Abstract
It remains unclear whether differences in gut microbiota noted between HIV-infected and uninfected individuals are driven by HIV or sexual behavior. We evaluated rectal swab microbiota of HIV-infected and uninfected women with similar demographic, neighborhood, and diet characteristics enrolled in the Chicago Women's Interagency HIV Study (WIHS). DNA was amplified for sequencing of fragments of bacterial small subunit (SSU or 16S) ribosomal RNA (rRNA) genes. HIV-infected and uninfected women did not differ by Shannon diversity index (p = .14), non-metric multidimensional scaling (NMDS) plot of Bray–Curtis indices (p = .488, r = 0.0027), or copy number of individual taxa. Both groups demonstrated marked microbiome stability over time (p = .889).
Keywords: HIV, microbiome, women, mucosal immunology
Background
Individuals with controlled HIV have excess morbidity and mortality that appear to be driven, in part, by immune activation related to gut microbial translocation.1,2 A shift in the gut microbiome or “dysbiosis” has been reported in HIV as well as other systemic inflammatory disorders such as metabolic syndrome, inflammatory bowel disease, and cardiovascular disease.3 Several researchers have found correlations between specific “dysbiotic” microbes and gut barrier function in HIV-infected individuals.4,5 This difference appeared to persist even when those with suppressed HIV were compared with HIV negative controls.6 These studies, however, included mostly male HIV-infected subjects. Recent publications suggest that differences in microbiota between HIV-infected and uninfected individuals are specific to men who have sex with men (MSM) rather than HIV infection.7 Noguera-Julian et al. demonstrated that the Prevotella-predominant enterotype is associated with MSM rather than HIV status. Kelley et al. also showed a shift toward Prevotellaceae in HIV-uninfected MSMs who reported condomless receptive anal intercourse.8 Currently there is a paucity of published data comparing the gut and/or rectal microbiome of HIV-infected and sociodemographically similar uninfected women, particularly from the same geographic locale. Diet, environmental, behavioral, and mental health factors also appear to be determinants of gut microbiome composition, making it difficult to tease out HIV-driven changes.9
Data from simian immunodeficiency virus (SIV)-infected nonhuman primates in controlled settings with standardized environment and diet have failed to show persistent changes in the gut microbiome associated with SIV infection.10 Studies of chimpanzees in their natural habitat, however, have suggested that SIV infection leads to greater instability of the gut microbiome over time.11 Gut mucosal CD4 cells often do not recover after antiretroviral therapy and this may lead to inability to select beneficial microbes and eliminate harmful ones.12 The makeup of the gut microbiome of HIV-infected individuals, particularly those with low CD4 may thus be more strongly influenced by the environment and less stable over time. We sought to determine whether the rectal microbiota composition and stability of HIV-infected women was similar to uninfected women.
Methods
Study design and setting
This was a nested study of rectal microbiota within the Women's Interagency HIV Study (WIHS). All women from the Cook County site of the Chicago WIHS were approached for recruitment during visit 41 (October 2014 to March 2015) and again 1 year later at visit 43. The WIHS is an ongoing observational study of HIV-infected and demographically similar uninfected women enrolled during one of three recruitment waves in 1994–1995, 2001–2002, and 2011–2013. Recruitment, retention, study procedures, and cohort characteristics have been previously described.13 In brief, WIHS women are evaluated semiannually with an in-depth interview to collect sociodemographic, behavioral, and clinical data and undergo phlebotomy and a targeted physical and gynecologic examination with specimen collection. This study included women who were undergoing a routine WIHS study visit with genital examination during visit 41 who agreed to participate. The Cook County Health and Hospitals System's Institutional Review Board reviewed and approved all study procedures. All participants provided informed consent.
Rectal swab sample collection
Clinicians collected rectal swabs (FLOQSwabs™ 552C, Copan, CA) during WIHS anogenital examinations of all participants who had a visit and consented 191/202 (95%), including 136/141 (96%) HIV-infected and 55/61 (90%) uninfected women. Ninety-seven HIV-infected and 42 uninfected women provided rectal swabs at both visits 41 and 43, which were 1 year apart. A FLOQSwabs moistened in sterile saline was inserted into the anal canal beyond the anal verge (+3 cm), rotated 360°, and then withdrawn. The FLOQSwabs was transferred to a 1.8 mL vial containing PowerBeads and 750 μL of PowerSoil buffer from the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA). The swabs were snap frozen in liquid nitrogen and immediately placed in a −80°C freezer. DNA isolation was subsequently performed using the PowerSoil DNA Isolation Kit. Rectal swabs were lysed using the proprietary kit lysis buffer in conjunction with mechanical lysis by vortexing the PowerBead tubes to release the microbial DNA. On completion of lysis step, the tubes were centrifuged and the supernatant was removed for further purification. Impurities and inhibitor proteins were precipitated out through various wash steps and the DNA-containing supernatant was loaded onto a spin filter for further purification. Once the purification steps were complete the filter-bound DNA was eluted off the filter and stored at −80°C until the samples were sent for genomic testing.
Basic microbiota sequence processing
Forward and reverse reads were merged using paired read end merger (PEAR).14 Ambiguous nucleotides were trimmed from the ends and reads with internal ambiguous nucleotides were discarded. Primer sequences were identified using Smith–Waterman alignment and trimmed from the sequence. Reads that lacked either primer sequence were discarded. Sequences were then trimmed based on quality scores using a modified Mott algorithm with Phred quality threshold of p = .01. After trimming any sequences <325 bp were discarded. Chimeric sequences were identified using the USEARCH algorithm with the GreenGenes 13_8 reference sequences.15 QIIME was then used to generate the OTU (operational taxonomic unit) table and taxonomic summaries.16 In brief, the resulting sequence files were then merged with sample information. OTU clusters were generated in a de novo manner using the UCLUST algorithm with a 97% similarity threshold. Taxonomic annotations for each OTU were determined using the UCLUST algorithm and GreenGenes 13_8 reference with a minimum similarity threshold of 90%.16 Taxonomic and OTU abundance data were merged into a single OTU table and summaries then rarefied to a depth of 5,600 counts per sample.16 The rarefied table was then used to generate taxonomic summaries that were used for subsequent analyses.
Demographics and other measures
Demographics, medical, and behavioral characteristics were obtained at study visit, including HIV status, age, race/ethnicity, income, education, body mass index, alcohol use, and CD4+ T-lymphocyte count and nadir, HIV RNA level, highly active antiretroviral therapy use and adherence, and type of antiretroviral treatment for HIV seropositive women.
Block 2014 food frequency questionnaires (FFQ) were collected at one of two yearly visits. The FFQ was done separately from the visits where rectal swabs were collected but within 12 months of the rectal swab visit. Data collected from the FFQ included the frequency and type of foods eaten, which were transformed into estimated amounts (in grams) of whole grains, dietary protein, carbohydrate, fat, fiber, saturated fat, monounsaturated fat, and polyunsaturated fat.17
Statistical analyses
Chi-square statistics and independent sample t-tests were used to assess the association between HIV serostatus and demographic, medical, and behavioral characteristics and food quality questions. Shannon and Bray–Curtis indices were calculated with default parameters in R using the vegan library.18 The rarefied genus data, taxonomic level 6, were used to calculate both indices. For Bray–Curtis indices, the rarefied genus data were filtered to remove any taxon with an abundance of <1% of the total abundance in the data set. Plots were generated in R using the ggplot2 library.19 Significant difference among tested groups was determined using the Kruskal–Wallis one-way analysis of variance using the group_significance.py script within the QIIME v1.8 package. Before group significance testing, all taxonomic summaries were filtered to remove any taxon with an abundance of <1% of the total abundance in the data set.
Correlations were tested between HIV serostatus, HIV treatment status (prescribed or not prescribed antiretrovirals), viral load, CD4 stratum, and dietary intake with all taxonomic units using Kendall Tau test of correlation in R. Before correlation testing, all taxonomic summaries were filtered to remove any taxon with an abundance of <1% of the total abundance in the data set.
Results
Of 202 women enrolled in the Cook County WIHS site who were approached for inclusion, 95% (191: 136 HIV-infected and 55 HIV uninfected) women agreed to enroll and were included in analyses for visit 41, 70 HIV-infected and 30 HIV-uninfected women provided samples on both visit 41 and 43. Demographics were similar between HIV-infected and uninfected women, although the proportion of HIV-uninfected women who were African American was slightly higher (Table 1). HIV-infected and uninfected women reported similar consumption of all food/nutrient groups except for whole grains, which HIV-uninfected women reported eating more frequently (p = .01) (Table 2). Of the HIV-infected women, 84% were prescribed antiretroviral treatment and 71% had a current undetectable HIV viral load at the time of rectal swab collection.
Table 1.
Demographic and Medical Characteristics of Women with Microbiome Data (n = 191)
| HIV+, N = 136 | HIV−, N = 55 | pa | |
|---|---|---|---|
| Age at interview (years), mean (SD) | 49.7 (8.0) | 49.9 (9.4) | .92b |
| Ethnicity, N (%) | |||
| Non-Hispanic African American | 102 (75.0) | 48 (87.3) | .05 |
| Non-Hispanic white | 17 (12.5) | 0 (0) | |
| Hispanic African American | 1 (0.7) | 1 (1.8) | |
| Hispanic white | 7 (5.2) | 5 (9.1) | |
| Hispanic other | 7 (5.2) | 1 (1.8) | |
| Other | 2 (1.5) | 0 (0) | |
| Average household income, N (%) | |||
| ≤$18,000 | 93 (68.4) | 39 (70.9) | .73 |
| $18,001+ | 43 (31.6) | 16 (29.1) | |
| Education level, N (%) | |||
| Less than high school | 55 (40.4) | 15 (27.3) | .09 |
| Completed high school or more | 81 (59.6) | 40 (72.7) | |
| Body mass index category, N (%) | |||
| <18.5 | 6 (4.4) | 1 (1.8) | .12 |
| 18.5–24.9 | 43 (31.6) | 9 (16.4) | |
| 25–29.9 | 37 (27.2) | 19 (34.6) | |
| 30+ | 50 (36.8) | 26 (47.3) | |
| Alcohol use, N (%) | |||
| Abstainer | 76 (56.7) | 24 (43.6) | .4 |
| 1–7 Drinks/week | 42 (31.3) | 22 (40.0) | |
| 8–12 Drinks/week | 4 (3.0) | 3 (5.5) | |
| >12 Drinks/week | 12 (9.0) | 6 (10.9) | |
P-value obtained by using the chi-square test unless otherwise specified.
P-value obtained by using the t-test for means.
SD, standard deviation.
Table 2.
Reported Dietary Intake
| Overall, N (%) | HIV+ (n = 113) | HIV− (n = 53) | p | ||
|---|---|---|---|---|---|
| Whole grains | |||||
| ≥3 Servings/day | 1 | 13 (7.8) | 9 (8.0) | 4 (7.6) | .01 |
| 1–2 Servings/day | 0.5 | 35 (21.1) | 16 (14.1) | 19 (35.8) | |
| <1 Serving/day | 0 | 118 (71.1) | 88 (77.9) | 30 (56.6) | |
| Green leafy vegetables | |||||
| ≥6 Servings/week | 1 | 34 (20.5) | 23 (20.4) | 11 (20.8) | .57 |
| 3–5 Servings/week | 0.5 | 48 (28.9) | 30 (26.5) | 18 (33.9) | |
| ≤2 Servings/week | 0 | 84 (50.6) | 60 (53.1) | 24 (45.3) | |
| Other vegetables | |||||
| ≥1 Serving/day | 1 | 80 (48.2) | 58 (51.3) | 22 (41.5) | .33 |
| 5–6 Servings/week | 0.5 | 23 (13.9) | 13 (11.5) | 10 (18.9) | |
| <5 Servings/week | 0 | 63 (37.9) | 42 (37.2) | 21 (39.6) | |
| Berries | |||||
| ≥2 Servings/week | 1 | 31 (18.7) | 21 (18.6) | 10 (18.9) | .88 |
| 1 Serving/week | 0.5 | 19 (11.4) | 12 (10.6) | 7 (13.2) | |
| <1 Serving/week | 0 | 116 (69.9) | 80 (70.8) | 36 (67.9) | |
| Fish | |||||
| ≥1 Serving/week | 1 | 58 (35.0) | 40 (35.4) | 18 (34.0) | .73 |
| 1–3 Servings/month | 0.5 | 56 (33.7) | 36 (31.9) | 20 (37.7) | |
| Rarely (<1 serving/month) | 0 | 52 (31.3) | 37 (32.7) | 15 (28.3) | |
| Poultry | |||||
| ≥2 Servings/week | 1 | 43 (25.9) | 26 (23.0) | 17 (32.1) | .12 |
| 1 Serving/week | 0.5 | 41 (24.7) | 25 (22.1) | 16 (30.2) | |
| <1 Serving/week | 0 | 82 (49.4) | 62 (54.9) | 20 (37.7) | |
| Beans | |||||
| >3 Servings/week | 1 | 31 (18.7) | 23 (20.4) | 8 (15.1) | .53 |
| 1–3 Servings/week | 0.5 | 48 (28.9) | 34 (30.1) | 14 (26.4) | |
| <1 Serving/week | 0 | 87 (52.4) | 56 (49.6) | 31 (58.5) | |
| Nuts | |||||
| ≥5 Servings/week | 1 | 48 (28.9) | 33 (29.2) | 15 (28.3) | .99 |
| 1 Serving/month to <5 servings/week | 0.5 | 105 (63.3) | 71 (62.8) | 34 (64.2) | |
| <1 Serving/month | 0 | 13 (7.8) | 9 (8.0) | 4 (7.5) | |
| Olive oil | |||||
| Primary oil used | 1 | 39 (23.5) | 25 (22.1) | 14 (26.4) | .54 |
| Not primary oil | 0 | 127 (76.5) | 88 (77.9) | 39 (73.6) | |
| Butter, margarine | |||||
| <1 Serving/day | 1 | 96 (57.8) | 64 (56.6) | 32 (60.4) | .21 |
| 1–2 Servings/day | 0.5 | 39 (23.5) | 15 (28.3) | 24 (21.2) | |
| >2 Servings/day | 0 | 31 (18.7) | 25 (22.1) | 6 (11.3) | |
| Red meats and products | |||||
| <4 Servings/week | 1 | 68 (41.0) | 42 (37.2) | 26 (49.1) | .35 |
| 4–6 Servings/week | 0.5 | 41 (24.7) | 30 (26.5) | 11 (20.7) | |
| ≥7 Servings/week | 0 | 57 (34.3) | 41 (36.3) | 16 (30.2) | |
| Fast/fried foods | |||||
| <1 Serving/week | 1 | 70 (42.2) | 46 (40.7) | 24 (45.3) | .84 |
| 1–3 Servings/week | 0.5 | 58 (34.9) | 40 (35.4) | 18 (34.0) | |
| ≥4 Servings/week | 0 | 38 (22.9) | 27 (23.9) | 11 (20.7) | |
| Cheese | |||||
| <1 Serving/week | 1 | 59 (35.5) | 39 (34.5) | 20 (37.7) | .76 |
| 1–6 Servings/week | 0.5 | 70 (42.2) | 47 (41.6) | 23 (43.4) | |
| ≥7 Servings/week | 0 | 37 (22.3) | 27 (23.9) | 10 (18.9) | |
| Pastries, sweets | |||||
| <5 Servings/week | 1 | 44 (26.5) | 28 (24.8) | 16 (30.2) | .22 |
| 5–6 Servings/week | 0.5 | 11 (6.6) | 10 (8.8) | 1 (1.9) | |
| ≥7 Servings/week | 0 | 111 (66.9) | 75 (66.4) | 36 (67.9) | |
| Alcohol/wine | |||||
| 1 Serving/day | 1 | 2 (1.2) | 1 (0.9) | 1 (1.9) | .81 |
| 1 Serving/month to 6 servings/week | 0.5 | 23 (13.9) | 15 (13.3) | 8 (15.1) | |
| >1 Serving/day or never | 0 | 141 (84.9) | 97 (85.8) | 44 (83.0) | |
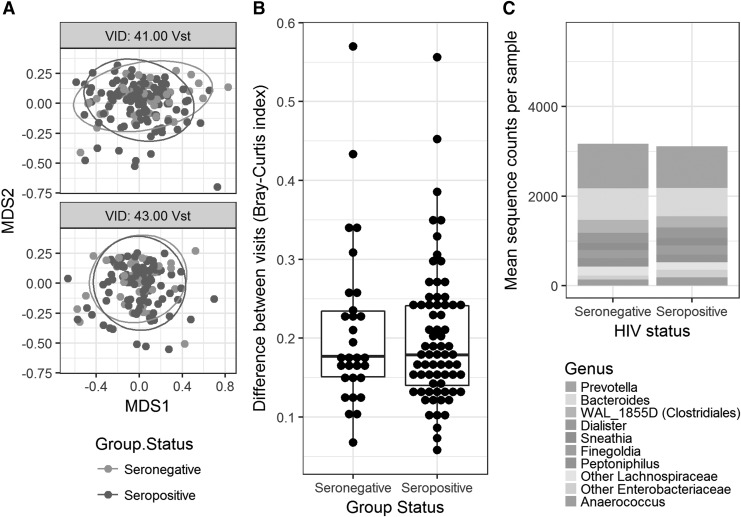
Shannon diversity index (Mann–Whitney, p = .14) and Bray–Curtis dissimilarity indices (analysis of similarity, R = 0.0027, p = .488) did not differ by HIV status (Fig. 1A). Bray–Curtis analysis of samples from the same individuals collected 1 year apart demonstrated similar rectal microbiome over time in both HIV-infected and HIV-uninfected women (Mann–Whitney, p = .889) (Fig. 1B). There were no statistically significant taxa that differed by HIV status (HIV infected vs HIV uninfected) (Fig. 1C). After correction for false discovery rate (FDR), HIV treatment status did not appear to influence overall microbial community structure but did impact the prevalence of several genera. Staphylococcus (logFC = 6.66, q = 5xE−10) and Corynebacterium (logFC = 3.69, q = 1.67xE−9) were statistically significantly enriched and Lactobacillus (logFC = −4.75, q = 7.62xE−3) were depleted in those (n = 22) with untreated HIV infection compared with HIV-uninfected women. Staphylococcus (logFC = 6.31, q = 8.51E−15) and Corynebacterium (logFC = 4.45, q = 2.48E−19) were also enriched in untreated HIV-infected versus treated HIV-infected. Prevotella were not enriched in untreated women compared with those on treatment (0.95).
FIG. 1.
(A) NMDS plot of Bray–Curtis indices compared with HIV status. VID: 41 represents Visit 41 (ANOSIM R = 0.0027, p = .49) and VID: 43 is Visit 43 (ANOSIM R = −0.021, p = .67). (B) Bray–Curtis similarity between visits 41 and 43 (Mann–Whitney U test, p = .89). (C) Stacked histogram of 10 most common genera by HIV status. ANOSIM, analysis of similarity; NMDS, non-metric multidimensional scaling.
The previously noted significant genera-specific differences were not seen when HIV-infected women on treatment were compared with HIV-uninfected women for Staphylococcus (q = 0.09) and for Corynebacterium (q = 0.95). Prevotella abundance was not different between HIV-infected women on treatment and HIV-uninfected women (p = .95). However, Actinomyces (logFC = −1.81, q = 4.43E−6) and Clostridium (logFC = −1.03, q = 5.44E−4) were depleted in HIV-infected women on treatment compared with HIV-uninfected women. Clostridium was also depleted in HIV-infected women on versus off therapy (logFC = −2.54, q = 9.32E−11). The family Streptococcaceae and the genus Streptococcus were slightly more enriched, although with borderline significance, in HIV-infected women with undetectable versus detectable viral load (Kruskal–Wallis, p = .001, FDR = 0.05 for both). Diversity measured by Shannon and Bray–Curtis indices were not associated with HIV viremia status. HIV-infected women were categorized by current CD4 into three groups: <200, 201–350, and >350 cells/mm3. CD4 group did not correlate with Shannon index, Bray–Curtis index, or any individual taxa.
Discussion
This study of sociodemographically similar HIV-infected and uninfected Chicago women suggests that HIV status is not associated with significant community shifts in rectal microbiota. There were no differences in the diversity, complexity, or stability of rectal microbiota by HIV status. These findings are similar to those of Noguera-Julian et al. who found that HIV status was not associated with differences in microbiota but rather that the MSM enterotype was likely driving the difference between HIV-infected and uninfected individuals.7 Our data demonstrate that change in gut microbiome as measured by rectal swabs in HIV-infected women for a 1-year period is low and similarly stable to that of matched HIV-uninfected women. The stability of the gut microbiome seen in HIV-infected women, including those with low CD4, suggests that the gut mucosal immune system may play a smaller role than previously believed in managing gut ecology, specifically at the rectum.
Gut immune function may focus on excluding organisms with high pathogenic potential or only affect mucosa-associated microbes.20 The HIV-associated Prevotella-enriched “dysbiosis” reported by others but not found in our relatively large sample may reflect differences in sampling site (stool/biopsy) as opposed to rectal swabs. The majority of prior publications have used stool or colonic samples, making direct comparisons difficult.3 McHardy et al. did, however, use rectal samples and found changes in microbiota composition associated with HIV in men.21 Several studies suggest that rectal swab microbiota may closely resemble samples from upstream sites.22,23 Kelley et al., using rectal biopsies, also found a similar Prevotellaceae-enriched enterotype associated with condomless receptive anal intercourse in HIV-uninfected MSM, suggesting that rectal sampling is sufficient to identify this Prevotella-enriched enterotype.8 Mounting evidence, including study by Noguera-Julian et al. and Kelley et al., suggests differences between HIV-infected and uninfected are due to behavioral or environmental confounders rather than HIV infection.
We cannot, however, rule out involvement by the gut immune system in selecting organisms associated with the mucosa or more proximal bowel. The enrichment of the family Streptococcaceae and genus Streptococcus in HIV-infected aviremic versus viremic participants has not previously been reported. If it were related to immunosuppression we would expect to see depletion associated with lower CD4 as well, which we did not, suggesting that viral replication and its direct effects on the gut may be responsible.
Among HIV-infected women, untreated status was strongly associated with the abundance of the genera Staphylococcus and Corynebacterium, which does not seem to be explained by persistent HIV viral replication since we did not see these same genera-specific correlations in comparisons between HIV viremic versus aviremic women. The relatively low abundance of Clostridium in HIV-infected women on antiretroviral treatment may suggest that antiretrovirals directly impact the gut microbiome.
This study has several strengths. Our large sample size affords adequate power to find HIV-associated microbial shifts should they exist. The use of the Chicago WIHS cohort, which includes HIV-infected and uninfected women with similar demographic and behavioral characteristics living in the same urban neighborhoods, allowed us to minimize several known environmental, social, dietary, and behavioral confounders. Lastly, examining two samples collected 1-year apart allowed us to address microbiome stability overall and by HIV status. A systems-based approach to integrate measurements of gut microbial metabolites, metagenomics, gut permeability, gut mucosal immune recovery, systemic immune activation, and behavior will be necessary to begin uncovering the effects that gut microbes have on clinical outcomes in HIV. Further data from this cohort will be presented in future articles, including further dietary analysis and serum markers of immune activation. Hopefully, this study will better inform design of those studies by demonstrating the limited role that HIV infection plays in modulating the gut microbiome.
Acknowledgments
Data in this article were collected by the WIHS. WIHS (Principal Investigators): Chicago WIHS (A.F.; Mardge Cohen), U01-AI-034993. The WIHS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute on Mental Health (NIMH).
This study was presented at the International AIDS Society in Paris, July 2017, abstract A-854-0029-04135.
Disclaimer
The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH).
Author Disclosure Statement
The authors report no conflicts of interests in regard to this article.
References
- 1. Tenorio AR, Zheng Y, Bosch RJ, et al. : Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 2014;210:1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hunt PW, Sinclair E, Rodriguez B, et al. : Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 2014;210:1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J, Williams B, Frank D, Dillon SM, Wilson CC, Landay AL: Inside out: HIV, the gut microbiome, and the mucosal immune system. J Immunol 2017;198:605–614 [DOI] [PubMed] [Google Scholar]
- 4. Paquin-Proulx D, Ching C, Vujkovic-Cvijin I, et al. : Bacteroides are associated with GALT iNKT cell function and reduction of microbial translocation in HIV-1 infection. Mucosal Immunol 2017;10:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dillon SM, Lee EJ, Kotter CV, et al. : An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal immunol 2014;7:983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dinh DM, Volpe GE, Duffalo C, et al. : Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015;211:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noguera-Julian M, Rocafort M, Guillén Y, et al. : Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016;5:135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelley CF, Kraft CS, de Man TJB, et al. : The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: Implications for HIV transmission and prevention. Mucosal immunol 2017;10:996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Filippo C, Cavalieri D, Di Paola M, et al. : Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klase Z, Ortiz A, Deleage C, et al. : Dysbiotic bacteria translocate in progressive SIV infection. Mucosal immunol 2015;8:1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moeller AH, Shilts M, Li Y, et al. : SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe 2013;14:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mehandru S, Poles MA, Tenner-Racz K, et al. : Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med 2006;3:e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bacon MC, von Wyl V, Alden C, et al. : The Women's Interagency HIV Study: An observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol 2005;12:1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J, Kobert K, Flouri T, Stamatakis A: PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014;30:614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDonald D, Price MN, Goodrich J, et al. : An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caporaso JG, Kuczynski J, Stombaugh J, et al. : QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dyett P, Rajaram S, Haddad EH, Sabate J: Evaluation of a validated food frequency questionnaire for self-defined vegans in the United States. Nutrients 2014;6:2523–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oksanen J, Blanchet FG, Friendly M, et al. : Vegan: Community Ecology Package. 2016 [Google Scholar]
- 19. Wickham H, Sievert C: In ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing, Cham, 2016 [Google Scholar]
- 20. Yang L, Poles MA, Fisch GS, et al. : HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS 2016;30:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McHardy IH, Li X, Tong M, et al. : HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013;1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Budding AE, Grasman ME, Eck A, et al. : Rectal swabs for analysis of the intestinal microbiota. PLoS One 2014;9:e101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bassis CM, Moore NM, Lolans K, et al. : Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol 2017;17:78. [DOI] [PMC free article] [PubMed] [Google Scholar]