Abstract
Current estimates report that approximately 25% of U.S. adults use dietary supplements for medicinal purposes. Yet, regulation and transparency within the dietary supplement industry remains a challenge, and economic incentives encourage adulteration or augmentation of botanical dietary supplement products. Undisclosed changes to the dietary supplement composition could impact safety and efficacy; thus, there is a continued need to monitor possible botanical adulteration or mis-identification. Goldenseal, Hydrastis canadensis L. (Ranunculaceae), is a well-known botanical used to combat bacterial infections and digestive problems and is widely available as a dietary supplement. The goal of this study was to evaluate potential adulteration in commercial botanical products using untargeted metabolomics, with H. canadensis supplements serving as a test case. An untargeted ultraperformance liquid chromatography-mass spectrometry (LC-MS) metabolomics analysis was performed on 35 H. canadensis commercial products. Visual inspection of the chemometric data via principal component analysis (PCA) revealed several products that were distinct from the main groupings of samples, and subsequent evaluation of contributing metabolites led to their confirmation of the outliers as originating from a non-goldenseal species or a mixture of plant materials. The obtained results demonstrate the potential for untargeted metabolomics to discriminate between multiple unknown products and predict possible adulteration.
Keywords: metabolomics, goldenseal, adulteration, dietary supplements, mass spectrometry
Graphical Abstract
1. Introduction
Dietary supplements have become a focal point in personal medicinal care, with natural products becoming increasingly prevalent within the industry. Approximately 25% of Americans take a dietary supplement as part of their everyday health regimen [Asher, 2017; Newman, 2016; Smith, 2016]. In the United States, the prevailing regulatory structure at the Food and Drug Administration (FDA) views herbal supplements as food rather than pharmaceuticals [Dietary Supplement Health and Education Act, 1994]. As such, the evaluation and reporting of adverse events remains with the manufacturer; the FDA does not generally perform pre-market testing on dietary supplements [Dietary Supplement Health and Education Act, 1994]. However, the FDA can act to remove any adulterated supplements from the market if proof of adulteration has been established [Dietary Supplement Health and Education Act, 1994]. As an example of some of the regulatory challenges surrounding dietary supplements, four stimulants - two 1,3-dimethylamylamine (DMAA) analogs and two banned stimulants (1,3-dimethylamylamine and 1,3-dimethylbutylamine) - were found in a study analyzing weight lose supplements [Cohen, 2017]. The FDA had banned the 1,3-DMAA stimulant and removed any supplements containing the compound in August 2016 due to increased incidence of correlated emergency room visitations and because the conditions for it to be legally marketed had not been met [Cohen, 2017]. However, analysis performed on other weight loss products still on the market after 2016 revealed analogs of 1,3-DMAA in five out of six commercial products tested [Cohen, 2017]. This is just one case that illustrates how dietary supplements can be adulterated with potentially harmful compounds.
Goldenseal, Hydrastis canadensis L. (Ranunculaceae), is among the top 40 herbal supplements sold in the United States [Smith, 2016]. It is used to treat gastrointestinal disturbances, eye infections, and inflammation [Cicero, 2016; Le, 2013; Leyte-Lugo, 2017]. Root extracts of this botanical have demonstrated antimicrobial and antibacterial [Cicero, 2016; Le, 2013; Leyte-Lugo, 2017], as well as cytotoxic properties in vivo [Karmakar, 2010; Le, 2013]. The root contains several benzylisoquinoline alkaloids, including berberine, hydrastine, and canadine [Cicero, 2016; Karmakar, 2010; Le, 2013; Le, 2014; Leyte-Lugo, 2017]. Berberine is the most abundant alkaloid in H. canadensis roots and the antimicrobial activity of H. canadensis has generally been attributed to this compound [Brown, 2008; Junio, 2011]. Several flavonoids have also shown to contribute to the antimicrobial activity of goldenseal, working synergistically with the alkaloid berberine [Britton, 2017; Junio, 2011].
Goldenseal dietary supplements are often harvested from wild populations and those are available in limited quantities; a cultivated plant takes years to fully mature and is expensive to farm [McGraw, 2003; Tims, 2016]. Thus, there is an economic incentive to adulterate goldenseal dietary supplements, and adulteration has become a pertinent issue with the dietary supplement industry. Adulteration can involve spiking plant material with synthetic compounds, using a different species in the same genus, or substituting a completely different species in place of the stated one [Tims, 2016]. Several plants that have been used to adulterate goldenseal supplements are barberry, Berberis vulgaris L. (Berberidaceae), Chinese goldthread, Coptis chinensis Franch. (Ranunculaceae), and Oregon grape, Mahonia aquifolium (Pursh) Nutt. (Berberidaceae). These plants also produce berberine in high quantities, yet possess distinctly different metabolic profiles from that of goldenseal [McGraw, 2003; Tims, 2016; Weber, 2003]. Material from berberine-producing plants has been known to have been incorporated into dietary supplement capsules in lieu of goldenseal [Pengelly, 2012; Tims, 2016; Weber, 2003]. Goldenseal, specifically the alkaloid hydrastine, has been shown to inhibit two major metabolic enzymes, CYP2D6 and CYP3A4, which metabolize approximately half of the drugs currently on the market [Gupta, 2015]. Goldenseal dietary supplements could thus affect the absorption, distribution, metabolism, and excretion of certain drugs taken concomitantly, and the presence of an unknown species could precipitate additional drug interactions that would not otherwise be expected [Gupta, 2015; Gurley, 2008]. The adulterated supplement could also demonstrate different biological activity, or have other side effects [Cicero, 2016].
There have been several published methodologies used to detect adulteration of goldenseal specifically. One method used HPLC-UV to analyze and quantify analytes in the plant material, finding non-goldenseal constituents palmatine, coptisine, and jatrorrhizine [Tims, 2016]. The other method utilized GC-MS to quantify metabolites in three different commercial root products [Weber, 2003] using various extraction solvents (hexane, chloroform, methanol, ethanol, and water). Palmatine, jatrorrhizine, and coptisine, all compounds found in plant species that also contain berberine, were found in one out of three of the commercial samples tested [McGraw, 2003]. Utilizing FT-IR data in combination with different chemometric techniques an in silico limit of detection was established at 5% adulteration, depending on the adulterant species [Liu, 2018]. Comparing botanical material can also be achieved via genomic methods, e.g., DNA barcoding. Barcoding has been shown to be an effective tool in authentication of botanical and dietary supplements that are comprised of fresh, dried, or powdered material, where intact DNA sequences are still present [Coutinho, 2015; Little, 2014; Raja, 2017]. However, the DNA barcoding approach is more difficult when applied to botanical extracts, as the manufacturing process often leads to removal or degradation of DNA or contamination with rice filler. DNA barcoding is not feasible for processed botanical products where the DNA is either not present or potentially highly degraded, or where there are two or more species present [Coutinho, 2015; New York State Office of the Attorney General, 2015; Parveen, 2016].
Metabolomics approaches are applied to characterize multiple small molecule metabolites in a biological sample set simultaneously, typically involving spectroscopic or spectrometric analyses, with nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS) being the most common analytical inputs [Sun, 2018]. Untargeted metabolomics can be employed to analyze datasets when little is known about the composition of the sample set and when the variance between samples could be attributed to several sources [Kellogg, 2016; Tao, 2018; Cappello, 2018]. Metabolomics can be used to distinguish one group of samples from another based on unique chemical profiles, and has been applied to a wide scope of biological and chemical applications, including identification of toxicological or disease biomarkers [Sun, 2018], natural product drug discovery [Kellogg, 2016], identification of secondary metabolites in Gram negative bacteria [Depke, 2017] and characterization of botanicals (black tea, green tea, ginseng, coffee) [Guo, 2018; Kellogg, 2017; Lu, 2013; Souard, 2018; Zhang, 2018].
In this study, liquid chromatography-mass spectrometry (LC-MS) data were utilized to perform untargeted metabolomics analysis on a range of commercial goldenseal products, comprised of either the aerial portion (leaves, stem), root, or rhizome. A variety of plant material references (both goldenseal and other berberine-producing species) were utilized to identify possible adulterated products. Though several targeted techniques have been used to find adulterants in dietary supplements [Simmler, 2017; Steuer, 2017], there has not yet been a study performed to detect adulteration in a sample set of commercial products, ostensibly derived from the same botanical, with an untargeted approach. Untargeted metabolomics approaches have been employed to discriminate between different botanical species as well as variations in the geographic origin of materials [Kang, 2008; Mncwangi, 2014], and studies adulterating pure botanical material have shown discriminating patterns that can be discerned using untargeted techniques [Dowlatabadi, 2017; Geng, 2015; Geng, 2017]. However, little attention has been paid to commercial botanical products, where neither the geographical provenance, cultivation conditions, nor the harvesting and production specifications are known. This unknown information introduces a degree of variability in product composition and could complicate efforts to discern patterns and identify outliers, while an untargeted approach to commercial products facilitates analysis without any a priori hypotheses on the nature of possible adulterants. All samples employed in this study were commercial products marketed as Hydrastis canadensis dietary supplements, and the only information regarding their composition was the label provided on the package by the manufacturer. Additionally, studying commercial supplements highlights the direct connection between possible adulteration and naïve consumption by the consumer. The goal of these studies was to employ untargeted metabolomics analyses to distinguish potential adulteration in a batch of 35 samples of commercial H. canadensis products simultaneously.
2. Materials and Methods
2.1. General Methods
All solvents and chemicals used were of reagent or spectroscopic grade, as required, and obtained from ThermoFisher Scientific (Waltham, MA, USA). Berberine and hydrastine standards were purchased from Sigma-Aldrich (St. Louis, MO, USA) and were found to have a purity of 99% and 98% respectively. A canadine reference was isolated and purified from H. canadensis as described previously [Leyte-Lugo, 2017] and demonstrated purity of 79%. Purity was determined via LC-UV.
2.2. Sample Selection and Reference Materials
Commercial goldenseal products were selected based on their popularity in online consumer sales reports [Amazon.com]. The 35 products included 19 capsules, six tinctures, eight powdered bulk materials, and two bagged teas (Table 1). Each sample was randomly coded with an internal reference number (beginning with the letters GS) to maintain manufacturer anonymity (Supplemental, Table S1).
Table 1:
Botanical and physical characteristics of the 35 commercial goldenseal products. Preparation refers to the post-harvest treatment a sample received (drying, extraction followed by drying, or freeze-drying); formulation represents how the final material was packaged for the consumer. Botanical source relates to the physiological portion of the plant which was harvested and incorporated into the final product.
| Preparation | Products (%) | Formulation | Products (%) | Botanical Source* | Products (%) |
|---|---|---|---|---|---|
| Dried material | 24 (69) | Capsule | 19 (54) | Root | 25 (72) |
| Botanical extract | 9 (26) | Tincture | 6 (17) | Rhizome | 4 (11) |
| Freeze-dried material | 2 (5) | Powder/loose material | 8 (24) | Herb/leaf | 4 (11) |
| Tea | 2 (5) | Aerial parts | 1 (3) | ||
| Not specified | 1 (3) |
Botanical source as reported by manufacturer.
Reference materials were obtained from commercial suppliers as well as harvested by the investigators. Hydrastis canadensis leaf (GS-35) and root (GS-36) samples were purchased from ChromaDex (Irvine, CA, USA). In addition, H. canadensis material was collected in August 2016 from William Burch in Hendersonville, North Carolina (NC, N 35°24.277′, W 082°20.993′, 702.4 m elevation), and a voucher specimen was deposited with the Herbarium of the University of North Carolina at Chapel Hill (accession: NCU583414) and authenticated by Dr. Alan S. Weakly. Leaf (GS-37) and root (GS-38) samples were dried in air at room temperature for several weeks prior to extraction.
Reference material from common goldenseal adulterants was also purchased from ChromaDex. These samples included Coptis chinensis rhizome (GS-39) and root (GS-40) samples, Mahonia aquifolium leaf (GS-41) and root (GS-42) references, and Berberis vulgaris root (GS-43) samples. All reference materials were obtained as dried powders, and extracted using the same methods applied for the H. canadensis samples.
2.3. Sample Extraction
Samples were weighed into scintillation vials (200 mg of material per sample) and 20 mL of methanol were added. Extractions were performed in triplicate. Samples were shaken for 24 hours then filtered with 13 mm Puradisc Whatman (GE Healthcare, Chicago, IL, USA) syringe filters. Drying of extracts was accomplished under N2 gas, and they were stored at room temperature prior to analysis.
2.4. Mass Spectrometry Analysis
Liquid chromatography tandem to mass spectrometry (LC-MS) data were acquired utilizing a Q Exactive Plus quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific) with an electrospray ionization (ESI) source coupled to an Acquity UPLC system (Waters, Milford, MA, USA). Samples were resuspended in CH3OH to a concentration of 1 mg/mL (expressed as mass of extract per volume solvent). Injections of 3 μL were performed on an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm, Waters) with a flow rate of 0.3 mL/min using a binary solvent gradient of H2O (0.1% formic acid added) and CH3CN (0.1% formic acid added): initial isocratic composition of 95:5 (H2O:CH3CN) for 1.0 min, increasing linearly to 0:100 over 7 minutes, followed by an isocratic hold at 0:100 for 1 min, gradient returned to starting conditions of 95:5 and held isocratic again for 2 min. The positive/negative switching ionization mode of the mass spectrometer was utilized over a full scan of m/z 150−2000 with the following settings: capillary voltage, 5 V; capillary temperature, 300 °C; tube lens offset, 35 V; spray voltage, 3.80 kV; sheath gas flow and auxiliary gas flow, 35 and 20 units, respectively. Extracted ion chromatographs were obtained from the XCalibur software (ThermoFisher Scientific).
2.5. Metabolomic Analysis
The LC-MS data were analyzed, aligned, and filtered using MZmine 2.28 software (http://mzmine.github.io/) with a slightly modified version of a previously reported method [Kellogg, 2017]. The following parameters were used for peak detection: noise level (absolute value), 1×105 counts; minimum peak duration 0.5 min; tolerance for m/z intensity variation, 20%. Peak list filtering and retention time alignment algorithms were performed to refine peak detection. The join algorithm was used to integrate all the chromatograms into a single data matrix using the following parameters: the balance between m/z and retention time was set at 10.0 each, m/z tolerance was set at 0.001, and retention time tolerance was defined as 0.5 min. The peak areas for individual ions detected in triplicate extractions were exported from the data matrix for further analysis. Samples that did not contain a particular marker ion were coded with a peak area of 0 for that variable to maintain a consistent number of variables throughout the dataset. Chemometric analysis was completed using Sirius version 10.0 (Pattern Recognition Systems AS, Bergen, Norway). Transformation from hetereoscedastic to homoscedasatic noise was carried out by a fourth root transform of spectral variables. Principal component analysis (PCA) was used for untargeted metabolomics profiling of the goldenseal samples with Sirius software. The 95% confidence interval was calculated using Hoetelling’s T2 in R with the R package ‘car’ [Fox, 2011]. Heatmap construction was performed on the log-transformed, mean-scaled peak area data for each relevant metabolite, using the R package ‘gplots’ [Warnes GR, 2016].
2.6. Compound Identification
Variables, unique m/z value and retention time (m/z-RT) pairs, present in the loadings plot were used to confirm and explain the variance in the corresponding scores plot. These ions were identified by using exact mass (< 5 ppm) and retention time. The compounds (berberine [1], canadine [2], hydrastine [3], coptisine [4], palmatine [5], jatrorrhizine [6], and dihydrocoptisine [7]) are all known and well documented (Figure 3). The m/z-RT pairs were compared and confirmed with literature values.
Fig 3:
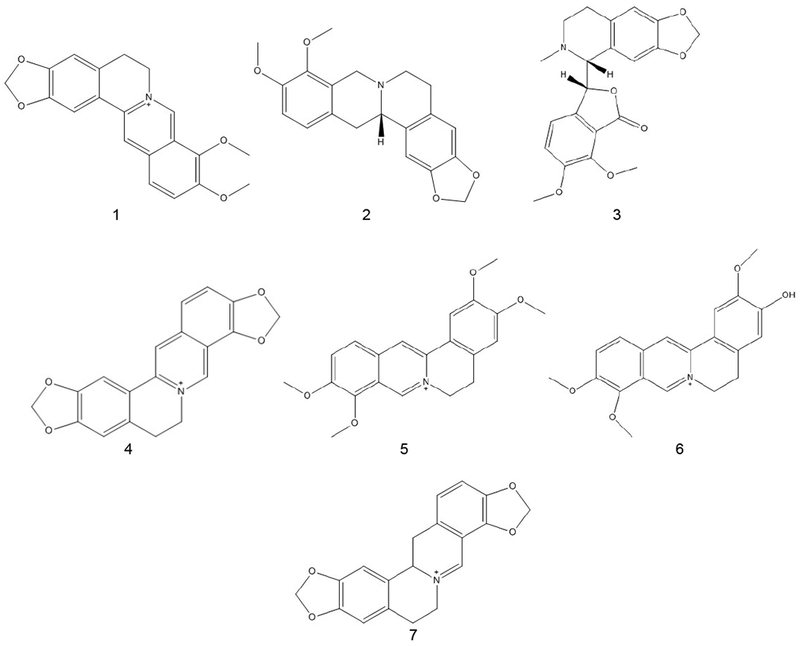
Structures of marker compounds in goldenseal and other berberine containing species. These compounds were confirmed using exact mass and retention time. The compounds are berberine (1), canadine (2), hydrastine (3), coptisine (4), palmatine (5), jatrorrhizine (6), and 13,14 dihydrocoptisine (7).
3. Results and Discussion
3.1. Identification of Goldenseal Outliers by Untargeted Mass Spectrometry Metabolomics
Untargeted metabolomic analysis of the goldenseal samples using LC-MS yielded 5,423 marker ions (unique retention time−m/z ion pairings) for 117 objects (35 commercial goldenseal samples and four goldenseal reference materials extracted in triplicate), which were statistically modeled using PCA. The extraction replicates of each goldenseal product were overlaid on the PCA plot (Supplemental, Figure S5), indicating repeatability of the extraction technique and subsequent LC-MS analysis. The three extractions were averaged for subsequent PCA analysis. The 4-component PCA model accounted for 68.0% of the variance in the sample set.
Inspection of the data based upon the botanical source of each sample (e.g., leaf, aerial portion, root, rhizome, whole plant) indicated two distinct sample clusters located in different regions of the two-dimensional space prescribed by principal component 1 (PC1, 25.0% variability explained) and principal component 2 (PC2, 17.3% variability explained) (Figure 1). The authenticated goldenseal reference samples (commercial and vouchered leaf and rhizome material) also clustered with their commercial counterparts (aerial (green) and root/rhizome (orange) samples, respectively). Sample variation was observed with the goldenseal root/rhizome and aerial supplements clustering together while three samples were located beyond the 95% confidence interval. Distinct clustering between the plant parts was shown in the scores plot of principal component two (PC2) versus principal component three (PC3) (Supplemental, Figure S2). The variability in spatial locations for the root and aerial samples in the PCA plot could represent variations in growth location, genetic differences, and/or processing methods; these differences have been shown to be detectable via metabolomics analyses [Ghatak, 2018; Kellogg, 2017; Pinasseau, 2017]. However, without more information from the supplier, it was not possible to determine the biological source of the observed variation.
Fig. 1:
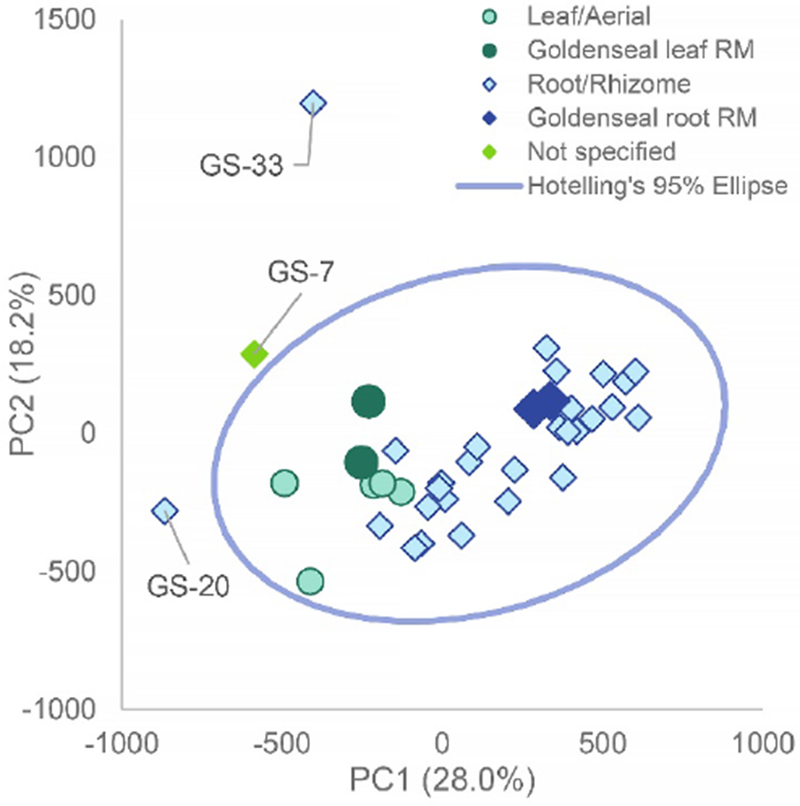
Principal component analysis (PCA) scores plot from untargeted mass spectrometry metabolomics analysis of commercial goldenseal samples. PC1 versus PC2 (25.0% and 17.3% explained variance, respectively) allowed for visualization of three distinct outliers: GS-20, GS-7, and GS-33. These outliers were distinct from the root and aerial samples (blue and green respectively) and fell outside the 95% Hotelling’s T2 confidence interval represented by the blue circle. The abbreviation “RM” represents “reference material” which are the standards that have been vouchered or purchased by Chromadex.
The untargeted metabolomics analysis of commercial goldenseal products revealed three samples which were distinct outliers from the goldenseal samples: GS-07, GS-20, and GS-33; all three fell outside the 95% confidence interval. While these three products were labeled as “goldenseal”, their distinct metabolomic location in the PCA scores plot raised suspicion that the three could be partially or completely adulterated with other botanical products. This hypothesis prompted further investigation into the chemical and botanical identity of these samples.
3.2. Tentative identification of unique compounds in outlier samples based upon PCA loadings plot
The separation observed in the PCA scores plot of PC1 versus PC2 (Figure 1) can be explained through metabolites highlighted in the corresponding PCA loading plots (Figure 2). The loadings plot graphically estimates the degree to which each variable (RT-m/z pair) contributes to the separation of three samples in the PCA scores plot; the greater the magnitude of a variable’s loading score, the more it contributes to that principal component. Graphing a corresponding PCA loadings plot highlights marker ions that are associated with the observed clustering [Lever, 2017].
Fig. 2:
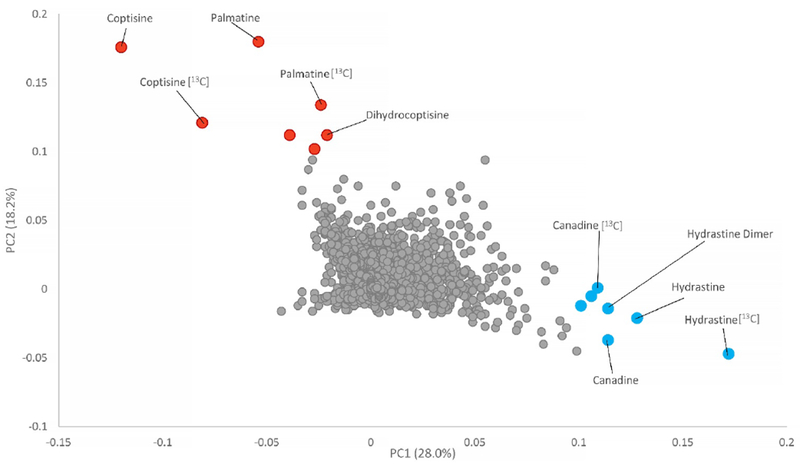
Loadings plots from untargeted MS-based PCA of goldenseal samples (PC1 vs PC2). Metabolites with greater positive loadings values along the y-axis (PC2, red labels) were present in higher concentrations in the outlier samples (GS-7, GS-20, and GS-33) and contribute to the separation observed along the vertical axis of Figure 1. Metabolites with greater positive values in the x-axis direction (PC1, blue labels) were more heavily represented in goldenseal supplements comprised of the aerial portions or the root/rhizome as well as the goldenseal reference material.
The PCA loadings plot highlighted hydrastine (m/z 384.1459 [M+H]+) and canadine (m/z 340.1537 [M+H]+) as having significant contributions towards the first principal component (PC1) due to their large magnitude in the x-direction. The loadings plot also included the 13C isotope peaks for each of the metabolites, lending additional confidence to the identification and significance of these compounds. A dimer of hydrastine was also present. Hydrastine and canadine are two of the main alkaloids present in goldenseal, root and leaf, and were found to be missing in the outlier samples [Le, 2013; Le, 2014]. The large cluster of goldenseal supplements, both of leaf and root/rhizome, were separated from the outliers along the positive x-axis due to the presence of these major alkaloids. These data suggest that the outliers could possess a possible mixture of plant material and/or a lack of Hydrastis canadensis.
A series of ions were observed to lie along the y-axis direction, contributing to the observed variance along the second principal component (PC2). PC2 was also responsible for discriminating outlier samples from goldenseal supplements (Figure 1). The loadings plot revealed several metabolites that were present in higher concentrations in the adulterated samples, and, thus were dominant peaks in the positive direction. These included coptisine (m/z 321.0954 [M+H]+), palmatine (m/z 352.1535 [M+H]+), their 13C isotopes (m/z 322.1072 [M+H]+ and 353.1571 [M+H]+, respectively), and dihydrocoptisine (m/z 323.1121 [M+H]+) (Figure 3). All of these compounds have been previously shown to be present in other berberine containing species, specifically Coptis chinensis, Berberis vulgaris, and Mahonia aquifolium [Ivanovska, 1996; Pengelly, 2012; Račková, 2004; Weber, 2003; Yang, 2017]. Their presence in the outliers (GS-07, GS-20, and GS-33) and absence in other goldenseal supplements and reference material was supported by the heat map (Figure 5) and the corresponding stacked mass spectrometry chromatograms (Figure 6).
Fig. 5:
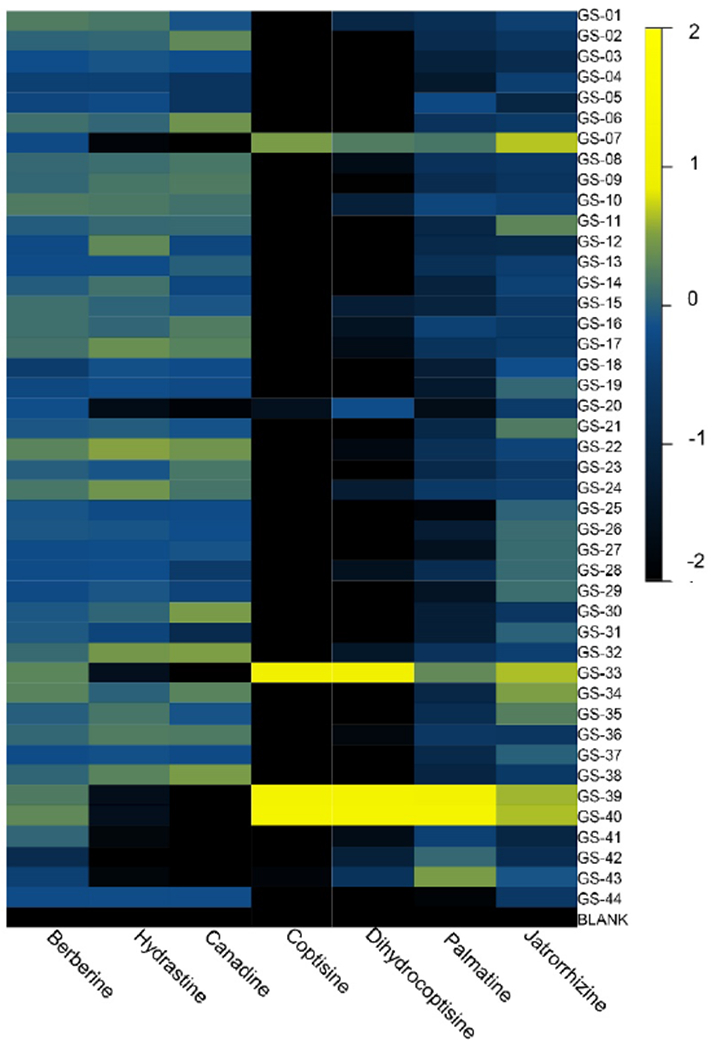
Heat map of the relative concentration of characteristic alkaloids in the goldenseal supplements and reference materials tested. The log of the mean-scaled peak area was used to determine the relative amount of these components present. Black represents low relative concentration; yellow corresponds to high relative quantities present. The outliers from the PCA, GS-07, GS-20, and GS-33 all yielded an alkaloid distribution pattern similar to that of the non-goldenseal reference materials and serve as an indication of possible adulteration.
Figure 6:
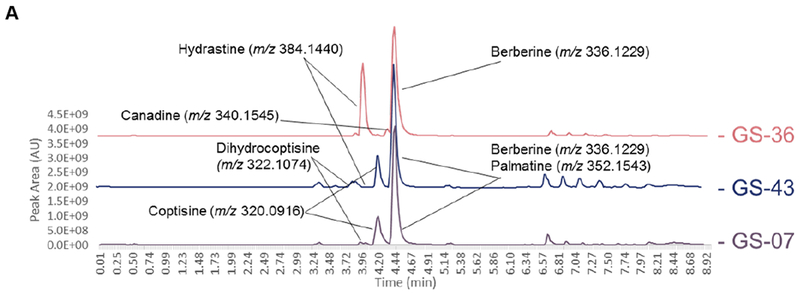
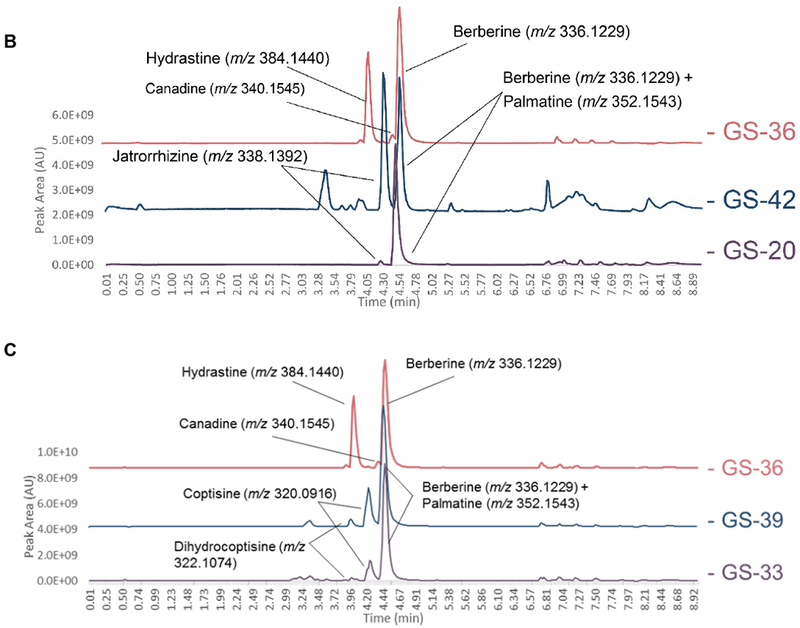
Liquid chromatography-mass spectrometry (LC-MS) chromatograms from supplement samples GS-07 (A), GS-33 (B), and GS-20 (C) (suspected to be altered compared against reference material profiles for H. canadensis (GS-36), C. chinensis (GS-39), B. vulgaris (GS-43), and M. Aquifolium (GS-42). Highlighted alkaloid peaks are identified through comparison of accurate mass from high-resolution mass spectrometry in the positive ionization mode. GS-07 and GS-33 revealed peaks corresponding to berberine, palmatine, coptisine, and dihydrocoptisine, the last three of which are not found in the H. canadensis reference material. The M. aquifolium standard possessed berberine, palmatine, and jatrorrhizine; all of which were also present in GS-20 but the latter two were found in much lower concentrations than what was observed in the M. aquifolium reference material.
3.3. Adulteration Analysis with Reference Materials
Reference materials for non-H. canadensis species were extracted and incorporated into the metabolomics analysis. These included Coptis chinensis rhizome (GS-39) and root (GS-40), Mahonia aquifolium leaf (GS-41) and root (GS-42), and Berberis vulgaris root (GS-43). The resulting dataset contained 5,573 marker ions for 135 objects (i.e., 35 goldenseal products, nine reference materials, and a process blank, all prepared by extraction in triplicate). After analysis of the reproducibility of the extraction method the average response of the triplicate extractions was taken to yield a final 5,423 × 45 dataset (RSD < 5%).
PCA analysis yielded a 4-component model explaining 62.6% of the variance contained within the dataset. Examining the position of the non-goldenseal reference material within the PCA scores plot, the standards for M. aquifolium herb and root (GS-41 and GS-42, respectively) clustered closely to sample GS-07. Sample GS-33 was located in close proximity to the C. chinensis reference materials (GS-39 and GS-40) and all three were shifted from the H. canadensis clusters, suggesting GS-33 was a dietary supplement formulated from C. chinensis instead of the labeled H. canadensis. Additionally, GS-07 was found in a similar region to the B. vulgaris (GS-43) reference material, implying there was a mix of plant material present. After examining the position of the samples in the PCA scores plot, the corresponding loadings plot and mass spectrometry chromatograms provided additional substantive evidence as to the variables (ions) responsible for the observed variation.
Liquid chromatography-mass spectrometry (LC-MS) chromatograms provided corroborating evidence as to the nature of the three adulterated samples (Figure 6). The base peak chromatogram for the H. canadensis reference material (GS-36 and GS-38) contained three main peaks in the positive ionization mode: berberine, m/z 336.1229 [M]+, hydrastine, m/z 384.1440 [M+H]+, and canadine, m/z 340.1545 [M+H]+, all of which are characteristic marker compounds for goldenseal [29–30]. Berberine was consistently present across all root/rhizome samples regardless of putative botanical origin (Figure 5); however, hydrastine and canadine levels were found to vary considerably. In the base peak chromatogram for GS-07, GS-20 and GS-33 (Figure 6A, 6B, and 6C, respectively), the peaks for canadine and hydrastine were not present. Three additional peaks – tentatively identified as palmatine (m/z 352.1543 [M+H]+), coptisine (m/z 320.0917 [M+H]+), and dihydrocoptisine (m/z 322.1074 [M+H]+) based on accurate mass determination – were present in GS-07 (Figure 6A), which coincided with ions having the same m/z (< 5.0 ppm) and retention time as the corresponding ions in reference material from B. vulgaris (GS-43) [Ivanovska, 1996; Weber, 2003]. The second outlier supplement, GS-20, had a large berberine peak present but was lacking hydrastine and canadine peaks (Figure 6B). The high berberine content was believed to be responsible for the clustering of GS-20 with GS-41 and GS-42 (M. aquifolium leaf and root, respectively). As shown in the extracted ion chromatograms, M. aquifolium root also contains jatrorrhizine (m/z 338.1392 [M]+) and palmatine (m/z 352.1543 [M+H]+) both of which were found in GS-20 (Figure 6C) [Račková, 2004; Weber, 2003]. The final outlier sample, GS-33, also displayed a spectral profile distinct from that of goldenseal [Weber, 2003; Yang, 2017] (Figure 6C). Canadine and hydrastine were not present, but palmatine, coptisine, and dihydrocoptisine were also detected in the sample. This was consistent with C. chinensis, commonly known as Chinese goldthread. The overlap observed in these chromatograms with non-goldenseal alkaloids, and the absence of two principal goldenseal marker compounds (hydrastine and canadine), supported the hypothesis from the untargeted metabolomic analysis that samples GS-07, GS-20, and GS-33 were adulterated.
4. Conclusion
Currently “there is a need to develop or extend existing analytical approaches to identify unexpected adulterants”, specifically in the dietary supplement industry [Pawar, 2017]. Untargeted metabolomics analysis of commercial goldenseal dietary supplements efficiently identified three samples as potentially being adulterated, without prior knowledge of composition or suspicion of adulteration. The altered status of samples GS-07, GS-20, and GS-33 was substantiated by incorporating standard reference materials of related botanical supplements into the metabolomics data analysis. This second, more detailed study revealed that these outliers possessed few goldenseal metabolites, instead containing other, non-goldenseal components. GS-07, appeared to contain B. vulgaris botanical material based upon the metabolite correlation found in the PCA scores plot (Figure 4) and mass spectrometry chromatograms of relevant reference materials (Figure 6A). GS-20, clustered closely to GS-42 (M. aquifolium root) and GS-41 (M. aquifolium herb) and shared several of the same non-goldenseal compounds (Figure 6B), showing the supplement may contain M. aquifolium rather than H. canadensis, or perhaps as a mixture of plant species. The last outlier, GS-33 was believed to contain C. chinensis root and rhizome, due to the high correlation with the references in the PCA plot (Figure 4), similar concentrations of metabolites (Figure 5) and comparative mass spectrometry chromatograms (Figure 6C).
Fig. 4:
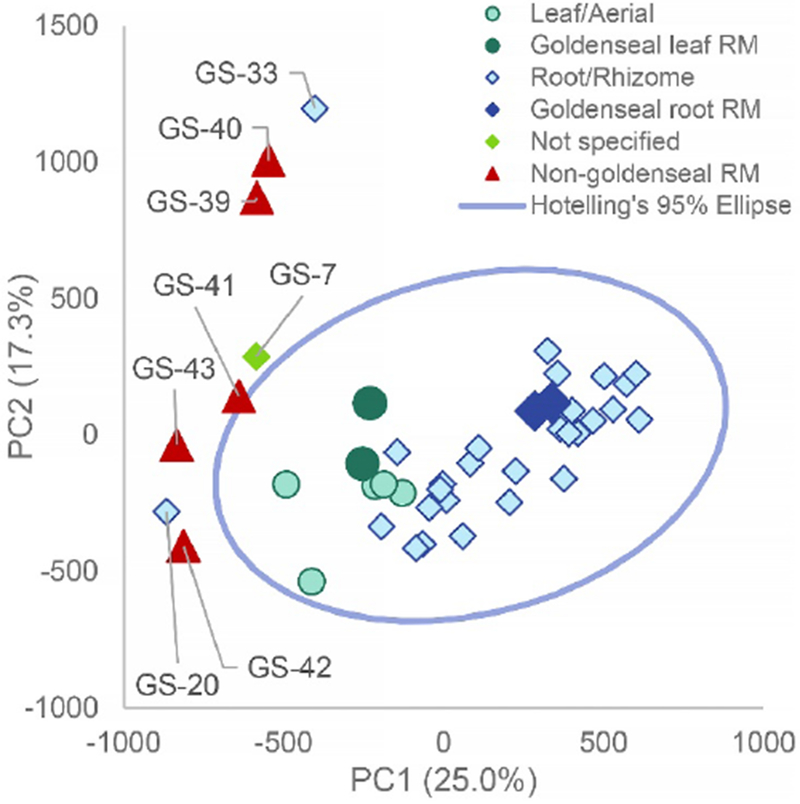
Principal component analysis (PCA) scores plot from untargeted mass spectrometry metabolomics analysis of commercial goldenseal samples with non-goldenseal reference material added for confirmation. Graphically comparing PC1 versus PC2 (25.0% and 17.3% explained variance, respectively) allowed for similar visualization as the original PCA analysis (Figure 1), showing clear discrimination between possibly adulterated supplements and goldenseal root/rhizome and aerial supplements. The blue line is a 95% confidence interval calculated using Hoetelling’s T2. The non-goldenseal reference materials (Coptis chinensis rhizome (GS-39), Coptis chinensis root (GS-40), Mahonia aquifolium leaf (GS-41), Mahonia aquifolium root (GS-42), and Berberis vulgaris root (GS-43)) were positioned outside of the 95% confidence interval and closely aligned with the previously identified outlier samples. GS-07 correlated with Berberis vulgaris root and herb standards, GS-20 correlated with Mahonia aquifolium root and herb standards, and GS-33 clustered with Coptis chinensis root and rhizome standards. The abbreviation “RM” represents “reference material” which are the standards that have been vouchered or purchased by Chromadex.
Analyzing similarity and variation within commercial botanical supplement products remains a challenge due to their innate phytochemical complexity. Hydrastis canadensis supplements possess a variety of bioactive secondary metabolites, which vary depending on the plant portion used for formulation [Le, 2013; Leyte-Lugo, 2017]. This study illustrates the effectiveness of untargeted metabolomics methodologies to analyze this variability and differentiate outlier samples that could indicate possible adulteration in a large sample set. Multivariate analysis of the metabolomics dataset effectively modeled the variance between the goldenseal supplements based upon physiological origin of the product, as well as differentiating three potentially adulterated samples from the other supplements via the PCA scores plot (Figure 1). Moreover, the distinction between the goldenseal supplements was achieved without any prior knowledge of the composition of the samples, nor of the identity of the possible adulterants. Subsequent investigation employing the PCA loadings plot (Figure 2) and the stacked mass spectrometry chromatograms (Figure 6) yielded discriminating features (ions) that were responsible for the differentiation between sample groups, providing information that suggests the identities of the botanical adulterants. These marker ions were discovered from a large sample set comprised of commercial botanical dietary supplements (reported on the labels to contain only goldenseal). One potential disadvantage to untargeted metabolomics is the possibility of ion suppression and matrix effects, where co-eluting components can affect the ionization efficiency of one another [Jorge, 2016; Lei, 2011]. This predominantly impacts lower abundant compounds or compounds with poor ionization efficiency, but it can also enhance ionization. Both of these effects of ion suppression and matrix effects have the potential to compromise accurate quantitation of the analytes across a sample set [Jorge, 2016; Lei, 2011]. Steps can be taken to minimize the effect of ion suppression, including more refined chromatographic separation of analytes, an alteration to the ionization mode, or inclusion of an appropriate internal standard [Antignac, 2005]. However, targeted techniques would have required an a priori understanding of the identity of the adulterating botanicals and the relevant marker ions associated with each species, and untargeted metabolomics effectively provided a simultaneous comparison, which included more of the complex chemical profile and is much more efficient than a pair-wise comparison.
There is a continued need to ascertain variability in complex botanical products, especially within the dietary supplement industry [Dietary Supplement Health and Education Act, 1994] to monitor quality control of products for adulteration [Tims, 2016], authenticate botanical samples [Smillie, 2010], or select samples from a broad range of commercial products [Kellogg, 2017]. The untargeted mass spectrometry-based metabolomics approach described herein has the potential to provide a versatile data acquisition tool for comparisons of multiple products with complex constituents. The collection of thousands of secondary metabolites represents a robust analytical technique that is capable of differentiating between closely-related samples, [Dowlatabadi, 2017; Geng, 2015; Geng, 2017; Kang, 2008; Mncwangi, 2014; Simmler, 2017; Steuer, 2017] and employing untargeted follow up analyses to discern potential adulteration from multiple complex botanical matrices representing a potentially valuable application of this analytical methodology.
Supplementary Material
Table 2:
The marker ions in Hydrastis canadensis, Berberis vulgaris, Mahonia aquifolium, and Coptis chinensis with [M+H]+ values. Check marks represent whether or not the ions were present in the following goldenseal samples: commercial goldenseal supplements GS-07, GS-20, GS-33, GS-36 (Hydrastis canadensis root standard), GS-39 (Coptis chinensis rhizome standard), GS-42 (Mahonia aquifolium root material), and GS-43 (Berberis vulgaris root material).
| compound | m/z value | GS-07 | GS-20 | GS-33 | GS-36 | GS-39 | GS-42 | GS-43 |
|---|---|---|---|---|---|---|---|---|
| Reference Material | Hydrastis canadensis | Coptis chinensis | Mahonia aquifolium | Berberis vulgaris | ||||
| Berberine | 336.1229 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Hydrastine | 384.1440 | ✓ | ||||||
| Canadine | 340.1545 | ✓ | ✓ | |||||
| Palmatine | 352.1543 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Coptisine | 320.0916 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Dihydrocoptisine | 322.1074 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Jatrorrhizine | 338.1392 | ✓ | ✓ | |||||
| Tentative botanical Identity | Berberis vulgaris | Mahonia aquifolium | Coptis chinensis | |||||
Highlights:
The composition of commercial goldenseal supplements was assessed by mass spectrometry-based profiling and untargeted metabolomic analysis.
Untargeted metabolomics analysis identified three outliers from a multi-sample dataset of goldenseal supplements.
These outliers were compared to non-goldenseal species to support the conclusion of possible adulteration.
Untargeted approaches did not require any a priori knowledge of the adulterating botanicals nor relevant marker ions.
5. Acknowledgements
This project was supported by the National Institutes of Health National Center for Complementary and Integrative Health (NIH NCCIH), specifically the Center of Excellence for Natural Product Drug Interaction Research (NaPDI, U54 AT008909). Mass spectrometry analyses were conducted in the Triad Mass Spectrometry Facility at the University of North Carolina at Greensboro (https://chem.uncg.edu/triadmslab/). The authors would like to thank TN Graf for his assistance with sample preparation and analytical guidance.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
7. References
- Amazon.com Amazon Best Sellers. https://www.amazon.com/Best-Sellers-Health-Personal-Care-Goldenseal-Herbal-Supplements/zgbs/hpc/3765771/ref=zg_bs_nav_hpc_3_3764461 (January, 2017).
- Antignac J-P, de Wasch K, Monteau F, De Brabander H, Andre F, Le Bizec B. The ion suppression phenomenon in liquid chromatography-mass spectrometry and its consequences in the field of residue analysis. Anal Chim Acta. 2005; 529: 129–36. [Google Scholar]
- Asher G, Corbett A, Hawke R. Common Herbal Dietary Supplement-Drug Interactions. Am Fam Physician. 2017; 96 (2): 101–07. [PubMed] [Google Scholar]
- Britton ER, Kellogg JJ, Kvalheim OM, Cech NB. Biochemometrics to Identify Synergists and Additives from Botanical Medicines: A Case Study with Hydrastis canadensis (Goldenseal). J Nat Prod. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, Roman MC. Determination of hydrastine and berberine in goldenseal raw materials, extracts, and dietarysupplements by high-performance liquid chromatography with UV: collaborative study. J AOAC Int. 2008; 91 (4): 694–701. [PMC free article] [PubMed] [Google Scholar]
- Cappello T, Giannetto A, Parrino V, De Marco G, Mauceri A, Maisano M. Food safety using NMR-based metabolomics: Assessment of the Atlantic bluefin tuna, Thunnus thynnus, from the Mediterranean Sea. Food Chem Toxicol. 2018; 115: 391–7. [DOI] [PubMed] [Google Scholar]
- Choen PA, Travis JC, Keizers PHJ, Deuster P, Venhuis BJ. Four experimental stimulants found in sports and weight loss supplements: 2-amino-6-methylheptane (octodrine), 1,4-dimethylamylamine (1,4-DMAA), 1,3-dimethylamylamine (1,3-DMAA) and 1,3-dimethylbutylamine (1,3-DMBA). Clin Toxicol. 2017. [DOI] [PubMed] [Google Scholar]
- Cicero AG, Baggioni A. Berberine and Its Role in Chronic Disease. Adv Exp Med Biol. 2016; 928: 27–45. [DOI] [PubMed] [Google Scholar]
- Coutinho Moraes DF, Still DW, Lum MR, Hirsch AM. DNA-Based Authentication of Botanicals and Plant-Derived Dietary Supplements: Where Have We Been and Where Are We Going? Planta Med. 2015; 81(09): 687–95. [DOI] [PubMed] [Google Scholar]
- Depke T, Franke R, Bronstrup M. Clustering of MS2 spectra using unsupervised methods to aid the identification of secondary metabolites from Pseudomonas aeruginosa. J Chromatogr. 2017; 1071: 19–28. [DOI] [PubMed] [Google Scholar]
- Dietary Supplement Health and Education Act, Public Law 103–417, 108 U. S. Stat. 4325, 25 October 1994, Sec. 2–13.
- Dowlatabadi R, Farshidfar F, Zare Z, Pirali M, Rabiei M, Khoshayand MR, Vogel HJ. Detection of adulteration in Iranian saffron samples by 1 H NMR spectroscopy and multivariate data analysis techniques. Metabolomics. 2017; 13: 1–11.27980501 [Google Scholar]
- Fox J, Weisberg S. An R Companion to Applied Regression. 2nd ed Thousand Oaks, CA: Sage; 2011. [Google Scholar]
- Geng P, Harnly JM, Chen P. Differentiation of Whole Grain from Refined Wheat (T. aestivum) Flour Using Lipid Profile of Wheat Bran, Germ, and Endosperm with UHPLC-HRAM Mass Spectrometry. J Agric Food Chem. 2015; 63 (27): 6189–211. [DOI] [PubMed] [Google Scholar]
- Geng P, Harnly JM, Sun J, Zhang M, Chen P. Feruloyl dopamine-O-hexosides are efficient marker compounds as orthogonal validation for authentication of black cohosh (Actaea racemosa)—an UHPLC-HRAM-MS chemometrics study. Anal Bional Chem. 2017; 409 (10): 2591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak A, Chaturvedi P, Weckwerth W. Metabolomics in Plant Stress Physiology. Berlin, Heidelberg, Springer Berlin Heidelberg: 1–50. [Google Scholar]
- Guo X, Long P, Meng Q, Ho C-T, Zhang L. An emerging strategy for evaluating the grades of Keemun black tea by combinatory liquid chromatography-Orbitrap mass spectrometry-based untargeted metabolomics and inhibition effects on alpha-glucosidase and alpha-amylase. Food Chem. 2018; 246: 74–81. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Barone G, Gurley BJ, Fifer EK, Hendrickson HP. Hydrastine pharmacokinetics and metabolism after a single oral dose of goldenseal (Hydrastis canadensis) to humans. Drug Metab Dispos. 2015; 43 (4): 534–52. [DOI] [PubMed] [Google Scholar]
- Gurley BJ, Swain A, Hubbard MA, Williams DK, Barone G, Hartsfield F, Tong Y, Carrier DJ, Cheboyina S, Battu SK. Clinical assessment of CYP2D6-mediated herb-drug interactions in humans: effects of milk thistle, black cohosh, goldenseal, kava, St. John’s wort, and Echinacea. Mol Nutr Food Res. 2008; 52 (7): 755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska N, Philipov S. Study on the anti-inflammatory action of Berberis vulgaris root extract, alkaloid fractions and pure alkaloids. Int J Immunopharmaco. 1996; 18 (10): 553–61. [DOI] [PubMed] [Google Scholar]
- Jorge TF, Mata AT, António C. Mass spectrometry as a quantitative tool in plant metabolomics. Philos Trans A Math Phys Eng Sci. 2016; 374 (2079): 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junio HA, Sy-Cordero AA, Ettefagh KA, Burns JT, Micko KT, Graf TN, Richter SJ, Cannon RE, Oberlies NH, Cech NB. Synergy-directed fractionation of botanical medicines: a case study with goldenseal (Hydrastis canadensis). J Nat Prod. 2011; 74 (7): 1621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lee S, Kang S, Kwon HN, Park JH, Kwon SW, Park S. NMR-based metabolomics approach for the differentiation of ginseng (Panax ginseng) roots from different origins. Arch Pharm Res. 2008; 31 (3): 330–36. [DOI] [PubMed] [Google Scholar]
- Karmakar SR, Biswas SJ, Khuda-Bukhsh AR. Anti-carcinogenic Potentials of a Plant Extract (Hydrastis canadensis): I. Evidence from In Vivo Studies in Mice (Mus musculus). Asian Pac J Cancer P. 2010; 11: 545–51. [PubMed] [Google Scholar]
- Kellogg JJ, Graf TN, Paine MF, McCune JS, Kvalheim OM, Oberlies NH, Cech NB. Comparison of Metabolomics Approaches for Evaluating the Variability of Complex Botanical Preparations: Green Tea (Camellia sinensis) as a Case Study. J Nat Prod. 2017; 80 (5): 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg JJ, Todd DA, Egan JM, Raja HA, Oberlies NH, Kvalheim OM, Cech NB. Biochemometrics for Natural Products Research: Comparison of Data Analysis Approaches and Application to Identification of Bioactive Compounds. J Nat Prod. 2016; 79(2): 376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le PM, et al. Characterization of the alkaloids in goldenseal (Hydrastis canadensis) root by high resolution Orbitrap LC-MSn. Anal Bioanal Chem. 2013; 405(13): 4487–98. [DOI] [PubMed] [Google Scholar]
- Le PM, McCooeye M, Windust A. Application of UPLC-QTOF-MS in MS(E) mode for the rapid and precise identification of alkaloids in goldenseal (Hydrastis canadensis). Anal Bioanal Chem. 2014; 406 (6): 1739–49. [DOI] [PubMed] [Google Scholar]
- Lei Z, Huhman DH, Sumner LW. Mass Spectrometry Strategies in Metabolomics. J Biol Chem. 2011; 286: 25435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever j, Krzywinski M, Altman N. Points of Significance: Principal component analysis. Nat Methods. 2017; 14: 641–2. [Google Scholar]
- Leyte-Lugo M, Britton ER, Foil DH, Brown AR, Todd DA, Rivera-Chavez J, Oberlies NH, Cech NB. Secondary Metabolites from the Leaves of the Medicinal Plant Goldenseal (Hydrastis canadensis). Phytochem Lett. 2017; 20: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DP. Authentication of Ginkgo biloba herbal dietary supplements using DNA barcoding. Genome. 2014; 57(9): 513–16. [DOI] [PubMed] [Google Scholar]
- Liu Y, Finley J, Betz JM, Brown P. FT-NIR characterization with chemometric analyses to differentiate goldenseal from common adulterants. Fitoterapia. 2018. [DOI] [PubMed] [Google Scholar]
- Lu Y, Sun J, Petrova K, Yang X, Greenhaw J, Salminen WF, Beger RD, Schnackenberg LK. Metabolomics evaluation of the effects of green tea extract on acetaminophen-induced hepatotoxicity in mice. Food Chem Toxicol. 2013; 62: 707–21. [DOI] [PubMed] [Google Scholar]
- McGraw JB, Sanders SM, Van der Voort M. Distribution and abundance of Hydrastis Canadensis L. (Ranunculaceae) and Panax quinquefolius L. (Araliaceae) in the central Appalachian region. J Torrey Bot Soc. 2003; 130 (2): 62–69. [Google Scholar]
- Mncwangi NP, Viljoen AM, Zhao J, Vermaak I, Chen W, Khan I. What the devil is in your phytomedicine? Exploring species substitution in Harpagophytum through chemometric modeling of 1H-NMR and UHPLC-MS datasets. Phytochemistry. 2014; 106: 104–15. [DOI] [PubMed] [Google Scholar]
- New York State Office of the Attorney General. A.G. Schneiderman asks major retailers to halt sales of certain herbal supplements as DNA tests fail to detect plant materials listed on majority of products tested. 2015.
- Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016; 79 (3): 629–61. [DOI] [PubMed] [Google Scholar]
- Parveen I, Gafner S, Techen N, Murch SJ, Khan IA. DNA Barcoding for the Identification of Botanicals in Herbal Medicine and Dietary Supplements: Strengths and Limitations. Planta Med. 2016; 82(14):1225–35. [DOI] [PubMed] [Google Scholar]
- Pawar SR, Handy SM, Cheng R, Shyong N, Grundel E. Assessment of the Authenticity of Herbal Dietary Supplements: Comparison of Chemical and DNA Barcoding Methods. Planta Med. 2017; 83(11): 921–936. [DOI] [PubMed] [Google Scholar]
- Pengelly A, Bennett K, Spelman K, Tims M. An Appalachian Plant Monograph: Goldenseal Hydrastis Canadensis L. Appalachian Center for Ethnobotanical Studies. 2012. [Google Scholar]
- Pinasseau L , Vallverdú-Queralt A 1, Verbaere A , Roques M , Meudec E , Cunff LL , Péros J-P, Ageorges A, Sommerer N, Boulet J-C, Terrier N, Cheynier V. Cultivar Diversity of Grape Skin Polyphenol Composition and Changes in Response to Drought and Investigated by LC-MS Based Metabolomics. Front Plant Sci. 2017; 8: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Račková L, Májeková M, Košt’álová D, Štefek M. Antiradical and antioxidant activities of alkaloids isolated from Mahonia aquifolium. Structural aspects. Bioorgan Med Chem. 2004; 12 (17): 4709–715. [DOI] [PubMed] [Google Scholar]
- Raja HA, Baker TR, Little JG, Oberlies NH. DNA barcoding for identification of consumer-relevant mushrooms: A partial solution for product certification? Food Chemistry. 2017; 214: 383–92. [DOI] [PubMed] [Google Scholar]
- Simmler C, Graham JG, Chen S-N, Pauli GF. Integrated analytical assets aid botanical authenticity and adulteration management. Fitoterapia. 2017; 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie TJ, Khan IA. A Comprehensive Approach to Identifying and Authenticating Botanical Products. Clin Pharmacol Ther. 2010; 87: 175–86. [DOI] [PubMed] [Google Scholar]
- Smith T, Kawa K, Eckl V, Johnson J. Sales of Herbal Dietary Supplements in US Increased 7.5% in 2015 Consumers spent $6.92 billion on herbal supplements in 2015, marking the 12th consecutive year of growth. Herbalgram. 2016; (111): 67–73. [Google Scholar]
- Souard F, Delporte C, Stoffelen P, Thevenot EA, Noret N, Dauvergne B, Kauffmann J-M, Van Antwerpen P, Stevigny C. Metabolomics fingerprint of coffee species determined by untargeted-profiling study using LC-HRMS. Food Chem. 2018; 245: 603–612. [DOI] [PubMed] [Google Scholar]
- Steuer AE, Arnold K, Schneider TD, Poetzsch M, Kraemer T. A new metabolomics-based strategy for identification of endogenous markers of urine adulteration attempts exemplified for potassium nitrate. Anal Bioanal Chem. 2017; 409 (26): 6235–44. [DOI] [PubMed] [Google Scholar]
- Sun L-M, Zhu B-J, Cao H-T, Zhang X-Y, Zhang Q-C, Xin G-Z, Pan L-M, Liu L-F, Zhu H-X. Explore the effects of Huang-Lian-Jie-Du-Tang on Alzheimer’s disease by UPLC-QTOF/MS-based plasma metabolomics study. J Pharmaceut Biomed. 2018; 151: 75–83. [DOI] [PubMed] [Google Scholar]
- Tao H-X, Xiong W, Zhao G-D, Peng Y, Zhong Z-F, Xu L, Duan R, Tsim KWK, Yu H, Wang Y-T. Discrimination of three Siegesbeckiae Herba species using UPLC-QTOF/MS-based metabolomics approach. Food Chem Toxicol. 2018:1–7. [DOI] [PubMed] [Google Scholar]
- Tims M On Adulteration of Hydrastis Canadensis root and rhizome. Botanical Adulterants Bulletin [Online], 2016. [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Moeller AMS, Schwartz M, Venables B. gplots: Various R Programming Tools for Plotting Data R package version 3.0.1. 2016. https://CRAN.R-project.org/package=gplots [Google Scholar]
- Weber HA, Zart MK, Hodges AE, Molloy HM, O’Brien BM, Moody LA, Clark AP, Harris RK, Overstreet JD, Smith CS. Chemical Comparison of Goldenseal (Hydrastis Canadensis L.) Root Powder from Three Commercial Suppliers. J Agri Food Chem. 2003; 51: 7352–58. [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng J, Li F, Liu X, Deng M, Wu H. Determination of Alkaloid Contents in Various Tissues of Coptis Chinensis Franch. by Reversed Phase-High Performance Liquid Chromatography and Ultraviolet Spectrophotometry. J Chromatogr Sci. 2017; 55 (5): 556–563. [DOI] [PubMed] [Google Scholar]
- Zhang L, Shen H, Xu J, Xu J-D, Li Z-L, Wu J, Zou Y-T, Liu L-F, Li S-L. UPLC-QTOF-MS/MS-guided isolation and purification of sulfur-containing derivatives from sulfur-fumigated edible herbs, a case study on ginseng. Food Chem. 2018; 246: 202–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


