changes. The preliminary associations are being investigated longitudinally with a larger sample size. In addition, relative abundance measures may vary depending on sampling techniques and sequencing depth. This bias was minimized by using the same amounts of template DNA and equimolar concentrations of PCR amplicons across samples. As bacteria may have functional redundancy, further studies on the genetic redundancy and/or metabolites would help elucidate whether clustering on the taxa level parallels functionality. Next, we allowed hierarchical clustering for the top 15 most dominant genera instead of all the genera to be consistent with our earlier work.4 The top 15 most abundant genera represented a median of 73% (interquartile range: 68%–78%) of all genera observed in the samples. Since the majority of the observed genera were included, potential bias in the formation of clusters was minimized. Lastly, the ART treatment difference by cluster membership may be confounded by duration of HIV infection, although for those who knew their status, their infection duration was within the past 5 years.
In summary, we have demonstrated the potential of clustering of microbial data as an analytic approach that offers a more comprehensive look at multiplexed microbial patterns. The results presented here suggest that a microbial cluster enriched with a member of the Fusobacteria Phylum, Sneathia, was associated with prevalent HPV-16 among MSM at risk of or living with HIV. Participants with an anal microbiota belonging to that cluster were more likely to be virally suppressed and have no vaginal sex partners. These findings should be validated in larger study populations to establish whether HPV-16 and other HR-HPVs are associated with certain anal microbial communities of ART-treated MSM.
Supplementary Material
Acknowledgments
The authors thank Dr. Patti Gravitt in her thoughtful review of the article, Mike Humphrys for his assistance in sequencing, the clients who willingly engaged in the TRUST/RV368 study regardless of the difficult contexts, and the ceaseless commitment of the study staff to their clients and the study team. The TRUST/RV368 Study Group includes Principal Investigators: Manhattan Charurat (IHV, University of Maryland, Baltimore, MD), Julie Ake (MHRP, Walter Reed Army Institute of Research, Silver Spring, MD); Coinvestigators: Sylvia Adebajo, Stefan Baral, Erik Billings, Trevor Crowell, George Eluwa, Abiola Fasina, Charlotte Gaydos, Sosthenes Ketende, Afoke Kokogho, Hongjie Liu, Jennifer Malia, Olumide Makanjuola, Nelson Michael, Nicaise Ndembi, Jean Njab, Rebecca Nowak, Oluwasolape Olawore, Zahra Parker, Sheila Peel, Habib Ramadhani, Merlin Robb, Cristina Rodriguez-Hart, Eric Sanders-Buell, Sodsai Tovanabutra; Institutions: Institute of Human Virology at the University of Maryland School of Medicine (IHV-UMB), University of Maryland School of Public Health (UMD SPH), Johns Hopkins Bloomberg School of Public Health (JHSPH), Johns Hopkins University School of Medicine (JHUSOM), U.S. Military HIV Research Program (MHRP), Walter Reed Army Institute of Research (WRAIR), Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF), Henry M. Jackson Foundation Medical Research International (HJFMRI), Institute of Human Virology Nigeria (IHVN), International Centre for Advocacy for the Right to Health (ICARH), the Initiative for Equal Rights (TIERS), Population Council (Pop Council) Nigeria.
The research reported in this publication was supported by seed funding from MPower and salary support from UMGCC P30 grant (P30 CA134274-04) from the National Cancer Institute. Additional support was provided by the National Institutes of Health (R01 MH099001, R01 AI120913, R01 MH110358); the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (W81XWH-11-2-0174); Fogarty Epidemiology Research Training for Public Health Impact in Nigeria program (D43TW010051); and the President's Emergency Plan for AIDS Relief through a cooperative agreement between the Department of Health and Human Services/Centers for Disease Control and Prevention, Global AIDS Program, and the Institute for Human Virology-Nigeria (NU2GGH002099).
Contributor Information
Collaborators: on behalf of the TRUST/RV368 Study Group, Manhattan Charurat, Julie Ake, Sylvia Adebajo, Stefan Baral, Erik Billings, Trevor Crowell, George Eluwa, Abiola Fasina, Charlotte Gaydos, Sosthenes Ketende, Afoke Kokogho, Hongjie Liu, Jennifer Malia, Olumide Makanjuola, Nelson Michael, Nicaise Ndembi, Jean Njab, Oluwasolape Olawore, Zahra Parker, Sheila Peel, Habib Ramadhani, Merlin Robb, Cristina Rodriguez-Hart, Eric Sanders-Buell, and Sodsai Tovanabutra
Authors' Contributions
W.A.B. and M.E.C. designed the TRUST study. R.G.N., S.M.B., and M.E.C. conceived the analysis for the article. Data collection and management were facilitated by R.G.N., W.D., J.R., and B.M. R.G.N. conducted the data analysis with input from S.M.B., J.R., B.M., and H.L. R.G.N. drafted the article and S.M.B., J.R., T.A.C., W.D., B.M., H.L., W.A.B., S.D.B., and M.E.C. provided critical review and editing. All authors have seen and approved the article; the corresponding author had full access to the data and had final responsibility for the decision to submit for publication.
Disclaimer
The views expressed are those of the authors and should not be construed to represent the positions of the National Institutes of Health, U.S. Army, the Department of Defense, the Department of Health and Human Services, or other funders. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70.
Author Disclosure Statement
T.A.C. has received a speaker fee from Gilead Sciences. The other authors declare no relevant conflicts of interest.
Supplementary Material
References
- 1. Shi Z, Gewirtz AT: Together forever: Bacterial–viral interactions in infection and immunity. Viruses 2018;10:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Noguera-Julian M, Rocafort M, Guillén Y, et al. : Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016;5:135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ: Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS 2014;28:753–760 [DOI] [PubMed] [Google Scholar]
- 4. Nowak RG, Bentzen SM, Ravel J, et al. : Rectal microbiota among HIV-uninfected, untreated HIV, and treated HIV-infected in Nigeria. AIDS 2017;31:857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zevin AS, McKinnon L, Burgener A, Klatt NR: Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 2016;11:182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mutlu EA, Keshavarzian A, Losurdo J, et al. : A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014;10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McHardy IH, Li X, Tong M, et al. : HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013;1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelley CF, Kraft CS, de Man TJ, et al. : The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: Implications for HIV transmission and prevention. Mucosal Immunol 2017;10:996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pescatore NA, Pollak R, Kraft CS, Mulle JG, Kelley CF: Short communication: Anatomic site of sampling and the rectal mucosal microbiota in HIV negative men who have sex with men engaging in condomless receptive anal intercourse. AIDS Res Hum Retroviruses 2018;34:277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nowak P, Troseid M, Avershina E, et al. : Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 2015;29:2409–2418 [DOI] [PubMed] [Google Scholar]
- 11. Wu GD, Bushmanc FD, Lewis JD: Diet, the human gut microbiota, and IBD. Anaerobe 2013;24:117–120 [DOI] [PubMed] [Google Scholar]
- 12. Stanley M: HPV—Immune response to infection and vaccination. Infect Agents Cancer 2010;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gravitt PE: The known unknowns of HPV natural history. J Clin Invest 2011;121:4593–4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin C, Franceschi S, Clifford GM: Human papillomavirus types from infection to cancer in the anus, according to sex and HIV status: A systematic review and meta-analysis. Infect Dis 2018;18:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Serrano-Villar S, Vásquez-Domínguez E, Pérez-Molina JA, et al. : HIV, HPV, and microbiota: Partners in crime? AIDS 2017;31:591–594 [DOI] [PubMed] [Google Scholar]
- 16. Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE: HIV-induced alteration in gut microbiota: Driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014;5:562–570 [DOI] [PubMed] [Google Scholar]
- 17. Faust K, Sathirapongsasuti JF, Izard J, et al. : Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 2012;8:e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levy R, Carr R, Kreimer A, Freilich S, Borenstein E: NetCooperate: A network-based tool for inferring host–microbe and microbe-microbe cooperation. BMC Bioinformatics 2015;16:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morton JT, Sanders J, Quinn RA, et al. : Balance trees reveal microbial niche differentiation. mSystems 2017;2:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ravel J, Gajer P, Abdo Z, et al. : Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gajer P, Brotman RM, Bai G, et al. : Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012;4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nowak RG, Gravitt PE, He X, et al. : Prevalence of anal high-risk human papillomavirus infections among HIV-positive and HIV-negative men who have sex with men in Nigeria. Sex Transm Dis 2016;43:243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Charurat ME, Emmanuel B, Akolo C, et al. : Uptake of treatment as prevention for HIV and continuum of care among HIV-positive men who have sex with men in Nigeria. J Acquir Immune Defic Syndr 2015;68(Suppl 2):S114–S123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baral SD, Ketende S, Schwartz S, et al. : Evaluating respondent-driven sampling as an implementation tool for universal coverage of antiretroviral studies among men who have sex with men living with HIV. J Acquir Immune Defic Syndr 2015;68(Suppl 2):S107–S113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Federal Ministry of Health (FMOH): National Guidelines for HIV Prevention Treatment and Care. Federal ministry of Health Nigeria, 2016, pp. 19 Available at: http://apps.who.int/medicinedocs/documents/s23252en/s23252en.pdf [Google Scholar]
- 26. Edgar RC: Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 27. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R: UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Q, Garrity GM, Tiedje JM, Cole JR: Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDonald D, Price MN, Goodrich J, et al. : An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harwich MD, Jr., Serrano MG, Fettweis JM, et al. : Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics 2012;13(Suppl 8):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fredricks DN: Molecular methods to describe the spectrum and dynamics of the vaginal microbiota. Anaerobe 2011;17:191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lennard K, Dabee S, Barnabas SL, et al. : Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect immun 2017;86:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manhart LE, Khosropour CM, Liu C, et al. : Bacterial vaginosis-associated bacteria in men: Association of Leptotrichia/Sneathia spp with nongonococcal urethritis. Sex Transm Dis 2013;40:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS: Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol 2009;47:38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gosmann C, Anahtar MN, Handley SA, et al. : Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young south african women. Immunity 2017;46:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nawrot R, Kamieniarz K, Malinowska M, et al. : The prevalence of Leptotrichia amnionii in cervical swabs of HPV positive and negative women. Eur J Gynaecol Oncol 2010;31:425–428 [PubMed] [Google Scholar]
- 37. Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, et al. : Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PloS One 2016;11:e0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen-Vercoe E, Jobin C: Fusobacterium and enterobacteriaceae: Important players for CRC? Immunol Lett 2014;162:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Han YW, Shi W, Huang GT, et al. : Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 2000;68:3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jewett A, Hume WR, Le H, et al. : Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun 2000;68:1893–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee JE, Lee S, Lee H, et al. : Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PloS One 2013;8:e63514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brotman RM, Shardell MD, Gajer P, et al. : Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis 2014;210:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitra A, MacIntyre DA, Lee YS, et al. : Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 2015;5:16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor S, Bunge E, Bakker M, Castellsagué X: The incidence, clearance and persistence of non-cervical human papillomavirus infections: A systematic review of the literature. BMC Infect Dis 2016;16:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burk RD, Harari A, Chen Z: Human papillomavirus genome variants. Virology 2013;445:232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goedert JJ: The microbiota and HIV: Shedding light on dark matters. AIDS 2017;31:863–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jackson MA, Goodrich JK, Maxan ME, et al. : Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016;65:749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klase Z, Ortiz A, Deleage C, et al. : Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol 2015;8:1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. : Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013;5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doléans-Jordheim A, Bergeron E, Bereyziat F, et al. : Zidovudine (AZT) has a bactericidal effect on enterobacteria and induces genetic modifications in resistant strains. Eur J Clin Microbiol Infect Dis 2011;30:1249–1256 [DOI] [PubMed] [Google Scholar]
- 51. Mcgowan I, Cranston RD, Duffill K, et al. : A phase 1 randomized, open label, rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of three formulations of tenofovir 1% gel (the CHARM-01 study). PloS One 2015;10:e0125363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klatt NR, Cheu R, Birse K, et al. : Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 2017;356:938–945 [DOI] [PubMed] [Google Scholar]
- 53. Heffron R, McClelland RS, Balkus JE, et al. : Efficacy of oral pre-exposure prophylaxis (PrEP) for HIV among women with abnormal vaginal microbiota: A post-hoc analysis of the randomised, placebo-controlled partners PrEP study. Lancet 2017;4:e449–e456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.