Abstract
Ex vivo explant models are used to characterize in vitro efficacy of preexposure prophylaxis (PrEP) agents. Tissue is challenged with virus in culture and HIV-1 p24 levels are quantified with enzyme-linked immunosorbent assay (ELISA) on supernatants collected throughout a 14–21-day incubation. Due to the narrow dynamic range of HIV-1 p24 kits, we evaluated whether droplet digital PCR (ddPCR) provides an alternative method to quantify HIV-1 replication in supernatant samples. We used samples from the MWRI-01 study, which evaluated the pharmacokinetic/pharmacodynamic profile of long-acting rilpivirine using the explant model (McGowan et al. Lancet HIV 2016). HIV-1 pol RNA was measured with ddPCR, either directly with a one-step method or reverse transcribed to cDNA before ddPCR (two-step method) on supernatants from the MWRI-01 study. Previously analyzed HIV-1 p24 antigen levels (Alliance; Perkin-Elmer) were available for comparison purposes. Both ddPCR methods strongly correlated with HIV-1 p24 and displayed similar patterns of HIV-1 suppression before and after rilpivirine. Compared to the p24 ELISA, two-step and one-step ddPCR reduced the amount of hands-on time by approximately one-half and two-thirds, respectively. ddPCR also required less sample and based on p24 versus ddPCR correlation, could potentially reduce the explant culture time from 14 to 10 days (r2 = 0.78, p < .001) due to the increased sensitivity of ddPCR. We demonstrate that ddPCR is a suitable alternative to HIV-1 p24 ELISA to quantify HIV-1 infection in the explant model and has the potential to decrease explant culture time.
Keywords: HIV-1 p24, HIV-1, explant challenge, qPCR, ddPCR
Introduction
There is increasing interest in using ex vivo/in vitro tissue explant challenge models to generate preliminary efficacy data for novel preexposure prophylaxis (PrEP) agents.1–3 The ex vivo challenge assay exposes fresh biopsy tissue to HIV-1 and, after a washout, is cultured for 14–21 days with supernatant collections every 3–4 days. Quantification of supernatant HIV-1 p24 antigen with an enzyme-linked immunosorbent assay (ELISA) is a commonly used exploratory endpoint in several clinical studies.1,2,4–6
HIV-1 p24 antigen increases with time in culture with the cumulative HIV-1 p24 through day 14 representing the earliest time point where quantification of infection is most reliable.7 This, in addition to virus growth levels increasing up to 3 or 4 log10 in the ex vivo challenge assay,8 limits quantifying HIV infection with the narrow dynamic range of most commercial HIV-1 p24 ELISA kits. An alternative method for measuring HIV infection is the amplification of nucleic acid sequences by quantitative polymerase chain reaction (qPCR), which has been used extensively to diagnose HIV-1 infection and monitor antiretroviral regimens.9 Several qPCR assays have been developed that detect HIV-1 proviral DNA, RNA, integration by Alu-PCR, and the long terminal repeat (LTR) region.9–16
While it has been widely used in virology, qPCR requires a prevalidated standard curve or endogenous controls, which are highly dependent on reaction efficiency, to estimate concentration in unknown samples. Insufficient amplification in the standard curve limits the accuracy of qPCR.17,18 Target sequence variation, instrument and operator variability, and subjectivity in data analysis are also limitations of this method.19 Droplet digital PCR (ddPCR) is an alternative PCR technique that uses sample partitioning to obtain absolute quantification without the need for a standard curve.18 Amplification of target sequences occurs in ∼20,000 nL-sized oil droplets to estimate an absolute count of target DNA. The massive sample partitioning creates a dynamic range from a single copy up to 100,000 copies with Poisson corrections extending the range to multiple copies per droplet.17 Numerous studies published to date have measured HIV-1 DNA, RNA, and 2-LTR with ddPCR; several directly compared ddPCR to qPCR as reviewed by Trypsteen et al.18 Overall, ddPCR has shown to be superior in accuracy, precision, and reproducibility compared to qPCR, but not always more sensitive (in measuring the smallest concentration of an analyte). ddPCR is also more resilient to mismatches between the primers/probes and the target sequence, which is often observed in HIV quantification.18
Studies have directly compared HIV-1 DNA or RNA quantification with qPCR to HIV-1 p24 measurement with ELISA in the rectal ex vivo challenge assay,7,20–23 demonstrating that qPCR produces similar trends as HIV-1 p24 antigen assays and can be used as an alternative endpoint in measuring HIV-1 infection. However, direct comparisons of ddPCR to HIV-1 p24 ELISA have yet to be performed. We therefore hypothesized that ddPCR can be used to quantify HIV-1 infection in the culture supernatants from the ex vivo challenge assay and with greater sensitivity at earlier time points than the HIV-1 p24 ELISA, which would potentially shorten the required culture time.
Thus, we tested three ddPCR methods on previously measured HIV-1 p24 samples to compare HIV-1 detection. First, viral RNA was extracted and subsequently reverse transcribed (RT) to cDNA in a separate step before the PCR (two-step ddPCR). Second, the RT step and the PCR occurred in the same step (one-step ddPCR). Third, one-step ddPCR was performed again, but droplet formation and transfer were done by an automated droplet generator (Bio-Rad, Hercules, CA) instead of manually (one-step automated ddPCR).
Materials and Methods
Sample collection
Archived samples from the MWRI-01 study, a phase 1 open-label clinical trial of long-acting rilpivirine performed at the University of Pittsburgh, were chosen to test the different ddPCR methodologies. After obtaining informed consent, participants underwent flexible sigmoidoscopy to obtain rectal biopsies before drug (referred subsequently as baseline) and on days 28, 56, 84, 112, and optional 140 and 168 days after rilpivirine injection. Four biopsies were subjected to the ex vivo challenge assay.1
Ex vivo challenge assay
Rectal biopsies were weighted before being placed into culture and exposed to the common viral stock of HIV-1BaL (105 TCID50) as described previously.7,21,24 Briefly, biopsies were incubated with virus for 2 h at 37°C at 5% CO2, and then washed with D-PBS (ThermoFisher Scientific, Grand Island, NY) before a 14-day culture period. Supernatants were collected at days 3, 7, 10, and 14 postinfection and stored at −80°C until HIV-1 p24 analysis.
HIV-1 p24 ELISA
Quantification of HIV-1 p24 was measured on the supernatants as per the manufacturer's instructions (Alliance ELISA; Perkin-Elmer Life Sciences, Boston, MA) with the assay's Lower Limit of Quantification (LLOQ) being 12.5 pg/mL. Nondetectable HIV-1 p24 values were converted to half LLOQ. Results were reported as cumulative HIV-1 p24, which is the biopsy weight-adjusted sum of days 3, 7, 10, and 14.
ddPCR methods: two step, one step, and one-step automated
RNA was extracted from 503 rectal explant supernatant samples from 6 participants (1 participant out of the 6 had the optional day 140 and 168 biopsies collected) of the MWRI-01 study using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA concentrations were measured on a NanoVue Plus spectrophotometer (GE Healthcare Life Sciences, Piscataway, NJ). Approximately 1 μg of RNA was reverse transcribed to cDNA using the QuantiTect Reverse Transcription Kit, according to the manufacturer's instructions (Qiagen), before amplification by ddPCR (two-step ddPCR). RNA was also used in the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad) for direct RNA quantification in the ddPCR (one-step ddPCR).
HIV-1 primers designed to bind to conserved regions of HIV pol (Hxb2 positions 2536–2662) were obtained from Integrated DNA Technologies (Coralville, IA). The two-step ddPCR consisted of 2.5 μL of undiluted cDNA in a total reaction volume of 25 μL containing 12.5 μL ddPCR Supermix for probes (no dUTP) (Bio-Rad), 900 nM primers, and 250 nM FAM-labeled double-quencher probe. For one-step and one-step automated ddPCR, 1 μL of undiluted RNA was used in a total reaction volume of 25 μL containing 6.25 μL Supermix, 20 U/μL Reverse Transcriptase, 15 mM DTT (materials provided in the One-Step RT-ddPCR Advanced Kit for Probes from Bio-Rad), and the same concentration of primers and probe as previously mentioned. The sequences are as follows: HIV pol forward primer (5′-GCA CTT TAA ATT TTC CCA TTA GTC CTA-3′), HIV pol reverse primer (5′-CAA ATT TCT ACT AAT GCT TTT ATT TTT TC-3′), and HIV pol double-quencher probe (5′-/56-FAM/AAG CCA GGA/ZEN/ATG GAT GGC C/3IABkFQ/-3′).25
The ddPCR reaction was prepared in a white/clear hard-shell 96-well PCR plate with samples and controls, mixed, and 20 μL was transferred to an 8 channel DG8 cartridge with 70 μL droplet generation oil for droplet formation in the QX200 droplet generator (all materials and equipment from Bio-Rad). Forty μL of droplets were transferred to a semiskirted green 96-well PCR plate (Eppendorf, Hauppauge, NY), sealed with a pierceable foil heat sealer in the PX1 PCR Plate Sealer, and placed into the C1000 Touch Thermal Cycler (Bio-Rad) for amplification. The cycling conditions were as follows for two-step ddPCR: 10 min at 95°C, 40 cycles each of 30 s at 94°C followed by 1 min at 58°C, and 10 min at 98°C. A 2°C/s ramp rate was set for each cycling step. Samples were transferred immediately to the QX200 Droplet Reader (Bio-Rad) or held overnight at 4°C in the thermal cycler. The cycling conditions for one-step and one-step automated ddPCR were the exact same except for a reverse transcription step of 60 min at 50°C before amplification.
For one-step automated ddPCR, the reaction was prepared as described above with samples and controls in a semiskirted green 96-well PCR plate, but placed into the automated droplet generator (AutoDG) with the appropriate consumables: automated droplet generation oil, DG32 cartridges, pipet tips, and a new semiskirted plate for the droplets. The AutoDG combined the reaction mix and oil to form droplets and transferred them to a new plate. Following completion, the plate was heat sealed, amplified in the thermal cycler, and read on the droplet reader.
Results from the QX200 droplet reader were recorded in Bio-Rad's QuantaSoft software (version 1.7.4) using absolute quantification. The software counts the number of positive and negative droplets and uses Poisson statistics to calculate copies/μL in the final 20 μL 1 × ddPCR reaction. A manual threshold of 4,000 was chosen based on preliminary experiments with the plasmid control to accurately separate positive and negative clusters and eliminate false positive droplets (data not shown).
Nontemplate and plasmid controls for two-step ddPCR
Nontemplate controls consisting of all components of the reaction, except nuclease-free water (VWR, Radnor, PA) in place of cDNA or RNA template, were included with each ddPCR run to verify samples were free of contamination. A plasmid control sample derived from amplifying the HIV-1 pol region was used with each cDNA plate (two-step ddPCR only) as a positive control to test assay efficiency and not used as a standard curve. DNA samples derived from biopsy tissue using the AllPrep DNA/RNA mini kit (Qiagen) that were positive on ddPCR were amplified with the same primers and probe using the AmpliTaq Gold Kit (ThermoFisher Scientific) and run on endpoint PCR on a Veriti thermal cycler (Applied Biosystems, Foster City, CA) with the following conditions: 10 min at 95°C, 40 cycles each of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C, followed by 1 cycle of 7 min at 72°C.
The amplification of the PCR products was verified by agarose electrophoresis on an ethidium bromide gel, cut out of the gel and purified with the QIAquick Gel Extraction Kit (Qiagen) before ligation with pCR™2.1-TOPO® vector and transformation with Subcloning Efficiency™ DH5α™ Competent Cells (Invitrogen, Carlsbad, CA), which were plated on LB agar plates with antibiotics ampicillin and kanamycin (Sigma, St. Louis, MO). Several positive bacterial colonies were individually grown overnight in LB broth (Sigma), and then isolated DNA obtained from the Qiagen QIAprep Spin Mini Kit was loaded onto an ethidium bromide gel to test for positive clones. Two positive colonies were each expanded into 100 mL of LB media overnight and DNA subsequently isolated with the Qiagen QIAprep Miniprep kit.
DNA yield was determined with the NanoVue spectrophotometer and converted to copy number. Serial dilutions of one plasmid DNA from 5,000 copies/μL down to 5 copies/μL were used in the two-step ddPCR reaction. An RNA sample that was positive for HIV pol in two-step ddPCR was used for every one-step ddPCR plate as a positive control.
Data analysis
To ensure 20 μL of mixture was transferred to the DG8 cartridge, the initial reaction volume was 25 μL. After threshold setting, copies/μL reported by the software was converted to number of copies in the starting sample according to Bio-Rad's ddPCR Applications Guide. The resulting copies/μL was converted to copies/mL since HIV-1 p24 is reported as pg/mL. Cumulative copy number was calculated through day 14, biopsy weight adjusted, and log transformed. Linear regression and repeated measures ANOVA tests were performed using GraphPad Prism software with p < .05 set for significance.
Results
HIV infection patterns are similar over 14 days in the ex vivo challenge assay when measured by ddPCR and p24
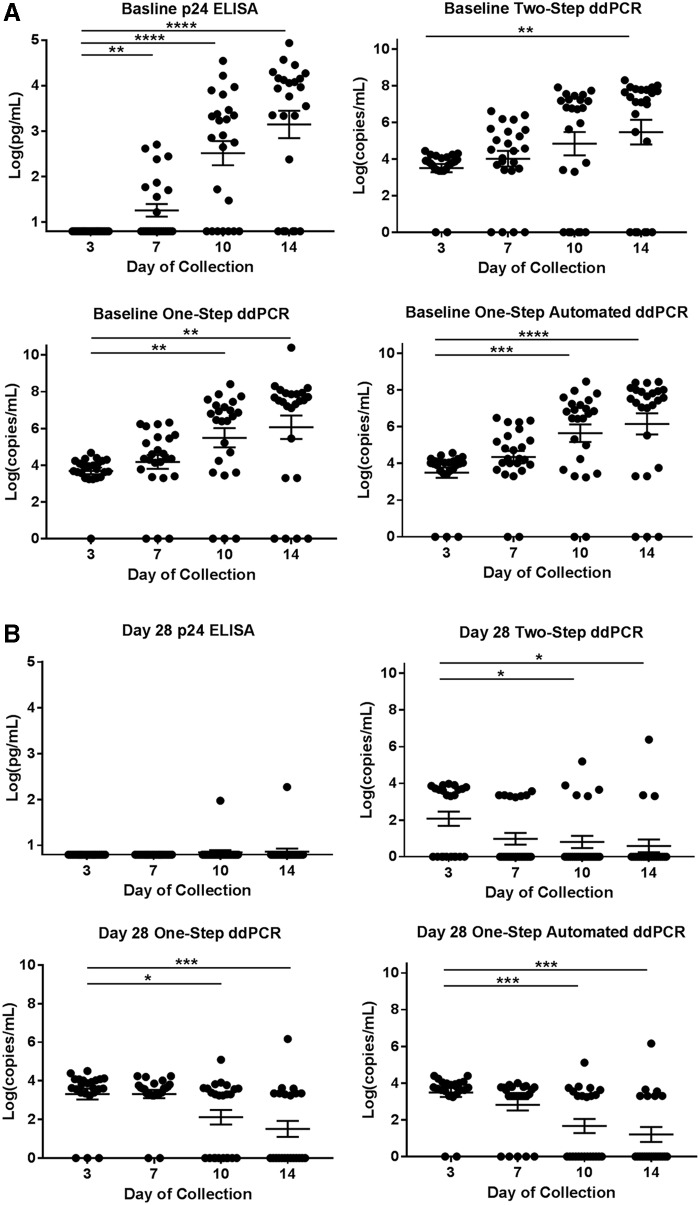
We directly compared HIV-1 p24 ELISA, one-step ddPCR, one-step automated ddPCR, and two-step ddPCR in baseline and day 28 (post rilpivirine exposure) supernatants derived from rectal explant tissue challenged with HIV-1BaL. As expected, HIV-1 p24 and RNA at baseline (before rilpivirine exposure) increased throughout days 3, 7, 10, and 14 of collection for all assays (Fig. 1A). However, no samples on day 3 had detectable HIV-1 p24, whereas most had detectable RNA with all ddPCR assays (Table 1). By day 7 of collection, only 38% of samples had detectable HIV-1 p24 compared to 83%–92% of detectable RNA with the various ddPCR methods. The percentages were comparable between all methods for days 10 and 14 of collection in the ex vivo challenge assay.
FIG. 1.
HIV-1 RNA and p24 antigen levels throughout the ex vivo challenge assay. Scatter plots with mean ± SE bars displaying HIV-1 p24 antigen in pg/mL from the Alliance ELISA and HIV-1 RNA copies/mL from each ddPCR method: one step, one-step automated, and two step for each day of collection in the ex vivo challenge assay. (A) Baseline data and (B) day 28 after rilpivirine injection are shown for each assay type. All data are log transformed. Repeated measures one-way ANOVA test with the mean of day 3 set as the control. Significance set to 0.05. *p ≤ .05, **p ≤ .01, ***p ≤ .001, ****p ≤ .0001. ddPCR, droplet digital PCR; ELISA, enzyme-linked immunosorbent assay.
Table 1.
Number of HIV-1 RNA or p24 Positive Samples for Each Day of Collection in the Ex Vivo Challenge Assay Determined by ddPCR and ELISA
| Baseline positive/total (%) | Day 28 positive/total (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Assay | Day 3 | Day 7 | Day 10 | Day 14 | Day 3 | Day 7 | Day 10 | Day 14 |
| One-step ddPCR | 23/24 (96) | 21/24 (88) | 21/24 (88) | 20/24 (83) | 20/23 (87) | 22/24 (92) | 14/24 (58) | 9/22 (41) |
| One-step automated ddPCR | 21/24 (88) | 22/24 (92) | 22/24 (92) | 21/24 (88) | 22/24 (92) | 19/24 (79) | 11/24 (46) | 7/22 (32) |
| Two-step ddPCR | 22/24 (92) | 20/24 (83) | 18/24 (75) | 18/24 (75) | 13/23 (57) | 7/24 (29) | 5/24 (21) | 3/22 (14) |
| HIV-1 p24 ELISA | 0/24 (0) | 9/24 (38) | 17/24 (71) | 18/24 (75) | 1/23 (4) | 0/24 (0) | 1/24 (4) | 1/22 (5) |
ddPCR, droplet digital PCR; ELISA, enzyme-linked immunosorbent assay.
On day 28, the first sampling after drug exposure, the differences in detectable HIV-1 p24 versus RNA were more evident (Table 1). Almost all samples had undetectable HIV-1 p24 for all days of collection, while RNA was detected in 63%–70% of samples with one-step and 30% with two-step ddPCR with a decreasing trend throughout time (Fig. 1B).
HIV-1 p24 and ddPCR have similar trends of drug-mediated viral suppression of HIV-1
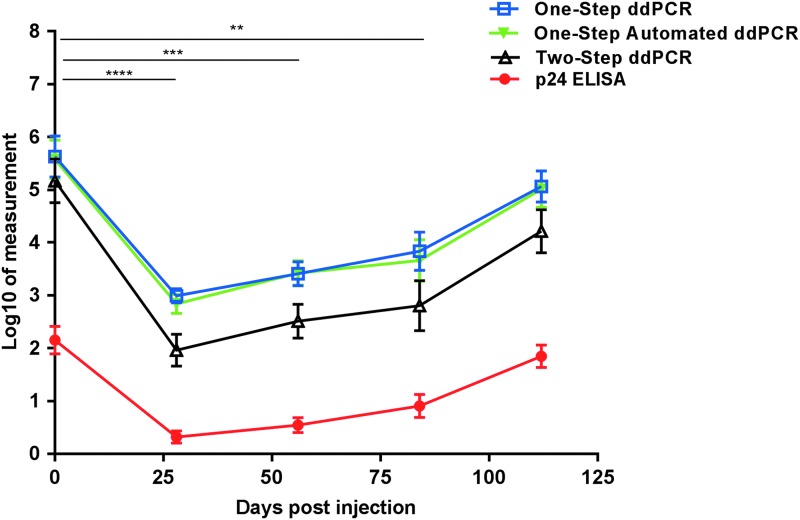
Cumulative HIV-1 p24 and RNA were calculated for each biopsy in the explant challenge. Compared to baseline levels, cumulative HIV-1 p24 antigen levels were significantly suppressed on day 28 after rilpivirine injection (p ≤ .0001), which persisted to day 56 (p ≤ .001) and day 84 (p ≤ .005) postdose. The same pattern occurred for cumulative HIV-1 RNA in the supernatant with all ddPCR methods, with equivalent levels of significance (Fig. 2). Two-step ddPCR measured significantly less cumulative HIV-1 RNA at each study visit than one-step (day 0 = p ≤ .05, days 28 and 56 = p ≤ .001, and days 84 and 112 = p ≤ .01) and one-step automated ddPCR (day 0, day 84, and day 112 = p ≤ .05, days 28 and 56 = p ≤ .005). There were no significant differences in the cumulative HIV-1 RNA between the one-step and one-step automated ddPCR methods at any time point in the study.
FIG. 2.
ddPCR and p24 pattern throughout the MWRI-01 study. Each symbol represents the cumulative mean ± SE at each study visit (n = 24; 6 patients each with 4 biopsy replicates). Day 0 (baseline, before rilpivirine injection), 28, 56, 84, and 112 after rilpivirine for p24 and each ddPCR method are displayed. Cumulative values were calculated by taking the sum of days 3 through 14 of explant culture, biopsy weight adjusted, and log transformed. Units: Log10[(pg/mL)/mg] for p24 and Log10 [(copies/mL)/mg] for all ddPCR methods. Repeated measures ANOVA test with the mean of day 0 set as the control was performed for each method with significance set to 0.05. **p ≤ .005, ***p ≤ .001, ****p ≤ .0001. Each significance annotation between study visits applies to all four methods.
Correlations between assays
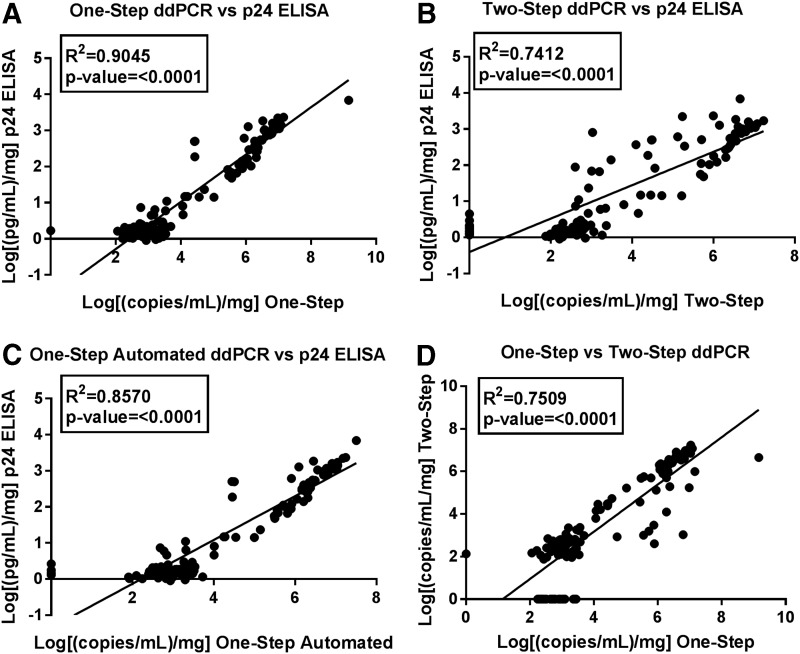
ddPCR and HIV-1 p24 significantly correlated for both one-step and two-step ddPCR (r2 = 0.5386, p < .001 and r2 = 0.5426, p < .001 respectively). There was a stronger correlation after calculating the cumulative values (one step vs. HIV-1 p24 r2 = 0.9045, p < .001 and two step vs HIV-1 p24 r2 = 0.7412, p < .001) (Fig. 3A, B) and the average of the cumulative values among the four biopsy replicates for each study visit (one step vs. HIV-1 p24 r2 = 0.9035, p < .001 and two step vs. HIV-1 p24 r2 = 0.8063, p < .001). As expected, weight-adjusted cumulative copy number correlated between one-step and two-step ddPCR (Fig. 3D).
FIG. 3.
Linear relationships between p24 and ddPCR assays. Each data point is the cumulative value of (A) one-step ddPCR or (B) two-step ddPCR, or (C) one-step automated ddPCR on x-axis versus Alliance p24 on y-axis. (D) One-step ddPCR on x-axis versus two-step ddPCR on y-axis. Cumulative values calculated by taking the sum of days 3 through 14 of explant culture, adjusted for biopsy weight, and log transformed.
Shortening the 14-day explant challenge with one-step ddPCR
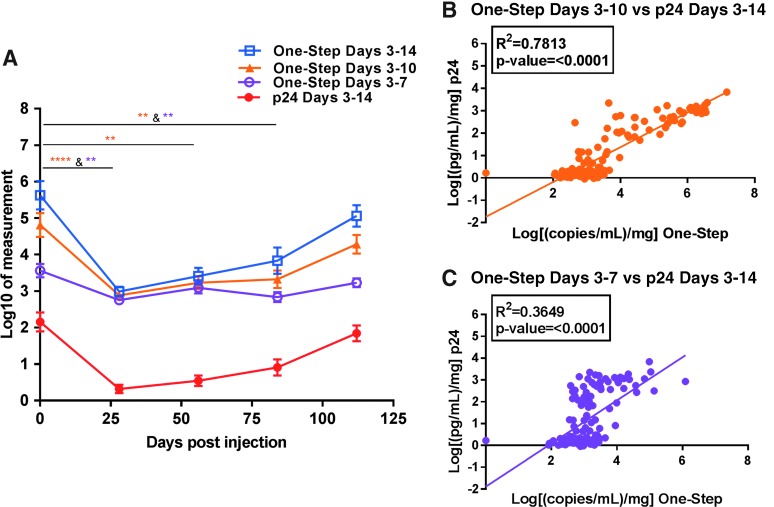
To determine if the 14-day explant challenge could be shortened to 7 or 10 days, cumulative one-step ddPCR and HIV-1 p24 values were calculated with days 10 and 14 omitted, or day 14 only omitted, with biopsy weight adjustment and log transformation. Figure 4A displays the HIV-1 RNA pattern with one-step ddPCR in a manner similar to Figure 2, before and after rilpivirine injection, with the shortened windows included (ddPCR only).
FIG. 4.
Shortening the ex vivo challenge assay with one-step ddPCR compared to p24. (A) Pattern of cumulative one-step ddPCR with shortened explant windows. Cumulative values were calculated by taking the sum of days 3 through 14 (open blue squares for ddPCR and solid red circles for p24), days 3 through 10 (solid orange triangles; ddPCR only), and days 3 through 7 (open purple circles; ddPCR only) of explant culture, adjusted for biopsy weight, and log transformed. Each symbol represents the mean ± SE at each study visit (n = 24; 6 patients each with 4 biopsy replicates). (B) Linear relationship between cumulative p24 (days 3–14) versus cumulative one-step ddPCR days 3–10 only or versus (C) one-step ddPCR days 3–7 only. Units and statistical test for (A) are the same as those presented in Figure 2.
Shortening the sampling period from 14 to 10 days produced equivalent responses at all time points tested for both methods, with only a small decrease in statistical power on day 56 for one-step ddPCR. However, further reducing the sampling period down to 7 days caused a loss of observed significance on day 56 for one-step ddPCR (Fig. 4A). HIV-1 p24 was only significant between baseline and day 28 (p ≤ .05) for cumulative days 3–7 (data not shown). Shortening the culture time also decreased the degree of correlation between one-step ddPCR and HIV-1 p24 despite still being statistically significant (linear relationship compared to cumulative HIV-1 p24 days 3–14: ddPCR days 3–10 only r2 = 0.7813, p < .0001 and days 3–7 only r2 = 0.3649, p < .0001) (Fig. 4B, C).
Discussion
The ex vivo challenge assay is a commonly used exploratory endpoint in numerous phase 1 clinical trials to test the efficacy of multiple PrEP agents.1,2,4–6 The measurement of HIV-1 p24 antigen in explant supernatants with commercial ELISA kits is a well-established method to test infection levels before and after drug exposure. While HIV-1 p24 ELISA kits are the gold standard, most have a narrow dynamic range and low sensitivity resulting in limited quantification. ddPCR has a much greater dynamic range and the potential to be more accurate and precise than qPCR,25 which has been shown to be an effective alternative to HIV-1 p24 ELISA.7,20–23 We determined that HIV-1 RNA in the supernatant can be quantified by ddPCR with more sensitivity at earlier time points than the HIV-1 p24 ELISA. The one-step ddPCR method had the strongest correlation to the HIV-1 p24 results and was the most time- and cost-efficient assay in measuring HIV-1 infection.
In baseline supernatants, HIV-1 RNA measured by ddPCR increased throughout the ex vivo challenge assay with similar kinetics as HIV-1 p24 antigen, but with more sensitivity on days 3 and 7 after infection. This contrasts with two earlier studies that measured HIV-1 RNA with qPCR and HIV-1 p24 with ELISA, in which both methods had similar sensitivity throughout the ex vivo challenge assay.7,20 Nondetectable HIV-1 p24 tends to occur at earlier time points in the rectal ex vivo challenge assay irrespective of HIV-1 p24 kit used,8 an outcome also demonstrated in this study. A high virus titer of HIV-1BaL (104 or 105 TCID50) is used to ensure infectibility for most baseline tissues due to intersubject and intrasubject variability.24 Although a washout step occurs after infection, some virus will remain adhered to the target cells. While this residual virus may or may not be replication competent, it combines with any viral growth to contribute to the HIV-1 RNA present at early time points postinfection.7 The greater precision of ddPCR at low HIV-1 template frequencies25 also likely explains the observed early time point RNA measurements, which are too low to be measured within the detection limits of the HIV-1 p24 ELISA.
Despite the magnitude log differences of the cumulative values, ddPCR and HIV-1 p24 had a strong positive correlation and displayed similar patterns of HIV-1 suppression throughout the MWRI-01 study. This finding agrees with previous studies that have measured both HIV-1 RNA with qPCR and HIV-1 p24 antigen with ELISA,7,20,22,23 including our previous work that found qPCR did not appreciably shorten the time necessary to detect HIV-1 infection despite good matches between assay primers and probes and the infecting sequence.7 Since HIV-1 RNA was detected in more day 3 and 7 samples than HIV-1 p24, the HIV-1 replication pattern measured with one-step ddPCR was assessed if later collection days were omitted. Shortening the explant challenge would increase the feasibility of performing this assay in a multisite clinical trial.
Cumulative HIV-1 p24, and thus HIV-1 RNA, is directly affected by the number of collection time points in the ex vivo challenge assay since more HIV-1 will likely accumulate as time increases.8 Reducing the culture time to 7 or 10 days still resulted in a significant reduction in HIV-1 infection by rilpivirine at day 28 postinjection compared to baseline; however, the statistical differences between baseline and later study visits were less evident as more collection days were omitted. In Richardson-Harman et al.'s retrospective analysis using nonlinear growth curve models, the active virus growth period for nontreated (baseline) rectal tissues has been reported to be short (3 days total) and occurs early at 6–8 days after infection compared to the longer and/or later viral growth in cervical and vaginal tissues.8 While they did not assess how PrEP treatments affected viral growth patterns, this may explain the inconsistent pattern of cumulative days 3–7 throughout the study compared to cumulative days 3–14, and how a significant increase in HIV-1 RNA was not seen until day 10 or 14 in baseline tissues with ddPCR. Shortening the culture period to 7 days may not capture enough viral growth distinguishable from day 3 of culture when using ddPCR. Thus, reducing the time of the explant challenge to 10 days would be sufficient to assess the viral growth pattern before and after rilpivirine injection.
Since HIV-1 replication kinetics were similar between ddPCR and HIV-1 p24, cost and hands-on technician time were also considered between methods. The cost per test for any of the ddPCR methods was about three times the cost of the HIV-1 p24 assay (Table 2; equipment costs not included). However, the length of the HIV-1 p24 assay coupled with the standard curve requirements that limit the number of samples that can be measured on one plate render it very time consuming. ddPCR was much more time efficient in assay setup; however, additional steps of RNA isolation and cDNA synthesis are required before running ddPCR. Despite the extra steps, the total amount of hands-on technician time was reduced by half using two-step ddPCR and by nearly two-thirds using either one-step ddPCR method compared to the HIV-1 p24 ELISA.
Table 2.
Cost and Time Analysis for Each ddPCR Method Compared to HIV-1 p24 ELISA
| HIV-1 p24— ELISA | ddPCR—two-step | ddPCR—one-step | ddPCR—one-step automated | |
|---|---|---|---|---|
| Assay costs | ||||
| I. Cost per test | $4.15 | $13.90 | $12.30 | $12.30 |
| II. Cost for n = 500 samples | $4,773a | $6,950 | $6,150 | $6,150 |
| Technician time/costs | ||||
| III. Samples loaded per plate | 32 | 88 | 88 | 88 |
| IV. Number of total plates = (500/III) | 19a | 6 | 6 | 6 |
| V. Technician time (h) for RNA isolation (n = 500) | — | 26 | 26 | 26 |
| VI. Technician time (h) for separate cDNA RT (n = 500) | — | 25 | — | — |
| VII. Technician time per HIV-1 p24 or ddPCR plate (h) | 6 | 2 | 2 | 1.3 |
| VIII. Total technician time (h) = (IV×VII)+V+VI | 114 | 63 | 38 | 34 |
| IX. Cost for technician time @$25/HR with benefits | $2,850 | $1,575 | $950 | $850 |
| Total cost | ||||
| X. Total cost for 500 samples = II+IX | $7,623 | $8,525 | $7,100 | $7,000b |
Costs are estimates and do not include equipment costs.
Includes 15% of samples that required dilutions and additional measurements due to the dynamic range of the kit; samples ran in duplicate.
Does not include cost of the automated droplet generator, the additional piece of ddPCR equipment needed to perform this specific method.
Shortening the length of the ex vivo challenge assay would also save on costs. Using one-step ddPCR and shortening the assay to 10 days for this sample set would have saved 8 h of hands-on time and approximately $1,480 compared to the full 14 days. Overall, one-step ddPCR was less expensive and more time efficient over two-step ddPCR and HIV-1 p24 after considering technician costs (Table 2).
In this study, one-step ddPCR measured higher levels of HIV-1 infection compared to two-step ddPCR. Previous reports comparing one-step ddPCR to one-step qPCR have found the ddPCR platform to be more accurate in quantifying human rhinovirus RNA with high sequence diversity,26 and had higher precision, repeatability, and lower susceptibility to inhibition in low waterborne virus RNA samples.27 Conversely and unexpectedly, Aizawa et al.28 found two-step ddPCR-detected human parechovirus type 3 RNA in more cerebrospinal fluid samples than did one-step ddPCR. The authors suggested that different reverse transcription efficiency and/or the double-quencher probe in the one-step ddPCR reaction could explain this finding. While the same probe was used, the reverse transcription enzyme and reaction conditions differed between one-step and two-step ddPCR, which is one limitation of this study. Nonetheless, using gene-specific primers in one-step ddPCR may have led to more efficient cDNA synthesis compared to using random hexamers and oligo dT primers in two-step ddPCR, especially for samples with low copy numbers.29
ddPCR may circumvent other sensitivity issues of detecting HIV-1 p24, such as lowering viral titer exposures in the ex vivo challenge assay. Kordy et al.30 were unable to detect new infections with low viral titers (100–103 TCID50 of HIV-1BaL) in experiments that tested colorectal biopsy infectibility in the presence of human semen and/or seminal plasma. The inability to detect HIV-1 p24 early in the ex vivo challenge assay was a limitation in that study since there are conflicted reports of whether semen enhances or has no effect on HIV-1 infection.30 The high viral titers necessary to ensure infectibility in the ex vivo challenge assay is much greater than what is predicted to be in semen4; therefore, a more sensitive assay such as ddPCR could enhance our understanding of semen's role on HIV-1 infection with or without PrEP drug exposure.
While ddPCR has potential to be a better alternative to qPCR, there are limits to ddPCR sensitivity regarding false positive droplets in nontemplate control wells reported in other studies.25,31,32 In this study, there were no positive droplets in any nontemplate control wells that measured above the threshold placed at the fluorescent amplitude of 4,000. This was the intermediate fluorescence between the positive and negative clusters that was the most accurate in measuring our positive control (discussed in Methods section 2.5). Related is the issue involving threshold setting as Bio-Rad uses a proprietary method or allows a user-defined cutoff, which can become problematic with the presence of “rain,” droplets in the region between positive and negative clusters.33 Different threshold determination methods have been developed as discussed by Trypsteen et al.,18 which is a topic needing further attention in quantifying low levels of HIV infection in this study.
In summary, ddPCR is a suitable alternative to HIV-1 p24 ELISA to quantify HIV-1 infection in the ex vivo challenge assay and has the potential to reduce the required infection time to 10 days instead of 14 days. ddPCR had greater sensitivity at earlier time points in the 14 days of culture yet displayed strong linear relationships with HIV-1 p24 antigen before and after rilpivirine drug exposure. It should be noted, however, that additional p24 ELISA kits that are available may lead to a different HIV-1 infection profile found in this study. The strong correlation with HIV-1 p24, shorter assay time, and cost of the one-step ddPCR method verified its superiority over two-step ddPCR. Both HIV-1 p24 ELISA and ddPCR are effective tools in measuring HIV-1 infection in explant supernatants and one-step ddPCR would be advantageous to explore further in future studies.
Acknowledgments
The authors would like to acknowledge both the study participants for their commitment to clinical research and the dedicated staff and physicians of the University of Pittsburgh Magee Women's Hospital Clinical Trials Research Center for their assistance in obtaining clinical samples. This study was funded by the Bill and Melinda Gates Foundation, grant no. OPP1045325, and was also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award UM1AI106707.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. McGowan I, Dezzutti CS, Siegel A, et al. : Long-acting rilpivirine as potential pre-exposure prophylaxis for HIV-1 prevention (the MWRI-01 study): An open-label, phase 1, compartmental, pharmacokinetic and pharmacodynamic assessment. Lancet HIV 2016;3:e569–e578 [DOI] [PubMed] [Google Scholar]
- 2. McGowan I, Cranston RD, Duffill K, et al. : A phase 1 randomized, open label, rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of three formulations of tenofovir 1% gel (the CHARM-01 Study). PLoS One 2015;10:e0125363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dezzutti CS, Russo J, Wang L, et al. : Development of HIV-1 rectal-specific microbicides and colonic tissue evaluation. PLoS One 2014;9:e102585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anton PA, Saunders T, Elliott J, et al. : First phase 1 double-blind, placebo-controlled, randomized rectal microbicide trial using UC781 gel with a novel index of ex vivo efficacy. PLoS One 2011;6:e23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anton PA, Cranston RD, Kashuba A, et al. : RMP-02/MTN-006: A phase 1 rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of tenofovir 1% gel compared with oral tenofovir disoproxil fumarate. AIDS Res Hum Retroviruses 2012;28:1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen BA, Panther L, Marzinke MA, et al. : Phase 1 safety, pharmacokinetics, and pharmacodynamics of dapivirine and maraviroc vaginal rings: A double-blind randomized trial. J Acquir Immune Defic Syndr 2015;70:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janocko L, Althouse AD, Brand RM, Cranston RD, McGowan I: The molecular characterization of intestinal explant HIV infection using polymerase chain reaction-based techniques. AIDS Res Hum Retroviruses 2015;31:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richardson-Harman N, Parody R, Anton P, et al. : Analytical advances in the ex vivo challenge efficacy assay. AIDS Res Hum Retroviruses 2017;33:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Désiré N, Dehée A, Schneider V, et al. : Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J Clin Microbiol 2001;39:1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malnati MS, Scarlatti G, Gatto F, et al. : A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc 2008;3:1240–1248 [DOI] [PubMed] [Google Scholar]
- 11. Li P, Ruel T, Fujimoto K, et al. : Novel application of Locked Nucleic Acid chemistry for a Taqman assay for measuring diverse human immunodeficiency virus type 1 subtypes. J Virol Methods 2010;170:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong F, Aga E, Cillo AR, et al. : Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 2016;54:902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rouet F, Chaix ML, Nerrienet E, et al. : Impact of HIV-1 genetic diversity on plasma HIV-1 RNA Quantification: Usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr 2007;45:380–388 [DOI] [PubMed] [Google Scholar]
- 14. Brussel A, Delelis O, Sonigo P: Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. In: Human Retrovirus Protocols: Virology and Molecular Biology (Zhu T, ed.) Humana Press, Totowa, NJ, 2005, pp. 139–154 [DOI] [PubMed] [Google Scholar]
- 15. Liszewski MK, Yu JJ, O'Doherty U: Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 2009;47:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drosten C, Panning M, Drexler JF, et al. : Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin Chem 2006;52:1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hindson BJ, Ness KD, Masquelier DA, et al. : High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trypsteen W, Kiselinova M, Vandekerckhove L, De Spiegelaere W: Diagnostic utility of droplet digital PCR for HIV reservoir quantification. J Virus Erad 2016;2:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bustin SA, Nolan T: Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 2004;15:155–166 [PMC free article] [PubMed] [Google Scholar]
- 20. Klein SA, Karsten S, Rüster B, et al. : Comparison of TaqMan real-time PCR and p24 Elisa for quantification of in vitro HIV-1 replication. J Virol Methods 2003;107:169–175 [DOI] [PubMed] [Google Scholar]
- 21. Fletcher PS, Elliott J, Grivel JC, et al. : Ex vivo culture of human colorectal tissue for the evaluation of candidate microbicides. AIDS 2006;20:1237–1245 [DOI] [PubMed] [Google Scholar]
- 22. Grivel JC, Elliott J, Lisco A, et al. : HIV-1 pathogenesis differs in rectosigmoid and tonsillar tissues infected ex vivo with CCR5- and CXCR4-tropic HIV-1. AIDS 2007;21:1263–1272 [DOI] [PubMed] [Google Scholar]
- 23. Dezzutti CS, Park SY, Marks KM, et al. : Heterogeneity of HIV-1 replication in ectocervical and vaginal tissue ex vivo. AIDS Res Hum Retroviruses 2018;34:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richardson-Harman N, Mauck C, McGowan I, Anton P: Dose–response relationship between tissue concentrations of UC781 and explant infectibility with HIV type 1 in the RMP-01 rectal safety study. AIDS Res Hum Retroviruses 2012;28:1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strain MC, Lada SM, Luong T, et al. : Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 2013;8:e55943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sedlak RH, Nguyen T, Palileo I, Jerome KR, Kuypers J: Superiority of digital reverse transcription-PCR (RT-PCR) over real-time RT-PCR for quantitation of highly divergent human rhinoviruses. J Clin Microbiol 2017;55:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rački N, Morisset D, Gutierrez-Aguirre I, Ravnikar M: One-step RT-droplet digital PCR: A breakthrough in the quantification of waterborne RNA viruses. Anal Bioanal Chem 2014;406:661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aizawa Y, Koyama A, Ishihara T, Onodera O, Saitoh A: Performance of a real-time PCR-based approach and droplet digital PCR in detecting human parechovirus type 3 RNA. J Clin Virol 2016;84:27–31 [DOI] [PubMed] [Google Scholar]
- 29. Wacker MJ, Godard MP: Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J Biomol Tech 2005;16:266–271 [PMC free article] [PubMed] [Google Scholar]
- 30. Kordy K, Elliott J, Tanner K, Johnson EJ, McGowan IM, Anton PA: Human semen or seminal plasma does not enhance HIV-1BaL ex vivo infection of human colonic explants. AIDS Res Hum Retrovir 2018;34:459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L: Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 2014;9:e85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bosman KJ, Nijhuis M, van Ham PM, et al. : Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir. Sci Rep 2015;5:13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones M, Williams J, Gärtner K, Phillips R, Hurst J, Frater J: Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, ‘definetherain’. J Virol Methods 2014;202:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]