Abstract
Growth factor receptor-bound protein 2 (Grb2) is an adaptor protein that plays a critical role in cellular signal transduction. It contains a central Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains. Binding of Grb2 SH2 to the cytoplasmic region of CD28, phosphorylated Tyr (pY) containing the peptide motif pY-X-N-X, is required for costimulatory signaling in T cells. In this study, we purified the dimer and monomer forms of Grb2 SH2, respectively, and analyzed their structural and functional properties. Size exclusion chromatography analysis showed that both dimer and monomer exist as stable states. Thermal stability analysis using circular dichroism showed that the dimer mostly dissociates into the monomer around 50°C. CD28 binding experiments showed that the affinity of the dimer to the phosphopeptide was about three fold higher than that of the monomer, possibly due to the avidity effect. The present crystal structure analysis of Grb2 SH2 showed two forms; one is monomer at 1.15 Å resolution, which is currently the highest resolution analysis, and another is dimer at 2.00 Å resolution. In the dimer structure, the C-terminal region, comprising residues 123–152, was extended towards the adjacent molecule, in which Trp121 was the hinge residue. The stable dimer purified using size exclusion chromatography would be due to the C-terminal helix “swapping”. In cases where a mutation caused Trp121 to be replaced by Ser in Grb2 SH2, this protein still formed dimers, but lost the ability to bind CD28.
Keywords: binding kinetics, crystal structure, swapped dimer, thermal stability
Significance.
We purified the stable form of Grb2 SH2 dimer and monomer, and analyzed their structural and functional properties. The dimer mostly dissociated into the monomer around 50°C. The CD28 binding affinity of dimer was about three fold higher than that of monomer. The stable dimer would be swapped dimer, in which the C-terminal region is extended towards the adjacent molecule. Because Trp121 was considered to be the key residue for dimer formation, W121S mutant was generated and its structural and functional properties were analyzed. The mutant still formed dimers, but lost the ability to bind CD28.
Growth factor receptor-bound protein 2 (Grb2) is an adaptor protein consisting of an Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains, that contributes to signal transduction of the Ras/mitogen-activated protein kinase pathway [1]. Grb2 exhibits a monomer-dimer equilibrium, which could determine normal versus oncogenic function [2–4]. The SH2 domain of Grb2 (Grb2 SH2) specifically binds to the consensus sequence pY-X-N-X (where X is any amino acid) present in phosphorylated Tyr (pY) containing proteins [5]. The selective inhibition of Grb2 SH2 binding to phosphorylated proteins is expected to disrupt protein-tyrosine kinase signaling, and this inhibition might prevent the occurrence of hyperproliferative diseases.
Signaling via CD28 receptors is required for complete activation of T cells and differentiation into effector T cells, in addition to signals via T cell receptors [6,7]. When CD28 binds to its ligands such as CD80 (B7-1) and CD86 (B7-2), protein tyrosine kinases are recruited to the CD28 cytoplasmic tail, which consists of 41 amino acid residues including four Tyr residues, and they phosphorylate the tail. The YMNM motif of CD28 cytoplasmic tail plays an important role in the co-stimulatory signaling pathway, and its Tyr residue is critical for phosphorylation [8]. We previously reported the crystal structure of Grb2 SH2 in complex with the CD28 phosphopeptide, SDpYMNMTP, at 1.35 Å resolution, and showed that the recognition mechanism of Grb2 SH2 is different from those of SH2 domains in the p85 sub-unit of phosphoinositide 3-kinase (PI3K), N-terminal (nSH2) and C-terminal (cSH2) SH2 domains [9,10]. The 3D structures of Grb2 SH2 in either peptide-unbound or -bound states have been reported previously [11–24]. There are also reports showing that Grb2 SH2 forms a swapped dimer using the protein with GST-tag or His-tag [25,26].
In this study, we purified the stable form of the dimer from Grb2 SH2 without additional tag under physiological conditions, and analyzed its structural and functional properties using size-exclusion chromatography (SEC), circular dichroism (CD), and surface plasmon resonance (SPR). We obtained the crystal of the Grb2 SH2 dimer in the peptide-unbound state, in addition to that of monomer, and determined their structures at high resolution. It was observed that a dimer formed through C-terminal helix “swapping”. Because Trp121 was considered to be the key residue for dimer formation based on the structural information, the mutant W121S was generated, and its structural and functional properties were also analyzed. The CD28 binding abilities of Grb2 SH2 dimer and its W121S mutant were first evaluated in this study, and compared with that of Grb2 SH2 monomer reported previously [9]. We discussed the role of dimer formation in signal transduction by comparing our results to those obtained with other SH2 domains from Grb2-related adaptor downstream of Shc (Gads) and PI3K [10].
Materials and Methods
Materials
Grb2 SH2 and its mutant, W121S, were overexpressed in Escherichia coli and purified, as described previously [9]. Briefly, the SH2 domain of human Grb2 (residues 60–152) was expressed in Escherichia coli BL21 (DE3) cells as a glutathione S-transferase (GST)-fusion protein. The expressed SH2 was firstly purified using a glutathione column (glutathione-Sepharose 4B fast flow, GE Healthcare). To isolate SH2 from GST, the thrombin protease was used, followed by application onto a benzamidine-Sepharose column (HiTrap benzamidine fast flow, GE Healthcare) to remove thrombin. The SH2 monomer and dimer were purified using a gel filtration column (HiPrep Sephacryl S-100 column, GE Healthcare). A DNA fragment encompassing W121S was generated by site-directed mutagenesis using Grb2 SH2 as the template. The protein concentrations were determined from UV absorbance at 280 nm and were calculated by using the molar absorption coefficients of 1.43×104 M−1 cm−1 and 8.48×103 M−1 cm−1 for Grb2 SH2 and W121S, respectively. CD28 derived phosphopeptides were chemically synthesized, as described previously [10].
SEC analysis
Gel-filtration HPLC was performed on a Cosmosil 5Diol-300-II column (7.5 mm×30 cm, Nacalai Tesque, Japan). HPLC was performed with PBS (pH 7.4) at the flow rate of 1.0 mL min−1 at room temperature. The loading volume was 50 μL, and the eluate was monitored at 280 nm.
CD measurements
Far-UV CD spectra were measured on a Jasco J-725 or J-820 spectropolarimeter at 20°C equipped with Peltier-type temperature control system. The spectra were obtained for the protein concentration, 0.04 mg mL−1, in PBS (pH 7.4) using quartz cell with 1.0 cm path-length. CD spectra were obtained using scanning speed of 20 nm min−1, a time response of 1 sec, a bandwidth of 1 nm, and an average over 4 scans.
The melting curves were recorded in temperature scanning mode at 222 nm, from 20°C to 80°C with a heating rate of 1.0°C min−1. The analysis of the transition curve to determine Tm was performed on the basis of a two-state transition model assuming that the heat capacity change is zero.
SPR measurements
A Biacore T200 (GE Healthcare) was used to measure interaction between SH2 proteins and CD28. Biotinylated CD28 peptide, 41-residue phosphopeptide corresponding to cytoplasmic region of CD28, biotin-RSKRSRLLHSDpYM NMTPRRPGPTRKHYQPYAPPRDFAAYRS, was immobilized to Biacore sensor chip SA surfaces with streptavidin pre-immobilized to dextran, as described previously [27]. To measure interactions, Grb2 SH2 and its mutant, W121S, were injected over the immobilized CD28 at 20 μL min−1 for 180 sec at 25°C, as described previously [27]. Kinetic data were analyzed using the BIAevaluation program 3.3, which was supplied with the BIAcore system. In this program, a global fitting method was used for determination of the kinetic rate constants, kon and koff. The Ka values were also determined by Scatchard analysis using the steady-state response unit, RUeq [28].
Crystallization, data collection, model building and refinement
All crystallization experiments were performed using monomer fraction of the purified Grb2 SH2. Crystallization conditions were screened by the sparse matrix method using commercially available screening kits (Hampton Research) with the hanging-drop vapor-diffusion method at 20°C. Prior to data collection, all the crystals were soaked in cryo-protectant solutions containing 20% (v/v) glycerol along with their respective reservoir buffers and flash-frozen using nitrogen gas stream at −183°C.
X-ray diffraction experiments were performed at the beamline BL-5A at KEK-PF. All data was processed and scaled using XDS [29] and truncated by the CCP4 software suite [30]. The initial phases were determined by the molecular replacement method using the program PHASER [31], and the structures of the Grb2 SH2 domain complexed with CD28 derived peptide (3WA4) was used as the search models. Several cycles of manual model rebuilding by using Coot [32] and refinement by using PHENIX [33] and REFMAC [34] were performed. The refined models were validated by MOLPROBITY [35]. The statistics for data collection and refinement are summarized in Table 1. The figures were prepared using the PyMOL software (http://www.pymol.org/).
Table 1.
Data collection and refinement statistics. Values in parentheses are for the highest resolution shell
| Data collection | monomer | dimer |
| Wavelength, Å | 1.0000 | 1.0000 |
| Space group | P41212 | I422 |
| Unit-cell parameters, Å | ||
| a/c (Å) | 79.6/69.6 | 80.4/73.6 |
| Resolution, Å | 50–1.15 (1.22–1.15) | 50–2.00 (2.12–2.00) |
| No. of observations | 1,072,294 | 118,431 |
| No. of unique reflections | 77,235 | 8,418 |
| Completeness, % | 97.2 (88.9) | 99.9 (99.8) |
| Average I/σ(I) | 27.9 (2.7) | 30.4 (4.0) |
| Redundancy | 13.8 (11.8) | 14.1 (14.2) |
| Rsym, % | 5.9 (93.5) | 5.8 (84.5) |
| CC1/2, % | 100 (78.7) | 100 (92.3) |
| Refinement | ||
| R, % | 14.4 | 21.9 |
| Rfree, % | 16.9 | 24.2 |
| Number of molecule in asymmetric unit | 2 | 1 |
| Average B factor, Å2 | 16.2 | 45.0 |
| r.m.s. deviation from ideal | ||
| Bonds, Å | 0.013 | 0.004 |
| Angles, ° | 1.4 | 0.7 |
| Ramachandran plot | ||
| Favored region (%) | 97.85 | 98.9 |
| Allowed region (%) | 2.15 | 1.1 |
| Outlier region (%) | 0 | 0 |
Results
SEC analyses of Grb2 SH2 showed three elution peaks, corresponding to the monomer, the dimer, and the oligomer, respectively. The eluted fractions of monomer and dimer including oligomer were collected, and SEC was performed again. The results showed that the respective fractions were eluted as the same peak, respectively (Fig. 1A). These results indicate that Grb2 SH2 dimer and monomer exist as stable states and support the notion that Grb2 SH2 can form the stable “swapped” dimer [25,26].
Figure 1.
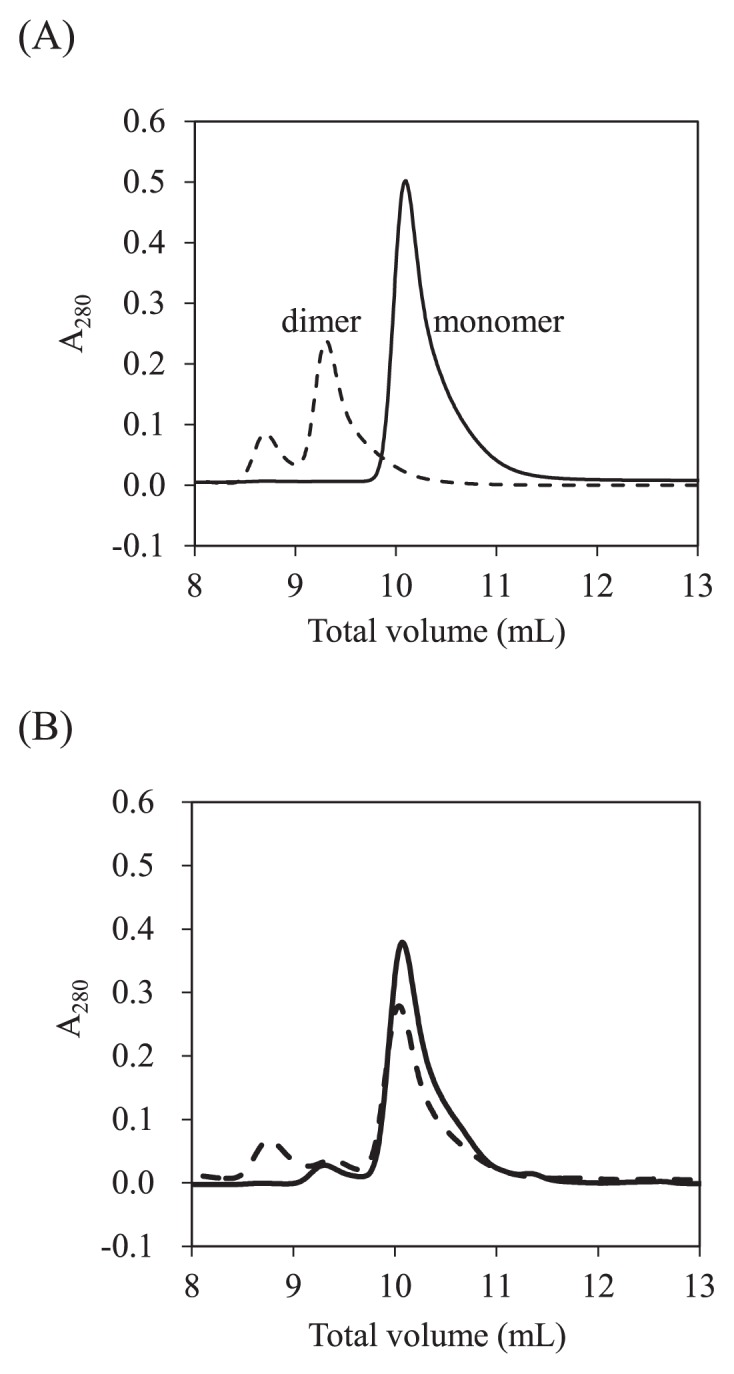
SEC analysis of Grb2 SH2 monomer and dimer. (A) Elution profiles of the monomer (solid line) and the dimer (broken line). (B) Elution profiles of the monomer (solid line) and the dimer (broken line) after heating up to 50°C. The eluates corresponding to monomer, dimer, and oligomer are observed at total volumes of 10.1, 9.4, and 8.7 mL, respectively.
Figure 2A shows the far-UV CD spectra of the Grb2 SH2 dimer and monomer purified using SEC. Figure 2B shows the thermal transition curves of the dimer and the monomer analyzed using CD. The transition temperature, Tm, of the monomer was determined to be 58.5°C. The transition curve of the dimer showed unique profile with two transitions; the first occurred between 40°C and 45°C, Tm,1 = 43.0°C, and the second between 55°C and 65°C, Tm,2 = 58.4°C, similar to that of monomer. After heating up to 80°C, both the monomer and the dimer aggregated, and the thermal transitions were irreversible. The CD spectrum of the monomer, after heating up to 50°C, was similar to that of the original monomer, while that of dimer was different from that of the original dimer and similar to that of monomer (Fig. 2C). In addition, when the original dimer was heated up to 50°C, it was eluted at the retention time corresponding to the monomer, except for a minor peak corresponding to an oligomer (Fig. 1B). These results suggest that the dimer mostly dissociates into the monomer around 50°C, which is when the first thermal transition observed in our CD experiments (Fig. 2B).
Figure 2.
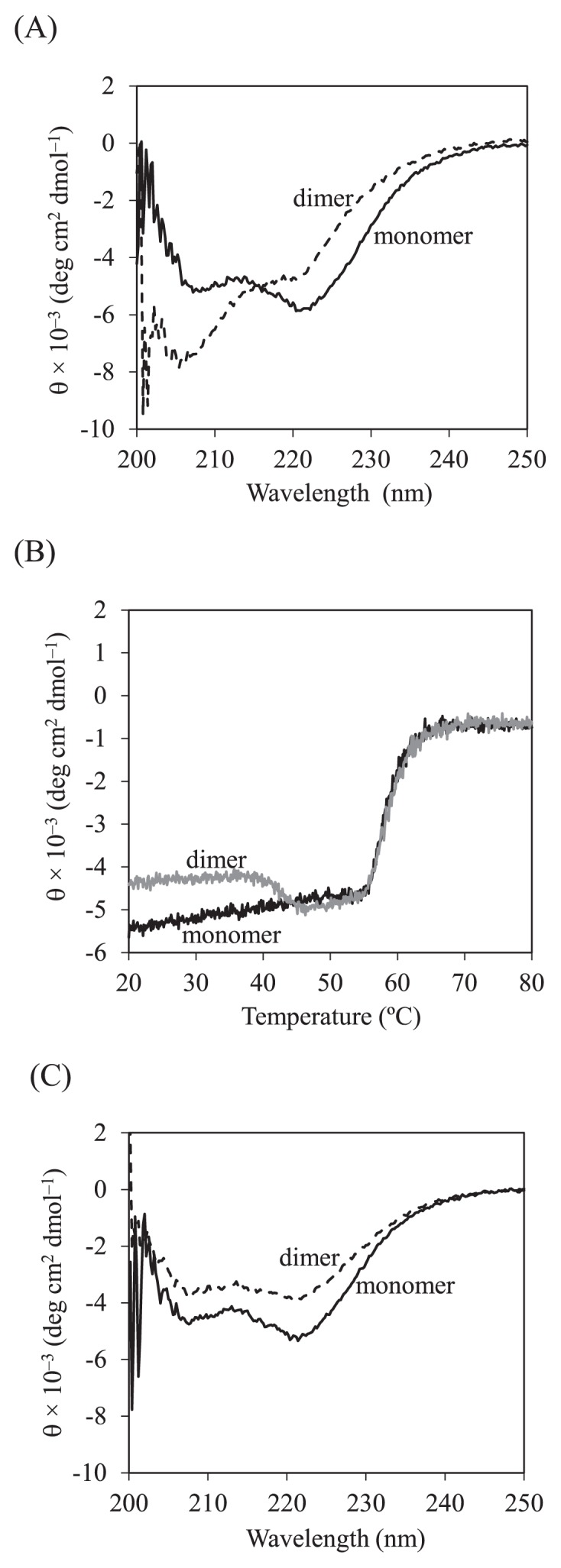
CD analysis of Grb2 SH2 monomer and dimer. (A) Far-UV CD spectra of the monomer (solid line) and the dimer (broken line) at 20°C. (B) Thermal transition curves of the monomer (black line) and the dimer (gray line). (C) Far-UV CD spectra of the monomer (solid line) and the dimer (broken line) at 20°C after heating up to 50°C.
The CD28 binding properties of dimer and monomer were analyzed using SPR biosensor. Figures 3A and 3B show sensorgrams for binding of the Grb2 SH2 monomer and dimer, and the binding kinetics are summarized in Table 2. The binding kinetics, association rate (kon) and dissociation rate (koff) constants, were determined using the model of 1:1 Langmuir binding. In the analyzed protein concentrations, 400, 800, 1600, 3200, and 6400 nM, this simple model could be applied; this was supported by the linear fitting in the Scatchard plots (Fig. 3C). The Ka values determined from binding kinetics were similar to those determined from binding equilibrium using Scatchard analysis (Table 2), showing that the binding affinity of the dimer increases about three fold compared to that of the monomer. The increased binding affinity of the dimer was mainly due to the decreased dissociation rate (Table 2).
Figure 3.
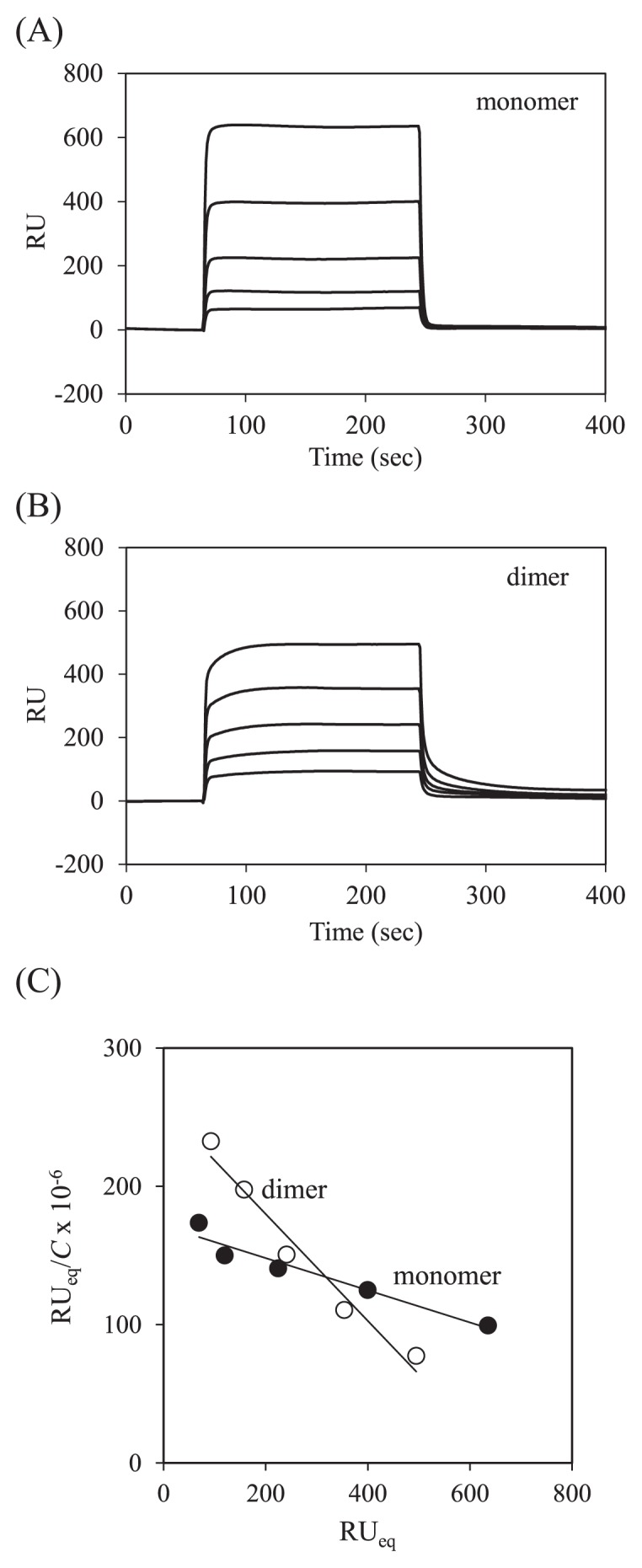
SPR analysis of Grb2 SH2 monomer and dimer. (A) Typical sensorgrams for binding of the monomer (400, 800, 1600, 3200, and 6400 nM) to immobilized CD28. (B) Typical sensorgrams for binding of the dimer (400, 800, 1600, 3200, and 6400 nM) to immobilized CD28. (C) Scatchard plots for binding of the monomer (closed circle) and the dimer (open circle).
Table 2.
Binding kinetics parameters for Grb2 SH2 binding to CD28 phosphopeptide at 25°C
| kon (M−1s−1) | koff (s−1) | Ka,kina (M−1) | Ka,Scab (M−1) | |
|---|---|---|---|---|
| monomer | 5.77×104 | 0.493 | 1.17×105 | 1.16×105 |
| dimer | 4.02×104 | 0.129 | 3.11×105 | 3.86×105 |
Ka,kin was calculated from the two kinetic rate constants.
Ka,Sca was calculated from a Scatchard analysis.
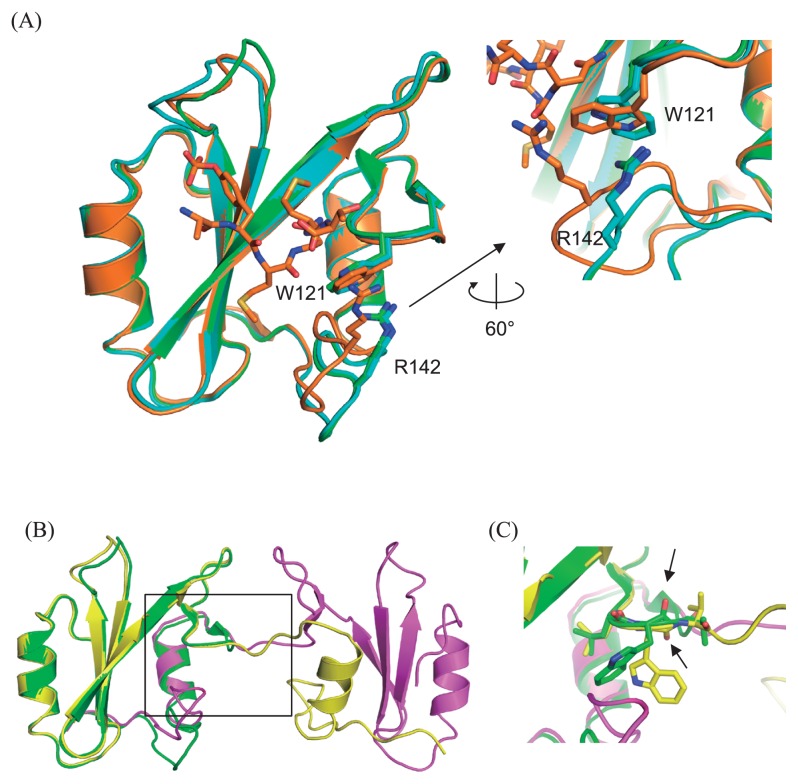
Crystals of the monomeric Grb2 SH2 in the absence of the CD28-derived phosphopeptide were obtained in the screening buffer, 0.1 M sodium chloride, 0.1 M HEPES (pH 7.5), 1.6 M ammonium sulfate in a concentration of 0.89 mM. The structure was determined at 1.15 Å resolution (Table 1), which is the highest resolution analysis among over 30 structures of the peptide bound/unbound Grb2 SH2 currently reported. The asymmetric unit contains two molecules of Grb2 SH2, and the overall structure of each monomer (Fig. 4A) is similar to that of the CD28-bound form, reported previously [9]. Remarkable structural change is observed at the loop region containing Arg142 and the interaction between Trp121 and Arg142 is altered. In the CD28-unbound form of present study, the indole ring of Trp121 is flipped and interacts more tightly with Arg142 by a cation-π interaction (Fig. 4A). This conformational change of the rotamer of Trp121 is due to the tight crystal contact around the loop region around Arg142.
Figure 4.
Overall structures of Grb2 SH2 monomer and dimer. (A) Left: Two monomers in the asymmetric unit (green and cyan) are superimposed on the structure of the complex with CD28 (orange, 3WA4). The root-mean-square-distance (rmsd) are 0.47 (between the monomer shown as green and 3WA4) and 0.54 (between the monomer shown as cyan and 3WA4) Å, respectively. Right: Close-up view of the interaction mode between W121 and R142. (B) Overall structures of the domain swapped dimer (yellow and magenta) superposed on that of one of the monomer (green, same molecule in panel A). (C) Close-up view around the hinge residue W121 (black square in (B)), whose peptide plane flips compared between that of monomer (green) and the domain swapped dimer (yellow), as highlighted by the black allows indicating the oxygen atoms of the main chain carbonyls.
Crystals of the peptide-unbound and domain-swapped dimer form of Grb2 SH2 were unexpectedly obtained from the monomer fraction of the purified protein in the presence of 1.5 mM of a CD28-derived 8-residue phosphopeptide, SDpYMNMTP. The protein concentration was 0.89 mM and the reservoir solution contains 0.1 M MES monohydrate (pH 6.5), 12% (w/v) polyethylene glycol 20,000. The structure was determined at 2.00 Å resolution (Fig. 4B), and the obtained electron density and refined structure clearly confirmed that Grb2 SH2 bound no CD28 peptide, despite the presence of CD28 peptide in the crystallization condition, and formed domain swapped dimer as same as previously reported uncomplexed swapped dimers of Grb2 SH2 [17,26]. The unit cell parameters of our structure are almost the same as previously reported two structures [17,26], so that the asymmetric unit contained one molecule of Grb2 SH2 with the root-mean-square-distance (rmsd) of 0.54 and 0.43 Å, respectively. Trp121 was the hinge residue of this extended, domain swapped form compared to the monomeric globular structure. The peptide plane flipping at this residue (Fig. 4C) seemed to cause the large conformational change for the C-terminal extended form (discussed later).
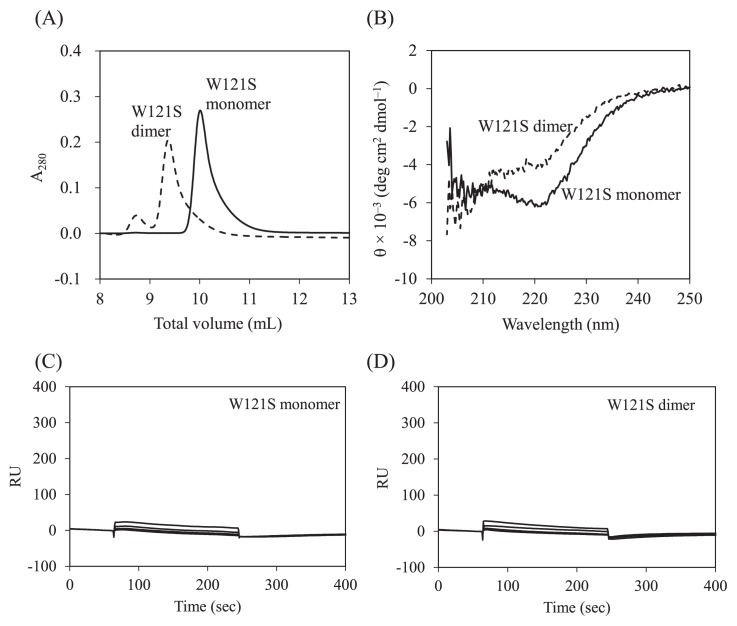
Comparison of the crystal structure of the Grb2 SH2 dimer with that of the monomer showed that the dihedral angles in the main-chain of Trp121 were largely different; (φ, ψ) angles of the dimer and monomer are (−153°, 147°) and (−133°, −82°), respectively. To analyze the role of Trp121 for dimer formation, the mutant W121S was overexpressed and purified, and its structural and functional properties were analyzed. Both SEC and CD analyses showed that W121S forms a stable dimer, like the wild-type (Figs. 5A and 5B). The SPR experiments showed that W121S loses the ability to bind to CD28 (Figs. 5C and 5D).
Figure 5.
SEC, CD, and SPR analysis of Grb2 SH2 mutant, W121S. (A) Elution profiles of the monomer (solid line) and the dimer (broken line). (B) Far-UV CD spectra of the monomer (solid line) and the dimer (broken line). (C) Typical sensorgrams for binding of the monomer (800, 1600, 3200, and 6400 nM) to immobilized CD28. (D) Typical sensorgrams for binding of the dimer (800, 1600, 3200, and 6400 nM) to immobilized CD28.
Discussion
The present crystal structure analysis at high resolution clearly showed that the overall structure of Grb2 SH2 monomer in the CD28-unbound state is similar to that in the CD28-bound state [9]. When comparing the monomer to the dimer, the main difference is the dihedral angle of Trp121, as described above, but overall the structures are similar. The binding site for phosphopeptide in the Grb2 SH2 dimer is exposed to solvent, possibly keeping the binding ability of each site, comparing to that of the monomer.
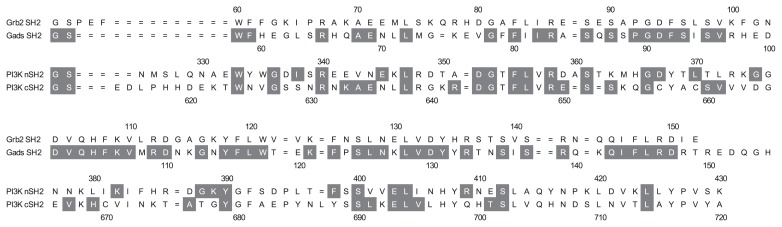
The dihedral angle in the main-chain of Trp121 closely correlated with two different forms of Grb2 SH2, that is the monomeric and dimeric form. In the monomer structure, the (φ, ψ) angles (−133°, −82°) are energetically disadvantageous because they are located at the edge of the allowed region in the Ramachandran plot [36]. In contrast, the angles (−153°, 147°) of the dimer are energetically preferred as they are in the favored region of Ramachandran plot. Interactions between Trp121 and Arg142 (CD28-unbound form) or between Trp121 and CD28 peptide (CD28-bound form) might compensate the energetically disadvantageous conformation of Trp121 in the monomeric state. The corresponding residues of Trp121 in Gads SH2, PI3K nSH2 and cSH2 are Trp118, Ser393, and Ala682, respectively (Fig. 6). Trp118 of Gads also shows energetically disadvantageous dihedral angles, but a similar compensation mechanism should exist because the Arg142 in Grb2 is conserved as Arg139 in Gads, and Gads binds CD28 in almost the same manner [10]. In contrast, Ser393 and Ala682 of PI3K nSH2 and cSH2 show energetically preferred angles. An insertion of a residue in the loop around this region would also contribute to energetically preferred geometry of PI3K nSH2 and cSH2 (Fig. 6). Indeed, SEC analysis showed that Gads SH2 exists as a monomer with a small population of dimers, similar to Grb2 SH2, while both nSH2 and cSH2 exist exclusively as monomers (data not shown).
Figure 6.
The sequence alignments of Grb2 SH2, Gads SH2, PI3K nSH2, and PI3K cSH2. The gray boxes indicate the sequence similarity to Grb2 SH2.
The mutation of Trp121 to Ser in Grb2 did not change the structural property of dimer formation; similar results were reported previously in the W121G mutant of Grb2 SH2 [37]. These results indicate that the dihedral angle of residue at 121 in the mutant is different in the monomer and the dimer. Our crystal structural analysis indicates that the bulky side-chains of Trp121 in Grb2 SH2 and Trp118 in Gads SH2 could force the peptide into the bent conformation observed in CD28 binding [9,10]. The mutation of W121S in Grb2 SH2 would cause a loss of ability to force the unique conformation of CD28 via direct interaction between Asn193 of CD28 and the side-chain of Trp121, resulting in a significant decrease in CD28 binding ability (Figs. 5C and 5D). Also, the SPR experiments showed that W121S mutant could not bind to the intracellular tail of inducible T-cell co-stimulator, to which PI3K nSH2 and cSH2 could bind (data not shown). The residue Trp121 could well contribute to the phosphopeptide binding [37,38]. This is in good agreement with the result that Trp121 in the monomeric state of Grb2 SH2 is stabilized by the interaction with CD28 peptide, but seems to contradict to the assumption that Trp121 is only the determining factor to form the dimer [37].
The SEC and CD analyses showed that the Grb2 SH2 dimer exists in stable form under physiological conditions and dissociates into monomers at 40–45°C (Figs. 1 and 2). A similar transition was recently reported in Gads SH2, analyzed using fluorescence and SEC [39]. After heating up to 50°C and cooling down to 20°C, most of Grb2 SH2 dimer dissociates into monomer while keeping the native structure (Figs. 1B and 2C). Together with the results that the monomer population is larger than the dimer population when Grb2 SH2 is expressed in Escherichia coli, these results suggest local energy minimum of the dimer is higher than that of the monomer. The cation-π interaction between the side-chains of Trp121 and Arg142 in the monomer could contribute to the energy stabilization [9]. The Grb2 SH2 monomer was purified using a gel filtration column as reported previously [9], which formed the swapped dimer in crystallization, indicating that the minor populated dimer is in equilibrium with the monomer, or interconversion between monomer and dimer might be occurred upon the concentration procedure [26], and the crystal of the swapped dimer grew up under the crystallization conditions used in this study.
The swapped dimer did not bind the phosphopeptide in the crystal, probably due to the contact of the two protein molecules near the binding site, which would promote the dissociation of peptide. In addition, it is noteworthy that the crystals of the swapped dimer were obtained from the crystallization solution containing a high density of polyethylene glycol, in contrast, the crystals of CD28 peptide bound Grb2 SH2 were obtained from the solution of high concentrated salt of sodium acetate [9]. However, the dimer should still be able to bind the phosphopeptide in solution, because the structure around the peptide binding site is similar to that of complex with CD28 peptide, and the peptide binding site is exposed to the solvent in the swapped dimer structure. This can explain the present results of CD28 binding experiments. The Grb2 SH2 dimer, possibly swapped dimer, possesses the two binding sites for the phosphopeptide. In the SPR system, the CD28 cytoplasmic tail was immobilized on the flexible dextran surface via its biotin at N-terminus on the sensor chip; this is similar to intracellular conditions where CD28 binds to adaptor SH2 signaling molecules. Grb2 SH2 monomer could bind to CD28, monovalently, while the dimer could mainly bind to CD28 divalently. Interestingly, the apparent binding affinity of the dimer was higher than that of the monomer (Table 2). The increased binding affinity of the dimer contrasts to previous results of decreased binding affinity of the dimer to pYVN tripeptide [26]. The divalent binding affinity might be partially compensated by the decreased binding affinity due to steric crowding, resulting in totally increased binding affinity. Similar to the antigen binding avidity of an antibody [28], the Grb2 SH2 dimer would bind two CD28 molecules immobilized on the sensor chip, simultaneously, resulting in slow dissociation rates.
Both Grb2 and Gads contain one SH2 domain, while PI3K p85 contains two SH2 domains, nSH2 and cSH2. The former SH2 domain forms a dimer and a monomer, while the latter SH2 domain only forms a monomer as revealed by our SEC analysis (data not shown). The SH2 dimer could bind to the phosphopeptide with increased affinity, similar to the tandem SH2 domains in PI3K. This could be closely correlated with the signaling event. The possible changes in binding affinity could regulate the signal in cells. The crystal structure analysis of intact Grb2 indicates that the respective domains SH3-SH2-SH3 could behave independently [2], suggesting it is possible to form the swapped dimer via SH2 interaction as observed in this study. In addition to the monomer-dimer equilibrium observed in intact Grb2 [2–4], the stable dimer observed in this study, possibly swapped dimer, might regulate the signaling event. Ahmed et al. demonstrated that monomeric Grb2 could bind to its pY-containing ligand and activate signal transduction, but dimer formation of Grb2 through SH2-C-terminal SH3 impairs the function as an adaptor molecule [4]. In this study, we found that Grb2 forms SH2-SH2 swapped dimer and this swapped dimer could strongly bind to its ligand, CD28, compared to monomer SH2 domain of Grb2. Therefore, it is possible that Grb2 swapped dimer may preferentially bind to cross-linked CD28, effectively transmitting CD28-mediated signals different to Grb2 monomer. SH2 domain-mediated dimerization of Gads could discriminate between dual and single phosphorylated linker for activation of T cells (LAT) and dimerized Gads-LAT interaction might increase antigen receptor sensitivity by increasing concentration of dually phosphorylated LAT at the antigen receptor signalosome [39], Grb2 SH2 also binds to phosphorylated LAT and contributes to the oligomerization of multivalent ligands, such as LAT, SOS, and Cbl downstream of antigen receptor [40]. Therefore, swapped dimer formation of Grb2 may be one of the critical mechanisms for effective activation of signaling molecules downstream of not only CD28 but also T-cell receptors.
Conclusions
We determined the crystal structures of Grb2 SH2 monomer and dimer at resolution of 1.15 Å and 2.00 Å, respectively. The 3D structure of Grb2 SH2 monomer is similar to that in the CD28-bound state, reported previously [9]. The Grb2 SH2 dimer forms through C-terminal helix swapping, in which Trp121 is the hinge residue. It should be noted that the dihedral angles in the main-chain of Trp121 in the monomer structure, φ = −133° and ψ = −82°, are energetically dis-advantageous, while those in the dimer structure, φ = −153° and ψ = 147°, are energetically preferred. The SEC analysis showed that Grb2 SH2 dimer and monomer exist as stable states. The thermal denaturation experiments using CD showed that the Grb2 SH2 dimer mostly dissociates into the monomer around 50°C, followed by denaturation of monomer around 60°C. The CD28 binding experiments using SPR showed that the binding affinity of Grb2 SH2 dimer increases about three fold compared to that of the monomer, mainly due to the decreased dissociation rate. The possible changes in binding affinity of Grb2 SH2 monomer and dimer could be closely correlated with the signaling event in cells. Because Trp121 was considered to be the key residue for dimer formation, structural and functional properties of W121S mutant were also analyzed. Both SEC and CD analyses showed that W121S forms a stable dimer, like the wild-type, and the SPR experiments showed that W121S loses the ability to bind to CD28.
The atomic coordinates and structure factors (codes 6ICG and 6ICH) have been deposited in the Protein Data Bank (pdbj.org).
Acknowledgements
This study was supported by Nanken-Kyoten, Tokyo Medical and Dental University. The X-ray crystal structure analyses were performed with the approval of the Photon Factory Program Advisory Committee (Proposal No. 2015G075 and 2016G638). The CD experiments were partly performed at the Okinawa Institute Science and Technology Graduate University (OIST); MO is a visiting researcher. The SPR experiments were performed at the Kyoto Integrated Science & Technology Bio-Analysis Center (KIST-BIC).
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions
R. A., N. I., and M. O. directed the project. Y. H. and S. I. purified Grb2 SH2 proteins. H. M. synthesized phosphopeptides. Y. H. carried out SEC, CD, and SPR measurements. Y. H, N. N., S. I. carried out X-ray crystal structure analysis. N. N., S. O., and M. O. wrote the manuscript.
References
- 1.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 2.Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 3.Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, et al. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed Z, Timsah Z, Suen KM, Cook NP, Lee GR, 4th, Lin CC, et al. Grb2 monomer-dimer equilibrium determines normal versus oncogenic function. Nat Commun. 2015;6:7354. doi: 10.1038/ncomms8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNemar C, Snow ME, Windsor WT, Prongay A, Mui P, Zhang R, et al. Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2-SH2. Biochemistry. 1997;36:10006–10014. doi: 10.1021/bi9704360. [DOI] [PubMed] [Google Scholar]
- 6.Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev. 2009;229:12–26. doi: 10.1111/j.1600-065X.2009.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA. CD28 costimulation: from mechanism to therapy. Immunity. 2016;44:973–988. doi: 10.1016/j.immuni.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa S, Watanabe M, Sakurai Y, Inutake Y, Watanabe S, Tai X, et al. CD28 signaling in primary CD4+ T cells: identification of both tyrosine phosphorylation-dependent and phosphorylation-independent pathways. Int Immunol. 2013;25:671–681. doi: 10.1093/intimm/dxt028. [DOI] [PubMed] [Google Scholar]
- 9.Higo K, Ikura T, Oda M, Morii H, Takahashi J, Abe R, et al. High resolution crystal structure of the Grb2 SH2 domain with a phosphopeptide derived from CD28. PLoS One. 2013;8:e74482. doi: 10.1371/journal.pone.0074482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inaba S, Numoto N, Ogawa S, Morii H, Ikura T, Abe R, et al. Crystal structures and thermodynamic analysis reveal distinct mechanisms of CD28 phosphopeptide binding to the Src homology 2 (SH2) domains of three adaptor proteins. J Biol Chem. 2017;292:1052–1060. doi: 10.1074/jbc.M116.755173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverría C, Furet P, et al. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 12.Thornton KH, Mueller WT, McConnell P, Zhu G, Saltiel AR, Thanabal V. Nuclear magnetic resonance solution structure of the growth factor receptor-bound protein 2 Src homology 2 domain. Biochemistry. 1996;35:11852–11864. doi: 10.1021/bi952615s. [DOI] [PubMed] [Google Scholar]
- 13.Rahuel J, García-Echeverría C, Furet P, Strauss A, Caravatti G, Fretz H, et al. Structural basis for the high affinity of amino-aromatic SH2 phosphopeptide ligands. J Mol Biol. 1998;279:1013–1022. doi: 10.1006/jmbi.1998.1790. [DOI] [PubMed] [Google Scholar]
- 14.Senior MM, Frederick AF, Black S, Murgolo NJ, Perkins LM, Wilson O, et al. The three-dimensional solution structure of the SRC homology domain-2 of the growth factor receptor-bound protein-2. J Biomol NMR. 1998;11:153–164. doi: 10.1023/A:1008250309874. [DOI] [PubMed] [Google Scholar]
- 15.Furet P, García-Echeverría C, Gay B, Schoepfer J, Zeller M, Rahuel J. Structure-based design, synthesis, and X-ray crystallography of a high-affinity antagonist of the Grb2-SH2 domain containing an asparagine mimetic. J Med Chem. 1999;42:2358–2363. doi: 10.1021/jm991013u. [DOI] [PubMed] [Google Scholar]
- 16.Ogura K, Tsuchiya S, Terasawa H, Yuzawa S, Hatanaka H, Mandiyan V, et al. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J Mol Biol. 1999;289:439–445. doi: 10.1006/jmbi.1999.2792. [DOI] [PubMed] [Google Scholar]
- 17.Nioche P, Liu WQ, Broutin I, Charbonnier F, Latreille MT, Vidal M, et al. Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor. J Mol Biol. 2002;315:1167–1177. doi: 10.1006/jmbi.2001.5299. [DOI] [PubMed] [Google Scholar]
- 18.Phan J, Shi ZD, Burke TRJr, Waugh DS. Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain. J Mol Biol. 2005;353:104–115. doi: 10.1016/j.jmb.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 19.Benfield AP, Teresk MG, Plake HR, DeLorbe JE, Millspaugh LE, Martin SF. Ligand preorganization may be accompanied by entropic penalties in protein-ligand interactions. Angew Chem Int Ed Engl. 2006;45:6830–6835. doi: 10.1002/anie.200600844. [DOI] [PubMed] [Google Scholar]
- 20.DeLorbe JE, Clements JH, Teresk MG, Benfield AP, Plake HR, Millspaugh LE, et al. Thermodynamic and structural effects of conformational constraints in protein-ligand interactions. Entropic paradoxy associated with ligand preorganization. J Am Chem Soc. 2009;131:16758–16770. doi: 10.1021/ja904698q. [DOI] [PubMed] [Google Scholar]
- 21.Delorbe JE, Clements JH, Whiddon BB, Martin SF. Thermodynamic and structural effects of macrocyclization as a constraining method in protein-ligand interactions. ACS Med Chem Lett. 2010;1:448–452. doi: 10.1021/ml100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myslinski JM, DeLorbe JE, Clements JH, Martin SF. Protein-ligand interactions: thermodynamic effects associated with increasing nonpolar surface area. J Am Chem Soc. 2011;133:18518–18521. doi: 10.1021/ja2068752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Raychaudhuri M, Sen U, Mukhopadhyay D. Functional implications of the conformational switch in AICD peptide upon binding to Grb2-SH2 domain. J Mol Biol. 2011;414:217–230. doi: 10.1016/j.jmb.2011.09.046. [DOI] [PubMed] [Google Scholar]
- 24.Myslinski JM, Clements JH, Martin SF. Protein-ligand interactions: Probing the energetics of a putative cation-pi interaction. Bioorg Med Chem Lett. 2014;24:3164–3167. doi: 10.1016/j.bmcl.2014.04.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiering N, Casale E, Caccia P, Giordano P, Battistini C. Dimer formation through domain swapping in the crystal structure of the Grb2-SH2-Ac-pYVNV complex. Biochemistry. 2000;39:13376–13382. doi: 10.1021/bi0012336. [DOI] [PubMed] [Google Scholar]
- 26.Benfield AP, Whiddon BB, Clements JH, Martin SF. Structural and energetic aspects of Grb2-SH2 domain-swapping. Arch Biochem Biophys. 2007;462:47–53. doi: 10.1016/j.abb.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higo K, Oda M, Morii H, Takahashi J, Harada Y, Ogawa S, et al. Quantitative analysis by surface plasmon resonance of CD28 interaction with cytoplasmic adaptor molecules Grb2, Gads and p85 PI3K. Immunol Invest. 2014;43:278–291. doi: 10.3109/08820139.2013.875039. [DOI] [PubMed] [Google Scholar]
- 28.Oda M, Azuma T. Reevaluation of stoichiometry and affinity/avidity in interactions between anti-hapten antibodies and mono- or multi-valent antigens. Mol Immunol. 2000;37:1111–1122. doi: 10.1016/s0161-5890(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 37.Papaioannou D, Geibel S, Kunze MB, Kay CW, Waksman G. Structural and biophysical investigation of the interaction of a mutant Grb2 SH2 domain (W121G) with its cognate phosphopeptide. Protein Sci. 2016;25:627–637. doi: 10.1002/pro.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marengere LE, Songyang Z, Gish GD, Schaller MD, Parsons JT, Stern MJ, et al. SH2 domain specificity and activity modified by a single residue. Nature. 1994;369:502–505. doi: 10.1038/369502a0. [DOI] [PubMed] [Google Scholar]
- 39.Sukenik S, Frushicheva MP, Waknin-Lellouche C, Hallumi E, Ifrach T, Shalah R, et al. Dimerization of the adaptor Gads facilitates antigen receptor signaling by promoting the cooperative binding of Gads to the adaptor LAT. Sci Signal. 2017;10:eaal1482. doi: 10.1126/scisignal.aal1482. [DOI] [PubMed] [Google Scholar]
- 40.Houtman JC, Yamaguchi H, Barda-Saad M, Braiman A, Bowden B, Appella E, et al. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat Struct Mol Biol. 2006;13:798–805. doi: 10.1038/nsmb1133. [DOI] [PubMed] [Google Scholar]