Abstract
Here we report the fragmentation of disulfide linked intact proteins using activated-ion electron transfer dissociation (AI-ETD) for top-down protein characterization. This fragmentation method is then compared to the alternative methods of HCD, ETD, and EThcD. We analyzed multiple precursor charge states of the protein standards bovine insulin, α-lactalbumin, lysozyme, β-lactoglobulin, and trypsin inhibitor. In all cases we found that AI-ETD provides a boost in protein sequence coverage information and the generation of fragment ions from within regions enclosed by disulfide bonds. AI-ETD shows the largest improvement over the other techniques when analyzing highly disulfide linked and low charge density precursors ions. This substantial improvement is attributed to the concurrent irradiation of the gas phase ions while the electron-transfer reaction is taking place, mitigating non-dissociative electron transfer, helping unfold the gas phase protein during the electron transfer event, and preventing disulfide bond reformation. We also show that AI-ETD is able to yield comparable sequence coverage information when disulfide bonds are left intact relative to proteins that have been reduced and alkylated. This work demonstrates that AI-ETD is an effective fragmentation method for the analysis of proteins with intact disulfide bonds, dramatically enhancing sequence ion generation and total sequence coverage compared to HCD and ETD.
Keywords: activated-ion electron transfer dissociation, electron transfer dissociation, mass spectrometry, proteomics, top down proteomics, disulfide bonds, ion/ion reactions
Graphical Abstract
Top-down mass spectrometry allows researchers to interrogate proteins and protein modifications without the need for protein digestion or derivatization1,2. The potential benefits to avoid these steps are myriad and include investigation of genetic variants, alternative splicing, and site occupancy of post-translational modifications – information that is often lost upon enzymatic digestion3–5. One limitation to the top down approach is that intact protein cations do not dissociate as completely or readily as peptides during tandem mass spectrometry (MS/MS). MS/MS methods such as collision-activated dissociation (CAD) and infrared multiple photon dissociation (IRMPD) often selectively cleave the most labile bonds, limiting sequence coverage and PTM localization6–8. Offering more extensive dissociation and the ability to preserve most PTMs, the electron based dissociation methods, i.e., electron-capture dissociation (ECD) and electron-transfer dissociation (ETD), have become particularly important for top-down mass spectrometry9–11.
Accessing information about PTMs is indeed one of the most attractive strengths of the top-down approach. Disulfide bonds are among the most common PTMs and have fundamental roles in protein stabilization, structure, and function12–14. That said, they are challenging to study as they necessitate extensive fragmentation of interlinked peptide backbone sequences. To improve fragmentation, most top-down MS/MS methodologies reduce and alkylate disulfide bonds prior to analysis. Some methods use online disulfide bond reduction just prior to electrospray ionization in both shotgun and top-down proteomic regimes to improve precursor ion fragmentation, eliminating the reduction and alkylation step in peptide or protein sample preparation15–18. Similarly, matrix-assisted laser desorption/ionization in-source decay (MALDI-ISD) can be performed in a reducing matrix, such as 1,5-diaminonaphthalene (1,5-DAN), to reduce disulfide bonds and fragment intact proteins19–21. This method has been shown to yield high sequence coverage for proteins, but is limited to producing singly charged fragment ions, necessitating a wide m/z range mass analyzer or pseudo-MS3 analysis for the interrogation of intact proteins22,23. Other studies have left disulfide bonds intact but performed enzymatic digestion so that disulfide bridged peptides can be detected. That approach, however, suffers the same disadvantages of all shotgun approaches, namely that combinatorial patterns of modification cannot be discerned24–30. Producing sequence informative fragment ions from disulfide-bridge peptides or proteins with intact disulfide bonds remains challenging, as collision-based methods, the most prevalent fragmentation type used in proteomics experiments, do not efficiently cleave disulfide bonds, limiting sequence coverage within the region contained by disulfide bridges.
Twenty years ago McLafferty and co-workers demonstrated that disulfide bonds can be cleaved in the gas phase by ECD31–33. Since that time many dissociation methods have been examined for their potential application to disulfide bond characterization – e.g., electron transfer dissociation (ETD)29, electron detachment dissociation (EDD)34, ultraviolet photodissociation (UVPD)35,36, infrared multiple photon dissociation (IRMPD)34, metal-cationization37–39, excitation energy transfer (EET)40, electron transfer and higher-energy collision dissociation (EThcD)24, and radical induced dissociation41. Recently, Loo and colleagues described that pre-activation of ribonuclease A with UV and IR photons followed by ECD improved fragmentation over ECD alone, even allowing cleavage of multiple disulfide bonds42. In this same work, fragmentation of porcine insulin cleaved all disulfide bonds, yielding 73% sequence coverage, an improvement over previous studies31,37,42,43. Another important finding was that lengthening the time between UV and IR laser pulses allowed close proximity disulfide bonds to reform, revealing that the disulfide bond reformation of radical thiols was between 10 and 100 ms42. Despite these advancements, no one method can fragment disulfide intact proteins nearly as well as the same protein with disulfide bonds reduced and alkylated prior to analysis.
Activated-ion ETD (AI-ETD) is a form of ETD where ions are concurrently irradiated with infrared photons during the ion-ion reaction44–46. This supplemental energy has been shown to increase peptide and protein fragmentation by mitigating non-dissociative electron transfer. Additionally, the concurrent nature of AI-ETD minimizes hydrogen abstraction events between product ions while also incurring no additional time costs to the ETD scan sequence. Our lab recently showed that AI-ETD greatly improves the sequence coverage of intact protein standards as compared to HCD, ETD, and EThcD47–50 and improves analysis of post-translationally modified intact proteins, namely phosphoproteins51. Here we explore the utility of AI-ETD for interrogating proteins with intact disulfide bonds. Specifically, we compared the fragmentation of five protein standards (bovine insulin, bovine β-lactoglobulin, soybean trypsin inhibitor, α-lactalbumin, and chicken egg lysozyme) with molecular weights ranging from 5.7 to 20 kDa that have two to four intact disulfide bonds which enclose varying degrees of the protein backbone. Having dissociated these species using HCD, ETD, EThcD, and AI-ETD, we conclude that AI-ETD yields greater sequence coverage, fragment ion generation, and disulfide bond cleavage for all precursor charge states studied, with the greatest benefit arising from low charge density precursors. Following from previous studies regarding gas phase protein structure and electron transfer mechanics, we hypothesize that the condensed state of the proteins with intact disulfide bonds greatly enhances the abundance of non-dissociative electron transfer and prevents electron transfer from occurring in the interior region of the gas phase protein44,48,52,53. The benefit of concurrent supplemental infrared irradiation in AI-ETD is two-fold: 1) non-dissociative electron transfer product ions are converted to sequence informative product ions and 2) protein cations are unfolded to allow electron transfer to occur in the interior regions of the protein cation and prevent disulfide bond reformation. These two phenomenon work in tandem to allow for a significant improvement in fragment ion yield and therefore protein structure elucidation. Furthermore, because AI-ETD is able to fragment proteins effectively across the entire charge state envelope, the necessity to select high charge density precursors for successful ETD reactions is eliminated. This makes AIETD amenable to a wide range of proteins, which is especially valuable for disulfide-bonded proteins that trend toward low charge density precursor ions.
EXPERIMENTAL SECTION
Materials and Sample Preparation.
The proteins bovine insulin, bovine β-lactoglobulin, and soybean trypsin inhibitor were purchased from Sigma-Aldrich (St. Louis, MO, USA) and α-lactalbumin and chicken egg lysozyme were obtained from Protea Biosciences (Morgantown, WV, USA). Formic acid ampules and acetonitrile were obtained from Thermo Fisher Scientific (Rockford, IL, USA). Solutions were prepared with Milli-Q water (Millipore Corporation, Billerica MA). Samples were prepared for infusion by suspending each protein in 49.9:49.9:0.2 acetonitrile/water/formic acid to a final concentration of 10 pmol per μL. For comparison to the disulfide intact protein, lysozyme was also reduced and alkylated. Lysozyme was suspended in buffer (8 M urea, 50 mM Tris, pH 8) and incubated with 5 mM diothiothreiol for 45 minutes at 58° C, then alkylated with 15 mM iodoacetamide for 45 minutes at room temperature in the dark. The sample was then desalted with a C2 SepPak (Waters, Milford, MA), evaporated, and resuspended in 49.9:49.9:0.2 acetonitrile/water/formic acid to a final concentration of 10 pmol per μL.
ESI-MS/MS Analysis.
Each protein standard was infused via syringe pump at a flow rate of 5 μL per minute and electrosprayed with a spray voltage of 4 to 5 eV and inlet capillary temperature of 275° C. All mass spectrometry experiments were performed on a Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) modified with a Firestar T-100 Synrad 60-W CO2 continuous wave laser (Mukilteo, WA) for performing AI-ETD, as previously described54. Survey scans using intact protein mode for each protein were performed at 240,000 resolution and averaged over 100 scans. MS/MS experiments were also performed using intact protein mode at 240,000 resolution with a precursor AGC target of 800,000 and averaged over 400 scans. For each protein, three precursor charge states spanning the protein envelope were selected for analysis. For HCD, normalized collision energies of 15, 20, and 25 were used to find the optimal energy for fragmentation. For ETD, EThcD, and AI-ETD, the reagent anion AGC target was set to 300,000 and the ETD reaction time was varied to optimize fragment ion generation and sequence coverage, from 20 to 38 milliseconds. Normalized collision energies of 8, 10, 12, and 15 were used for EThcD and laser powers of 18, 24, 30, and 36 Watts were used for AI-ETD to determine optimal fragmentation.
Data Analysis.
Raw MS/MS spectra were deconvoluted using the Xtract algorithm (Thermo Fisher Scientific). The spectra were then compared against all possible b, y, c, and z•-type fragment ions which could be formed from that protein. Modifications were allowed for fragment ions containing a cysteine involved in a disulfide bond to consider all possible cleavage positions of the disulfide bond (S-S and C-S cleavage) and for hydrogen rearrangement products. Cleavage of all disulfide bonds were allowed but only one peptide backbone bond cleavage was considered. Internal fragment ions were not considered because they have been shown to be significantly less prevalent than terminal fragment ions and would greatly increase the fragment ion search space, leading to the false identification of fragment ions55–57. Fragment ions were matched within a mass tolerance of 10 parts per million.
RESULTS AND DISCUSSION
Dissociate proteins with intact disulfide bonds.
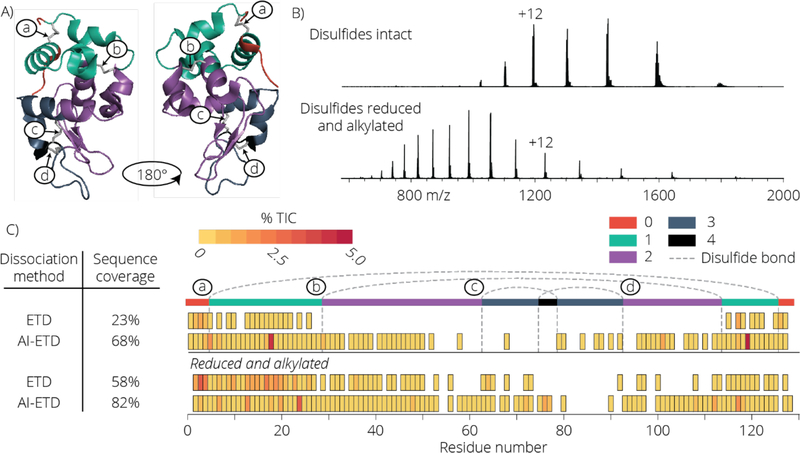
AI-ETD improves the fragmentation of intact proteins when compared to ETD47,48. This boost is realized by the absorption of infrared photons by the protein cations – a process that ultimately induces gas-phase protein unfolding and concomitantly a boost in ETD efficiency. To investigate the potential of AI-ETD for dissociation of disulfide linked protein cations, we selected lysozyme, a protein that contains four disulfide bonds (Table 1, Figure 1 panel A) enclosing 94% of the protein backbone. We analyzed the +12 charge state precursor of lysozyme using ETD and AI-ETD both with the disulfide bonds intact and cleaved (i.e., reduced and alkylated prior to analysis). The precursor ion charge distributions for the lysozyme cations are shown in Figure 1 panel B. Panel C of Figure 1 presents a sequence coverage map afforded by each fragmentation method. When the disulfide bonds are reduced and cysteines are alkylated, ETD and AI-ETD cleave 58% and 82% of the backbone residues, respectively. Dissociating proteins with intact disulfides, however, presents a much greater challenge. While both methods have reduced sequence coverage, ETD provides only 23% coverage while AI-ETD achieves a much higher 58% coverage. The coverage map shown in panel C of Figure 1 illuminates the underlying cause for this discrepancy. Here the coverage map is divided into five regions (0, 1, 2, 3, and 4) where the region number corresponds to the number of disulfide bonds that must be cleaved in addition to a protein backbone bond in order to produce a sequence-informative product ion. For example, any observed fragment resulting from dissociation of the backbone between residues 31 and 63 (region 2) can only be formed if three bonds are broken – one protein backbone and two disulfides. Note that ETD does not generate any fragments where more than two dissociations are required – i.e., one backbone and one disulfide. This indicates a clear relationship between the number of disulfide bonds enclosing a region and the amount of sequence informative fragment ions formed. Infrared photoactivation of protein cations during ETD (AI-ETD) disrupts non-covalent interactions and helps to reduce non-dissociative electron transfer (ETnoD)58,59. These data demonstrate that the concurrent photoactivation used in AI-ETD can open the precursor gas-phase structure and expose the interior of a disulfide linked protein cation so that multiple dissociative electron transfer events can occur. In fact, for lysozyme we detect many fragments that result from three bond cleavages (one backbone and two disulfides) and several that result from four (one backbone and three disulfides).
Table 1.
Summary of proteins
| Protein | MW (Da) | Length | # disulfides |
|---|---|---|---|
| Bovine insulin | 5729 | 51 | 3 |
| Alpha-lactalbumin | 14169 | 123 | 4 |
| Lysozyme | 14296 | 129 | 4 |
| Beta-lactoglobulin | 18263 | 162 | 2 |
| Trypsin Inhibitor | 19965 | 180 | 2 |
Figure 1.
Panel A shows the 3 dimensional structure of lysozyme with all 4 disulfide bonds intact in its native state. The regions are color coded based on the number of disulfide bond breakages required to generate a fragment ion for that region. The four disulfide bonds are labeled a, b, c, and d to illustrate where they occur along the protein backbone. The electrospray ionization MS1 spectra for lysozyme with disulfide bonds intact and disulfide bonds reduced and alkylated is shown (B). Panel C compares the sequence coverage of lysozyme precursor charge state +12 using ETD and AI-ETD with disulfide bonds intact or reduced and alkylated. Fragment ion intensities are reported as a percentage of the total ion current (TIC), calculated by taking the sum of the intensity of all fragment ions where a specific residue cleavage occurred and dividing by the total signal of all peaks in the mass spectrum. Sequence coverage of the highly disulfide linked region is hindered greatly using ETD, while AI-ETD shows pronounced coverage of this portion of the protein.
Comparison of AI-ETD and other methods for dissociation of proteins entirely enclosed by disulfide bonds.
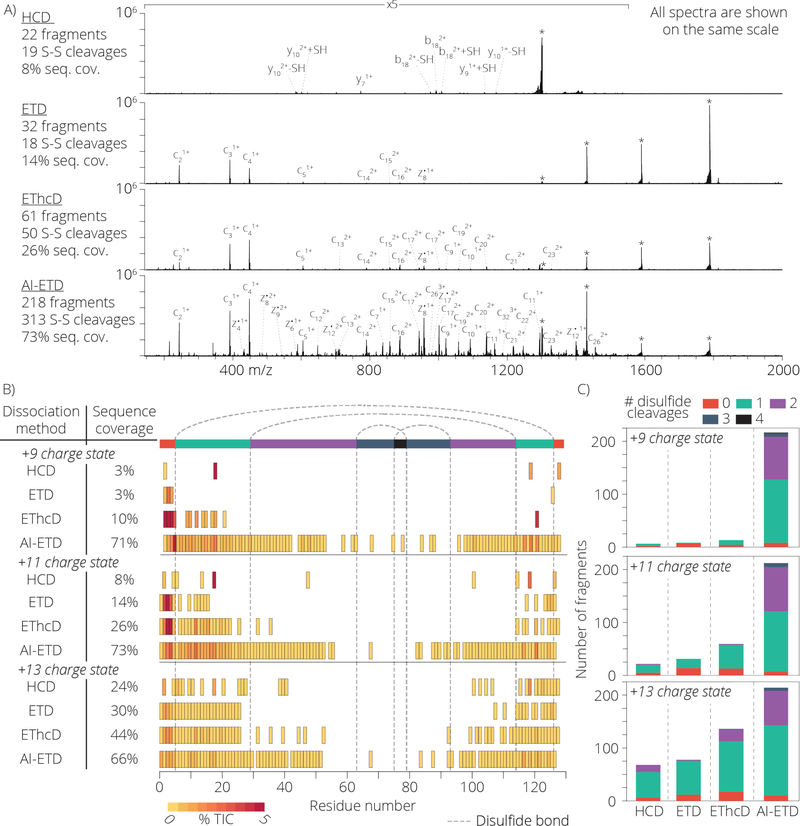
Having established the efficacy of AI-ETD to dissociate multiple disulfide linkages, we next sought to characterize performance for various charge states of the same protein (i.e., z = +9, z = +11, and z = +13 precursor ions of lysozyme) and benchmark this performance to other common dissociation methods including beam-type collisional activation (HCD), ETD, and electron transfer dissociation with HCD as supplemental collisional activation (EThcD). Figure 2 summarizes the results for lysozyme. Figure 2 panel A compares the MS/MS spectra using each dissociation method for the z = +11 charge state. The percentage of the total product ion signal contained in sequence-informative fragment ion channels is 10%, 14%, 25%, and 49% for HCD, ETD, EThcD, and AI-ETD, respectively. From these data we conclude AI-ETD induces much more extensive fragmentation than any of the other tested methods. To see how these fragment ions facilitate sequence analysis, we generated sequence coverage maps for each dissociation method for all three precursor ion charge states (Figure 2, panels B and C). As with the example above, AI-ETD generates substantially more sequence informative fragment ions, especially for those ions requiring disulfide bond cleavages. Again AI-ETD allows for the observation of fragment ions that result from cleavage of up to five bonds (i.e., one backbone and four disulfide linkages). For the z = +9 charge state precursor, only AI-ETD produced any fragmentation within the region enclosed by two or more disulfide bonds (show in purple, blue, and black along the top of the plot). As the charge density of the precursor increased, the sequence coverage for HCD, ETD, and EThcD is improved, while AI-ETD stays constant. This is consistent with previous work showing that higher charge states improve fragmentation for ETD while AI-ETD is more or less indifferent to precursor charge density. We next conducted a similar study but with a different protein – α-lactalbumin (14.2 kDa). The sequence coverage for the z = +10 charge state precursor of this protein reveals AI-ETD yields 64% sequence coverage while HCD, ETD, and EThcD show 6%, 12%, and 26%, respectively (Figure S1). The results of the fragmentation of these two proteins reveal that AI-ETD is very effective at fragmenting highly disulfide bonded proteins across all charge states, but also that vibrational activation is sufficient to cleave both disulfide bonds and peptide bonds, as noted by the performance of HCD, which cleaved 1 and sometimes 2 disulfide bonds for higher charge state precursors.
Figure 2.
Panel A shows a comparison of HCD, ETD, EThcD, and AI-ETD fragmentation of lysozyme with precursor charge state +11 with all 4 disulfide bonds intact. All spectra are 400 scan averages and are shown on the same scale. The scan range of 150 – 1550 m/z are shown at 5 times magnification. Peaks annotated with an asterisk (*) show the unreacted precursor and charge reduced precursor ions. A summary of the number of detected fragment ions, number of 22 disulfide cleavages (either S-S or C-S) amongst identified fragments, and percentage of inter-residue bond cleavages (referred to as sequence coverage) is shown to the right for each dissociation method. Panel B illustrates the sequence coverage achieved for each dissociation method tested for the precursor charge states +9, +11, and +13. Notably, AI-ETD is able to generate fragment ions within the region of the protein requiring 2 or more disulfide bond cleavages far more often than the other dissociation methods. Panel C illustrates the number of identified fragment ions and the amount of disulfide bond cleavages necessary to form that ion.
Comparison of AI-ETD and other methods for dissociation of proteins partially enclosed by disulfide bonds.
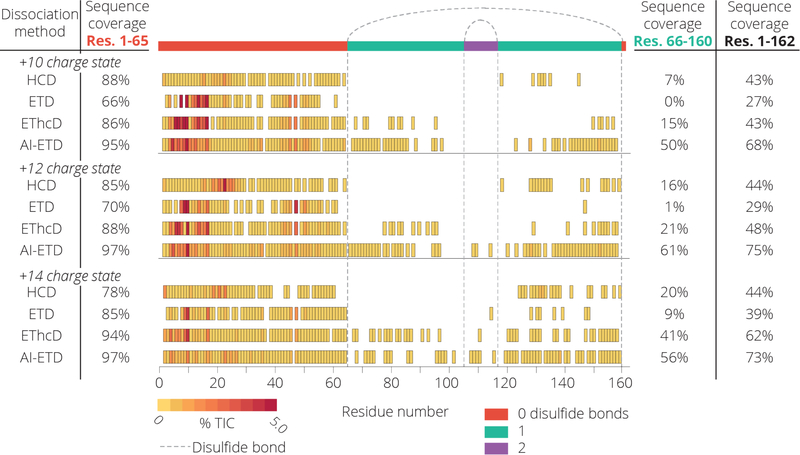
To contrast the highly disulfide linked proteins lysozyme and α-lactalbumin, we next analyzed β-lactoglobulin which has two disulfide bonds enclosing 59% of the protein. Figure 3 presents sequence coverage maps following dissociation of the +10, +12, and +14 precursor charge states of β-lactoglobulin with either HCD, ETD, EThcD, or AI-ETD. To isolate the effect of the disulfide bond, we also calculate sequence coverage percentages between residues 1–65 (not enclosed by a disulfide bond) and residues 66–160 (enclosed by one or two disulfide bonds). Not surprisingly the region containing no disulfide bonds has high sequence coverage regardless of precursor charge state or dissociation method. The region enclosed by disulfide bonds, however, is much less accessible. AI-ETD cleaves at least 50% of the bonds in this region while ETD and HCD produce very few fragment ions. In order to account for the extent of fragmentation due to infrared irradiation, we compared the fragmentation of the +12 charge state precursor of β-lactoglobulin using IRMPD, ETD, and AI-ETD (Figure S2). This experiment revealed that at the laser power and reaction time used for AI-ETD, very little fragmentation occurs. However, of the three low intensity fragment ions that were identified, two (z•14 and b138) required the cleavage of a disulfide bond, while the 48 identified fragments using ETD only contained one which cleaved a disulfide bond. This experiment clearly demonstrates the synergistic effect of concurrent IR irradiation and ETD, with AI-ETD providing drastically more sequence informative fragmentation than ETD and IRMPD alone. In addition, the structurally similar protein trypsin inhibitor was examined. This protein has two disulfide bonds enclosing 31% of the protein backbone. AI-ETD successfully sequenced 60% of the protein for all charge states examined (Figure S3). These data demonstrate that the benefit of AI-ETD is most pronounced in regions that are enclosed by disulfide bonds.
Figure 3.
The sequence coverage cleavage map for the +10, +12, and +14 charge state precursors of β-lactoglobulin are shown. The left sequence coverage values represent the region of the protein which does not contain any disulfide bonds and the left values show the sequence coverage which contains 1 or 2 disulfide bonds. All fragmentation methods perform well in the open region, while AI-ETD shows significant increase in coverage for the region enclosed by disulfide bonds.
Comparison of AI-ETD and other methods for dissociation of a protein with interpeptide disulfide bonds.
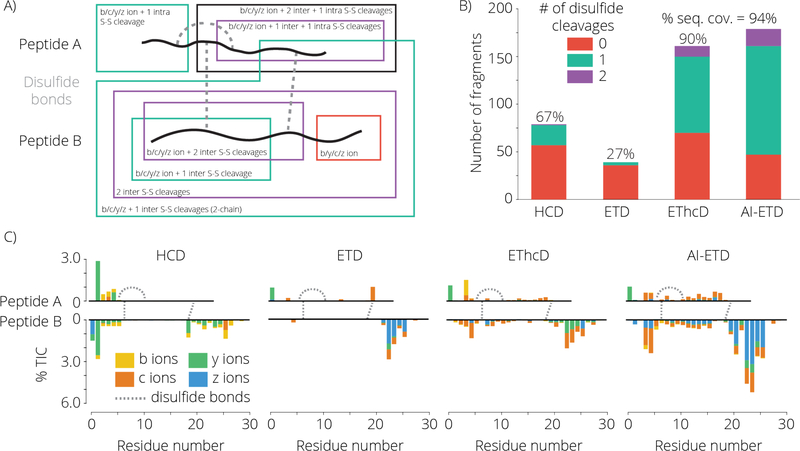
Lastly we investigated the fragmentation of bovine insulin with intact disulfides. Insulin comprises of two separate peptide chains linked by two disulfide bonds (Figure 4 panel A). There is an additional intrapeptide disulfide bond on the A-chain. Figure 4 panel A highlights the different types of fragment ions which can be formed, classifying each fragment ion by the number of disulfide bond cleavages necessary for the formation of the fragment. The results of the MS/MS fragmentation of insulin with HCD, ETD, EThcD, and AI-ETD for the z = +5 precursor charge state of insulin is shown in Figure 4 panels B and C. Both EThcD and AI-ETD yield near complete sequence coverage of the protein; however, the intensity of fragment ions in AI-ETD make up a significantly larger percent of the total ion current (Figure S4). Interestingly, while EThcD and AI-ETD generate fragments of all types, the c- and z•-type fragment ions comprise most of the fragment ions which required disulfide cleavage. This suggests that electron-driven dissociation is predominantly responsible for the formation of these fragment ions for this protein and charge state. Fragment ion generation within the region of the protein enclosed by disulfide bonds is noticeably low with ETD. While ETD preferentially cleaves disulfide bonds, the highly compact and charge dense characteristics of bovine insulin likely causes substantial ETnoD product ion formation.
Figure 4.
A cartoon representation of bovine insulin with intact disulfide bonds is shown (A). Representative fragment ions are illustrated and are color coded based on the number of disulfide bond cleavages necessary to generate that fragment. Unlike the other proteins analyzed in this study, insulin consists of 2 separate disulfide linked peptides, generating a complex assortment of fragment ion possibilities. Panel B shows the number of fragment ions observed using each dissociation method and the number of disulfide cleavages necessary for each ion. The relative signal intensity and position of each ion is shown in panel C.
CONCLUSION
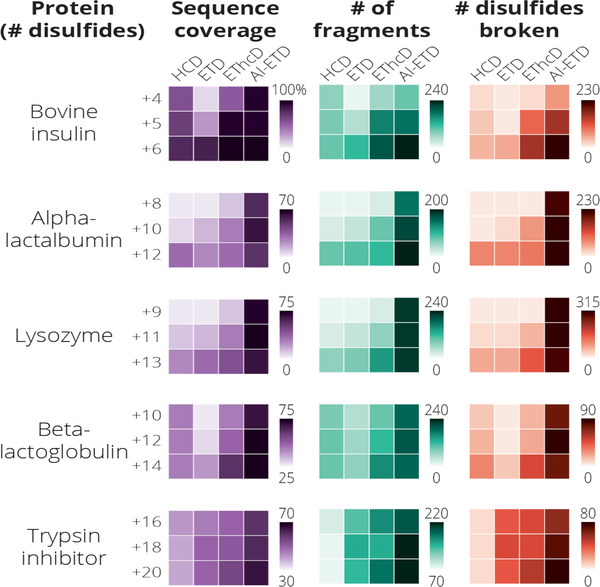
We demonstrate that AI-ETD is an extremely effective fragmentation method for five protein standards which contain intact disulfide linkages. The results for all proteins investigated are summarized in Figure 5 where we show that the total sequence coverage for each protein, the number of fragments ions generated, and the number of total disulfide bonds broken across all fragment ions for all precursor ion charge states examined is optimal when AI-ETD as the fragmentation method. Note, when calculating the number of total disulfide bonds broken, a fragment requiring two disulfide cleavages would be counted twice. These results build upon previous observations that 1) ETD preferentially fragments disulfide bonds but can suffer from ETnoD and 2) AI-ETD improves fragmentation of intact proteins by reducing the amount of ETnoD product ion formation. Concurrent irradiation allows for the unfolding of the gas phase protein while the electron transfer reaction is occurring, exposing regions of the protein inaccessible to the ETD reagent otherwise and preventing disulfide bond reformation. The method is particularly effective with proteins which are highly compact in the gas phase, such as lysozyme and insulin, where a majority of the protein backbone is enclosed by disulfide linkages. Furthermore, AI-ETD shows effective fragmentation across the precursor ion charge state envelope, allowing the interrogation of low charge density precursor ions which generally offer poor fragmentation by ETD alone.
Figure 5.
A summary of the sequence coverage, number of identified fragments, and number of disulfide bonds cleaved for each protein and precursor charge state analyzed in this study. In all cases tested AI-ETD yields the greatest sequence coverage, number of fragment ions formed, and number of disulfide bond cleavages within the fragment ions.
The ability to effectively fragment highly disulfide linked intact proteins with AI-ETD will likely advance efforts towards the structural characterization of many types of proteins such as intact antibodies, toxins, native proteins, and protein complexes. Additionally, AI-ETD was recently shown to benefit top-down characterization of intact proteins in LC-MS/MS analyses,60 and the work described here lays the groundwork for future experiments that could be conducted on a chromatographic timescale to screen complex mixtures of proteins with intact disulfide bonds. In all, AI-ETD is a superior fragmentation technique for proteins with intact disulfide bonds and will continue to be explored as a tool for disulfide bond analysis in a variety of applications.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge support from Thermo Fisher Scientific and NIH grants R35 GM118110. N.M.R. was funded through a NIH Predoctoral to Postdoctoral Transition Award (F99 CA212454).
Footnotes
SUPPORTING INFORMATION
Supporting Information Available: Sequence coverage map of the z = +8, z = +10, and z = +12 precursor charges states for α-lactalbumin (Figure S1). Comparison of IRMPD, ETD, and AI-ETD for z=+12 β-lactoglobulin (Figure S2). Sequence coverage map of the z = +16, z = +18, and z = +20 precursor charge states for trypsin inhibitor (Figure S3). Bovine insulin fragmentation grouped by fragment type for the z = +5 precursor charge state (Figure S4).
REFERENCES
- (1).Toby TK; Fornelli L; Kelleher NL Annu. Rev. Anal. Chem 2016, 9 (1), 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chen B; Brown KA; Lin Z; Ge Y Anal. Chem 2018, 90 (1), 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Siuti N; Kelleher NL Nat. Methods 2007, 4 (10), 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Smith LM; Kelleher NL Nat. Methods 2013, 10 (3), 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Catherman AD; Skinner OS; Kelleher NL Biochem. Biophys. Res. Commun 2014, 445 (4), 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Little DP; Speir JP; Senko MW; O’Connor PB; McLafferty FW Anal. Chem 1994, 66 (18), 2809–2815. [DOI] [PubMed] [Google Scholar]
- (7).Raspopov SA; El-Faramawy A; Thomson BA; Siu KWM Anal. Chem 2006, 78 (13), 4572–4577. [DOI] [PubMed] [Google Scholar]
- (8).Ahlf DR; Compton PD; Tran JC; Early BP; Thomas PM; Kelleher NL J. Proteome Res 2012, 11 (8), 4308–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zubarev RA; Kelleher NL; McLafferty FW J. Am. Chem. Soc 1998, 120 (13), 3265–3266. [Google Scholar]
- (10).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF In Proceedings of the Nationalal Academy of Sciences; 2004; Vol. 101, pp 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Riley NM; Coon JJ Anal. Chem 2018, 90 (1), 40–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Thornton JM J. Mol. Biol 1981, 151, 261–287. [DOI] [PubMed] [Google Scholar]
- (13).Matsumura M; Signor G; Matthews BW Nature 1989, 342 (6247), 291–293. [DOI] [PubMed] [Google Scholar]
- (14).Wedemeyer WJ; Welker E; Narayan M; Scheraga HA Biochemistry 2000, 39 (15), 4207–4216. [DOI] [PubMed] [Google Scholar]
- (15).Zhang Y; Dewald HD; Chen HJ Proteome Res. 2011, 10 (3), 1293–1304. [DOI] [PubMed] [Google Scholar]
- (16).Zhang Y; Cui W; Zhang H; Dewald HD; Chen H Anal. Chem 2012, 84 (8), 3838–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nicolardi S; Giera M; Kooijman P; Kraj A; Chervet JP; Deelder AM; Van Der Burgt YEM J. Am. Soc. Mass Spectrom 2013, 24 (12), 1980–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhao DS; Gregorich ZR; Ge Y Proteomics 2013, 13 (22), 3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Brown RS; Lennon JJ Anal. Chem. 1995, 67 (21), 3990–3999. [DOI] [PubMed] [Google Scholar]
- (20).Asakawa D Mass Spectrom. Rev 2016, 35 (4), 535–556. [DOI] [PubMed] [Google Scholar]
- (21).Fukuyama Y; Iwamoto S; Tanaka KJ Mass Spectrom. 2006, 41 (2), 191–201. [DOI] [PubMed] [Google Scholar]
- (22).Nicolardi S; Switzar L; Deelder AM; Palmblad M; van der Burgt YE M. Anal. Chem 2015, 87 (6), 3429–3437. [DOI] [PubMed] [Google Scholar]
- (23).Schroeder MJ; Shabanowitz J; Schwartz JC; Hunt DF; Coon JJ Anal. Chem 2004, 76 (13), 3590–3598. [DOI] [PubMed] [Google Scholar]
- (24).Liu F; van Breukelen B; Heck AJ R. Mol. Cell. Proteomics 2014, 13 (10), 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ni W; Lin M; Salinas P; Savickas P; Wu SL; Karger BL J. Am. Soc. Mass Spectrom 2013, 24 (1), 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wu SL; Jiang H; Hancock WS; Karger BL Anal. Chem 2010, 82 (12), 5296–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang Y; Lu Q; Wu SL; Karger BL; Hancock WS Anal. Chem 2011, 83 (8), 3133–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lu S; Fan SB; Yang B; Li YX; Meng JM; Wu L; Li P; Zhang K; Zhang MJ; Fu Y; Luo J; Sun RX; He SM; Dong MQ Nat. Methods 2015, 12 (4), 329–331. [DOI] [PubMed] [Google Scholar]
- (29).Wu S; Jiang H; Lu Q; Dai S; Hancock WS; Karger BL Anal. Chem 2009, 81 (1), 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Clark DF; Go EP; Desaire H Anal. Chem 2013, 85 (2), 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zubarev RA; Kruger NA; Fridriksson EK; Lewis MA; Horn DM; Carpenter BK; McLafferty FW J. Am. Chem. Soc 1999, 121 (8), 2857–2862. [Google Scholar]
- (32).Ganisl B; Breuker K ChemistryOpen 2012, 1 (6), 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ge Y; ElNaggar M; Sze SK; Oh H. Bin; Begley TP; McLafferty FW; Boshoff H; Barry CE J. Am. Soc. Mass Spectrom 2003, 14 (3), 253–261. [DOI] [PubMed] [Google Scholar]
- (34).Kalli A; Håkansson K Int. J. Mass Spectrom 2007, 263 (1), 71–81. [Google Scholar]
- (35).Fung YME; Kjeldsen F; Silivra OA; Chan TWD; Zubarev RA Angew. Chemie - Int. Ed 2005, 44 (39), 6399–6403. [DOI] [PubMed] [Google Scholar]
- (36).Agarwal A; Diedrich JK; Julian RR Anal. Chem 2011, 83 (17), 6455–6458. [DOI] [PubMed] [Google Scholar]
- (37).Mentinova M; McLuckey SA Int. J. Mass Spectrom 2011, 308 (1), 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lioe H; Duan M; O’Hair RAJ Rapid Commun. Mass Spectrom 2007, 21, 2727–2733. [DOI] [PubMed] [Google Scholar]
- (39).Gunawardena HP; O’Hair RAJ; McLuckey SA J. Proteome Res 2006, 5 (9), 2087–2092. [DOI] [PubMed] [Google Scholar]
- (40).Hendricks NG; Lareau NM; Stow SM; McLean JA; Julian RR J. Am. Chem. Soc 2014, 136 (38), 13363–13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lee M; Lee Y; Kang M; Park H; Seong Y; Sung BJ; Moon B; Oh H. Bin. J. Mass Spectrom 2011, 46 (8), 830–839. [DOI] [PubMed] [Google Scholar]
- (42).Wongkongkathep P; Li H; Zhang X; Ogorzalek Loo RR; Julian RR; Loo JA Int. J. Mass Spectrom 2015, 390 (2015), 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Liu J; Gunawardena HP; Huang TY; McLuckey SA Int. J. Mass Spectrom 2008, 276 (2–3), 160–170. [Google Scholar]
- (44).Ledvina AR; McAlister GC; Gardner MW; Smith SI; Madsen JA; Schwartz JC; Stafford GC; Syka JEP; Brodbelt JS; Coon JJ Angew. Chemie Int. Ed 2009, 48 (45), 8526–8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ledvina AR; Beauchene NA; McAlister GC; Syka JEP; Schwartz JC; Griep-Raming J; Westphall MS; Coon JJ Anal. Chem 2010, 82 (24), 10068–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ledvina AR; Rose CM; McAlister GC; Syka JEP; Westphall MS; GriepRaming J; Schwartz JC; Coon JJ J. Am. Soc. Mass Spectrom 2013, 24 (11), 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Riley NM; Westphall MS; Coon JJ Anal. Chem 2015, 87 (14), 7109–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Riley NM; Westphall MS; Coon JJ J. Proteome Res 2017, 16 (7), 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Riley NM; Westphall MS; Coon JJ J. Am. Soc. Mass Spectrom 2018, 29 (1), 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Riley NM; Mullen C; Weisbrod CR; Sharma S; Senko MW; Zabrouskov V; Westphall MS; Syka JEP; Coon JJ J. Am. Soc. Mass Spectrom 2016, 27 (3), 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Riley NM; Hebert AS; Dürnberger G; Stanek F; Mechtler K; Westphall MS; Coon JJ Anal. Chem 2017, 89 (12), 6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Breuker K; Oh H; Horn DM; Cerda BA; McLafferty FW J. Am. Chem. Soc 2002, 124 (22), 6407–6420. [DOI] [PubMed] [Google Scholar]
- (53).Neff D; Sobczyk M; Simons J Int. J. Mass Spectrom 2008, 276 (2–3), 91–101. [Google Scholar]
- (54).Riley NM; Westphall MS; Hebert AS; Coon JJ Anal. Chem 2017, 89 (12), 6358–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Durbin KR; Skinner OS; Fellers RT; Kelleher NL J. Am. Soc. Mass Spectrom 2015, 26 (5), 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Xiao K; Yu F; Fang H; Xue B; Liu Y; Li Y; Tian Z J. Proteomics 2017, 160, 21–27. [DOI] [PubMed] [Google Scholar]
- (57).Lyon YA; Riggs D; Fornelli L; Compton PD; Julian RR J. Am. Soc. Mass Spectrom 2018, 29 (1), 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Pitteri SJ; Chrisman PA; McLuckey SA Anal. Chem 2005, 77 (17), 5662–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pitteri SJ; Chrisman PA; Hogan JM; McLuckey SA Anal. Chem 2005, 77 (6), 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Riley NM, Sikora JW, Seckler HS, Greer JB, Fellers RT, Westphall MS, Thomas PM, Kelleher NL, C. J. Anal. Chem 2018, submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.