Abstract
Background
RNA-Seq technology is routinely used to characterize the transcriptome, and to detect gene expression differences among cell types, genotypes and conditions. Advances in short-read sequencing instruments such as Illumina Next-Seq have yielded easy-to-operate machines, with high throughput, at a lower price per base. However, processing this data requires bioinformatics expertise to tailor and execute specific solutions for each type of library preparation.
Results
In order to enable fast and user-friendly data analysis, we developed an intuitive and scalable transcriptome pipeline that executes the full process, starting from cDNA sequences derived by RNA-Seq [Nat Rev Genet 10:57-63, 2009] and bulk MARS-Seq [Science 343:776-779, 2014] and ending with sets of differentially expressed genes. Output files are placed in structured folders, and results summaries are provided in rich and comprehensive reports, containing dozens of plots, tables and links.
Conclusion
Our User-friendly Transcriptome Analysis Pipeline (UTAP) is an open source, web-based intuitive platform available to the biomedical research community, enabling researchers to efficiently and accurately analyse transcriptome sequence data.
Keywords: NGS, Transcriptome, RNA-Seq, Sequence analysis pipeline, Bioinformatics workflow, Differentially expressed genes, Genome mapping, Bulk MARS-Seq, UMI (unique molecular identifier), Gene expression profile, Normalization
Background
Next-generation sequencing (NGS) technologies are the most advanced molecular tools currently available to interrogate the complexities of the transcriptome[1, 5], with proven efficient and cost-effective mechanisms for studying gene expression and reliably predicting differential gene expression [6]. Many methods for preparing the libraries have emerged, including Poly A or RiboZero for mRNA enrichment, complete transcript sequencing, strand-specific sequencing [2] and 3′ UTR sequencing [7]. In addition, in cases of initial low RNA levels, unique molecular identifiers (UMIs) are often incorporated in order to label individual cDNA molecules with a random nucleotide sequence before amplification. Advances in short-read sequencing instruments have yielded easy-to-operate machines, with high throughput, at a low price per base.
The massive amount of data created by NGS requires bioinformatics expertise to tailor specific solutions for each type of library preparation. Implementing the solutions typically requires scripting and running commands in the Linux environment. An example of such protocols can be seen at [8]. To address this challenge and simplify the analysis, we developed a transcriptome pipeline, with an intuitive user interface (Fig. 1; results in supplementary materials; demonstration).
Fig. 1.
An example of a page in the pipeline’s Web Graphical Interface. Demonstrates the information required from the user in order to run the pipeline
Implementation
Workflow
The UTAP system is composed of a Snakemake [9] workflow system backend, and Python (v2.7) and a Django (v1.11) - based web user interface (WUI) through which users can run analyses.
Snakemake bundles in-house scripts (written in Python and R) and public bioinformatics tools for completing stepwise processes. Sequence quality control is assessed by FastQC (v0.11.7), read-genome mapping by STAR [10] (v2.5.2b), gene count calculation by either STAR or HTSeq [11] (0.9.1) along with our specialized scripts for UMI counting. SAM and BAM file manipulation is accomplished by Samtools [12] (v1.6), and gene body coverage plotting is performed by ngsplot [13] (v2.61). Differentially expressed genes (DEG) detection and count normalization analysis are performed by DESeq2 [14] (1.18.1). The R package fdrtool [15] (1.2.15) is used to adjust p values when UTAP deduces that the raw p-value distribution is biased. The sva [16] (3.26.0) R package is used for batch correction of the counts when batch adjustments are required.
Web Interface
To increase usability, thereby broadening the potential audience of UTAP, the WUI was planned to be intuitive. Researchers select a pipeline type (demultiplexing or transcriptome), provide the Illumina sequence data (bcl or fastq files), and choose the relevant genome and its annotation source (GENCODE or RefSeq). When running DESeq2, samples should be grouped by category and can be assigned to batches, using a select and drag approach (Fig. 1; supplementary information; demonstration). Batches are sub-groups of measurements that might have qualitatively different behaviour across conditions, and are unrelated to the biological or scientific variables in the study.
Packaging
UTAP is available as a Docker image, which can run locally on one server, or integrated into LSF (Platform Load Sharing Facility, IBM) or PBS professional (OpenPBS; http://www.pbspro.org/) HTC (High-throughput computing) clusters.
Customization
We chose the various pipeline parameters based on our rich experience in transcriptome analysis. This works very well for users who are not deeply familiar with bioinformatics software, and who prefer to quickly benefit from these choices without having to delve into the pipeline’s architecture. On the other hand, many research groups have their own particular preferences, and can achieve system-wide and/or run-specific flexibility by making adjustments to the parameters or code (Snakefile, R scripts) as described in the guide.
Results
Our User-friendly Transcriptome Analysis Pipeline (UTAP) requires minimal user intervention. After providing the information described above (see demonstration), all steps required per library type are automatically executed. Upon completion, the system produces a rich and structured report as output. The transcriptome pipeline is designed for stranded or non-stranded TruSeq libraries, or, alternatively, for bulk RNA 3′ UTR MARS-Seq samples.
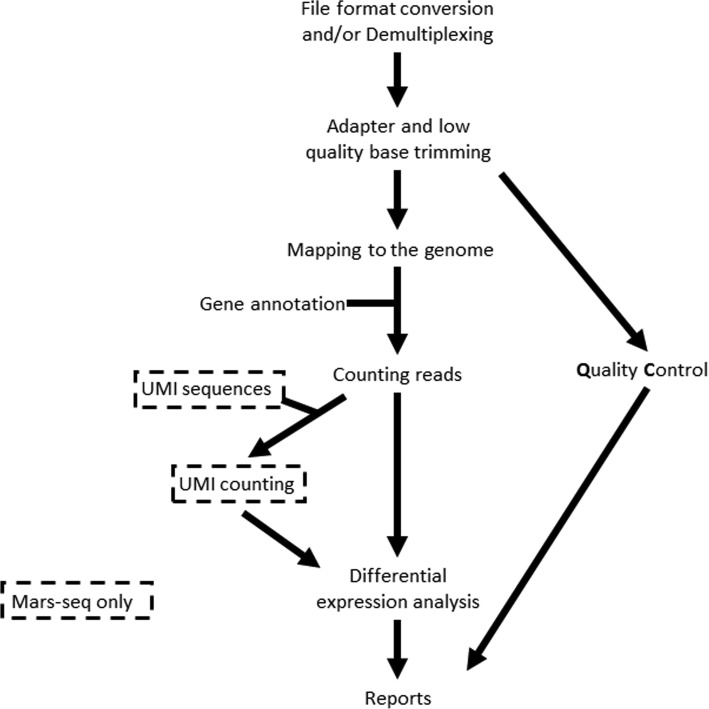
The pipeline runs the following steps (see Fig. 2 and examples in supplementary materials): demultiplexing, adapter and low-quality trimming, quality checks, mapping to a genome, gene quantification, UMI counting (if required), normalization, and detection of statistically significant differentially expressed genes (DEG) for pairwise comparisons of user-defined categories. Once a run has been completed, the user can redefine the samples and categories and rerun only DESeq2. If batches are defined, DESeq2 analyses take them into account.
Fig. 2.
Flow of analysis step performed by the UTAP pipeline. Note that steps that take place only in the MARS-Seq pipeline are shown within broken-line rectangles
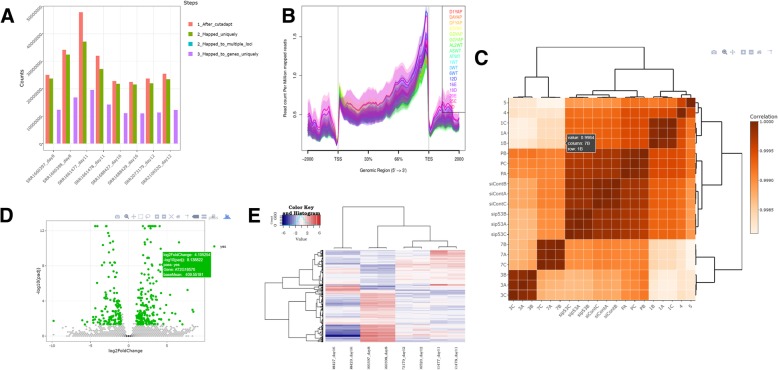
The comprehensive report (see Fig. 3 and examples in supplementary materials) contains dozens of figures for visual inspection, including statistical information, enabling one to explore the efficiency of the process. The figures contain details covering the number of reads per sample in the various steps of the process, the amount of similarity between the samples, and more. In addition, the report contains tables with information on the DEG in each category (up/down) as well as links to gene annotation at GeneCards [17] and submitting gene sets for pathway analysis on Intermine [18]. The report closes with a description of the databases, tools and parameters used, and links to additional results. All pipeline outputs, such as trimmed fastq files, mapped and indexed bam files, matrices of raw, normalized counts and statistical DEG values, are available in structured folders. R scripts containing code for plots and statistics and logs are also included, thus packaging the analysis into a reproducible format.
Fig. 3.
Selection of plots produced in a UTAP report. a Histogram with the number of reads for each sample in the various pipeline steps. b Sequence coverage on and near gene regions using ngs.plot [13] c. Heatmap of Pearson correlation between samples according to gene expression values. d. Scatter plot of significance (y axis) versus fold-change (x axis). e Hierarchical clustering heatmap of differentially expressed genes. Plots D and E are created when DESeq2 analysis is executed
The pipeline is scalable, utilizing the full power of the server or cluster. The Docker image has been tested on LSF and OpenPBS clusters. The scalability allows for fast processing of the data. When the pipeline runs in parallel on each sample with 20 threads per sample, the run time is ~ 1 h for MARS-Seq analysis and ~ 2.5 h for RNA-Seq analysis.
A collection of features that significantly differentiates UTAP from previously reported pipelines and platforms [19–25] is presented in Table 1. Specifically, the other platforms either lack a friendly graphical user interface, and/or are not scalable, and/or have complex installations, and/or do not provide predefined pipelines, and/or do not provide meticulous ways to detect differentially expressed genes, and/or do not have structured outputs. All of the other systems create reproducible results, but lack analysis for bulk MARS-Seq, and do not automatically create summaries via comprehensive reports.
Table 1.
Comparison of Transcriptome Analysis Pipelines
| Tool/platform | Graphical user interface (GUI) | Workflow | DEG detection | Scalable (cluster) | Hosting | Installation | Reproducible runs | Automatic comprehensive report with statistics | Structured output folders | Bulk MARS-Seq | NGS tools other than for DE | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chipster | Yes | user defined | Yes | Yes | Local, Remote server and cloud | Medium (requires virtualization sw) | Yes | No | No | No | Yes | 19 |
| RNACocktail | No | predefined | Yes | No | Local | Easy (Docker) | Yes | No | No | No | Yes | 20 |
| hppRNA-a | No | predefined | Yes | Yes | Local | Medium (Installation script) | Yes | No | Yes | No | Yes | 21 |
| aRNApipe | No | predefined | No | Yes | Local | Medium | Yes | Partial (no DEG) | Yes | No | Yes | 22 |
| Galaxy | Yes | user defined | Yes | Yes | Local, Remote server and cloud | Complex | Yes | No | No | No | Yes | 23 |
| Illumina BaseSpace | Yes | predefined | Yes | Yes | Remote (requires a fee) | NA | Yes | Partial | Yes | No | Yes | 24 |
| docker4seq | Yes | predefined | Yes | Yes | Local | Easy (Docker) | Yes | No | Yes | No | Yes | 25 |
| UTAP | Yes | predefined | Yes | Yes | Local | Easy (Docker) | Yes | Yes | Yes | Yes | No | NA |
Our future plans include improving customization by providing options to modify parameters via the web interface, adding NGS pipelines such as small RNAs, ChIP-Seq, ATAC-Seq, Ribo-Seq, SNP detection in RNA-Seq and single-cell RNA-Seq, and adapting the pipeline to run on other types of computing clusters and in the cloud.
Conclusions
UTAP is an open source, web-based intuitive, scalable and comprehensive platform available to the biomedical research community. It executes an efficient and accurate analysis of transcriptome sequence data, producing sets of differentially expressed genes and sophisticated reports, and requiring minimal user expertise.
Availability and requirements
Project name: UTAP: User-friendly Transcriptome Analysis.
Pipeline Installation manual: https://utap.readthedocs.io
Operating system(s): Linux.
Programming language: Python v2.7, R.
Other requirements: Docker v1.7, miniconda v2.
The pipeline consumes ~40GB RAM. The required disk space for the output files is ~1GB per sample for MARS-Seq analysis and ~6GB per sample for RNA-Seq analysis. In addition, ~135GB are required for storage of the genome files.
License: GNU GPL version 3.
Any restrictions to use by non-academics: License needed for commercial use.
Acknowledgements
We thank the reviewers for their insights and suggestions for improvements.
Funding
Not applicable.
Availability of data and materials
Information about where to download the UTAP Docker application can be found at https://utap.readthedocs.io.
Supplementary information
Examples of reports:
Published RNA-Seq data [3] report results - See https://bip.weizmann.ac.il/rna-seq
Published MARS-Seq data [4] report results – See https://bip.weizmann.ac.il/mars-seq
Explore our UTAP interface (demo site - See http://utap-demo.weizmann.ac.il/).
Abbreviations
- BAM
Binary alignment map
- DEG
Differentially expressed genes
- GB
Gigabyte
- NGS
Next generation sequencing
- RAM
Random access memory
- SAM
Sequence alignment map
- SNP
Single nucleotide polymorphism
- UMI
Unique molecular identifier
- WUI
Web user interface
Authors’ contributions
RK designed, implemented and developed UTAP and the Docker image. JB and GH took part in code development. DL conceived, designed and tested UTAP, and chose the list of competing tools. DL, EF and MS compared the tools. EF and GS helped in design and testing, and created the plots in the paper. DL and RK wrote the paper. RK wrote the manuals. MS edited the paper and manuals. KK helped in Docker image creation and testing. All of the authors read and approved the paper.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Refael Kohen, Email: refael.kohen@weizmann.ac.il.
Jonathan Barlev, Email: jonathan.barlev@gmail.com.
Gil Hornung, Email: gil.hornung@gmail.com.
Gil Stelzer, Email: gil.stelzer@weizmann.ac.il.
Ester Feldmesser, Email: ester.feldmesser@weizmann.ac.il.
Kiril Kogan, Email: kirkog10@gmail.com.
Marilyn Safran, Email: marilyn.safran@weizmann.ac.il.
Dena Leshkowitz, Email: dena.leshkowitz@weizmann.ac.il.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klepikova AV, Logacheva MD, Dmitriev SE, Penin AA. RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genomics. 2015;16:466. doi: 10.1186/s12864-015-1688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feigelson SW, Solomon A, Biram A, Hatzav M, Lichtenstein M, Regev O, Kozlovski S, Varol D, Curato C, Leshkowitz D, et al. ICAMs are not obligatory for functional immune synapses between naive CD4 T cells and lymph node DCs. Cell Rep. 2018;22(4):849–859. doi: 10.1016/j.celrep.2017.12.103. [DOI] [PubMed] [Google Scholar]
- 5.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17(1):4–11. doi: 10.1016/j.cbpa.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Zheng S, Papalexi E, Butler A, Stephenson W, Satija R. Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol. 2018;14(3):e8041. doi: 10.15252/msb.20178041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yalamanchili HK, Wan YW, Liu Z. Data analysis pipeline for RNA-seq experiments: from differential expression to cryptic splicing. Curr Protoc Bioinformatics. 2017;59:11.15.11–11.15.21. doi: 10.1002/cpbi.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 10.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strimmer K. Fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24(12):1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Łabaj PP, Zumbo P, Sykacek P, Shi W, Shi L, Phan J, Wu PY, Wang M, Wang C, et al. Detecting and correcting systematic variation in large-scale RNA sequencing data. Nat Biotechnol. 2014;32(9):888–895. doi: 10.1038/nbt.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.31–31.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 18.Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, Hu F, Smith R, Stěpán R, Sullivan J, et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42(Web Server issue):W468–W472. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallio MA, Tuimala JT, Hupponen T, Klemelä P, Gentile M, Scheinin I, Koski M, Käki J, Korpelainen EI. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8(1):59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D. hppRNA-a Snakemake-based handy parameter-free pipeline for RNA-Seq analysis of numerous samples. Brief Bioinform. 2018;19(4):622-626. 10.1093/bib/bbw143. [DOI] [PubMed]
- 22.Alonso A, Lasseigne BN, Williams K, Nielsen J, Ramaker RC, Hardigan AA, Johnston B, Roberts BS, Cooper SJ, Marsal S, et al. aRNApipe: a balanced, efficient and distributed pipeline for processing RNA-seq data in high-performance computing environments. Bioinformatics. 2017;33(11):1727–1729. doi: 10.1093/bioinformatics/btx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illumina BaseSpace [https://support.illumina.com/sequencing/sequencing_software/basespace.html]. Accessed 18 Mar 2019.
- 25.docker4seq [http://www.bioinformatica.unito.it/reproducibile.bioinformatics.html]. Accessed 18 Mar 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Information about where to download the UTAP Docker application can be found at https://utap.readthedocs.io.