Abstract
Objectives
Natural language processing (NLP) and machine learning approaches were used to build classifiers to identify genomic-related treatment changes in the free-text visit progress notes of cancer patients.
Methods
We obtained 5889 deidentified progress reports (2439 words on average) for 755 cancer patients who have undergone a clinical next generation sequencing (NGS) testing in Wake Forest Baptist Comprehensive Cancer Center for our data analyses. An NLP system was implemented to process the free-text data and extract NGS-related information. Three types of recurrent neural network (RNN) namely, gated recurrent unit, long short-term memory (LSTM), and bidirectional LSTM (LSTM_Bi) were applied to classify documents to the treatment-change and no-treatment-change groups. Further, we compared the performances of RNNs to 5 machine learning algorithms including Naive Bayes, K-nearest Neighbor, Support Vector Machine for classification, Random forest, and Logistic Regression.
Results
Our results suggested that, overall, RNNs outperformed traditional machine learning algorithms, and LSTM_Bi showed the best performance among the RNNs in terms of accuracy, precision, recall, and F1 score. In addition, pretrained word embedding can improve the accuracy of LSTM by 3.4% and reduce the training time by more than 60%.
Discussion and Conclusion
NLP and RNN-based text mining solutions have demonstrated advantages in information retrieval and document classification tasks for unstructured clinical progress notes.
Keywords: machine learning, natural language processing, electronic health records, cancer, genomics
INTRODUCTION
The advent of next generation sequencing (NGS) technologies and their continually declining costs have resulted in the accumulation of very large sets of genetic data and facilitated identification of actionable genetic alterations in different tumor types. Despite the dramatic growth of the availability and affordability of such testing, it has also brought challenges, including the need of evaluating the effectiveness and actionability of genetic testing that could be invaluable for assisting tumor diagnosis and prognosis to direct patient treatment.1
Additionally, with the widespread use of electronic health record (EHR) systems in clinical care, secondary use of clinically relevant information of cancer patients are available to biomedical research including comparative effectiveness, patient reported outcomes, clinical actionability of genomic profiling, and precision medicine.2 However, in contrast to structured available data, a sizable percentage of the patient data are unstructured (or semistructured), which makes them not easily parsable by the machines and software.3,4 Therefore, harnessing the potential of clinical narratives in the EHR requires strategies for efficient and automated information extraction and understanding.
Natural language processing (NLP) and machine learning techniques could map unstructured text into structured (semistructured) form as well as could enable automatic identification and extraction of relevant information. Additionally, such automated system would significantly reduce delays in EHR processing and allow more accurately extraction of embedded information.3 Many clinical NLP systems have been in development and widely adopted in biomedical settings, for example, the Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES),5 MetaMap,6 and Noble Tools.7 However, these approaches mainly focus on utilizing medical vocabularies such as unified medical language system (UMLS)8 to perform concepts recognition and information extraction.7
There are many conventional machine learning algorithms have been used in clinical text mining, however, these models require human experts to encode domain knowledge through feature engineering, and have so far had mixed results modeling sequential events or time dependencies.9 More recently, multilayer neural networks, or deep learning, have been applied to gain actionable insights from heterogeneous clinical data.9,10 The major differences between deep learning and conventional neural network (NN) are the number of hidden layers, as well as their capability to learn meaningful abstractions of the input.11 Deep learning has been applied to process aggregated EHR documents, including both structured (eg diagnosis, laboratory tests) and unstructured data (eg medical notes, images).9 Several studies used deep learning to predict diseases from the patient clinical notes, for example, Cheng et al12 used a 4-layer convolutional neural network (CNN) to predict congestive heart failure and chronic obstructive pulmonary disease and showed promising performance.
Word embedding, learned in an unsupervised manner, has seen a successful word representation method in numerous NLP tasks in recent years. Unlike traditional word representation methods, such as bag-of-words and one-hot encoding, word embedding can capture the semantic meanings of the words within numeric vectors.13 Words that are semantically similar are closer to each other in distance, while words that are semantically different are farther apart in distance. Word embedding has been utilized extensively in biomedical named entity recognition (NER) tasks14,15 such as medical synonym extraction,16 relation extraction including chemical-disease relations,17 drug-drug interactions,18,19 protein-protein interactions,20 biomedical IR,21,22 and medical abbreviation disambiguation.23
In this project, we explored how word embedding and deep learning techniques can help to efficiently extract information from free-text EHR documents (eg progress notes) and evaluate the effectiveness and actionability of genetic testing in assisting cancer patient treatment adjustment. A total of 5889 deidentified progress reports for 755 cancer patients who have undergone a clinical NGS testing in Wake Forest Baptist Comprehensive Cancer Center have been included in our data analyses. The primary goal of this project is to (1) identify the section of the progress report that discusses genomic testing results and treatment information, (2) predict if there is a treatment change (or not) based on the extracted information using deep learning and word embedding, and (3) compare the performance of 4 recurrent neural network (RNN)-based approaches and 5 conventional machine learning algorithms for text classification task using clinical progress reports.
METHODS
Progress reports and preprocessing
The progress reports for cancer patients were obtained from the Translational Data Warehouse at the Wake Forest Baptist Health upon the institutional review board approval for the study. The study corpus contains 5889 progress reports (2439 words on average) that were charted for the 755 NGS patients after their NGS tests. We excluded 28 patients who have NGS testing completed twice. A text preprocessing pipeline was implemented to perform cleaning and reformatting. All the English letters were converted to lowercase. We removed English stop words, special characters and punctuations, empty spaces, and strings with length <2. Numbers were also excluded since they usually do not carry relevant information in this type of analysis. Abbreviations were replaced with the full terms, for example, “‘ve” was replaced with “have,” “‘re” was replaced with “are,” and “‘ll” was replaced with “will.” We also performed word stemming for non-NN machine learning models.
We identified the section for each report that discusses genomic testing results based on keyword searches. A list of keywords including genes, mutations, and treatment names were populated from the information provided by the NGS vendors, namely Foundation Medicine (https://www.foundationmedicine.com/), Caris (https://www.carislifesciences.com/), Guardant (http://www.guardanthealth.com/), as well as our local database. A 400-word text window was extracted for each report surrounding the location of the first keyword. By extracting the target section, we reduced the size of the reports from an average of ∼5000 words to 289, which greatly eliminated the redundant content, as well as improved the efficiency of our training.
Establish true labels
A subset (44) of 755 cancer patients (452 reports) were manually classified as genomic-related treatment change and nontreatment-change groups by our precision medicine nurse. These annotations served as “true” labels to evaluate the performance of our classification task. Additionally, a “rule-based” annotation method was also implemented to label the group of the reports based on her experience, vendors’ name, cancer gene, mutation, as well as therapeutic-related keywords (Supplementary Material). We further evaluated the performance of the generated labels using a clustering algorithm to compare the natural separation and the labeled groups. The 452 manually annotated labels, as well as the generated labels using the “rule-based” method were used as “ground truth” to evaluate the machine learning algorithms.
Word representations
Two types of word representation techniques were used to convert word tokens in each report into numerical vectors, term frequency-inverse document frequency (TF-IDF) and word embedding (Word2vec), for conventional machine learning models and RNNs, respectively. TF-IDF weight is a statistical measure used to evaluate how important a word is to a document in a collection or corpus.24 Word2vec takes a large corpus of text as its input and produces a high-dimension vector space through which each unique word in the corpus being assigned a corresponding vector in the space. Word2vec can utilize a continuous bag-of-words (CBOW) architecture to predict the current word from a window of surrounding context words, therefore, the order of context words is not important.24 In this study, we applied 2 methods to generate word embeddings: (1) using Word2Vec with CBOW architecture to pretrain word embeddings on our entire corpus,25 (2) including an embedding layer in the network and train the word embeddings on the fly. We then compared the performance of the NNs with and without pretrained word embeddings.
Recurrent neural networks
RNNs are neural networks that add additional weights to the network to create cycles in the network, in an effort to model time dependencies and sequential events.15 Variations of RNN, long short-term memory (LSTM) networks and gated recurrent unit (GRU), have been invented to better handle gradient vanishing problems.26 LSTM was used to create DeepCare,18 which is an end-to-end deep dynamic network that infers current illness states and predicts future medical outcomes using EHR. Another variation of RNN, GRU,19 was used to develop Doctor AI, which is another model to use patient history to predict diagnoses and medications for subsequent encounters.20 A modified version of LSTM, bidirectional LSTM (LSTM_Bi), which allows analyzing sequential data from both directions, has been used to process medical text data and achieved elevated performances over nondeep learning tools in NER tasks.21,22 Since RNN architecture is designed to model the sequential events, such as word sequences, this architecture is specifically suitable for capturing meaningful linguistic patterns across long sequences of words within a document.27 We implemented 4 variations of RNN in this study: (1) LSTM with word embedding trained on the fly (LSTM_onFly); (2) LSTM with pretrained word embedding on the entire corpus (LSTM_Pre); (3) LSTM_Bi with pretrained word embedding (LSTM_Bi); and (4) a simplified version of LSTM, GRU with pretrained word embedding. We also evaluated the performance of these 4 RNN models for information extraction and text classification in this study.
Convolutional layer
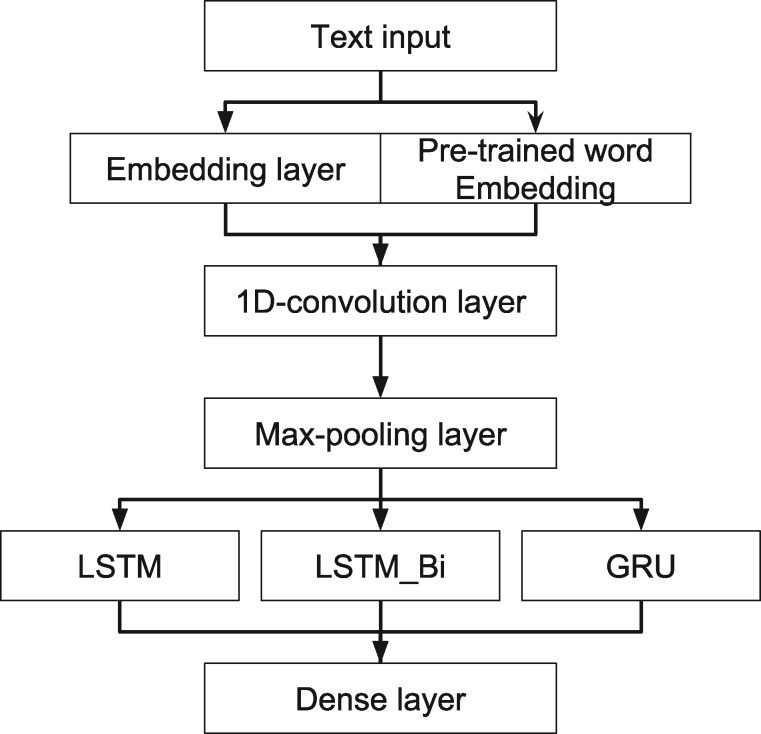
CNN has been successfully applied in image processing and NLP.28,29 We incorporated a 1D-convolution layer with 32 filters, a kernel size of 3, and stride of 1 word, followed by a max-pooling layer, in our RNNs. The convolutional layer, as well as the max-pooling layer, can help to learn useful word representations and reduce the dimensions of the input corpus.
Non-neural network models
We compared the performance of the RNNs against the performance of several conventional predictive models that can also be used for text classification. These include Naive Bayes (NB),30 K-nearest Neighbor (KNN),31 Support Vector Machine for classification (SVC),32 Random forest (RF),33 and logistic regression (LR).34 We generated TF-IDF vectors on the processed text using unigrams with a minimum document frequency of 5, and a maximum document frequency of 80%. Singular-value decomposition (SVD) was applied to reduce the dimension of the input matrix.
Hyperparameter optimization
We used a grid search technique to perform hyperparameter optimization for non-NN algorithms. Specifically, smoothing parameter alpha of NB, number of neighbors of KNN, penalty parameter C, kernel types (linear or radial basis function), and kernel coefficient gamma of SVC, the maximum depth of a tree, the minimum number of samples required to split an internal node, the minimum number of samples required to be at a leaf node of RF, and the L2 penalty parameter C of LR, were optimized using the grid search method. A 3-fold cross-validation was used during hyperparameter optimization to evaluate the performance of each version of the algorithms.
Model setup and evaluation metrics
To be consistent, we split the data into 0.66/0.33 train/test datasets, without any overlapping patients between train and test, for each model. In addition, we performed stratified 5-fold cross-validation during the training to evaluate the model performance. Binary cross entropy was used as the loss function for all the classifiers. For RNN algorithms, we implemented early stopping mechanism—the model stops training when the loss function does not improve for 5 epochs on the validation dataset. After training, the performance of each model was tested on the test set. We used 5 evaluation metrics to compare the performance of the models, including accuracy, precision, recall, and F1 score.
Open source platforms
We used high-level NN API Keras (https://keras.io/) running on top of Tensorflow (https://github.com/tensorflow/tensorflow) to set up our neural network structures. Non-NN models, as well as clustering and parameter search algorithms were derived from Scikit-Learn (http://scikit-learn.org/). Word embedding was performed using Gensim (https://radimrehurek.com/gensim/).
EXPERIMENTAL RESULTS
Study samples
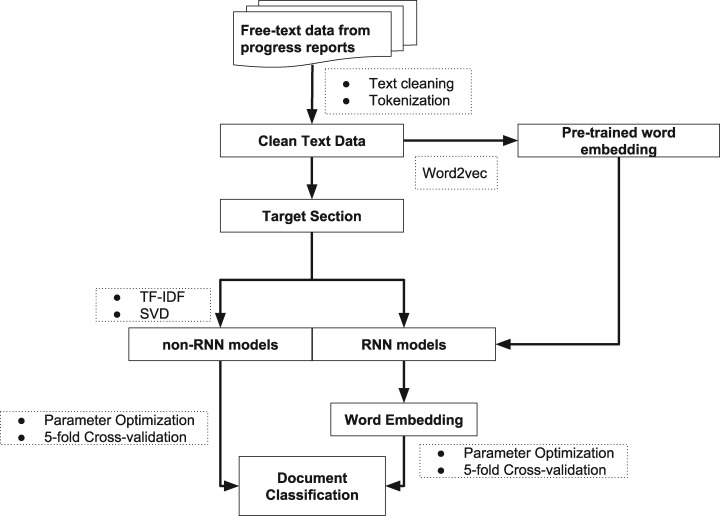
The flow chart of study design has been shown in Figure 1. Briefly, in this study, we processed 5889 free-text clinical reports from 755 patients. Target text windows from the reports were extracted for the subsequent classification task. We implemented a total of 9 classifiers, both RNN-based and traditional machine learning algorithms, to classify the treatment-change for each document. A word embedding matrix was pretrained based on the whole text corpus for some of the RNN-based models. A subset (44) of the cancer patients (452 reports) were annotated by clinical experts. These manually generated labels, along with the labels that generated by a rule-based keywords searching method, were used as “true” labels to evaluate the model performance. A total of 3736 documents being labeled as treatment-change and 2153 documents being labeled as no-treatment-change.
Figure 1.
Workflow of text processing and document classification using machine learning models.
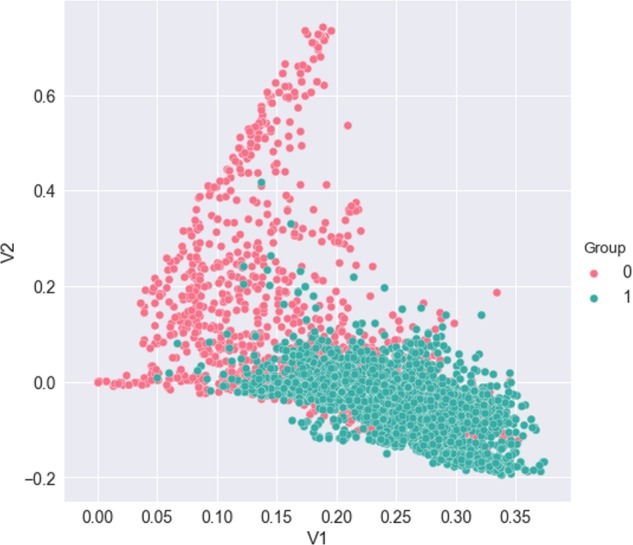
To explore additional insight about the progress reports and the separability of 2 labeled groups, we performed a SVD on the TF-IDF representation of the reports. The top 2 eigenvectors of SVD were used to plot the similarity between the 2 target groups (Figure 2). From the plot, we note that natural clustering occurs between progress reports corresponding to labeled groups. This technique can also help to better understand document misclassifications in our classification task.
Figure 2.
Dimensional reduction of term frequency-inverse document frequency (TF-IDF) representation of the documents via singular-value decomposition (SVD). Data points are colored by treatment-change (1) and nontreatment-change (0) groups.
Hyperparameter optimization
Key hyperparameters for each machine learning algorithm were optimized using a grid search method. For traditional models, smoothing parameter alpha of NB was optimized to be 0; the best number of neighbors for KNN was 7; linear kernel and a penalty parameter of 30 were selected for SVC; the maximum depth of a tree, the minimum number of samples required to split an internal node, and the minimum number of samples required to be at a leaf node were optimized as 6, 5, and 5, respectively, for RF; and an L2 penalty parameter of 10 was picked for LR (Table 1).
Table 1.
Best hyperparameters for the classifiers
| Classifier | Hyperparameters |
|---|---|
| Deep learning classifiers | |
| LSTM_onFly | Optimizer=Adam, batch size=64, dropout rate=0, word embedding=trained on the fly, recurrent layer=single directional LSTM |
| LSTM_Pre | Optimizer=Adam, batch size=64, dropout rate=0, word embedding=pretrained on the whole corpus, recurrent layer=single directional LSTM |
| LSTM_Bi | Optimizer=Adam, batch size=64, dropout rate=0, word embedding=pretrained on the whole corpus, recurrent layer=bidirectional LSTM |
| GRU | Optimizer=Adam, batch size=64, dropout rate=0, word embedding=pretrained on the whole corpus, recurrent layer=single directional LSTM |
| Conventional classifiers | |
| KNN | Number of neighbors=7 |
| LR | L2 penalty parameter=10 |
| NB | Smoothing parameter alpha=0 |
| RF | Maximum depth of a tree=6 Minimum number of samples required to split an internal node=5 Minimum number of samples required to be at a leaf node=5 |
| SVC | Kernel=linearL 2 penalty parameter=30 |
GRU: gated recurrent unit; KNN: K-nearest Neighbor; LR: logistic regression; LSTM: long short-term memory; NB: Naive Bayes; RF: random forest; SVC: Support Vector Machine for classification.
For RNN-based models (Figure 3), we selected Adam optimization algorithm as the default optimizer.35 Except for 1 LSTM model (LSTM_onFly), all the models used pretrained word embedding matrix as the input. Based on the parameter turning, we chose batch size of 64 and a dropout rate of 0.
Figure 3.
Architecture of RNN models. GRU: gated recurrent unit; LSTM: long short-term memory; LSTM_Bi: bidirectional LSTM; RNN: recurrent neural network.
Classification performance
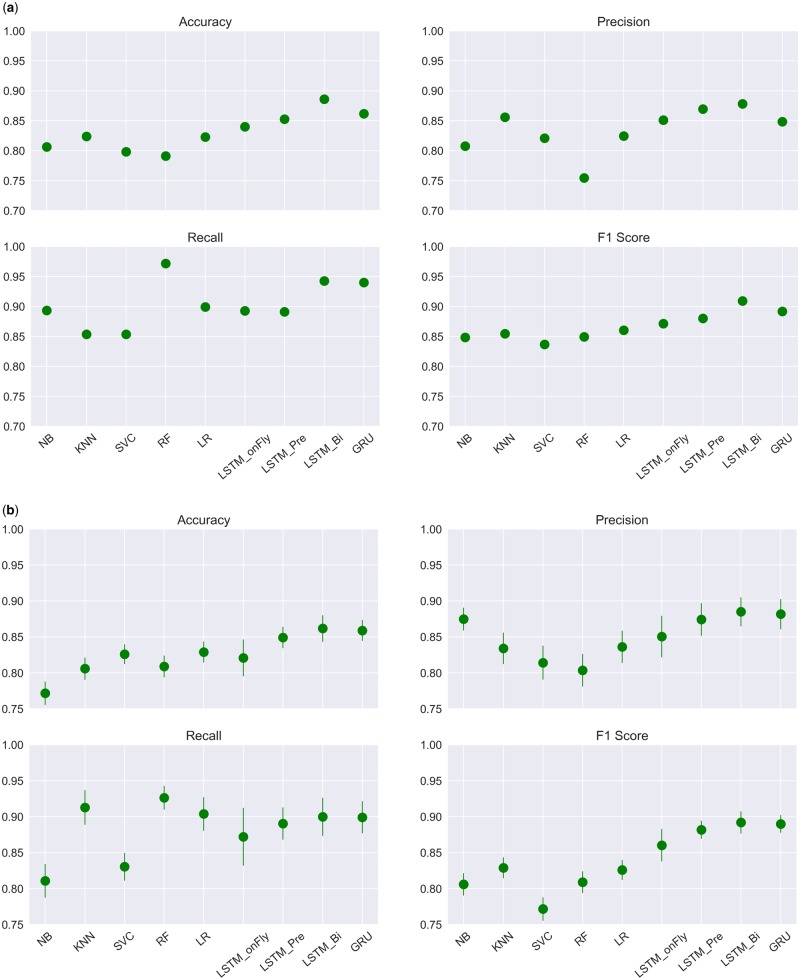
Four performance evaluation metrics on the task were included in Table 2 and Figure 4, including accuracy, precision, recall, and F1 score.
Table 2.
Performance of classifiers on the document classification repeated for 100 times
| Classifier | Accuracy (mean±SD) | Precision (mean±SD) | Recall (mean±SD) | F1 score (mean±SD) |
|---|---|---|---|---|
| Deep learning classifiers | ||||
| LSTM_onFly | 0.821±0.026 | 0.850±0.029 | 0.872±0.040 | 0.860±0.023 |
| LSTM_Pre | 0.849±0.015 | 0.874±0.023 | 0.890±0.022 | 0.882±0.013 |
| LSTM_Bi | 0.862±0.019 | 0.885±0.020 | 0.900±0.026 | 0.892±0.015 |
| GRU | 0.859±0.014 | 0.882±0.021 | 0.899±0.022 | 0.890±0.012 |
| Conventional classifiers | ||||
| KNN | 0.806±0.016 | 0.834±0.022 | 0.913±0.024 | 0.829±0.015 |
| LR | 0.829±0.015 | 0.836±0.022 | 0.904±0.023 | 0.826±0.014 |
| NB | 0.772±0.016 | 0.875±0.016 | 0.811±0.023 | 0.806±0.016 |
| RF | 0.809±0.015 | 0.804±0.023 | 0.926±0.017 | 0.809±0.015 |
| SVC | 0.826±0.014 | 0.814±0.024 | 0.830±0.019 | 0.772±0.016 |
GRU: gated recurrent unit; KNN: K-nearest Neighbor; LR: logistic regression; LSTM: long short-term memory; NB: Naive Bayes; RF: random forest; SD: standard deviation; SVC: Support Vector Machine for classification.
Figure 4.
Performance comparisons of 9 Machine Learning algorithms based on (A) a single run, and (B) models repeated for 100 times. Mean metrics (dots) and their standard deviations (bars) were included.
Overall, RNN-based classifiers outperformed the traditional machine learning algorithms. LSTM_Bi with pretrained word embedding and a 1D-convolution layer followed by max-pooling outperformed all other models in accuracy (0.886), precision (0.878), and the F1 score (0.909). RF had the highest recall, with a score of 0.972, followed by LSTM_Bi (0.943). Because of the stochastic nature of machine learning algorithms, we repeated each model for 100 times and calculated the average metrics and corresponding standard deviations. Again, LSTM_Bi outperformed all the others in accuracy (0.862 ± 0.019), precision (0.885 ± 0.02), and F1 score (0.892 ± 0.015), while recall was leading by RF (0.926 ± 0.017) (Figure 4B).
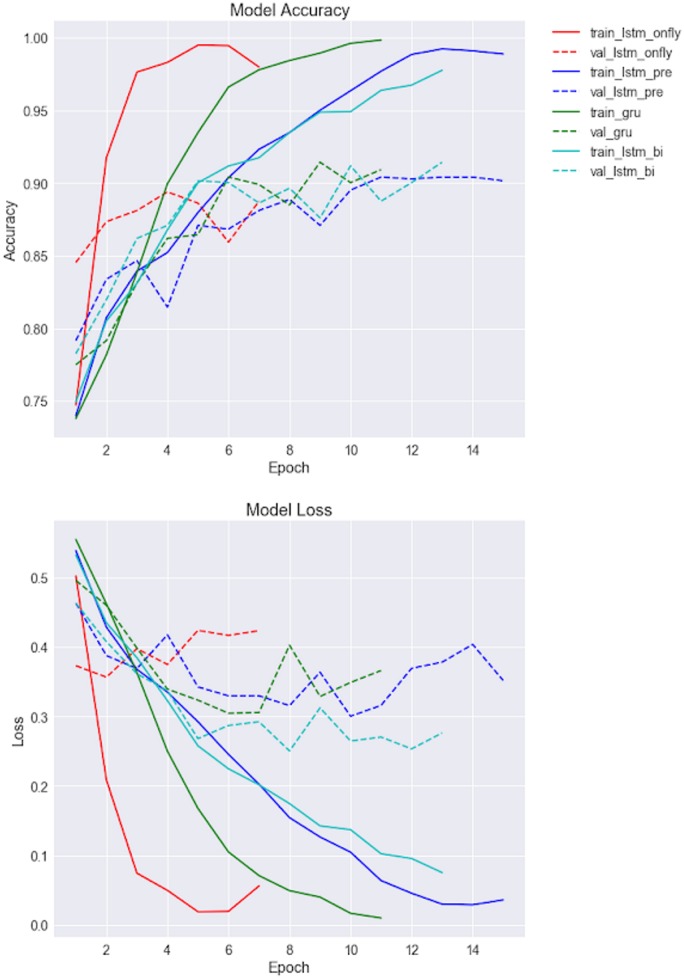
RNN-based models training
Accuracy and model loss-based training curves of RNN-based classifiers have been shown in Figure 5. As we can see, LSTM without pretrained word embedding (LSTM_onFly) revealed the fastest model convergence (the shorter learning curve was due to early stopping), followed by GRU. LSTMs with pretrained word embeddings had similar convergence curve. However, LSTM_onFly model quickly overfitted after the first epoch, it also has the largest discrepancy between training data and validation data, while the LSTM_Bi had the smallest discrepancy.
Figure 5.
Training curves of the first 15 epochs for RNN-based models, where the upper panel is the model accuracy for training and validation datasets, and lower panel is the model loss for training and validation dataset. RNN: recurrent neural network.
Error analysis
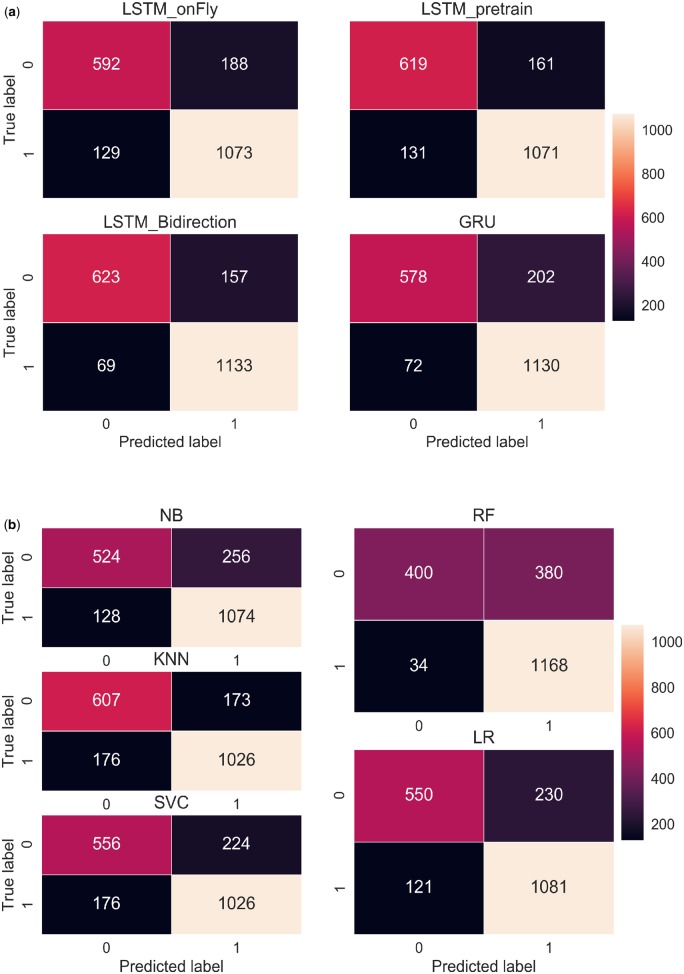
We analyzed the confusion matrices of 9 classifiers based on their classification performance on 1982 testing documents, with 1202 documents labeled as treatment-change, and 780 documents labeled as no-treatment-change. LSTM_Bi, which indicated the highest accuracy (0.886), had 69 false negatives and 157 false positives (Figure 6). The other 2 LSTM variations, LSTM_onFly and LSTM_Pre, resulted in significantly higher number misclassifications, especially for in the false negative category, where the misclassifications nearly doubled. The GRU model, on the other hand, had similar number of false negative classifications (72) comparing to LSTM_Bi, however, it mistakenly classified 202 no-treatment-change documents as treatment-change group (false positive).
Figure 6.
Confusion matrix of (A) RNN-based models, and (B) conventional machine learning models. GRU: gated recurrent unit; LSTM: long short-term memory; NB: Naive Bayes; RF: random forest; RNN: recurrent neural network; SVC: Support Vector Machine for classification.
For the conventional classifiers, KNN achieved the highest accuracy (0.824) as it correctly identified 1026 treatment-change documents, and 607 no-treatment-change documents, which was the highest among the conventional models. Notably, RF correctly classified 1168 treatment-change documents, which was the highest among all 9 models. However, it misclassified 380 no-treatment-change documents as treatment-change, which was also the highest. It is consistent with what we have observed from Table 2 and Figure 4, RF has the highest recall (0.972) but the lowest precision (0.755).
DISCUSSION
We have successfully applied NLP and machine learning methods to extract information from clinical progress reports and classify them into treatment-change and no-treatment-change groups. RNN-based algorithms with pretrained word embedding, especially LSTM_Bi, demonstrated significantly better performance on the classification task than conventional machine learning algorithms with TF-IDF features. It is most like because of the RNN structure that can capture linguistic patterns across long sequences of words and the pretrained word embeddings on the entire text corpus. In addition, we noticed that KNN and NB outperformed SVC in this study, possibly because the decision planes of SVC were not able to sufficiently separate classes due to the data structure.
We first compared the performance of LSTM with and without pretrained word embedding. Based on the results in Table 2 and Figure 4, LSTM_Pre outperformed LSTM_onFly in 3 of 4 evaluation metrics, except recall, where these 2 models had comparable results. This may be because of LSTM_onFly only trains word embedding based on a smaller extracted text window, which is not able to model linguistic patterns accurately. We then compared 3 RNN models with pretrained word embeddings, LSTM_Pre, LSTM_Bi, and GRU. LSTM_Bi outperformed the other 2 models in all 4 metrics. LSTM_Bi allows analyzing sequential data from both directions, and has been used to process medical text data and achieved elevated performances over nondeep learning tools in terms of NER.15,36 Interestingly, the simplified variation of LSTM, GRU,37 showed better results than LSTM_Pre in 3 of 4 evaluations, except precision. This observation is consistent with previous explorations, where GRU has yield similar performance compared with LSTM, however GRU could have better performance on smaller dataset.38,39
TF-IDF based non-NN classifiers overall had poorer performance in this study. One reason is that vector space word representations, such as TF-IDF and bag-of-words, cannot take the context of each word into account, instead, they rely on the ordering of words within a small text window. Cancer progress reports typically including complex information and structures, which are usually challenging to be sufficiently captured by vector space word representations. Our results suggest that pretrained word embeddings on a large related corpus can extract information more efficiently and improve the subsequent classifications tasks.
Furthermore, RNN architecture is designed to model the sequential events, such as word sequences. In our study, this architecture is able to capture meaningful linguistic patterns across long sequences of words within a document. It provides a method to extract higher-level information and make decisions based on the context of each word. Therefore, the combination of RNN and pretrained word embeddings further boosted the model performance.
Due to the stochastic nature of machine learning algorithms, evaluating their performances based on a single model is not always accurate. Randomness can be introduced at any stage of the study, such as data processing, data splitting, word representation methods, weight initialization, and random seeds. To reduce the randomness and evaluate the models more accurately, we repeated each model for 100 times. The ranges, means, and standard deviations of the evaluation metrics for each model were calculated. Our results in Table 2 and Figure 4B indicated that the performances of machine learning models are consistent and reproducible.
One goal of this study is to implement an automated system to reduce the time required for progress report annotation. However, NLP and machine learning models, especially for deep learning models, suffer from long processing and training time. We thus implemented several methods to improve our model efficiency. The first method was to extract a target text window from each document instead of using the whole progress report, which is usually very complex and redundant. Moreover, pretrained word embeddings significantly reduced the training time for RNN-based models, since they avoided training word embeddings on the fly. In addition, 2-dimensional reduction methods, 1D-convolution layer with max pooling and SVD, were used to further reduce the training time for RNN-based and non-NN classifiers, respectively. These methods ensure our model can make decisions more efficiently and reduce the burden of manually annotating the reports by medical experts.
One important limitation of our study is that most of the progress reports lack true labels. Reading progress reports and correctly labeling them is time-consuming and challenging even for human experts. However, we generated labels for a subset of the reports to validate and improve our rule-based labeling method. Another limitation is the small sample size of our dataset only 755 qualified cancer patients are available for this study. Although we included reports at multiple visits for each patient, 5889 documents are not likely to reach the full effectiveness of RNN models. In addition, using a dataset with a small number of samples but multiple documents for each sample would increase the risk of model overfitting. To reduce overfitting, we split training and test dataset based on unique samples, which prevented the classifier from seeing the reports from 1 patient in both training and testing phases.
To our knowledge, this is the first study extracting genomics-related information in clinical progress reports using NLP and deep learning. Our goal is to implement an automated annotation system for clinical progress reports that can improve the annotation accuracy, as well as reduce the time required. Moving forward, we will extend this NLP and RNN analysis pipeline to perform more tasks, for example, classify cancer stages, predict survival rate, deep phenotyping, and annotate unknown genomic mutations. Another important future direction is to generalize this pipeline to read data from multiple research facilities and multiple resources, such as pathology reports, radiology reports, medical images, as well as NGS results. In addition, during genomic testing, thousands of genetic alterations are generated with unknown pathogenic impacts on specific cancer types. Distinguishing the alterations that contribute to cancer risk from the neutral alterations is very challenging and time-consuming since it is mainly done manually. Thus, an automated genetic alteration interpretation system based on our NLP and RNN methods could be developed to incorporate relevant information from text-based sources such as pathology reports and progress notes.
CONCLUSIONS
An automated NLP and deep learning solution has demonstrated advantages and potentials in information retrieval and document classification tasks for unstructured clinical progress notes. It will help to evaluate the impact of genomic testing in the clinical practices.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONTRIBUTORS
MG designed data clean, processing, and analysis pipelines, drafted and revised the manuscript. UT contributed to data collection and conception of the work. SC participated in study design, revised analysis pipeline, and reviewed draft. RP contributed to data collection and ground truth generation. WZ and BP reviewed drafts and provided feedback on study design.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the study patients whose data have been used for the study. We thank the contributions of investigators and staff for data collection, management, and data analysis.
FUNDING
The work was partially supported by the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197). The authors acknowledge use of the services and facilities, funded by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (UL1TR001420).
Conflict of interest statement. None declared.
REFERENCES
- 1. National Academies of Sciences Engineering Medicine. An Evidence Framework for Genetic Testing. Washington, DC: National Academies Press (US); 2017. doi:10.17226/24632 [PubMed] [Google Scholar]
- 2. Manion FJ, Harris MR, Buyuktur AG, et al. Leveraging EHR data for outcomes and comparative effectiveness research in oncology. Curr Oncol Rep 2012; 146: 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen ES, Sarkar IN.. Mining the electronic health record for disease knowledge. Methods Mol Biol 2014; 1159: 269–86. [DOI] [PubMed] [Google Scholar]
- 4. Simmons M, Singhal A, Lu Z.. Text mining for precision medicine: bringing structure to EHRs and biomedical literature to understand genes and health. Adv Exp Med Biol 2016; 939: 139–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 175: 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 173: 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tseytlin E, Mitchell K, Legowski E, et al. NOBLE—flexible concept recognition for large-scale biomedical natural language processing. BMC Bioinformatics 2016; 17: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32: D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miotto R, Wang F, Wang S, et al. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 2018; 196: 1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ravì D, Wong C, Deligianni F, et al. Deep learning for health informatics. IEEE J Biomed Health Inform 2017; 21: 4–21. [DOI] [PubMed] [Google Scholar]
- 11. LeCun Y, Bengio Y, Hinton G.. Deep learning. Nature 2015; 5217553: 436–44. [DOI] [PubMed] [Google Scholar]
- 12. Cheng Y, Wang F, Zhang P, et al. Risk prediction with electronic health records: a deep learning approach. Soc Ind Appl Math 2016; 432–40. doi:10.1137/1.9781611974348.49 [Google Scholar]
- 13. Bengio Y, Ducharme R, Vincent P, et al. A neural probabilistic language model. J Mach Learn Res 2003; 3: 1137–55. [Google Scholar]
- 14. Liu S, Tang B, Chen Q, et al. Effects of semantic features on machine learning-based drug name recognition systems: word embeddings vs. manually constructed dictionaries. Information 2015; 64: 848–65. [Google Scholar]
- 15. Tang B, Cao H, Wang X, et al. Evaluating word representation features in biomedical named entity recognition tasks. Biomed Res Int 2014; 2014: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jagannatha A, Chen J, Yu H.. Mining and ranking biomedical synonym candidates from Wikipedia In: Proceedings of the Sixth International Workshop on Health Text Mining and Information Analysis. Lisbon, Portugal: Association for Computational Linguistics; 2015: 142–151. http://aclweb.org/anthology/W15-2619 (accessed June 18, 2018). [Google Scholar]
- 17. Xu J, Wu Y, Zhang Y, et al. CD-REST: a system for extracting chemical-induced disease relation in literature. Database (Oxford) 2016; 2016. pii: baw036. doi:10.1093/database/baw036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S, Tang B, Chen Q, et al. Drug-drug interaction extraction via convolutional neural networks. Comput Math Methods Med 2016; 2016: 6918381. doi:10.1155/2016/6918381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Liu S, Rastegar-Mojarad M, et al. Dependency and AMR embeddings for drug-drug interaction extraction from biomedical literature. In: Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics New York, NY: ACM; 2017: 36–43. doi:10.1145/3107411.3107426
- 20. Jiang Z, Li L, Huang D.. A general protein-protein interaction extraction architecture based on word representation and feature selection. Int J Data Min Bioinf 2016; 143: 276–91. [Google Scholar]
- 21. Jo S-H, Lee K-S.. CBNU at TREC 2016 clinical decision support track. Proc TREC 2016; 4. [Google Scholar]
- 22. Wang Y, Rastegar-Mojarad M, Komandur-Elayavilli R, et al. An ensemble model of clinical information extraction and information retrieval for clinical decision support. Proc TREC 2016; 10. [Google Scholar]
- 23. Wu y, Xu J, Zhang Y, et al. Clinical abbreviation disambiguation using neural word embeddings In: Proceedings of BioNLP 15. Beijing, China: Association for Computational Linguistics; 2015: 171–6. http://www.aclweb.org/anthology/W15-3822 (accessed June 18, 2018). [Google Scholar]
- 24. Salton G, Wong A, Yang CS.. A vector space model for automatic indexing. Commun ACM 1975; 1811: 613–20. [Google Scholar]
- 25. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. arXiv: 13013781 [cs] Published Online First: January 16, 2013. http://arxiv.org/abs/1301.3781 (accessed June 17, 2018).
- 26. Hochreiter S, Bengio Y, Frasconi P, et al. Gradient Flow in Recurrent Nets: The Difficulty of Learning Long-Term Dependencies. Wiley-IEEE Press; 2001.
- 27. Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. arXiv: 150600019 [cs] Published Online First: May 29, 2015. http://arxiv.org/abs/1506.00019 (accessed June 17, 2018).
- 28. Collobert R, Weston J.. A unified architecture for natural language processing: deep neural networks with multitask learning In: Proceedings of the 25th International Conference on Machine Learning. New York, NY: ACM; 2008: 160–7. [Google Scholar]
- 29. Krizhevsky A, Sutskever I, Hinton GE.. ImageNet classification with deep convolutional neural networks. Commun ACM 2017; 606: 84–90. [Google Scholar]
- 30. Kazmierska J, Malicki J.. Application of the Naïve Bayesian classifier to optimize treatment decisions. Radiother Oncol 2008; 862: 211–6. [DOI] [PubMed] [Google Scholar]
- 31. Trstenjak B, Mikac S, Donko D.. KNN with TF-IDF based framework for text categorization. Procedia Engineering 2014; 69: 1356–64. [Google Scholar]
- 32. Joachims T. Text categorization with support vector machines: learning with many relevant features In: Machine learning: ECML-98. Berlin, Heidelberg: Springer; 1998: 137–42. doi:10.1007/BFb0026683 [Google Scholar]
- 33. Breiman L. Random forests. Mach Learn 2001; 451: 5–32. [Google Scholar]
- 34. Speech and Language Processing. https://web.stanford.edu/∼jurafsky/slp3/ (accessed June 18, 2018).
- 35. Kingma DP, Ba LJ. Adam: a method for stochastic optimization. Published Online First: 2015. https://dare.uva.nl/search? identifier=a20791d3-1aff-464a-8544-268383c33a75 (accessed June 18, 2018).
- 36. Habibi M, Weber L, Neves M, et al. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 2017; 3314: i37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho K, van Merrienboer B, Gulcehre C, et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv: 14061078 [cs, stat] Published Online First: June 3, 2014. http://arxiv.org/abs/1406.1078 (accessed June 17, 2018).
- 38. Chung J, Gulcehre C, Cho K, et al. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv: 14123555 [cs] Published Online First: December 11, 2014. http://arxiv.org/abs/1412.3555 (accessed June 18, 2018).
- 39. Jozefowicz R, Zaremba W, Sutskever I. An empirical exploration of recurrent network architectures. In: Proceedings of the 32Nd International Conference on International Conference on Machine Learning - Volume 37 Lille, France: JMLR.org; 2015: 2342–50. http://dl.acm.org/citation.cfm? id=3045118.3045367 (accessed June 18, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.