Abstract
Extant genomes are largely shaped by the global transposition, copy number fluctuation, and rearrangement of DNA sequences, rather than by the substitutions of single nucleotides. Although many of these large-scale mutations have low probabilities and are unlikely to repeat, others are recurrent or predictable in their effects, leading to stereotyped genome architectures and genetic variation in both eukaryotes and prokaryotes. Such recurrent, parallel mutation modes can profoundly shape the paths taken by evolution, and undermine common models of evolutionary genetics. Similar patterns are also evident at the smaller scales of individual genes or short sequences. The scale and extent of this ‘non-substitution’ variation has recently come into focus through the advent of new genomic technologies; however, it is still not widely considered in genotype-phenotype association studies. In this review, we identify common features of these disparate mutational phenomena and comment on the importance and interpretation of these mutational patterns.
Keywords: mutation, short tandem repeats, transposons, repetitive DNA, rDNA, parallel mutation
The dominance of repetitive DNA in mutation
Substitution (see glossary) mutations do not substantially contribute to differences between species compared to other classes of mutations, and generally account for only a minority of new mutations (Table 1). As an example of the dominance of non-SNV variation, consider the difficulty of aligning whole genomes; most pairs of genomes are not syntenic enough or similar enough in size for a substantial role of substitutions in generating the observed diversity. Nevertheless, prominent reviews on the mutation rate almost exclusively focus on substitution rates [1,2].
Table 1.
Rate and genomic impact of various mutation types across eukaryotes.
| Mutation type | Mutation rate (per element) |
Mutation rate (per genome copy)a |
Mutation rate (bp/generation)b |
References |
|---|---|---|---|---|
| Substitution | 10^-9 to 10^-8 | ~30 (human) ~1 (weed) ~0.6 (fly) ~0.1 (yeast) |
~30 (human) ~1 (weed) ~0.6 (fly) ~0.1 (yeast) |
[3–6] |
| Transposition | 10^-6 to 10^-4 | ~0.05 (human) 0.001-0.2 (fly) |
~60 (human) 2.9-581 (fly) |
[7–9] |
| STR copy number change | 10^-5 to 10^-3 | ~40 (human) ~0.24 to 2.4 (weed) ~0.014 to 0.14 (yeast) |
>80 (human) >0.5 to >5 (weed) >0.03 to >0.3 (yeast) |
[4,10–13] |
Where available, estimates are taken from the literature. For weed and yeast, rates are estimated as the product of the element-wise mutation rate and the number of relevant elements (taken from the references).
Where available, estimates are taken from the literature. Estimates are made based on the product of element unit size and genome-wide mutation rate. Human transposition numbers are based on size and mutation rates of Alu, L1, and SVA elements reported in [7]; fly transposition numbers assume the size of the P element (2907 bp). As a lower bound on STR bp effects, we assume that all STR mutations are a one-unit change in a dinucleotide. Throughout, “human” is Homo sapiens, “fly” is Drosophila melanogaster “ yeast is Saccharomyces cerevisiae, and “weed” is Arabidopsis thaliana.
For example, the rate of spontaneous substitutions is lower than the rate of spontaneous short tandem repeat (STR) mutations in humans [10], and for decades transposable elements (TEs) have been thought to account for most spontaneous Drosophila mutations [14]. Such non-substitution mutational modes hold in common an idiosyncratic and high rate of per-locus mutation, and are sometimes referred to as “repetitive” DNA mutations, in that the affected DNA elements usually exist in high copy numbers in the genome. However, other mechanisms of high mutation rate are possible, as with plasmid acquisition and loss in prokaryotes. The importance of such mutational modes is illustrated by:
The above-cited numerical dominance of non-substitution mutations;
The large genomic footprint of many classes of non-substitution mutations such as large indels, ploidy changes, and chromosomal rearrangements;
The elaborate cellular machineries devoted to ameliorating or reducing the rate of devastating mutations (e.g. repeat-mediated deletion suppression in humans [15] and RNA-directed DNA methylation (RdDM) repression of TEs in plants [16])
The long-known overabundance in genomes of repetitive element families (particularly TEs) signifying past mutations [17].
In this review, we take these points of importance as largely self-evident, given their longstanding and uncontested nature (though we do touch on each as needed).
We focus instead on the common characteristics of highly mutable genetic elements that meet two criteria. First, we require that these mutations not be substitutions, as these are extremely well-studied and reviewed elsewhere [2]. Second, we require that the modes of mutation demonstrate parallel mutation; that is, that their rate of mutation is sufficiently high to repeatedly give rise to recurrent or repeated mutations at the same locus. More specifically, we require that these mutational modes violate the infinite-sites model (in many interesting cases the infinite-alleles model also will be violated) [18]. The infinite-sites model assumes that the number of possible sites is very large compared to the mutation rate, and the infinite alleles model assumes that the same allele never arises from mutation more than once; thus, both models assume no parallel evolution.
To illustrate some of the pertinent features of mutations fulfilling these two criteria, we begin by reviewing several important classes of genomic structural variation (including variation in copy number, satellite DNA, transposable elements, and others). We also discuss the example of STRs in some detail, as they are relatively simple and easy to study. We will continue by exploring some of the biological and evolutionary consequences of different mutational modes satisfying these criteria. We will additionally discuss cases of particular interest, including ribosomal DNA (rDNA) copy number variation, a fascinating and little-understood class of variation contributing to phenotypic variation.
The quantitative and qualitative preponderance of non-SNV variation.
The vast majority of variation in DNA sequences between organisms is due to differences in ploidy and in transposable element (TE) content. This is most well-described in plants [19], but is also marked in animal lineages [20]. Sister species/strains of maize [21,22], rice [23], or Arabidopsis [24–27] differ dramatically in their TE content. Moreover, it appears that these differences arise due to the preferential expansion and contraction of different TE families in closely related lineages [28]. Although qualitatively distinct from highly mutable, non-mobile elements in their mutational pattern and effects [28], TEs nonetheless indisputably evince parallel mutations of high rate. They additionally share other features, such as attenuated linkage with surrounding variation [25,29], limiting the power of SNP-based association approaches.
A further highly mutable class of variation is satellite DNA, one of the defining architectural features of eukaryotic genomes. Satellite DNA defines centromeres, telomeres, and other components of chromosomes. Such satellites consist of short motifs (usually less than 1000bp) arranged tandemly in very high copy number. These critically important elements, which participate in crucial genomic functions such as chromosome segregation and maintenance [30], evolve at remarkable speeds [31]. For example, Drosophila melanogaster centromeric repeats (which are generally 5-10bp elements) are dramatically different from closely related Drosophila simulans and Drosophila mauritania centromeric repeats (mostly ~500bp repeats) [32]. Noncentromeric satellite DNA follows similarly divergent patterns among Drosophila species [33,34]. In each case, as with TEs, it appears that different families of satellite repeats have expanded in different lineages of Drosophila by unknown mechanisms, leading to hotspots of diversification in the least-ascertainable portions of their genomes. Similar rapid evolutionary dynamics of satellite DNA have also been observed within and between primate lineages [35].
There are a large number of additional mechanisms for mutation that depend on specific aspects of genome architecture. For example, genomes with large families of closely related genes are amenable to gene conversion mutations, which occur by recombination between highly similar loci. Specifically, trypanosomes and some other human pathogens rely on recombination between high-copy host interaction genes as a mechanism to generate diversity in response to host selection [36,37], with consequences for public health. In these cases, these recombination events are frequently facilitated by nearby TEs.
These adaptive mechanisms can easily be observed in the laboratory. When budding yeast is grown under nutrient-limited conditions, genes encoding fitness-limiting transporters are frequently amplified to high copy number [38,39]. These adaptations are highly replicable across parallel continuous culture systems due to adjacent genomic features such as origins or inverted repeats, tandemly repeated homologous genes, or mobile elements, which all allow for elevated rates of amplification and local copy number expansion [4,38,39].
In summary, although these various sequence elements differ wildly in their mechanism of mutation, they hold in common the features of high rate, repeatability and even predictability. These features are also well illustrated by STR variation.
Lessons from short tandem repeat (STR) variation concerning genomic elements with high mutation rates.
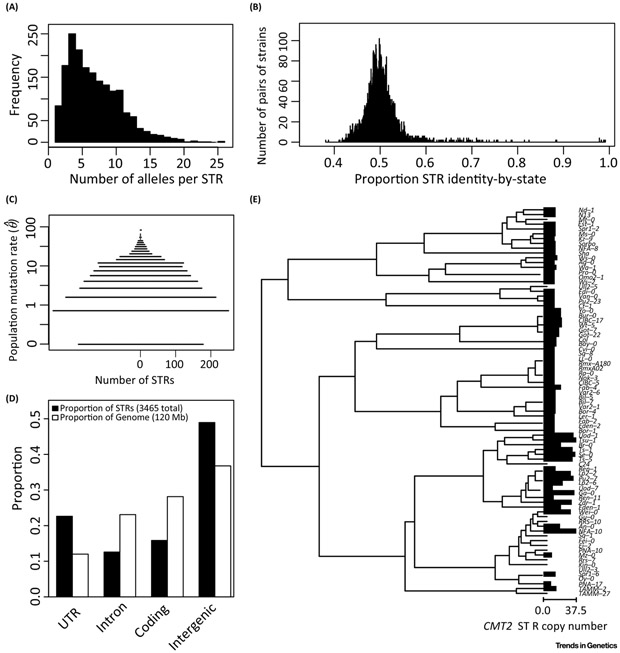
STRs (also known as microsatellites) provide a useful model for understanding the dynamics of elements with elevated mutation rates. Specifically, they are abundant, highly mutagenic, contribute to phenotypic variation, and more or less ignored in most population genomics. Thanks to technology advances coupled with long-standing theoretical work, we now have a fairly complete understanding of this class of variation, in terms of both its population variation and its molecular and phenotypic effects. Recent studies in humans [40–42] and Arabidopsis thaliana [13] provide high-accuracy genotypes and evidence for selective and phenotypic consequences of STR variation. We use some examples from A. thaliana STRs to illustrate the previously identified features of elements with high mutation rates (Figure 1):
Figure 1. STRs demonstrate features of high mutation rate elements.
STR loci demonstrate (A) multiallelism, (B) low allelic similarity between strains, (C) high mutation rate, (D) context-dependent mutation rate variation, and (E) parallelism. Data and figures adapted from [13]. (A): Number of alleles at each STR locus. (B): All pairs of strains were compared at all positions where both strains had STR allele calls, and the number of alleles in common was computed. (C): Population mutation rate was computed according to [52]. Observed mutation rates of zero had a small nonzero value added such that they could be shown on the log scale. (D): Gross localization of STRs in the A. thaliana genome. Annotations from Araport11 were compared to STR calls from [13]; UTR: untranslated regions. (E) Parallel expansions and contractions of the CMT2 STR across A. thaliana strains, adapted from [13]; tree represents UPGMA clustering of strains according to full CMT2 gene sequence, according to the Kimura 2-parameter model (which considers only transitions and transversions). Bars represent the relative copy number of the CMT2 STR, bars are omitted in cases of missing data for a strain; observed values ranged from 8.5 to 37.5 repeat units.
The expected number of mutations and segregating alleles from high-mutation-rate elements is very large (Figure 1A; and this variation has effects on phenotypic variation).
The genomic context of an element strongly influences its mutation rate (Figure 1C).
Several assumptions and qualitative expectations of classical evolutionary genetics are changed by high mutation rates (Figure 1E).
Most common population genomic methods and computer programs assume that loci are biallelic. This is true of less than 15% of 2,046 typed STRs across 96 strains of A. thaliana (Figure 1A). Moreover, there is not even a “major allele” for at least half of STR loci, because no single allele has a frequency above 50%. When comparing any two such A. thaliana strains, only half of STR loci will have the same allele (Figure 1B), whereas nucleotide positions will be identical at ~99% of ascertained sites in such comparisons. This demonstrates the massive population variation of these elements.
Substantial prior work has demonstrated the association of such STR variation with phenotypic variation in a variety of organisms [41,43–47]. Moreover, several studies have presented evidence that genic STRs are subject to substantial selective constraint [13,48], indicating that phenotypic effects of this STR variation contribute actively not only to evolutionary paths, but also to the mutational load afflicting populations.
STR mutation rates are strongly influenced by the genomic context of the STR. For example, transcribed STRs have a substantially higher mutation rate than comparable untranscribed STRs [49]. Indeed, STRs disproportionately tend to be located in otherwise nonrepetitive genic DNA [50], and specifically 5’ UTRs (Figure 1D), even though selection should remove STRs from genic DNA to avoid gene disruptions. Presumably, STRs are maintained in genic DNA by a higher rate of expansion or birth in these regions. Specifically, the mutagenic effect of transcription appears to increase the rate of STR unit insertions [49], which may lead to higher rates of STR “birth” in genic sequences (though they may subsequently be removed by selection from coding sequence).
Finally, STRs show parallel mutation. For example, nonsense mutations in the CMT2 gene in A. thaliana were previously described as subject to positive selection [51], but more recently we showed that an intronic STR in this gene shows repeated dramatic changes in copy number consistent with repeated mutation and selection (Figure 1E). Taking into account the local ancestry of this region in these A. thaliana strains, the most parsimonious explanation is multiple repeated mutations at this locus. The similarity of STR copy number between closely related strains suggests this may potentially (but not necessarily) occur via stepwise mutation of the STR.
High mutation rates and evolutionary genetics.
The genetic elements discussed in this paper all have high mutation rates. This is notable because the mutation rate is a key parameter in many evolutionary models. It is in fact a simplifying assumption in population genetics that the rate of evolution is equal to the mutation rate, as evolution itself is often assumed to be mutation-limited [53]. This is sometimes called the strong-selection weak-mutation (SSWM) model. However, when mutation is not limiting relative to selection (e.g. “strong mutation”), the dynamics of the evolutionary process change dramatically. For example, the seminal work on “soft” selective sweeps specifically noted recurrent mutation as a factor that would increase the frequency of soft sweeps from selected loci [54]. In distinction to “hard” sweeps, the rapid spread to fixation of a specific mutation on a distinctive haplotype, soft sweeps are characterized by the emergence of either multiple distinct adaptive mutations in a region, or the same adaptive mutation associated with heterogeneous haplotypes. Soft sweeps will manifest with population mutation rates (θ) greater than 0.01 even under very strong selection; the A. thaliana STRs discussed above have average population mutation rates on the order of one thousand times higher than this threshold (Figure 1C). Experimental evolution experiments in microbes (which often have very high population mutation rates due to large populations) frequently observe soft sweeps, with the emergence of multiple adaptive alleles leading to “clonal interference” between lineages carrying different adaptive alleles, frequently at the same locus [55]. One such example in Methylobacterium extorquens observed 17 distinct adaptive insertions into the same gene [56], a potent demonstration of the parallelism attainable with both large population sizes and high mutation rates.
High-mutation-rate loci show qualitatively different behavior from low-mutation-rate loci under selection (and their interaction with associated haplotypes), requiring different tools for detecting selection [54,57,58]. Therefore, vast differences in mutation rate within the genome and across mutational types can lead to dramatically different expectations for evolutionary outcomes. Simply, the SSWM model breaks down in the face of high-mutation-rate genetic elements. This is because the rate of adaptation is no longer limited by the rate of mutation due to the abundant supply of mutations at adaptive loci. Recent theoretical work suggests that this breakdown occurs with rates θ > 0.1 [59], leading to new dynamics such as population-size-dependence of the rate of evolution. Again, estimated average θ for STR mutations is approximately 100 times as large as this threshold (Figure 1C). This likely explains the observations of repeated mutations putatively contributing to adaptive variation for these loci, as observed for both STRs in A. thaliana (Figure 1E) and TEs in Drosophila [60]. Further theoretical work suggests that multi-mutation “jumps” become possible with elevated mutation rates relative to selection, changing dynamics of evolution on rugged fitness landscapes [43,61].
Even in the absence of selection, there are consequences of very high mutation rates and multiallelism for the dynamics of molecular evolution [62], some of which we discuss in more detail in Box 1. Overall, the large number and apparent impact of high-mutation-rate elements, combined with the proposition that adaptive evolution is mutation-limited, leads us to the natural conclusion that such elements contribute disproportionately to adaptation, even if presently available techniques are ill-suited to detecting this contribution.
High mutation rates in the human genome.
Mutation in the human genome is of inherent interest, and there exists a large body of work on this subject, reviewed elsewhere [63]. However, a few pertinent features of human mutation are worth noting here. First, much of the sequence difference between humans and great apes occurs in segmentally duplicated regions that are difficult to resolve due to high homology between duplicates [64,65]. Specifically, multiple human-specific genes with roles in neurodevelopmental processes appear to have arisen through such duplication events in the human lineage [65–67]. Second, copy number variation is major contributor to human genetic diversity [63], and tends to occur preferentially in repetitive regions such as the pericentromeres and peritelomeres that are difficult to analyze with traditional short read sequencing [68]. Some such regions consisting of low-copy repeats comprise 5% of the human genome, and show dramatic population variation consisting of rearrangements and large differences of copy number [69]. These variants are moreover nearly impossible to reconstruct without recently-developed methods such as proximity ligation or optical mapping. These observations highlight again the effects of genomic context and the importance of low-complexity genomic regions in generating genetic diversity.
rDNA variation and heritability.
The ribosomal RNA genes, which are organized into high copy regions known as the rDNA, are notable for their high level of sequence conservation and are universally present throughout cellular life. The copy numbers of these genes vary enormously. rDNA copy number variation in A. thaliana largely accounts for the size variation of the entire genome observed among strains [70]. Species estimates of rDNA copy number differ by orders of magnitude across eukaryotes [71], and natural isolates within a species may vary in rDNA copy number by as much as 10-fold [72–77]. Moreover, rDNA copy number is highly labile as an off-target mutation in yeast [78]. The expression and chromatin state of rDNA repeats are among the most tightly regulated features of the eukaryotic nucleus [79], and while only a subset of units are transcriptionally active, their gene products make up ~80% of total RNA in the cell [80]. Transcription from the rDNA (also termed nucleolar organizing regions) leads to formation of the nucleolus, the most obvious feature of gross nuclear morphology, and to such genetic phenomena as nucleolar dominance.
Perhaps due to such regulation, strong selection appears to maintain copy number, as observed in the large rDNA copy number fluctuations observed upon disruption and subsequent complementation of yeast orc2 mutants [63] and the return of yeast rDNA copy number back to the native ~150 copies after artificial reduction [82]. Reductions in germline rDNA copy number are heritable in Drosophila, yet rDNA copy number also recovers rapidly in those progeny that inherited reduced rDNA arrays [83].
Of relevance to understanding the consequences of rDNA copy number variation are the many different mechanisms proposed for its generation. In yeast, transcription-replication conflicts may contribute to rDNA instability, due in part to the presence of an origin of replication in the rDNA intergenic spacer [81,84,85], another reminder of the importance of genomic context in determining mutation rates. Intrachromatid recombination has similarly been proposed to produce copy number reduction [86], along with unequal meiotic recombination leading to changes in rDNA copy number between generations; in humans there is ~10% chance of a recombination in an rDNA array that will result in a change in rDNA copy number [87].
The potential phenotypic consequences of rDNA variation are vast and largely unexplored. Beyond the documented fitness consequences of catastrophic reductions in rDNA copy number [88,89], no causal relationships have yet been demonstrated between phenotype and rDNA copy number variation in the naturally-occurring range, although a weak positive association has recently been found between rDNA copy number and flowering time in maize [90]. Extrachromosomal circular rDNA sequences accumulate with age in yeast [86], and both their accumulation as well as instability of the rDNA locus itself have been proposed causative agents of aging in yeast [91,92]. Recently, interest in the relationship between rDNA stability and cancer has arisen, due to observations that rDNA copy number modestly decreases in some cancers [93–95] . Whether rDNA may act on cell physiology through ribosome biogenesis, maintaining genome integrity [96], balance of heterochromatin [97], influence on genome replication [84], or through some other mechanism remains to be resolved. One intriguing possibility is that, due to its centrality in cellular physiology and the processing of genetic information, rDNA copy number variation may affect not specific traits but the expressivity of other genetic variants [98]. rDNA copy number alteration has been reported to have genomewide impact on gene expression in Drosophila [97], and to influence position effect variegation [99]. Human studies have further revealed an association between rDNA copy number and genome-wide gene expression, as well as an inverse relationship with mitochondrial DNA abundance [72]. The potential central role of rDNA copy number variation in genomic structure and gene regulation puts rDNA at a critical point of research into human health and aging.
High mutation rates and parallel evolution in prokaryotes
While we chiefly focus on eukaryotes, prokaryotic genomes also highlight diverse recurrent mutational modes. Indeed, large-scale reorganization and gene gain and loss is probably even more biologically significant in prokaryotes than it is in eukaryotes [100]. For example, pathogenic organisms carrying the genus name Shigella are not a genus, and indeed not even monophyletic [101]. Each lineage of Shigella in fact arose independently from E. coli ancestors by a concerted and localized process of massive gene loss and acquisition [101–103]. In this example, recurrent large mutations follow a predictable path, due to the contextual influence of E. coli genome architecture, to yield the convergent outcome of the Shigella genome. A similar host-associated parallel mutation trajectory is known from the soil microbe Mesorhizobium ciceri, in the form of a large “symbiosis island” integration element that is broken up and integrated at three different genomic locations [104]. This element carries genes associated with diazotrophic symbiosis with plants, and its integration is repeatable, highly stereotyped, and can be recapitulated in the laboratory [105]. Moreover, it appears that this tripartite integration mechanism is conserved across at least the genus Mesorhizobium as a mechanism for facilitating the spread of beneficial mobile genetic elements [106]. More generally, adaptive horizontal transfer events are repeatable due to epistasis [107], and are specifically facilitated by the genomic context of mobile elements and associated cellular pathways and cellular features.
These well-trodden horizontal evolutionary pathways are superficially eye-catching, but in the context of microbial genomic evolution they are unremarkable. As seen in the above M. extorquens example [56], the large population sizes of microbes make them tractable systems for experimental evolution. Although the population sizes of experimentally evolved bacteria are sufficiently large that even substitutions are dominated by parallelism [108], these experiments emphasize the adaptive importance of non-SNV variation. Genomic optical mapping of parallel lab-evolved E. coli populations uncovered a dramatic diversity of rearrangements, generally mediated by recombination between distant IS elements [109]. Remarkably, these rearrangements were highly parallel, in that most such events were observed in more than one among only 12 populations. In the same populations, the most dramatic fitness increase over decades of evolution consisted of a highly repeatable tandem gene amplification that depended on a predisposing genomic context [110].
These mechanisms for yielding repeated high-impact adaptive mutations in prokaryotes highlight the prevalence, diversity, and adaptive significance of recurrent high-rate mutation events in the dominant clades of cellular life. The phenotypic consequences of this form of variation are vast and largely unexplored.
Concluding Remarks
We have discussed abundant cases of recurrently mutable DNA elements determining the architecture of genomes and variation in phenotypes. These highly abundant elements shape the direction of evolution through their large supply of ready genetic variation. In the last two decades, the vastly improved ascertainment of single nucleotide variants and substitutions prompted genetic and genomic researchers to focus on this much more tractable subject of study. This focus was driven largely by the advent of automated DNA sequencers and efficient computer programs for sequence alignment, which in those early iterations experienced difficulties with other classes of genetic variation. These technological difficulties are in some influential cases the explicit reason for ignoring other forms of variation [1], likely biasing both results and discourse. This bias is potentially reinforced by the common assumption of quantitative genetics that genome-wide SNP genotyping is sufficient to ascertain neighboring mutations due to linkage [111]. (Several studies of at least STRs and TEs indicate that this is unlikely to be true for multiallelic loci with high mutation rates [13,40,60,112].) However, we are encouraged that recent methodological advances such as optical mapping, proximity ligation, multiplexed sequence capture, and long-read sequencing have vastly expanded the pool of accessible variants [113].
Genomic elements with high mutation rates are intrinsically difficult to analyze by molecular methods. It is possible that methodological artifacts have influenced our understanding of these elements, just as we argue that past methods have biased us. For this reason we must evaluate results regarding these elements with more caution than substitutional variation. Nonetheless, we believe that the balance of evidence argues for important roles of genomic elements with high mutation rates. In the future, we must investigate whether the dizzying array of molecular variation in these elements has a commensurate effect on phenotype, or whether this variation is merely a genomic extravagance (See Outstanding Questions).
Outstanding Questions.
What are the relative contributions of different mutation classes (substitutions, transpositions, copy-number changes) to heritable variation in different organisms?
The number of substitutions per generation is well ascertained across many organisms, but what is the total number of mutations—including other mutation types that are more difficult to observe?
Are there general rules for the emergence of new families of elements such as transposons or satellites with very high mutation rates?
Are there generalizable effects of different genomic contexts (e.g., pericentromeres, peritelomeres, transcribed regions, plasmids) on the rate of different mutational modes?
If the rate of evolution is not mutation-limited, does this undermine other assumptions or models in currency?
BOX 1. Mutational modes and molecular evolution.
John Maynard Smith [114] proposed that molecular evolution might be understood with reference to a popular parlor game of his time, inferring the path of evolution by considering the most parsimonious number of single letter substitutions to transform one word into another:
word-->wore-->gore-->gone-->gene
However, this set of rules (parsimony, single letter substitution) does not necessarily describe the expected evolutionary path of a given DNA sequence. DNA sequences are altered according to rules allowing many more kinds of transitions. For example, one might consider the following scenario instead, allowing also duplications, inversions, and rearrangements of letters:
word-->drow-->brow-->brew-->brewer-->brewed-->breed-->breeder-->breed-->greed-->green-->greet-->great-->geat -->gent-->gene
Comparatively, this scenario is positively circuitous; multiple steps are redundant, with no effect on the outcome. Many transitions involve addition, subtraction, or rearrangement of existing sequences. Some words (“drow”, “geat”) may strain the dictionary. Nonetheless, we believe that many geneticists will (reluctantly) concede that it is a more familiar path than the simple one trod by Maynard Smith, while hastening to add that Maynard Smith’s has a higher tutelary value.
To defend this assertion, we can present some arguments, which are based on the empirical failures of parsimony as a criterion in phylogenetic inference [115], Firstly, while the most parsimonious path may be the most likely single path, it may have a lower probability than the summation of other paths. Second, we cannot assume that all transitions have equal probability [116]; or even that transition probabilities are constant along the path [117], These assumptions do not even hold for the single-letter substitutions in Maynard Smith’s simplified model. Indeed, cursory reference to biological experience argues that transition probabilities must change based on the sequence state, and that there is very large variation in transition probabilities (i.e. mutation rates) between sites and types of transitions. Overall, we must confront the possibility that intuitively obvious paths in molecular evolution may not be the true ones, given the observed dynamics of genome architectures and sequence variation throughout evolution.
HIGHLIGHTS.
Single nucleotide variants or mutations (e.g. point mutations) are less common than other variations and mutations, and cannot generate observed genomic diversity.
Genomic elements such as short tandem repeats, ribosomal RNA gene arrays, or transposable elements have extremely high mutation rates that likely contribute most mutations in eukaryotic genomes.
These high-rate-elements are very diverse and their importance depends on their biological context. For example, in prokaryotes the more important such elements are plasmids and integrative and conjugative elements.
Functional elements with very high mutation rates behave very differently than functional elements with low mutation rates in evolution. Specifically, the same mutation can occur multiple times in different lineages, and evolution is no longer mutation-limited.
Acknowledgments
We would like to thank members of the Queitsch lab for helpful conversations. MOP would like to thank attendees of the Population, Evolutionary, and Quantitative Genetics (PEQG) 2018 meeting, attendees of the Plasmid Biology 2018 meeting, and Emily Ebel for interesting scientific presentations and conversations. EM, ANH, and CQ are supported by R21AG052020 and R01GM122088 to CQ. ANH is supported by training grant T32GM007270.
GLOSSARY.
- Transposable element (TE):
DNA elements which reproduce themselves in genomes via “cut-and-paste” or “copy-and-paste” mechanisms, leading to large insertions and deletions of DNA. Sometimes called “selfish” DNA, or “jumping genes”.
- Satellite DNA:
Regions of DNA consisting of tandemly repeated DNA sequences at high copy number. This copy number mutates rapidly. Genomic regions such as telomeres or centromeres tend to consist of satellite DNA. Short tandem repeats, also called microsatellites, consist of very short repeat units (<10 nt).
- Parallel mutation:
A mutation that occurs at the same locus as another previous mutation, but independently from the same starting allele, usually in different genetic lineages. Mutations at the same locus in the same lineage are called “stepwise” mutations.
- Substitution:
A mutation that replaces a nucleotide at a single position (A, C, G, T) with one of the other three nucleotides.
- rDNA:
Regions of genomes consisting of many copies of ribosomal RNA genes, which vary dramatically in copy number across species and individuals while remaining conserved in the sequence of each gene.
- Population mutation rate:
The total number of mutations arising across the entire population of an organism. Either a larger population or a higher rate of per-locus mutation can increase this measure. Sometimes written as θ.
- Hard and soft selective sweeps:
When favorable mutations occur in populations, they tend to increase in frequency over time until they dominate the population due to positive selection. In hard sweeps, positive selection is very strong and the mutation goes to fixation very quickly. In soft sweeps, selection is weaker and multiple mutations are simultaneously under positive selection, leading to complex population dynamics which are more difficult to detect and interpret.
- Infinite sites model.
A model of molecular evolution under which it is assumed that all mutations happen at different sites. Under this assumption, parallel (or recurrent) mutation does not occur. This condition is satisfied by simply assuming that the number of sites in the genome is infinite while keeping a mutation rate constant, such that the probability of mutation at any specific site becomes infinitesimally small.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
MOP is an employee of Phase Genomics Inc.
References
- 1.Lynch M Evolution of the mutation rate. Trends in genetics: TIG. 2010;26: 345–52. doi: 10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, et al. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet. 2016;17: 704–714. doi: 10.1038/nrg.2016.104 [DOI] [PubMed] [Google Scholar]
- 3.Ossowski S, Schneeberger K, Lucas-Lledó Jl, Warthmann N, Clark RM, Shaw RG, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science (New York, NY). 2010;327: 92–4. doi: 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. PNAS. 2008;105: 9272–9277. doi: 10.1073/pnas.0803466105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keightley PD, Ness RW, Halligan DL, Haddrill PR. Estimation of the Spontaneous Mutation Rate per Nucleotide Site in a Drosophila melanogaster Full-Sib Family. Genetics. 2014;196: 313–320. doi: 10.1534/genetics.113.158758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44: 1277–1281. doi: 10.1038/ng.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns KH, Boeke JD. Human Transposon Tectonics. Cell. 2012;149: 740–752. doi: 10.1016/j.cell.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuzhdin SV, Mackay TF. The genomic rate of transposable element movement in Drosophila melanogaster. Mol Biol Evol. 1995;12: 180–181. doi: 10.1093/oxfordjournals.molbev.a040188 [DOI] [PubMed] [Google Scholar]
- 9.Bergman CM, Bensasson D. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc Natl Acad Sci US A. 2007;104: 11340–11345. doi: 10.1073/pnas.0702552104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willems T, Gymrek M, Poznik GD, Tyler-Smith C, 1000 Genomes Project Chromosome Y Group, Erlich Y. Population-Scale Sequencing Data Enable Precise Estimates of Y-STR Mutation Rates. Am J Hum Genet. 2016;98: 919–933. doi: 10.1016/j.ajhg.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nature genetics. 2005;37: 986–90. doi: 10.1038/ng1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324: 1213–6. doi: 10.1126/science.1170097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Press MO, McCoy RC, Hall AN, Akey JM, Queitsch C. Massive variation of short tandem repeats with functional consequences across strains of Arabidopsis thaliana. Genome Res. 2018; doi: 10.1101/gr.231753.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finnegan DJ. TRANSPOSABLE ELEMENTS In: Lindsley DL, Zimm GG, editors. The Genome of Drosophila Melanogaster. San Diego: Academic Press; 1992. pp. 1096–1107. doi: 10.1016/B978-0-12-450990-0.50010-1 [DOI] [Google Scholar]
- 15.Mendez-Dorantes C, Bhargava R, Stark JM. Repeat-mediated deletions can be induced by a chromosomal break far from a repeat, but multiple pathways suppress such rearrangements. Genes Dev. 2018; doi: 10.1101/gad.311084.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bousios A, Gaut BS. Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Current Opinion in Plant Biology. 2016;30: 123–133. doi: 10.1016/j.pbi.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 17.Britten RJ, Kohne DE. Repeated Sequences in DNA. Science. 1968;161: 529–540. doi: 10.1126/science.161.3841.529 [DOI] [PubMed] [Google Scholar]
- 18.Tajima F Infinite-allele model and infinite-site model in population genetics. J Genet. 1996;75: 27. doi: 10.1007/BF02931749 [DOI] [Google Scholar]
- 19.Wendel JF, Lisch D, Hu G, Mason AS. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Current Opinion in Genetics & Development. 2018;49: 1–7. doi: 10.1016/j.gde.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez F, Arkhipova IR. Transposable elements and polyploid evolution in animals. Current Opinion in Genetics & Development. 2018;49: 115–123. doi: 10.1016/j.gde.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vielle-Calzada J-P, Martínez de la Vega O, Hernández-Guzmán G, Ibarra-Laclette E, Alvarez-Mejía C, Vega-Arreguín JC, et al. The Palomero genome suggests metal effects on domestication. Science. 2009;326: 1078. doi: 10.1126/science.1178437 [DOI] [PubMed] [Google Scholar]
- 22.Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546: 524–527. doi: 10.1038/nature22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Saniyal A, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16: 1262–1269. doi: 10.1101/gr.5290206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legrand S, Caron T, Maumus F, Schvartzman S, Quadrana L, Durand E, et al. Differential retention of transposable element-derived sequences in outcrossing Arabidopsis genomes. bioRxiv. 2018; 368985. doi: 10.1101/368985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. In: eLife [Internet], 2 December 2016. [cited 4 Aug 2018], doi: 10.7554/eLife.20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z-W, Hou X-H, Chen J-F, Xu Y-C, Wu Q, González J, et al. Transposable elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol Evol. doi: 10.1093/gbe/evy171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quadrana L, Silveira AB, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, et al. The Arabidopsis thaliana mobilome and its impact at the species level. eLife. 2016;5: e15716. doi: 10.7554/eLife.15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arkhipova IR. Neutral Theory, Transposable Elements, and Eukaryotic Genome Evolution. Mol Biol Evol. 2018;35: 1332–1337. doi: 10.1093/molbev/msy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn A, Ong YM, Cheng C-Y, Wong TY, Quake SR, Burkholder WF. Linkage disequilibrium and signatures of positive selection around LINE-1 retrotransposons in the human genome. PNAS. 2014;111: 8131–8136. doi: 10.1073/pnas.1401532111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plohl M, Luchetti A, Meštrović N, Mantovani B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409: 72–82. doi: 10.1016/j.gene.2007.11.013 [DOI] [PubMed] [Google Scholar]
- 31.Lower SS, McGurk MP, Clark AG, Barbash DA. Satellite DNA evolution: old ideas, new approaches. Current Opinion in Genetics & Development. 2018;49: 70–78. doi: 10.1016/j.gde.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talbert PB, Kasinathan S, Henikoff S. Simple and Complex Centromeric Satellites in Drosophila Sibling Species. Genetics. 2018;208: 977–990. doi: 10.1534/genetics.117.300620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagannathan M, Warsinger-Pepe N, Watase GJ, Yamashita YM. Comparative Analysis of Satellite DNA in the Drosophila melanogaster Species Complex. G3 (Bethesda). 2016;7: 693–704. doi: 10.1534/g3.116.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei KH-C, Lower SE, Caldas IV, Sless TJS, Barbash DA, Clark AG. Variable Rates of Simple Satellite Gains across the Drosophila Phylogeny. Mol Biol Evol. 2018;35: 925–941. doi: 10.1093/molbev/msy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cechova M, Harris RS, Tomaszkiewicz M, Arbeithuber B, Chiaromonte F, Makova KD. High inter- and intraspecific turnover of satellite repeats in great apes. bioRxiv. 2018; 470054. doi: 10.1101/470054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talavera-Lopez C, Messenger LA, Lewis MD, Yeo M, Reis-Cunha JL, Bartholomeu DC, et al. Repeat-driven generation of antigenic diversity in a major human pathogen, Trypanosoma cruzi. bioRxiv. 2018; 283531. doi: 10.1101/283531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faino L, Seidl MF, Shi-Kunne X, Pauper M, van den Berg GCM, Wittenberg AHJ, et al. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 2016;26: 1091–1100. doi: 10.1101/gr.204974.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15: 931–942. doi: 10.1093/oxfordjournals.molbev.a026009 [DOI] [PubMed] [Google Scholar]
- 39.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai D a, Ward A, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genetics. 2008;4: e1000303. doi: 10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willems TF, Gymrek M, Highnam G, Mittelman D, Erlich Y. The landscape of human STR variation. Genome Research. 2014; doi: 10.1101/gr.177774.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nature Genetics. 2015;48: 22–29. doi: 10.1038/ng.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, et al. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 2016;44: 3750–3762. doi: 10.1093/nar/gkw219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Press MO, Carlson KD, Queitsch C. The overdue promise of short tandem repeat variation for heritability. Trends in Genetics. 2014;30: 504–512. doi: 10.1016/j.tig.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabib A, Vishwanathan S, Seleznev A, McKeown PC, Downing T, Dent C, et al. A Polynucleotide Repeat Expansion Causing Temperature-Sensitivity Persists in Wild Irish Accessions of Arabidopsis thaliana. Arabidopsis thaliana. 2016; 1311. doi: 10.3389/fpls.2016.01311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nature Reviews Genetics. 2018; doi: 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- 46.Bilgin Sonay T, Carvalho T, Robinson M, Greminger M, Krutzen M, Comas D, et al. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome research. 2015;25: 1591–1599. doi: 10.1101/gr.190868.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–8. doi: 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gymrek M, Willems T, Erlich Y, Reich DE. A framework to interpret short tandem repeat variation in humans. bioRxiv. 2016; 092734. doi: 10.1101/092734 [DOI] [Google Scholar]
- 49.Zavodna M, Bagshaw A, Brauning R, Gemmell NJ. The effects of transcription and recombination on mutational dynamics of short tandem repeats. Nucleic Acids Res. 2018;46: 1321–1330. doi: 10.1093/nar/gkx1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30: 194–200. doi: 10.1038/ng822 [DOI] [PubMed] [Google Scholar]
- 51.Shen X, Jonge JD, Forsberg SKG, Pettersson ME, Sheng Z, Hennig L, et al. Natural CMT2 Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality. PLOS Genetics. 2014;10: el004842. doi: 10.1371/journal.pgen.1004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haasl RJ, Payseur BA. The Number of Alleles at a Microsatellite Defines the Allele Frequency Spectrum and Facilitates Fast Accurate Estimation of θ. Mol Biol Evol. 2010;27: 2702–2715. doi: 10.1093/molbev/msq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura M Evolutionary Rate at the Molecular Level. Nature. 1968;217: 624–626. [DOI] [PubMed] [Google Scholar]
- 54.Pennings PS, Hermisson J. Soft Sweeps III: The Signature of Positive Selection from Recurrent Mutation. PLOS Genetics. 2006;2: e186. doi: 10.1371/journal.pgen.0020186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GI Lang, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500: 571–574. doi: 10.1038/nature12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M-C, Marx CJ. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics. 2013;193: 943–952. doi: 10.1534/genetics.112.148502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haasl RJ, Johnson RC, Payseur BA. The Effects of Microsatellite Selection on Linked Sequence Diversity. Genome Biol Evol. 2014;6: 1843–1861. doi: 10.1093/gbe/evu134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haasl RJ, Payseur BA. Fifteen years of genomewide scans for selection: trends, lessons and unaddressed genetic sources of complication. Mol Ecol. 2016;25: 5–23. doi: 10.1111/mec.13339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koning APJ de, Sanctis BDD. The Rate of Observable Molecular Evolution When Mutation May Not Be Weak. bioRxiv. 2018; 259507. doi: 10.1101/259507 [DOI] [Google Scholar]
- 60.Rech GE, Bogaerts-Marquez M, Barron MG, Merenciano M, Villanueva-Canas JL, Horvath V, et al. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. bioRxiv. 2018; 380618. doi: 10.1101/380618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katsnelson MI, Wolf YI, Koonin EV. On the feasibility of saltational evolution. bioRxiv. 2018; 399022. doi: 10.1101/399022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura M, Ohta T. Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci U S A. 1978;75: 2868–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends in Genetics. 2013;29: 575–584. doi: 10.1016/j.tig.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dennis MY, Eichler EE. Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev. 2016;41: 44–52. doi: 10.1016/j.gde.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dennis MY, Harshman L, Nelson BJ, Penn O, Cantsilieris S, Huddleston J, et al. The evolution and population diversity of human-specific segmental duplications. Nat Ecol Evol. 2017;1. doi: 10.1038/s41559-016-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, et al. Human-specific evolution of novel SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149: 912–922. doi: 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dougherty ML, Nuttle X, Penn O, Nelson BJ, Huddleston J, Baker C, et al. The birth of a human-specific neural gene by incomplete duplication and gene fusion. Genome Biology. 2017;18: 49. doi: 10.1186/s13059-017-1163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monlong J, Cossette P, Meloche C, Rouleau G, Girard SL, Bourque G. Human copy number variants are enriched in regions of low mappability. Nucleic Acids Res. doi: 10.1093/nar/gky538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demaerel W, Mostovoy Y, Yilmaz F, Vervoort L, Pastor S, Hestand MS, et al. The 22q11 low copy repeats are characterized by unprecedented size and structure variability. bioRxiv. 2018; 403873. doi: 10.1101/403873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45: 884–890. doi: 10.1038/ng.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46: 48–50. doi: 10.1139/g02-103 [DOI] [PubMed] [Google Scholar]
- 72.Gibbons JG, Branco AT, Yu S, Lemos B. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat Commun. 2014;5: 4850. doi: 10.1038/ncomms5850 [DOI] [PubMed] [Google Scholar]
- 73.Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23: 1749–1762. doi: 10.1101/gr.157651.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, et al. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv. 2018;4: eaao0665. doi: 10.1126/sciadv.aao0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritossa FM, Scala G. Equilibrium variations in the redundancy of rDNA in Drosophila melanogaster. Genetics. 1969; Available: http://agris.fao.org/agris-search/search.do;jsessionid=1E08D6134C19B3ACC4F456D8CE1F5E86?request_locale=ru&recordID=US201301233618&query=&sourceQuery=&sortField=&sortOrder=&agrovocString=&advQuery=¢erString=&enableField= [PubMed] [Google Scholar]
- 76.Mohan J, Ritossa FM. Regulation of ribosomal RNA synthesis and its bearing on the bobbed phenotype in Drosophila melanogaster. Dev Biol. 1970;22: 495–512. doi: 10.1016/0012-1606(70)90165-X [DOI] [PubMed] [Google Scholar]
- 77.James SA, O’Kelly MJT, Carter DM, Davey RP, Oudenaarden van A, Roberts IN. Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 2009;19: 626–635. doi: 10.1101/gr.084517.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwan EX, Wang XS, Amemiya HM, Brewer BJ, Raghuraman MK. rDNA Copy Number Variants Are Frequent Passenger Mutations in Saccharomyces cerevisiae Deletion Collections and de Novo Transformants. G3. 2016;6: 2829–2838. doi: 10.1534/g3.116.030296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: Insights into a chromosome-scale epigenetic on/off switch. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 2007;1769: 383–392. doi: 10.1016/j.bbaexp.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences. 1999;24: 437–440. doi: 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- 81.Sanchez JC, Kwan EX, Pohl TJ, Amemiya HM, Raghuraman MK, Brewer BJ. Defective replication initiation results in locus specific chromosome breakage and a ribosomal RNA deficiency in yeast. PLOS Genetics. 2017;13: e1007041. doi: 10.1371/journal.pgen.1007041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu KL, Nelson JO, Watase GJ, Warsinger-Pepe N, Yamashita YM. Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. In: eLife [Internet]. 13 February 2018. [cited 19 Aug 2018]. doi: 10.7554/eLife.32421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, et al. A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan. PLoS Genet. 2013;9. doi: 10.1371/journal.pgen.1003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17: 1497–1506. doi: 10.1101/gad.1085403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- 87.Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ritossa FM, Atwood KC, Spiegelman S. A molecular explanation of the bobbed mutants of Drosophila as partial deficiencies of “ribosomal” DNA. Genetics. 1966;54: 819–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23: 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li B, Kremling K, Wu P, Bukowski R, Romay M, Xie E, et al. Co-regulation of ribosomal RNA with hundreds of genes contributes to phenotypic variations. Genome Res. 2018; gr.229716.117. doi: 10.1101/gr.229716.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ganley ARD, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell. 2009;35: 683–693. doi: 10.1016/j.molcel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 92.Saka K, Ide S, Ganley ARD, Kobayashi T. Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr Biol. 2013;23:1794–1798. doi: 10.1016/j.cub.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 93.Wang M, Lemos B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017;13: e1006994.doi: 10.1371/journal.pgen.1006994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu B, Li H, Perry JM, Singh VP, Unruh J, Yu Z, et al. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017;13: e1006771. doi: 10.1371/journal.pgen.1006771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Udugama M, Sanij E, Voon HPJ, Son J, Hii L, Henson JD, et al. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. PNAS. 2018;115: 4737–4742. doi: 10.1073/pnas.1720391115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kobayashi T A new role of the rDNA and nucleolus in the nucleus--rDNA instability maintains genome integrity. Bioessays. 2008;30: 267–272. doi: 10.1002/bies.20723 [DOI] [PubMed] [Google Scholar]
- 97.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7: e1001376. doi: 10.1371/journal.pgen.1001376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waddington CH. CANALIZATION OF DEVELOPMENT AND THE INHERITANCE OF ACQUIRED CHARACTERS : Abstract : Nature. Nature. 1942;150: 563–565. [Google Scholar]
- 99.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA. 2009;106: 17829–17834. doi: 10.1073/pnas.0906811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. Genomes in turmoil: Quantification of genome dynamics in prokaryote supergenomes. BMC biology. 2014;12: 66. doi: 10.1186/s12915-014-0066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97: 10567–10572. doi: 10.1073/pnas.180094797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Lin K. A phylogenomic analysis of Escherichia coli / Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation. BMC Evolutionary Biology. 2012;12: 174. doi: 10.1186/1471-2148-12-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. Organised Genome Dynamics in the Escherichia coli Species Results in Highly Diverse Adaptive Paths. PLOS Genetics. 2009;5: e1000344. doi: 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haskett TL, Terpolilli JJ, Bekuma A, O’Hara GW, Sullivan JT, Wang P, et al. Assembly and transfer of tripartite integrative and conjugative genetic elements. PNAS. 2016;113: 12268–12273. doi: 10.1073/pnas.1613358113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haskett TL, Terpolilli JJ, Ramachandran VK, Verdonk CJ, Poole PS, O’Hara GW, et al. Sequential induction of three recombination directionality factors directs assembly of tripartite integrative and conjugative elements. PLOS Genetics. 2018;14: e1007292. doi: 10.1371/journal.pgen.1007292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haskett TL, Ramsay JP, Bekuma AA, Sullivan JT, O’Hara GW, Terpolilli JJ. Evolutionary persistence of tripartite integrative and conjugative elements. Plasmid. 2017;92: 30–36. doi: 10.1016/j.plasmid.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 107.Press MO, Queitsch C, Borenstein E. Evolutionary assembly patterns of prokaryotic genomes. Genome Research. 2016; gr.200097.115. doi: 10.1101/gr.200097.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. PNAS. 2006;103: 9107–9112. doi: 10.1073/pnas.0602917103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raeside C, Gaffé J, Deatherage DE, Tenaillon O, Briska AM, Ptashkin RN, et al. Large Chromosomal Rearrangements during a Long-Term Evolution Experiment with Escherichia coli. mBio. 2014;5: e01377–14. doi: 10.1128/mBio.01377-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489: 513–518. doi: 10.1038/nature11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bush WS, Moore JH. Chapter 11: Genome-Wide Association Studies. PLOS Computational Biology. 2012;8: e1002822. doi: 10.1371/journal.pcbi.1002822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sawaya S, Jones M, Keller M. Linkage disequilibrium between single nucleotide polymorphisms and hypermutable loci [Internet]. Cold Spring Harbor Labs Journals; 2015. June p. 020909. Available: http://biorxiv.org/content/early/2015/06/15/020909.abstract [Google Scholar]
- 113.Dijk van EL, Jaszczyszyn Y, Naquin D, Thermes C. The Third Revolution in Sequencing Technology. Trends in Genetics. 2018;34: 666–681. doi: 10.1016/j.tig.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 114.MAYNARD SMITH J Natural Selection and the Concept of a Protein Space. Nature. 1970;225: 563–564. doi: 10.1038/225563a0 [DOI] [PubMed] [Google Scholar]
- 115.Felsenstein J Cases in which Parsimony or Compatibility Methods Will be Positively Misleading. Systematic Zoology. 1978;27: 401–410. doi: 10.2307/2412923 [DOI] [Google Scholar]
- 116.Yang Z Among-site rate variation and its impact on phylogenetic analyses. Trends in Ecology & Evolution. 1996;11: 367–372. doi: 10.1016/0169-5347(96)10041-0 [DOI] [PubMed] [Google Scholar]
- 117.Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970;4: 579–593. doi: 10.1007/BF00486096 [DOI] [PubMed] [Google Scholar]