Abstract
Objectives
We aimed to gain a better understanding of how standardization of laboratory data can impact predictive model performance in multi-site datasets. We hypothesized that standardizing local laboratory codes to logical observation identifiers names and codes (LOINC) would produce predictive models that significantly outperform those learned utilizing local laboratory codes.
Materials and Methods
We predicted 30-day hospital readmission for a set of heart failure-specific visits to 13 hospitals from 2008 to 2012. Laboratory test results were extracted and then manually cleaned and mapped to LOINC. We extracted features to summarize laboratory data for each patient and used a training dataset (2008–2011) to learn models using a variety of feature selection techniques and classifiers. We evaluated our hypothesis by comparing model performance on an independent test dataset (2012).
Results
Models that utilized LOINC performed significantly better than models that utilized local laboratory test codes, regardless of the feature selection technique and classifier approach used.
Discussion and Conclusion
We quantitatively demonstrated the positive impact of standardizing multi-site laboratory data to LOINC prior to use in predictive models. We used our findings to argue for the need for detailed reporting of data standardization procedures in predictive modeling, especially in studies leveraging multi-site datasets extracted from electronic health records.
Keywords: hospital readmission, heart failure, logical observation identifiers names and codes, predictive modeling, medical informatics/standards
INTRODUCTION
The growing repository of available healthcare data has motivated the healthcare community to improve medical decision-making by integrating knowledge learned from data-driven analyses.1,2 Often, these analyses are geared toward enhancing clinical decision support (CDS) systems with models that predict events of clinical relevance, such as disease risk or progression.2 Laboratory data are particularly valuable information in predictive modeling as they can provide insight about a patient’s current and potential future clinical state. Unfortunately, the secondary use of laboratory data poses challenges due to the lack of enforced standardization.3 Currently, the only available standard for lab tests is the logical observation identifiers names and codes (LOINC), which provides a universal set of structured codes to identify laboratory and clinical observations.4,5 We have noticed in the literature, however, that most predictive modeling studies utilizing clinical laboratory data provide little to no information on the standardization processes used.
As an illustrative example of the lack of reporting in the literature, we considered readmission risk prediction models, which have grown increasingly popular since the introduction of financial penalties for excess readmissions by the Centers for Medicare and Medicaid Services (CMS).6 A number of studies on predicting readmission risk have utilized laboratory data7–24; however, most multi-site readmission prediction models using laboratory information provide limited details on the data standardization procedures used across sites.8,9,11–13,15,16,23,24 In particular, we found only 1 study that included any traceable record of standardizing to LOINC.12 Failing to report standardization procedures makes it challenging to accurately reproduce these multi-site predictive models and presents potential methodological issues in the modeling approach. For example, if a multi-site study failed to standardize laboratory test names across sites, it would result in incorrectly treating clinically comparable laboratory tests from different sites as unique tests in the model. This could result in poor overall model performance in addition to potentially mitigating the predictive power of laboratory data. This risk is especially high for data-driven modeling approaches, which are gaining popularity in the healthcare domain.25 The potential impact of standardizing laboratory data on prediction performance in multi-site datasets, however, has been largely ignored and under reported.
OBJECTIVES
In this study, we aimed to gain a better understanding of how the standardization of laboratory data can impact predictive model performance. We specifically focused on understanding how standardizing local laboratory test codes to LOINC impacts predictive model performance in multi-site datasets. We hypothesized that standardizing local laboratory codes to LOINC would produce predictive models that significantly outperform those learned utilizing local laboratory codes. To test our hypothesis, we performed a case study using 30-day readmission risk predictive models for adult heart failure patients, as this population is currently subject to financial penalties by the CMS.6 Findings from our study were used to construct an argument for the importance of reporting data standardization procedures in multi-site predictive modeling studies. The main contributions of this work included: (1) empirical evidence to support the need for data standardization in predictive modeling using multi-site datasets and (2) suggested recommendations for reporting laboratory data standardization.
METHODS
We extracted laboratory test results for adult heart failure patient visits from a large, multi-hospital health system. We then cleaned and standardized test results and mapped local laboratory test codes to LOINC. We constructed a set of features, and then learned several models to predict risk of 30-day hospital readmission using a variety of feature selection and modeling techniques. We compared the performance of models learned using local laboratory test codes to the same models learned using LOINC. These processes are described in detail in the following sub-sections. This study was reviewed and approved by the institutional review board (IRB) at the University of Pittsburgh (PRO18040108).
Dataset
We utilized an IRB certified honest broker to retrieve all electronic health records (EHRs) for in-patient visits to 13 individual hospitals within the University of Pittsburgh Medical Center (UPMC) Health System from 2008 to 2012. Heart failure-specific visits were identified using primary discharge ICD-9 codes [428 family (428.XX), 402.01, 402.11, 402.91, 404.01, 404.03, 404.11, 404.13, 404.91, 404.93]. Visits with in-hospital deaths and any visit without at least 1 valid laboratory test value available were excluded. If a patient returned to any UPMC hospital within 30 days following discharge from a visit, then the visit was classified as “Readmitted” (R); otherwise the visit was classified as “Not Readmitted” (NR). All visit information was then deidentified by the honest broker and provided to the research team for analysis. Laboratory test results from each visit were manually cleaned and standardized, and then were flagged as normal/abnormal (a detailed report of cleaning and standardization procedures is available in the Supplementary Material). Only data collected prior to discharge from the visit were used to predict whether the visit would result in a 30-day readmission, that is, only data from the initial visit were included in the prediction model.
Mapping to LOINC
As part of an ongoing effort to convert to LOINC, 1 UPMC hospital had previously mapped 456 of the most commonly ordered local laboratory test codes to LOINC. At the time of this study, this partial mapping was the only available mapping to LOINC across all 13 hospitals. The mapping process was completed manually by 3 coders from the Laboratory Information System (LIS) division who had more than 20 years of clinical laboratory experience and medical technologist certifications from the American Society for Clinical Pathology. Two coders independently mapped local laboratory codes to LOINC and discussed discrepancies. A third coder (T.G.) oversaw the process and reviewed discrepancies if the two coders could not come to an agreement. A supervisor of the UPMC core laboratory vetted the resulting list of LOINC assignments as a final technical review. Unfortunately, this list was not originally generated for research use, therefore the intercoder reliability was not captured, initial false positive mappings were not racked, and no formal validation of the mapping process was able to be performed.
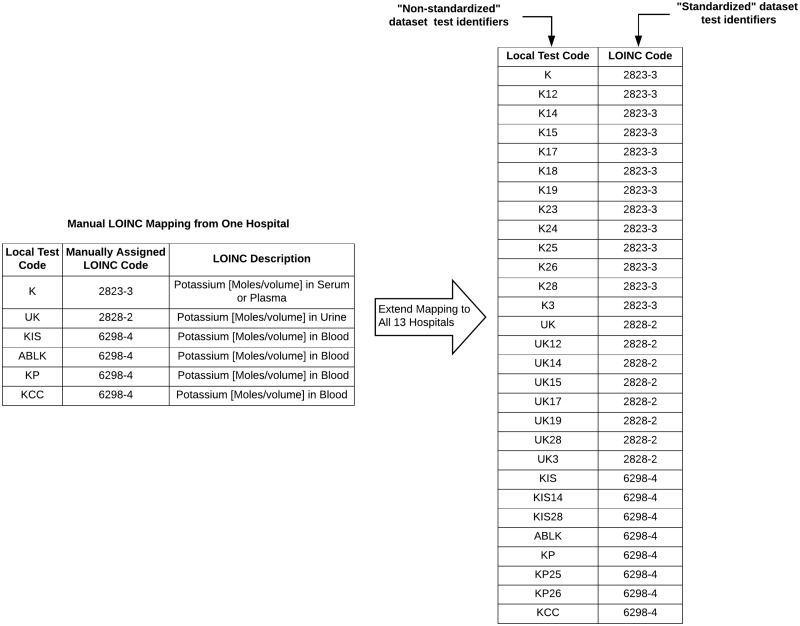
UPMC hospitals’ local laboratory test codes consist of a descriptive code for the test and a hospital ID tag indicating the source hospital (e.g, code “K14” represents a serum potassium test for hospital with ID 14). By removing the hospital ID tags, we were able to use the list of 456 mapped codes from a single hospital to map local laboratory codes to LOINC for all 13 hospitals. This process yielded 2 datasets for analysis: (1) a “non-standardized” dataset where tests were identified via the local laboratory codes (ie, no mapping of laboratory codes was performed) and (2) a “standardized” dataset where tests were identified via LOINC. An example of the mapping process is illustrated in Figure 1. As only a partial LOINC mapping was available, we discarded any tests that could not be mapped to LOINC from both the “non-standardized” and “standardized” datasets. This was done to ensure that we compared model performance across the same set of laboratory tests to get an unbiased estimate of the effect of standardizing laboratory codes to LOINC.
Figure 1.
Example of LOINC mapping for potassium laboratory tests. A manual mapping from 1 hospital (left) was extended to map local laboratory test codes from 13 hospitals to LOINC. After mapping, we had an “non-standardized” dataset, where laboratory tests were identified via the unmapped, local laboratory test codes and a “standardized” dataset, where laboratory tests were identified via a LOINC code.
Feature construction
Due to the asynchronous, time-series nature of laboratory data, we defined a fixed set of features to summarize test results for each patient visit. The features are listed in Table 1. Many of these features were part of a laboratory data feature set originally described by Hauskrecht et al.,26 but we also derived some new features. In Table 1, we have identified the Hauskrecht et al.26 features with superscript ‘H’s. To summarize the results for all laboratory tests that occurred during a patient visit, we defined 3 features (Table 1, column 1): (1) the average number of test results received per day (defined as number of tests divided by length of stay), (2) the percentage of most recent test results that were flagged as abnormal, that is, the percentage of abnormal results when considering only the most recently recorded result from each test, and (3) the percentage of all test results that were flagged as abnormal. For each categorical lab test, results were summarized using the results from the 2 most recent tests, the result from the first test, and the baseline result across all tests, which was defined as the mode of all test results excluding the most recent test (Table 1, column 2). For each continuous lab test, results were summarized using the percentage of all test results that were flagged as abnormal, the results from the 2 most recent tests, the result from the first test, the baseline result across all tests (defined as the mean of all test results excluding the most recent test), the nadir (min) and apex (max) results from all tests, and several features aimed to summarize result trends over time, such as the difference, percent change, and slope between the 2 most recent test results (Table 1, column 3). To reduce the amount of missing data generated in constructing the feature set, some features were only constructed for a given test if the median number of results per patient for that test was greater than 1 or 2. For example, for a categorical test for which most patients only have 1 test result, we would only use the most recent test result as a feature. Features were constructed for both the “non-standardized” and “standardized” datasets. All numeric constructed features were discretized using the minimum description length criterion discretization method.27
Table 1.
Features constructed to summarize laboratory test results each patient visit
| Included features | Summary of results for |
|||
|---|---|---|---|---|
| All lab tests | Each categorical lab test | Each continuous lab test | ||
| Average # of tests per day (# tests/length of stay) | X | |||
| % Abnormal tests for most recent testsa | X | |||
| % Abnormal testsa | X | X | ||
| Flag (normal/abnormal) for most recent test | X | |||
| Most recent test result | XH | XH | ||
| Second most recent test result (if median test count >1) | XH | XH | ||
| First test result (if median test count >2) | XH | X | ||
| Baseline result (mean/mode of values prior to most recent) (if median test count>1) | X | XH | ||
| Nadir (min) result (if median test count>2) | XH | |||
| Apex (max) result (if median test count>2) | XH | |||
| Difference between most recent test result and…. | Second most recent test result | XH | ||
| First test result | X | |||
| Apex result | XH | |||
| Nadir result | XH | |||
| Baseline result | XH | |||
| % change between most recent test result and… | Second most recent test result | XH | ||
| First test result | X | |||
| Apex result | XH | |||
| Nadir result | XH | |||
| Baseline result | XH | |||
| Slope between most recent test result and… | Second most recent test result | XH | ||
| First test result | X | |||
| Apex result | XH | |||
| Nadir result | XH | |||
| Baseline result | XH | |||
X: feature was derived for dataset; H: feature was originally described in Hauskrecht et al.26
Tests with “NA” flags were not included in these computations.
Model learning and evaluation
To learn and validate predictive models, we split each of the “non-standardized” and “standardized” datasets into a training dataset (data from 2008 to 2011) and a test dataset (data from 2012). We used the training datasets to learn models utilizing a variety of popular feature selection techniques and model types. We examined 2 popular strategies for feature selection: (1) correlation-based feature subset (CFS)28 selection which aims to find a set of features that have high correlation with the target class but low intercorrelation with each other, that is, a set of non-redundant, highly informative features and (2) information gain (IG) filter with a threshold greater than 0, which results in selecting features that contain at least some information with respect to the target class. For models, we examined logistic regression, naïve Bayes, and random forest classifiers, which are three popular models within the medical domain. We used the WEKA (Waikato Environment for Knowledge Acquisition) version 3.829 implementation of all algorithms. We adopted the default algorithm settings provided by WEKA, except for treating missing values as a separate category in our feature selection approaches, which had been previously shown to improve model performance,30 and learning a larger number of trees (500) in the random forest classifier. For each feature selection and classifier pair, we learned a predictive model based on the “non-standardized” and “standardized” datasets. The learned models are summarized in Table 2.
Table 2.
30-Day heart failure readmission model descriptions, evaluations, and comparisons. Prior to feature selection, there were 10,032 and 1881 features from non-standardized dataset (local codes) and standardized dataset (LOINC) respectively.
| # | Feature selection | Classifier | Dataset | Number of features | AUC (95% CI) | P-value |
|---|---|---|---|---|---|---|
| 1 | Information gain | Logistic regression | Non-standardized (Local codes) | 1154 | 0.538 (0.516–0.559) | 0.001 |
| 2 | Standardized (LOINC codes) | 388 | 0.573 (0.551–0.594) | |||
| 3 | Naïve Bayes | Non-standardized (Local codes) | 1154 | 0.560 (0.539–0.582) | 5.3e-5 | |
| 4 | Standardized (LOINC codes) | 388 | 0.603 (0.583–0.624) | |||
| 5 | Random forest | Non-standardized (Local codes) | 1154 | 0.590 (0.570–0.612) | 0.036 | |
| 6 | Standardized (LOINC codes) | 388 | 0.605 (0.585–0.626) | |||
| 7 | Correlation-based feature selection | Logistic regression | Non-standardized (Local codes) | 57 | 0.566 (0.545–0.587) | 2.3e-4 |
| 8 | Standardized (LOINC codes) | 46 | 0.601 (0.580–0.622) | |||
| 9 | Naïve Bayes | Non-standardized (Local codes) | 57 | 0.571 (0.550–0.592) | 8.9e-6 | |
| 10 | Standardized (LOINC codes) | 46 | 0.607 (0.586–0.628) | |||
| 11 | Random forest | Non-standardized (Local codes) | 57 | 0.561 (0.539–0.582) | 2.5e-4 | |
| 12 | Standardized (LOINC codes) | 46 | 0.602 (0.581–0.622) |
Note: Bolded P-values indicate significant differences in model performance.
We used the respective test datasets to evaluate the learned predictive models. All evaluation metrics were computed using the pROC package31 version 1.13.0 in R version 3.4.32 Evaluation metrics for each model included the area under the receiver-operating characteristic curve (AUC) and the 95% confidence interval (CI) computed using 2000 stratified bootstrap replicates (see pROC package documentation for details on bootstrapping approach).33 DeLong’s 1-sided comparisons34 with Bonferroni multiple-hypotheses correction35 were used to compare AUCs of the models based on the “non-standardized” and “standardized” datasets.
RESULTS
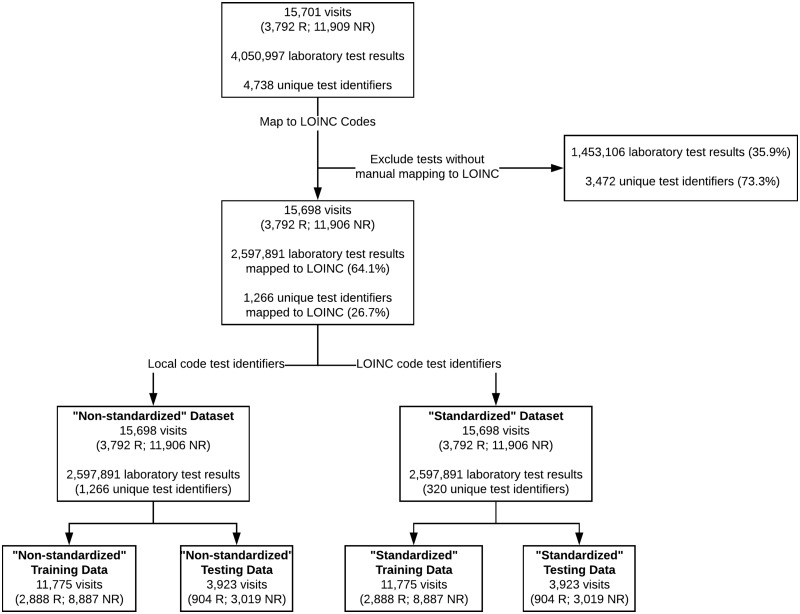
Figure 2 summarizes the coverage of the mapping process and provides a description of the training and test datasets. Table 2 summarizes the models learned to predict 30-day hospital readmission for adult heart failure patients, including the number of features used based on each feature selection technique, the AUC with 95% CI, and the P-values of the model comparisons. Complete lists of features selected by the CFS method for each dataset are provided in Table A1 of the Supplementary Material. As indicated by the bold-faced P-values in Table 2, nearly all models learned on the “standardized” dataset (ie, where tests were identified via LOINC) performed significantly better than models learned on the “non-standardized” dataset (ie, where tests were identified via local laboratory codes).
Figure 2.
LOINC mapping coverage and description of training and test datasets. “R” and “NR” stand for the classification as “Readmitted” or “Not Readmitted”, respectively.
DISCUSSION
We examined the effect of standardizing local laboratory test names to LOINC on predictive model performance in multi-site datasets. More specifically, we evaluated this effect in a case study on predicting 30-day hospital readmissions for a multi-site cohort of adult heart failure patients. To the best of our knowledge, this is the first study to examine this effect. Our results in Table 2 demonstrated that standardizing local laboratory codes to LOINC for multi-site datasets consistently resulted in models that achieved significantly higher predictive performance, regardless of the feature selection technique and classifier approach used. The final AUCs of our models were modest; however, the goal of this study was not to build a high-performing model, but rather to determine whether standardization of laboratory test names to LOINC improved model performance. We noticed significant improvement in performance even with the limited predictive ability of our models, and we believe that higher performing models using additional data would also benefit from standardization of laboratory data. This could lead to better overall predictive models to be used in CDS systems, especially since previous work has shown that standardization of data tends to lead to better outcomes for CDS systems.36 Given the potential impact standardizing laboratory data might have on predictive model performance, we find it alarming that many multi-site predictive modeling studies fail to include details on laboratory data standardization.
The low quality of reporting of prediction model studies in the healthcare domain has been previously identified as an issue, and it presents challenges in reproducing models and assessing the potential bias and usefulness of the models.37 Efforts have been made to develop recommendations for researchers when reporting the development and validation of models, such as the Transparent Reporting of a multivariate prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement.37 The TRIPOD statement is an excellent guideline for transparent model reporting and has been used to describe machine learning modeling approaches,38 but it provides limited consideration for data-driven approaches that utilize multi-site datasets. Specifically, it offers no guidance for reporting data standardization procedures. As our study has demonstrated, the standardization procedures used can have a profound impact on model performance and reproducibility when employing an EHR data-driven approach to prediction. Thus, detailed reporting on standardization procedures seems crucial to critically evaluate such models. These aspects will become an increasingly important part of predictive model reporting as EHR data-driven approaches to prediction gain popularity. Therefore, we argue that current predictive model reporting recommendations should be expanded to consider some of the unique challenges present when modeling with multi-site datasets extracted from EHRs. In particular, we argue for explicit recommendations pertaining to the reporting of data standardization procedures across sites.
Specific attention should be given to developing recommendations for reporting standardization procedures for laboratory data. Although LOINC is the accepted standard for reporting laboratory test names, it is a highly specific coding system and there is no standard procedure for mapping to LOINC. This presents a granularity problem when performing LOINC mapping.5,39–46 Therefore, wide variation in mapping specificity exists across institutions,39,47,48 which may pose significant challenges in predictive modeling on multi-site datasets. In a dataset with multiple mapping approaches performed by different institutions, effects on model performance due to varying levels of mapping specificity may be comparable to those observed in our study. We therefore recommend that multi-site studies evaluate and report on any differences in LOINC mapping processes used across sites. When possible, studies should report the level of agreement between LOINC mappings from different institutions.
Several prior studies specifically point out the need for lower resolutions of LOINC (eg, code groups or hierarchical structuring) to promote accurate data sharing and analysis across institutions.39,43,46,48 This need will become increasingly prevalent as more initiatives are undertaken to create and analyze networks of healthcare data across multiple institutions, such as the National Patient-Centered Clinical Research Network.49 The new LOINC Groups project by the Regenstrief Institute aims to address this need by creating sets of clinically similar codes. When completed, LOINC Groups could prove to be an invaluable tool for grouping the LOINC mappings in large multi-institutional datasets in a clinically meaningful way.50 As suggested by the findings of our study, these groupings may improve the quality and performance of predictive models learned from these networks of data. Without detailed reporting of the data standardization procedures used, however, it may be challenging to critically appraise and reproduce predictive models learned from these large, multi-institutional datasets. We therefore recommend that as part of laboratory data standardization reporting requirements, future studies should include any LOINC aggregation procedures used. In particular, we suggest that once the LOINC Groups project is completed, it should be recommended as the standard approach for aggregating codes.
Recently, an argument was made against the need for EHR data standardization and harmonization due to advancements in deep learning approaches to modeling, which are capable of achieving high performance when using large sets of messy data.51 Although our work did not explore deep learning approaches, it is worth discussing the idea as it contradicts our argument for the need for reporting data standardization procedures. The deep learning approach is a promising avenue for achieving high performance models based on raw EHR data, but these approaches have not yet been validated on multi-site datasets where the lack of data standardization presents significant challenges. Moreover, due to the demand for model interpretability in healthcare,52 it is likely that more traditional approaches to modeling will remain relevant. Thus, we assert that it is still essential to develop better recommendations for reporting data standardization procedures used when modeling with multi-site datasets extracted from EHRs.
Limitations
This study had several limitations that should be addressed in future work. First, as the LOINC mapping utilized was part of an ongoing project at UPMC, only a partial LOINC mapping was available at the time of this study. Therefore, we chose to exclude from our analysis any laboratory tests that did not have a LOINC mapping. This allowed for a fair comparison of model performance with and without standardization to LOINC across the same set of laboratory tests. Alternatively, we could have utilized the local laboratory test codes when a LOINC mapping was unavailable, but we felt that this approach would introduce too much bias against LOINC standardization due to the partially complete mapping. This alternative approach would be appropriate to utilize once the UPMC team has finished the LOINC mapping process. Thus, our conclusions are based only on a subset of laboratory data; however, this subset captured a large portion of all laboratory test results in our dataset (∼64% of all test results). We therefore believe that an analysis based on a complete LOINC mapping would yield similar results, but plan to evaluate this idea in future work when a complete mapping is available.
Additionally, as the partial mapping was not originally generated for research purposes, intercoder agreement and false positive mappings were not tracked. Thus, a formal validation of the mapping process was unable to be performed. It would be beneficial to validate our claims using more rigorously tested mapping approaches; however, it would take significant time and expertise to complete such mappings. Moreover, the two coders on the mapping team were highly qualified, thoughtfully selected subject matter experts and the accuracy of these individuals working together to map codes was expected to be high. The mapping team subjectively estimated that less than 5% of the initial codes resulted in discrepancies that needed to be reviewed, and they were confident in the accuracy of their approach (ie, it was unlikely that false positive mappings would have occurred).
Finally, our definition of readmission included both planned and unplanned visits and we only examined a single prediction task for a specific patient population. We note that our claims may not be valid for other patient populations or for other prediction tasks. Future studies examining the impact of standardizing to LOINC on prediction performance should include in a variety of population and prediction tasks and utilize all available laboratory test results. The impact of standardizing other EHR data types on predictive model performance should be also explored. Such studies could provide further support for the need for detailed reporting on standardization procedures in predictive modeling studies.
CONCLUSION
This study investigated the impact of standardizing local laboratory codes to LOINC on predictive model performance in a multi-site dataset. We quantitatively demonstrated that standardizing to LOINC significantly improves predictive performance across a variety of feature selection and modeling techniques. Based on our findings, we have argued for the need for detailed reporting of data standardization procedures in predictive modeling, especially in studies leveraging multi-site datasets extracted from EHRs.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONTRIBUTORS
AB and FT designed this study. AB executed the study design and prepared the manuscript. VR provided substantial assistance in executing the study design and provided revisions to the manuscript. TG provided the LOINC mappings and critical insights into the mapping process. FT oversaw the research and provided revisions to the manuscript. All authors reviewed and approved the final manuscript version.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the assistance of Jose Posada, Lingyun Shi, and Ye Ye.
FUNDING
This work was supported in part by the Richard King Mellon Foundation under award 5487 and the Innovation Works (2014W.DZ01621E-1) and the National Institutes of Health (NIH) through the National Library of Medicine (NLM) under award 5 T15 LM007059-27 and the Clinical and Translational Science Institute (CTSI) under award 5 UL1 TR000005-09. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Richard King Mellon Foundation, NIH, or Innovation Works.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tan SS-L, Gao G, Koch S.. Big data and analytics in healthcare. Methods Inf Med 2015; 54: 546–7. [DOI] [PubMed] [Google Scholar]
- 2. Simpao AF, Ahumada LM, Gálvez JA, et al. A review of analytics and clinical informatics in health care. J Med Syst 2014; 38:45. [DOI] [PubMed] [Google Scholar]
- 3. Hauser RG, Quine DB, Ryder A.. LabRS: a Rosetta stone for retrospective standardization of clinical laboratory test results. J Am Med Inform Assoc 2018; 25: 121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inf Assoc 1998; 53: 276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baorto DM, Cimino JJ, Parvin CA, et al. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC). Int J Med Inform 1998; 51: 29–37. [DOI] [PubMed] [Google Scholar]
- 6. CMS.gov. Readmissions Reduction Program (HRRP). 2016. https://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html Accessed April 18, 2016.
- 7. Walsh C, Hripcsak G.. The effects of data sources, cohort selection, and outcome definition on a predictive model of risk of thirty-day hospital readmissions. J Biomed Inform 2014; 52: 418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huynh QL, Saito M, Blizzard CL, et al. Roles of nonclinical and clinical data in prediction of 30-day rehospitalization or death among heart failure patients. J Card Fail 2015; 215: 374–81. [DOI] [PubMed] [Google Scholar]
- 9. Choudhry SA, Li J, Davis D, et al. A public-private partnership develops and externally validates a 30-day hospital readmission risk prediction model. Online J Public Health Inform 2013; 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Donzé J, Aujesky D, Williams D, et al. Potentially avoidable 30-day hospital readmissions in medical patients: derivation and validation of a prediction model. JAMA Intern Med 2013; 1738: 632–8. [DOI] [PubMed] [Google Scholar]
- 11. Hammill BG, Curtis LH, Fonarow GC, et al. Incremental value of clinical data beyond claims data in predicting 30-day outcomes after heart failure hospitalization. Circ Cardiovasc Qual Outcomes 2011; 41: 60–7. [DOI] [PubMed] [Google Scholar]
- 12. Hao S, Wang Y, Jin B, et al. Development, validation and deployment of a real time 30 day hospital readmission risk assessment tool in the Maine healthcare information exchange. PLoS One 2015; 10: e0140271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenzi J, Avaldi VM, Hernandez-Boussard T, et al. Risk-adjustment models for heart failure patients’ 30-day mortality and readmission rates: the incremental value of clinical data abstracted from medical charts beyond hospital discharge record. BMC Health Serv Res 2016; 16: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rubin DJ, Golden SH, McDonnell ME, et al. Predicting readmission risk of patients with diabetes hospitalized for cardiovascular disease: a retrospective cohort study. J Diabetes Complications 2017; 318: 1332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shadmi E, Flaks-Manov N, Hoshen M, et al. Predicting 30-day readmissions with preadmission electronic health record data. Med Care 2015; 533: 283–9. [DOI] [PubMed] [Google Scholar]
- 16. Tabak YP, Sun X, Nunez CM, et al. Predicting readmission at early hospitalization using electronic clinical data: an early readmission risk score. Med Care 2017; 553: 267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleming LM, Gavin M, Piatkowski G, et al. Derivation and validation of a 30-day heart failure readmission model. Am J Cardiol 2014; 1149: 1379–82. [DOI] [PubMed] [Google Scholar]
- 18. Amarasingham R, Moore BJ, Tabak YP, et al. An automated model to identify heart failure patients at risk for 30-day readmission or death using electronic medical record data. Med Care 2010; 4811: 981–8. [DOI] [PubMed] [Google Scholar]
- 19. Hebert C, Shivade C, Foraker R, et al. Diagnosis-specific readmission risk prediction using electronic health data: a retrospective cohort study. BMC Med Inform Decis Mak 2014; 14: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bradley EH, Yakusheva O, Horwitz LI, et al. Identifying patients at increased risk for unplanned readmission. Med Care 2013; 519: 761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. AbdelRahman SE, Zhang M, Bray BE, et al. A three-step approach for the derivation and validation of high-performing predictive models using an operational dataset: congestive heart failure readmission case study. BMC Med Inform Decis Mak 2014; 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rothman MJ, Rothman SI, Beals J.. Development and validation of a continuous measure of patient condition using the Electronic Medical Record. J Biomed Inform 2013; 465: 837–48. [DOI] [PubMed] [Google Scholar]
- 23. Cubbon RM, Woolston A, Adams B, et al. Prospective development and validation of a model to predict heart failure hospitalisation. Heart 2014; 10012: 923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amarasingham R, Velasco F, Xie B, et al. Electronic medical record-based multicondition models to predict the risk of 30 day readmission or death among adult medicine patients: validation and comparison to existing models. BMC Med Inform Decis Mak 2015; 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beam AL, Kohane IS.. Big data and machine learning in health care. JAMA 2018; 31913: 1317–8. [DOI] [PubMed] [Google Scholar]
- 26. Hauskrecht M, Batal I, Valko M, et al. Outlier detection for patient monitoring and alerting. J Biomed Inform 2013; 461: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fayyad UM, Irani KB Multi-interval discretization of continuous valued attributes for classification learning. In: Proceedings of the Thirteenth International Joint Conference on Artificial Intelligence San Francisco, CA: 1993. 1022–9. [Google Scholar]
- 28. Hall MA. Correlation-Based Feature Selection for Machine Learning [dissertation]. Hamilton, New Zealand: The University of Waikato; 1999. [Google Scholar]
- 29. Frank E, Hall MA, Witten IH.. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques.” 4th ed Morgan Kaufmann; 2016. [Google Scholar]
- 30. López Pineda A, Ye Y, Visweswaran S, et al. Comparison of machine learning classifiers for influenza detection from emergency department free-text reports. J Biomed Inform 2015; 58: 60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robin X, Turck N, Hainard A, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing 2017. https://www.R-project.org/.
- 33.Robin X, Turck N, Hainard A, et al. Package ‘pROC ’. 2018. https://cran.r-project.org/web/packages/pROC/pROC.pdf Accessed November 15, 2018.
- 34. DeLong ER, DeLong DM, Clarke-Pearson DL.. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 443: 837–45. [PubMed] [Google Scholar]
- 35. Dunn OJ. Multiple comparisons among means. J Am Stat Assoc 1961; 56293: 52–64. [Google Scholar]
- 36. Ahmadian L, van Engen-Verheul M, Bakhshi-Raiez F, Peek N, Cornet R, de Keizer NF.. The role of standardized data and terminological systems in computerized clinical decision support systems: Literature review and survey. Int J Med Inform 2011; 802: 81–93. [DOI] [PubMed] [Google Scholar]
- 37. Collins GS, Reitsma JB, Altman DG, Moons KGM.. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015; 350: g7594.. [DOI] [PubMed] [Google Scholar]
- 38. De Bari B, Vallati M, Gatta R.. Development and validation of a machine learning-based predictive model to improve the prediction of inguinal status of anal cancer patients: A preliminary report. Oncotarget 2017; 8: 108509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin MC, Vreeman DJ, McDonald CJ, et al. Auditing consistency and usefulness of LOINC use among three large institutions—using version spaces for grouping LOINC codes. J Biomed Inform 2012; 454: 658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim H, El-Kareh R, Goel A, et al. An approach to improve LOINC mapping through augmentation of local test names. J Biomed Inform 2012; 454: 651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vreeman DJ, Hook J, Dixon BE.. Learning from the crowd while mapping to LOINC. J Am Med Inform Assoc 2015; 226: 1205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khan AN, Russell D, Moore C, et al. The map to LOINC project. In: AMIA Annu Symp Proc 2003; 2003: 890. [PMC free article] [PubMed] [Google Scholar]
- 43. Lau LM, Johnson K, Monson K, et al. A method for the automated mapping of laboratory results to LOINC. Proc AMIA Symp 2000; 2000: 472–6. [PMC free article] [PubMed] [Google Scholar]
- 44. Khan AN, Griffith SP, Moore C, et al. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc 2006; 133: 353–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gamache RE, Dixon BE, Grannis S, et al. Impact of selective mapping strategies on automated laboratory result notification to public health authorities. AMIA Annu Symp Proc 2012; 2012: 228–36. [PMC free article] [PubMed] [Google Scholar]
- 46. Hauser RG, Quine DB, Ryder A, Campbell S. Unit conversions between LOINC codes. J Am Med Inform Assoc 2017; 25: 192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kume N, Suzuki K, Kobayashi S, et al. Development of unified lab test result master for multiple facilities. Stud Health Technol Inform 2015; 216: 1050. [PubMed] [Google Scholar]
- 48. Steindel S, Loonsk JW, Sim A, et al. Introduction of a hierarchy to LOINC to facilitate public health reporting. Proc AMIA Symp 2002; 2002: 737–41. [PMC free article] [PubMed] [Google Scholar]
- 49. Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. J Am Med Inform Assoc 2014; 214: 576–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Regenstrief Institute. LOINC Groups. https://loinc.org/groups/ Accessed January 27, 2018.
- 51. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning for electronic health records. NPJ Digit Med 2018; 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cabitza F, Rasoini R, Gensini GF.. Unintended consequences of machine learning in medicine. JAMA 2017; 3186: 517–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.