Abstract
Background
Microscopic detection of malaria parasites is the standard method for clinical diagnosis of malaria in Brazil. However, malaria epidemiological surveillance studies specifically aimed at the detection of low-density infection and asymptomatic cases will require more sensitive and field-usable tools. The diagnostic accuracy of the colorimetric malachite green, loop-mediated, isothermal amplification (MG-LAMP) assay was evaluated in remote health posts in Roraima state, Brazil.
Methods
Study participants were prospectively enrolled from health posts (healthcare-seeking patients) and from nearby villages (healthy participants) in three different study sites. The MG-LAMP assay and microscopy were performed in the health posts. Two independent readers scored the MG-LAMP tests as positive (blue/green) or negative (clear). Sensitivity and specificity of local microscopy and MG-LAMP were calculated using results of PET-PCR as a reference.
Results
A total of 91 participants were enrolled. There was 100% agreement between the two MG-LAMP readers (Kappa = 1). The overall sensitivity and specificity of MG-LAMP were 90.0% (95% confidence interval (CI) 76.34–97.21%) and 94% (95% CI 83.76–98.77%), respectively. The sensitivity and specificity of local microscopy were 83% (95% CI 67.22–92.66%) and 100% (95% CI 93.02–100.00%), respectively. PET-PCR detected six mixed infections (infection with both Plasmodium falciparum and Plasmodium vivax); two of these were also detected by MG-LAMP and one by microscopy. Microscopy did not detect any Plasmodium infection in the 26 healthy participants; MG-LAMP detected Plasmodium in five of these and PET-PCR assay detected infection in three. Overall, performing the MG-LAMP in this setting did not present any particular challenges.
Conclusion
MG-LAMP is a sensitive and specific assay that may be useful for the detection of malaria parasites in remote healthcare settings. These findings suggest that it is possible to implement simple molecular tests in facilities with limited resources.
Keywords: Malaria, Plasmodium, Malachite green loop-mediated isothermal amplification, Diagnosis
Background
Malaria is a devastating disease that remains a major global health burden. The illness arises from infection with parasites of the genus Plasmodium. Cases of the most significant morbidity and mortality in humans are caused by the most prevalent species, Plasmodium vivax and Plasmodium falciparum. Plasmodium ovale and Plasmodium malariae also cause human malaria, but the infections are typically associated with milder symptoms. In 2017, an estimated 219 million cases of malaria occurred worldwide [1]. Most malaria cases in 2017 were reported in sub-Saharan Africa (200 million, 92%). The WHO Region of the Americas recorded a rise, largely due to increases in malaria transmission in Brazil, Nicaragua, and Venezuela [1]. In Brazil, the vast majority of malaria cases are concentrated in the Brazilian Amazon Region. The State of Roraima in Brazil is located in the Amazon region in the far north on the border with Venezuela and Guyana. In 2017, Roraima reported 11,966 cases of malaria, which was a 44% increase compared to 2016 (8307) [2]. Based on data from the Brazilian Secretariat of Health Surveillance, 50% of patients seen in the State of Roraima were residents of Venezuela and Guyana. There is frequent movement of the population and vectors in the border region, and access to preventive healthcare in Venezuela and Guyana is limited. Control of malaria in Roraima is endangered and the border area is vulnerable to malaria outbreaks and epidemics.
One of the challenges for malaria surveillance and control programmes is the timely identification of low-density infections not detected by the routine diagnostic tests: microscopy and standard rapid diagnostic tests (RDTs). Currently, the primary method used in Brazil for the diagnosis of malaria is microscopy of a Giemsa-stained thick or thin blood smear, but there are limitations of microscopy, including inability to detect very low density (sub-microscopic) parasitaemia, occasional misdiagnosis of mixed-species infection, and the fact that it is time consuming [3–7]. A majority of sub-microscopic infections are asymptomatic. Individuals who are asymptomatic do not seek treatment resulting in a population of individuals with persistent infections, capable of transmitting malaria in the population, (reviewed in [8]). It is important to identify and treat persons with these low-level parasitaemia during malaria epidemiological surveys. Furthermore, the elimination of malaria will require active case detection in low transmission areas as well as the ability to detect sub-microscopic infections [9]. There is a need to develop and validate sensitive diagnostic tools. Molecular-based diagnostic tools provide more sensitive and specific methods for detecting Plasmodium infections than microscopy and RDTs. For a ‘significant improvement’ over expert microscopy, it is recommended that molecular tests be at least 1 log more sensitive than microscopy; preferably have a detection limit of 2 parasites/μL or fewer [10]. The use of molecular-based diagnostic tools in research and in epidemiological surveys has expanded in recent years. However, their use is limited to laboratories with more sophisticated facilities, due to the requirement for specialized equipment and technical expertise. Simpler molecular tests, such as the loop-mediated isothermal amplification (LAMP) assays, promise to facilitate the use of molecular tests even in facilities with limited resources [11–15].
As recently reviewed [16], several malaria LAMP-based assays have been described to date. Many of these have excellent diagnostic performance, e.g., detecting as few as 1 parasite/μL (illumigene LAMP), or 1–5 parasites/μL (EIKEN LAMP), however, they are not without limitations, which include the requirement for additional equipment for read-out, the limited number of samples tested per run, and the fact that some are capable of detecting malaria parasites at genus level only. Recently, the development of a malaria malachite green loop-mediated isothermal amplification (MG-LAMP) as a LAMP method for diagnosing Plasmodium infection was reported [17]. Three factors make the MG-LAMP assay appealing: (1) performance of the MG-LAMP assay requires only a small portable heat block and mini-centrifuge; (2) it is a colorimetric assay that does not require any special read-out equipment; and, (3) the heat block used has a 38-sample capacity allowing for the testing of many samples at once, with the potential for use in large-scale studies. To date, only two other high through-put (HTP) colorimetric malaria LAMP assays have been described [18, 19].
In this study, the performance of the MG-LAMP assay was tested in health posts of three municipalities of Roraima, Brazil using freshly isolated patient samples. The MG-LAMP diagnosis was compared to results provided by local microscopists at the sites of study. The sensitivity and specificity of MG-LAMP performed in these remote health posts, with limited laboratory infrastructure, were compared to that of a real-time PCR (PET-PCR) [20] assay.
Methods
Collection of clinical samples
This prospective study was carried out between July and August 2017 in malaria heath posts in three municipalities of Roraima, Brazil (Boa Vista, Pacaraima, Rorainopolis). All patients attending the health posts for malaria screening and treatment were eligible to be enrolled in the study. In addition, healthy controls were enrolled from houses near the health posts. Blood samples were obtained from all enrolled patients by venipuncture. Enrolled patients were tested for malaria by a trained local microscopist using 10% Giemsa-stained thick blood smear, and the diagnosis and parasitaemia level were recorded for each patient. Additionally, all consenting patients filled out a clinical questionnaire that addressed whether the patient had symptoms, their age, gender, residence, and whether they had prior Plasmodium infections.
LAMP logistics
Blood sample collection and processing, microscopy, DNA extraction, and MG-LAMP assays were all performed in the malaria health posts in Roraima by a USA-based graduate student with training in molecular biology but with no field experience. Two laboratory technicians with no previous experience with LAMP were trained to read the MG-LAMP results. To simplify the MG-LAMP procedure, a three-component ready-to-use kit was used: component I contained all the necessary reaction components for the assay (LAMP buffer: 40 mM Tris–HCL pH 8.8, 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)SO4, 0.2% Tween-20, 0.8 M Betaine, and 2.8 mM of dNTPs and the primers (stored in a 4 °C refrigerator); component II contained the Bst polymerase (stored at − 20 °C), and component III contained 0.2% malachite green dye. To perform the assay, 13.8 µL of Component I was mixed with 0.8 µL of the Bst polymerase and 0.4 μL of the malachite green dye for a final concentration of malachite green of 0.008%. Five µL of DNA template was added and the tubes were placed in the preheated heat block.
DNA extraction
The DNA extraction was performed in small rooms within the health posts. DNA was extracted from 200 μL of whole blood using the QIAamp DNA Mini Kit (Qiagen Inc, Chatsworth, CA, USA). The manufacturer’s provided DNA extraction protocol was slightly modified in that all of the spins were performed at 2000 g using a mini-centrifuge (Myfuge™) that was easily transported in the field setting.
LAMP method
All samples were screened for Plasmodium using the genus assay as described previously [17] in a final reaction volume of 20 μL. Samples were incubated for 1 h at 63 °C in a mini heat block (GeneMate, Bioexpress, Utah, USA) to amplify the DNA. Following the 1-h incubation, samples were removed from the heat block and allowed to cool for 15 min, the results were then scored by two independent readers as being positive (light blue/green) or negative (clear/colourless). Positive and negative controls were included during each run using P. falciparum 3D7 DNA or nuclease-free water, respectively. Plasmodium falciparum and P. vivax species-specific MG-LAMP assays were carried out on all samples that were positive by the genus assay. These assays were performed using the 3-component ready-to-use in-house kits prepared using previously published P. falciparum and P. vivax primers [21, 22]. Each reaction contained 5 μL of isolated DNA in a final reaction volume of 20 μL. Positive controls included a P. falciparum-positive sample and a P. vivax-positive sample. Nuclease-free water was included as a negative control.
PET-PCR method
DNA samples were shipped to the malaria branch laboratory at the CDC using cold packs. Plasmodium genus-specific PET-PCR was performed in duplicate as described previously except that 5 μL of DNA was used instead of 2 μL [20]. The reactions contained 2× TaqMan Environmental Master Mix 2.0 (Applied Biosystems, Foster City, CA, USA), 250 nM of Genus forward Primer and FAM-Genus reverse primer, and 5 μL of isolated DNA for a final volume of 20 μL. The PET-PCR reaction was run using an Agilent Mx3005pro thermocycler (Agilent Technologies, Santa Clara, CA, USA) using the following cycling parameters: 15 min initial hot-start at 95 °C followed by 45 cycles of denaturing at 95 °C for 20 s, annealing at 63 °C for 40 s, and an extension of 30 s at 72 °C. A positive and negative control, 3D7 and nuclease-free water, respectively, were included in each run. Samples were designated as positive if they had a threshold cycle (Ct) value below 40.0 and negative if they had no Ct value or Ct values above 40.0. Species-specific PET-PCR was performed in duplicate on all samples that were positive by the genus specific PET-PCR, using species-specific primers. P. falciparum and P. ovale PET-PCR primers have been used and verified previously [20, 23]. P. malariae and P. vivax PET-PCR primers can be found in Table 1. Two duplex reactions were set up to detect P. ovale together with P. falciparum and P. malariae together with P. vivax. The duplexed reactions were 20 μL containing 2× TaqMan Environmental Master Mix 2.0 (Applied Biosystems), 250 nM of FAM-P. ovale forward primer, 250 nM P. ovale reverse primer, 250 nM of P. falciparum forward primer, 125 nM of HEX-P. falciparum reverse primer, 250 nM P. malariae forward primer, 250 nM FAM-P. malariae, 125 nM P. vivax forward primer, 125 nM HEX-P. vivax reverse primer and 5 μL of isolated DNA. Reactions were run using the same cycling conditions as the Genus PET-PCR. Positive controls consisting of samples with known Plasmodium species and nuclease-free water as a negative control were included in each run.
Table 1.
PET-PCR primers utilized in the evaluation
| Primer | Sequence |
|---|---|
| P. vivax Forward | 5′-ACT GAC ACT GAT GAT TTA GAA CCC ATT T-3′ |
| HEX-P. vivax Reverse | 5′-agg cgc ata gcg cct ggT GGA GAG ATC TTT CCA TCC TAA ACC T-3′ |
| P. malariae Forward | 5′-AAGGCAGTAACACCAGCAGTA-3′ |
| FAM-P. malariae Reverse | 5′-agg cgc ata gcg cct ggTCCCATGAAGTTATATTCCCGCTC-3′ |
HEX-labelled: based on the plasmepsin gene; FAM-labelled: based on dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene
Statistical analyses
The percentage specificity and sensitivity were calculated as follows: Sensitivity = true positives/(true positives + false negatives) × 100. Specificity = true negatives/(true negatives + false positives) × 100. In addition, 95% Confidence Intervals (95% CI) for both sensitivity and specificity were calculated. The agreement between the human readers and diagnostic tests was assessed by calculating the Kappa coefficients. 95% CIs were calculated using MEDCALC® and GraphPad.
Results
Patient enrolment
A total of 91 participants were enrolled during the 2 months of the study: 65 patients presenting with malaria symptoms (axillary temperature ≥ 37.5 °C) at the health posts and 26 healthy participants from nearby villages (Fig. 1). None of the 26 healthy participants exhibited any symptoms of malaria. Of the 91 enrolled participants, 86 (94.5%) reported having had previous malaria infections while 4 (4.4%) had no previous malaria, and 1 (1.1%) did not provide this information.
Fig. 1.
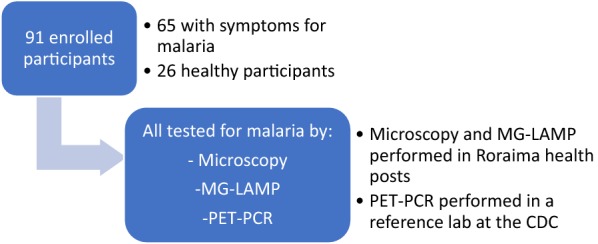
Summary of enrolled patients and sample processing
Agreement between human readers for the MG-LAMP assay
Overall, performing the MG-LAMP in this setting did not present any particular challenges. Two independent human readers scored the MG-LAMP tests as positive or negative. There was 100% agreement between the two readers (Kappa = 1).
Overall results of microscopy, MG-LAMP, and PET-PCR
Of the 91 samples, 33 (36%) were malaria positive by microscopy, 39 (43%) were positive by MG-LAMP, and 40 (44%) were positive by PET-PCR (Fig. 2). All samples were negative for P. malariae and P. ovale.
Fig. 2.
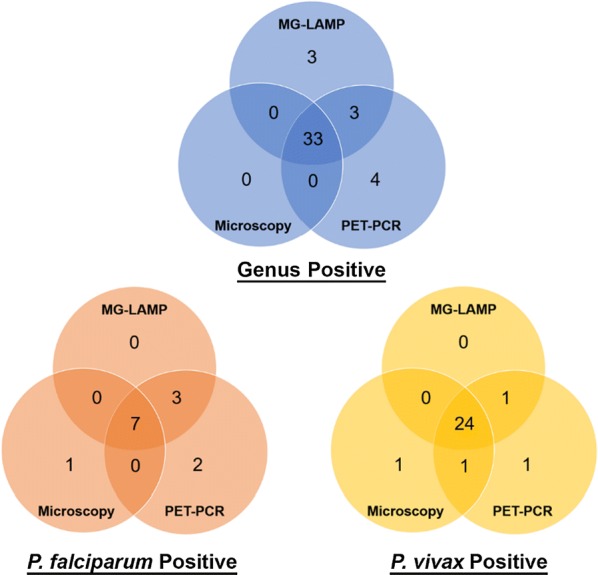
Summary of positive results by microscopy, MG-LAMP and PET-PCR
Specificity and sensitivity of MG-LAMP and microscopy compared to PET-PCR
The sensitivity and specificity of the MG-LAMP assays and microscopy were calculated using PET-PCR as a reference test (Table 2).
Table 2.
Sensitivity and specificity of MG-LAMP and microscopy using PET-PCR as a reference
| Method | Sensitivity | Specificity | |
|---|---|---|---|
| Genusa | Microscopy (n = 91) |
83% (95% CI 67.22–92.66%) | 100% (95% CI 93.02–100.00%) |
| MG-LAMP (n = 91) |
90% (95% CI 76.34–97.21%) | 94% (95% CI 83.76–98.77%) | |
| P. falciparum | Microscopy (n = 91) |
64% (95% CI 93.02–100.00%) | 99% (95% CI 93.23–99.97%) |
| MG-LAMP (n = 39) |
82% (95% CI 48.22–97.72%) | 100% (95% CI 95.49–100.00%) | |
| P. vivax | Microscopy (n = 91) |
83% (95% CI 65.28–94.36%) | 98% (95% CI 91.20–99.96%) |
| MG-LAMP (n = 39) |
90% (95% CI 73.47–97.89%) | 100% (95% CI 94.13–100.00%) |
aSamples which were negative for genus were considered to be negative for species in the sensitivity and specificity calculations
Agreement of MG-LAMP with PET-PCR
The data show that Plasmodium genus assay for MG-LAMP and PET-PCR agreed 92.3% of the time (Kappa = 0.84, 95% CI 0.732–0.955). When comparing P. falciparum and P. vivax MG-LAMP and PET-PCR assays, there was 97.8% (Kappa = 0.89, 95% CI 0.735–1.000) and 96.7% (Kappa = 0.92, 95% CI 0.839–1.000) agreement between the two tests, respectively.
Detection of mixed infections
Microscopy detected one mixed P. falciparum and P. vivax infection, which was detected to be a P. falciparum only infection by both the MG-LAMP and PET-PCR assays. There were six mixed infections detected by PET-PCR; two of these were also identified by MG-LAMP but none was identified by microscopy. In the four cases where the MG-LAMP did not detect the mixed infections identified by the PET-PCR, the Ct values were high, suggesting low parasite density infections (Table 3).
Table 3.
Detection of mixed infections by PET-PCR, MG-LAMP and microscopy
| Sample | Microscopy diagnosis | MG-LAMP diagnosis | PET-PCR diagnosis | PET-PCR CT value for P. falciparum | PET-PCR CT value for P. vivax |
|---|---|---|---|---|---|
| PC121 | P. vivax | Mixed | Mixed | 22.68 | 28.50 |
| PC123 | P. falciparum | Mixed | Mixed | 26.56 | 39.90 |
| BV237 | P. vivax | P. vivax | Mixed | 35.10 | 29.12 |
| BV217 | P. vivax | P. vivax | Mixed | 36.24 | 32.23 |
| BV239 | P. vivax | P. falciparum | Mixed | 31.37 | 35.38 |
| BV241 | P. falciparum | P. falciparum | Mixed | 29.43 | 39.92 |
| BV240 | Mixeda | P. falciparum | P. falciparum | 37.82 | No Ct |
aOnly one P. vivax parasite was seen by microscopy for this sample
Detection of parasitaemia in asymptomatic patients
Of the 26 enrolled healthy participants, five were positive for Plasmodium by MG-LAMP and three were positive for Plasmodium by PET-PCR assay. None of these was positive by microscopy (Table 4). Four of the five cases that were positive by MG-LAMP were positive only at genus level and the infecting species could not be determined (Table 4). Two of these samples were positive by both MG-LAMP and PET-PCR, one only at genus level.
Table 4.
Results of MG-LAMP and PET-PCR in asymptomatic patients
| Sample | Microscopy diagnosis | MG-LAMP diagnosis | PET-PCR genus (Ct value) | PET-PCR P. vivax (Ct value) | PET-PCR P. falciparum (Ct value) |
|---|---|---|---|---|---|
| RR09 | Negative | Genus only | Negative (40.70) | Negative (No Ct) | Negative (No Ct) |
| RR10 | Negative | Genus only | Negative (41.76) | Negative (No Ct) | Negative (No Ct) |
| RR37a | Negative | P. vivax | Positive (32.74) | Positive (35.96) | Negative (No Ct) |
| RR41a | Negative | Genus only | Positive (38.76) | Negative (41.99) | Negative (No Ct) |
| RR42 | Negative | Genus only | Negative (40.74) | Negative (41.69) | Negative (No Ct) |
| RR53 | Negative | Negative | Positive (34.99) | Positive (39.09) | Negative (No Ct) |
aSamples shown to be positive by both MG-LAMP and PET-PCR
Discordant results
Seven samples were found to have discordant results among the three tests (Table 5). Four of these samples were negative by microscopy and MG-LAMP but positive by PET-PCR. Three of these samples were positive by PET-PCR genus test and negative by species tests, while one was positive by PET-PCR P. vivax (Table 5). In these four cases, the Ct values by PET-PCR were all above 35.0. Three samples yielded a positive MG-LAMP genus test but were negative for the MG-LAMP P. falciparum and P. vivax tests and by both microscopy and PET-PCR (Table 5).
Table 5.
Summary of discordant results
| Sample | Microscopy diagnosis | MG-LAMP genus diagnosis | PET-PCR genus (Ct value) | PET-PCR P. vivax (Ct value) | PET-PCR P. falciparum (Ct value) |
|---|---|---|---|---|---|
| RR53 | Negative | Negative | Positive (34.99) | Positive (39.09) | Negative (No Ct) |
| BV235 | Negative | Negative | Positive (35.78) | Negative (No Ct) | Negative (No Ct) |
| RR01 | Negative | Negative | Positive (37.96) | Negative (No Ct) | Negative (No Ct) |
| BV236 | Negative | Negative | Positive (39.34) | Negative (No Ct) | Negative (No Ct) |
| RR09 | Negative | Positive | Negative (40.70) | Negative (No Ct) | Negative (No Ct) |
| RR10 | Negative | Positive | Negative (41.76) | Negative (No Ct) | Negative (No Ct) |
| RR42 | Negative | Positive | Negative (40.74) | Negative (41.69) | Negative (No Ct) |
Discussion
The findings presented in this study demonstrate the accuracy of the MG-LAMP as a malaria diagnostic test in remote health posts in a malaria-endemic country. Importantly, these data demonstrate that the MG-LAMP is sensitive at identifying infections not detectable by microscopy. Additionally, the results establish that this assay, like the PET-PCR assay used as a reference test in this study, is capable of detecting mixed infections that microscopy missed. However, the MG-LAMP assay missed four positive samples and four mixed infections detected by PET-PCR. These missed infections were all shown to be of much lower parasite densities (based on the high Ct values (between 35 and 39) in the PET-PCR assay. Extrapolation using previously obtained PET-PCR data shows that a Ct value of 35.0 corresponds to about 16 parasites/μL [20], therefore, the missed samples likely had parasite densities of about 16 parasites/μL (3 samples) and below (5 samples). While a detection limit of 16 parasites/μL is much better than that for routine microscopy, it is below the detection limits of many PCR-based assays and some previously published LAMP-based assays, which claim detection limits below 16 parasites/μL. Previously, the malaria MG-LAMP assay was shown to have a limit of detection of 1–8 parasites/μL [17] using quantified standard curves, however, this limit of detection did not hold when the assay was performed in a field setting. More sensitive MG-LAMP primers or a change in assay conditions may be required to achieve the same level of diagnostic accuracy as the PET-PCR assay in field settings. In addition, there were three cases where MG-LAMP yielded a positive genus result, while microscopy and PET-PCR were negative. It is likely that these are false positives by the MG-LAMP assay, however, one cannot rule out that these are indeed true positives missed by PET-PCR, a phenomenon that has been observed before in evaluation studies using low-density infection samples [11, 12].
While PCR-based assays, such as PET-PCR, have superior sensitivity for diagnosing low-density infections, they are far more complicated procedurally compared to the MG-LAMP, as they require costly equipment and supplies. The MG-LAMP assay evaluated in this study can be performed using a small portable heat block and mini-centrifuge and does not require any special read-out equipment since it is a colorimetric assay. It is an appealing test for use in resource-limited facilities. In addition, it has a 38-sample capacity allowing for HTP testing and therefore has the potential for use in large-scale studies. Further investment in refining simple molecular tests to increase sensitivity would allow them to be used in resource-limited settings for the detection of low-density infections.
A limitation of the current format of the MG-LAMP is the fact that the LAMP buffers and polymerase require cold chain, which is not ideal in more resource-limited settings. Currently, there are two available malaria LAMP assays that do not require a cold chain: the EIKEN LAMP and illumigene LAMP, but each of these have limitations, reviewed in [16]. For example, the illumigene LAMP assay is a genus-specific test only and is only capable of testing 10 samples per run. Elimination of the need for a cold chain will be required if the MG-LAMP assay is to be used in settings without a laboratory. However, in facilities similar to the health posts used in this study, the current format of MG-LAMP assay can be performed. The use of DNA extracted using commercially available blood kits should be avoided as this adds extra steps and cost to the test; the use of boil-and-spin DNA isolation should be further explored in future field studies.
Although healthy participants were enrolled in an effort to estimate the ability of the MG-LAMP to detect asymptomatic parasitaemia, the number of healthy participants was too low to draw firm conclusions.
Conclusion
Overall, MG-LAMP evaluated in this study provided a portable, sensitive and specific assay for the detection of malaria parasites in a remote health clinic in Brazil when compared to microscopy. However, the current format of the assay was not sensitive enough to be recommended for detection of low-density infections and improvements will be required to enhance its sensitivity and if possible, to make it a more field usable tool that does not require a cold-chain.
Authors’ contributions
HMK, JL, DL, KAK, JOF, and NWL made substantial contributions to the conception and design, acquisition of data, and analysis/interpretation of data. HMK, VM, JOF, and NWL were involved drafting the manuscript and revising it critically for important intellectual content. HMK, VM, JOF, and NWL agree to be accountable for all aspects of this work in ensuring that questions related to the accuracy or integrity of any part of this work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank all the patients who enrolled in this study and made this work possible and the Secretary of Health of Roraima and the local malaria control program team of Boa Vista, Pacaraima and Rorainopolis for their logistical support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Please contact the corresponding author for data requests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was part of a larger study approved by the Federal University of Roraima Ethical Committee (CAAE: 44055315.0.0000.5302). Written informed consent was obtained from all participants. The Centers for Disease Control and Prevention (CDC) investigators provided technical advice but did not have direct contact with study participants or access to any personally identifiable information and were considered not to be engaged in the research (Protocol 2017-105).
Funding
This work was supported by the US National Institutes of Health to H.M.K (T32AI060546). JOF is recipient of Research Productivity Fellowships from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- MG-LAMP
malachite green loop-mediated isothermal amplification
- RDT
rapid diagnostic test
- HTP
high through-put
- PET-PCR
photo-induced electron transfer-polymerase chain reaction
- CI
confidence interval
- CT
cycle threshold
Contributor Information
Vasant Muralidharan, Email: vasant@uga.edu.
Joseli Oliveira-Ferreira, Email: lila@ioc.fiocruz.br.
Naomi W. Lucchi, Email: frd9@cdc.gov
References
- 1.WHO . World malaria report. Geneva: World Health Organization; 2018. [Google Scholar]
- 2.PAHO . Epidemiological alert: increase of malaria in the Americas. Washington: Pan American Health Organization; 2018. [Google Scholar]
- 3.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am J Trop Med Hyg. 2007;77:119–127. doi: 10.4269/ajtmh.2007.77.119. [DOI] [PubMed] [Google Scholar]
- 4.Tajebe A, Magoma G, Aemero M, Kimani F. Detection of mixed infection level of Plasmodium falciparum and Plasmodium vivax by SYBR Green I-based real-time PCR in North Gondar, north-west Ethiopia. Malar J. 2014;3:411. doi: 10.1186/1475-2875-13-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehtesham R, Fazaeli A, Raeisi A, Keshavarz H, Heidari A. Detection of mixed-species infections of Plasmodium falciparum and Plasmodium vivax by nested PCR and rapid diagnostic tests in southeastern Iran. Am J Trop Med Hyg. 2015;93:181–185. doi: 10.4269/ajtmh.14-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishna S, Bharti PK, Chandel HS, Ahmad A, Kumar R, Singh PP, et al. Detection of mixed infections with Plasmodium spp. by PCR, India, 2014. Emerg Infect Dis. 2015;21:1853–1857. doi: 10.3201/eid2110.150678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth JM, Korevaar DA, Leeflang MM, Mens PF. Molecular malaria diagnostics: a systematic review and meta-analysis. Crit Rev Clin Lab Sci. 2016;53:87–105. doi: 10.3109/10408363.2015.1084991. [DOI] [PubMed] [Google Scholar]
- 8.Okell LC, Ghani AC, Lyons E, Drakeley CJ. Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis. 2009;200:1509–1517. doi: 10.1086/644781. [DOI] [PubMed] [Google Scholar]
- 9.WHO . A framework for malaria elimination. Geneva: World Health Organization; 2017. [Google Scholar]
- 10.WHO. Evidence review group on malaria diagnosis in low transmission settings. Geneva: World Health Organization; 2014. http://www.who.int/malaria/mpac/mpac_mar2014_diagnosis_low_transmission_settings_report.pdf.
- 11.Cook J, Aydin-Schmidt B, Gonzalez IJ, Bell D, Edlund E, Nassor MH, et al. Loop-mediated isothermal amplification (LAMP) for point-of-care detection of asymptomatic low-density malaria parasite carriers in Zanzibar. Malar J. 2015;14:43. doi: 10.1186/s12936-015-0573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins H, Gonzalez IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, et al. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis. 2013;208:645–652. doi: 10.1093/infdis/jit184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, et al. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS ONE. 2010;5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, Notomi T, et al. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–2871. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oriero EC, Jacobs J, Van Geertruyden JP, Nwakanma D, D’Alessandro U. Molecular-based isothermal tests for field diagnosis of malaria and their potential contribution to malaria elimination. J Antimicrob Chemother. 2015;70:2–13. doi: 10.1093/jac/dku343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucchi NW, Ndiaye D, Britton S, Udhayakumar V. Expanding the malaria molecular diagnostic options: opportunities and challenges for loop-mediated isothermal amplification tests for malaria control and elimination. Expert Rev Mol Diagn. 2018;18:195–203. doi: 10.1080/14737159.2018.1431529. [DOI] [PubMed] [Google Scholar]
- 17.Lucchi NW, Ljolje D, Silva-Flannery L, Udhayakumar V. Use of malachite green-loop mediated isothermal amplification for detection of Plasmodium spp. parasites. PLoS ONE. 2016;11:e0151437. doi: 10.1371/journal.pone.0151437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britton S, Cheng Q, Grigg MJ, William T, Anstey NM, McCarthy JS. A sensitive, colorimetric, high-throughput loop-mediated isothermal amplification assay for the detection of Plasmodium knowlesi. Am J Trop Med Hyg. 2016;95:120–122. doi: 10.4269/ajtmh.15-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Britton S, Cheng Q, Sutherland CJ, McCarthy JS. A simple, high-throughput, colourimetric, field applicable loop-mediated isothermal amplification (HtLAMP) assay for malaria elimination. Malar J. 2015;14:335. doi: 10.1186/s12936-015-0848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucchi NW, Narayanan J, Karell MA, Xayavong M, Kariuki S, DaSilva AJ, et al. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS ONE. 2013;8:e56677. doi: 10.1371/journal.pone.0056677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamura M, Makimura K, Ota Y. Evaluation of a new rapid molecular diagnostic system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification, and melting curve analysis. Jpn J Infect Dis. 2009;62:20–25. [PubMed] [Google Scholar]
- 22.Patel JC, Oberstaller J, Xayavong M, Narayanan J, DeBarry JD, Srinivasamoorthy G, et al. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS ONE. 2013;8:e54986. doi: 10.1371/journal.pone.0054986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akerele D, Ljolje D, Talundzic E, Udhayakumar V, Lucchi NW. Molecular diagnosis of Plasmodium ovale by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS One. 2017;12:e0179178. doi: 10.1371/journal.pone.0179178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact the corresponding author for data requests.


