Abstract
Moringa oleifera (moringa) has been traditionally used for the treatment of diabetes and in water purification. We previously showed that moringa seed extract (MSE), standardized to its primary bioactive isothiocyanate (MIC-1), modulated inflammatory and antioxidant signaling pathways in vitro. To understand the efficacy and mechanisms of action of MSE in vivo, we incorporated MSE into the diets of normal and obese C57Bl/6J male mice fed a standard low-fat diet or a very high-fat diet for 12 wk, respectively. MSE supplementation resulted in reduced body weight, decreased adiposity, improved glucose tolerance, reduced inflammatory gene expression, and increased antioxidant gene expression. 16S rRNA gene sequencing and quantitative PCR of fecal/cecal samples showed major modulation of the gut microbial community and a significantly reduced bacterial load, similar to an antibiotic response. This suggests that MSE improves metabolic health by its intracellular anti-inflammatory and antioxidant activities, and/or its antibiotic-like restructuring of the gut microbiota.
1. INTRODUCTION
Metabolic syndrome is a combination of symptoms (abdominal obesity, hypertension, high triglycerides, low HDL cholesterol and hyperglycemia) that increase the risk for cardiovascular disease and type 2 diabetes (Wilson et al., 2005). Chronic low-grade inflammation and oxidative stress have been linked to obesity-induced insulin resistance, which can lead to the development of type 2 diabetes and other conditions associated with metabolic syndrome (Carrier, 2017; Olefsky & Glass, 2010). Obesity-induced insulin resistance is associated with elevated inflammatory markers, such as inducible nitric oxide synthase (iNOS), interleukins-1β and −6 (IL-1β and IL-6), and tumor necrosis factor-α (TNF-α). These markers can be mediated by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), a transcription factor that regulates the inflammatory response (Das & Mukhopadhyay, 2011; Lawrence, 2009). Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a key player in mediating oxidative stress through activation of the phase II detoxification enzymes, including NAD(P)H quinone oxidoreductase (NQO1), which scavenge reactive oxygen species. Molecular crosstalk between NF-κB and Nrf2 has been identified linking the inflammatory and antioxidant response through a complex network of molecular interactions, capable of functioning at both the transcriptional and post-transcriptional level (Wardyn et al., 2015). Nrf2 has also been linked to other metabolic pathways associated with obesity and metabolic syndrome, including adipogenic pathways, lipid metabolism pathways and insulin signaling pathways, though some of these interactions remain unclear (Chartoumpekis & Kensler, 2013).
More recently, the development of obesity and insulin resistance in response to a high-fat diet (HFD) has been strongly correlated with localized intestinal inflammation and gut microbial community structure (Cani, Bibiloni, et al., 2008; Cani & Delzenne, 2011; Ding & Lund, 2011). Germ-free mice fed a HFD did not develop obesity and insulin resistance, whereas germ-free mice colonized with the gut microbiota from obese mice showed increased intestinal inflammation and total body fat (Cani, Delzenne, et al., 2008; Turnbaugh et al., 2006). In response to a HFD, other studies have shown major changes in the composition of the gut microbial community that were correlated with changes in gut barrier function, enhanced inflammatory response, and the development of insulin resistance (Cani et al., 2012). Several mediators have been identified as markers of intestinal inflammation, such as lipopolysaccharide (LPS), which may affect intestinal permeability leading to ‘metabolic endotoxemia’ and subsequently obesity-induced insulin resistance (Cani et al., 2007; Cani, Bibiloni, et al., 2008; Cani, Delzenne, et al., 2008; Cani et al., 2012).
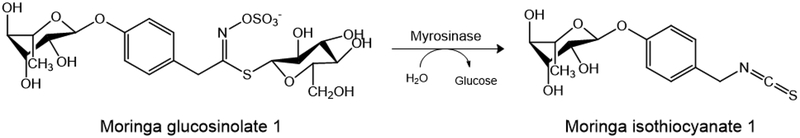
Moringa oleifera Lam. (moringa) is a tropical plant that is widely cultivated in Africa, Asia and the Americas. All parts of the plant have been traditionally used in the diet and as medicine for the treatment of various inflammatory-mediated diseases including cardiovascular diseases and diabetes. However, limited scientific studies have shown the hypoglycemic and anti-diabetic properties of moringa preparations from the leaves and seeds (Ajibola et al., 2014; Al-Malki & El Rabey, 2015; Anwar et al., 2007; Leone et al., 2015; Omodanisi et al., 2017; Tuorkey, 2016; Waterman et al., 2015). Additionally, moringa seeds have been used in water purification due to their antimicrobial properties (Eilert et al., 1981; Galuppo et al., 2013; Padla et al., 2012; Rim Jeon et al., 2014). In support of its traditional use, moringa has been shown to contain bioactive phytochemicals associated with anti-inflammatory, antioxidant, and antimicrobial properties. Moringa isothiocyanates (ITCs) are most abundant in the seeds and are formed by the enzymatic bioconversion of glucosinolates by the endogenous myrosinase (Fig 1), and are well documented for their anti-inflammatory and antioxidant activities in vitro and in vivo (Brunelli et al., 2010; Cheenpracha et al., 2010; Jaja-Chimedza et al., 2017; Tumer et al., 2015; Waterman et al., 2014). Moringa ITCs are similar to those of cruciferous vegetables (e.g. sulforaphane derived from broccoli and phenethyl isothiocyanate derived from wintercress) (Traka & Mithen, 2009). However, moringa ITCs are more stable than cruciferous ITCs due to the presence of a sugar moiety, thereby opening opportunities for the development of moringa ITCs as shelf-stable therapeutics. The most abundant ITC in the moringa plant is 4-[(α-L-rhamnosyloxy)benzyl] isothiocyanate (MIC-1; Fig 1), which we have shown to be the primary anti-inflammatory and antioxidant compound in moringa seeds, downregulating the gene expression of inflammatory markers iNOS, IL-1β and IL-6 while upregulating the antioxidant gene expression of NQO1, HO1 and GST in vitro (Jaja-Chimedza et al., 2017).
Figure 1:
The bioconversion of moringa glucosinolate 1 to moringa isothiocyanate 1 (MIC-1) by the endogenous enzyme myrosinase.
We showed earlier, that a dietary administration of an ITC-enriched moringa leaf concentrate (1–3% ITCs) reduced weight gain and insulin resistance in obese mice on a 60% HFD (Waterman et al., 2015). This bioactivity was likely due to the ITCs, however additional components of the extract, such as polyphenols, may have contributed to the observed bioactivities. For this study we prepared a standardized moringa seed extract (MSE), containing 47% of its primary bioactive compound, MIC-1, and lacking polyphenols. This well-defined extract was evaluated for efficacy in promoting metabolic resilience and tissue-specific expression of anti-inflammatory and antioxidant markers in the C57BL/6J mouse model of diet-induced obesity and insulin resistance, as well as for its effects on the gut microbiome.
2. RESEARCH DESIGN AND METHODS
2.1. Preparation of MSE and rodent diets
MSE was prepared as previously reported (Jaja-Chimedza et al., 2017). Briefly, moringa seeds were ground and incubated with water at a 1:3 ratio for 2 h at 37 °C. Ethanol was then added to the mixture (4x the volume of water), which was filtered and dried using a rotary evaporator and freeze-dryer. Dried extract was stored at −20 °C until needed. MSE was prepared from multiple 1 kg batches for a total yield of 400 g, resulting in 47% MIC-1 by weight. Custom diets were prepared by Research Diets (New Brunswick, NJ) as follows: a very-high-fat diet (VHFD, D12492; 60% fat) supplemented with 0.25% MIC-1 (0.54% MSE), and a low-fat diet (LFD, D12450J; 10% fat) supplemented with 0.34% MIC-1 (0.73% MSE) (Supplementary Table 1).
2.2. Animals and treatment (12-week study)
All animal studies were carried out according to the approved Rutgers University Institutional Animal Care and Use Committee (IACUC) protocol (#04–023). Forty-eight C57BL/6J mice were obtained from Jackson Laboratory (Bar Habour, ME) at 5 weeks old. Animals were housed on a light/dark cycle of 12 h at 22 ± 2 °C, and allowed access to food and water ad libitum. Prior to all studies, animals were acclimated for 1 week on the LFD.
Animals were subsequently split into 2 groups of 24 with 1 group on the LFD and the other group on a VHFD for 12 weeks to induce obesity and insulin resistance. Animals from each group were then randomized into 2 experimental groups (2 mice per cage); 1 group on the control diet and 1 group supplemented with MSE for a total of 4 groups (VHFD, VHFD+MSE, LFD, LFD+MSE). Body weight and food consumption were measured weekly. Oral glucose tolerance test (OGTT) was performed prior to MSE supplementation followed by every 2 weeks for the first 6 weeks and every 3 weeks until the end of the study using a glucometer (AlphaTRAK 32004–02; Abbott Animal Health). Body mass composition was measured every 3 weeks by quantitative nuclear MRI (EchoMRI 3-in-1 Analyzer). At the end of the study, tissues (liver, ileum, jejunum and colon) were collected, flash frozen in liquid nitrogen, and stored at −80 °C until processed for gene expression analyses.
2.3. Lipid liver analysis
Liver tissue was collected from each mouse and weighed at the time of necropsy. Evaluation of the total lipid content from the liver samples was performed using the Folch’s method with modifications (Cequier-Sanchez et al., 2008; Folch et al., 1957). Briefly, 250–350 mg of liver tissues was pre-weighed and homogenized in CH2Cl2 : MeOH (2:1) mixture at a 1:20 ratio (g tissue: mL solvent) using a tissue processor (Tissumizer, TEKMAR) until all the material in the vial was finely pulverized. The homogenized tissue was filtered through Whatman paper #1 and collected in pre-weighed glass tubes. The lipid extract was washed with saline solution (0.2x the volume of filtrate), vortexed and centrifuged at 1500 rpm for 5 min. The upper phase was then carefully removed and the interface was rinsed twice with 400 μL of the previously prepared CH2Cl2:MeOH:NaCl pure solvent upper phase. The lipid extracts were evaporated to dryness using a speed vacuum and freeze-dryer and subsequently weighed to determine the yield.
2.4. mRNA extraction and gene expression
Gene expression assays were performed as described previously with some modifications (Jaja-Chimedza et al., 2017). Briefly, total RNA was extracted from tissues using the RNeasy Plus Universal Mini Kit (QIAGEN), followed by cleanup with NucleoSpin RNA Clean-up XS (Macherey-Nagel). RNA quality was assessed on the NanoDrop 1000 (NanoDrop Technologies). cDNA synthesis was performed using the ABI High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with RNAse I inhibitor, according to the manufacturer’s instructions. cDNAs were diluted 3-fold for qRT-PCR analysis on the QuantStudio 3® Real-Time PCR System (Applied Biosystems) with Taqman Fast Advanced Master Mix. Inventoried Taqman primer sets (ThermoFisher Scientific) were used for analyses of HMBS (Mm01143545_m1), IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), TNF-α (Mm00443258_m1), iNOS (Mm00440502_m1), GAPDH (Mm99999915_g1), and NQO1 (Mm01253561_m1). Gene expression was quantified by the comparative ΔCt method (Schmittgen & Livak, 2008), normalizing to HMBS for inflammatory markers and GAPDH for the antioxidant markers. Tissues from eight mice were randomly selected for the gene expression and all analyses were performed in triplicates.
2.5. 16S rRNA Gene Sequencing
Genomic DNA was extracted from fecal pellets and cecal content using PowerSoil-htp 96 Well Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA). Amplification of the V4 region of the bacterial 16S rRNA gene was performed based on a previously established protocol (Kozich et al., 2013). PCR reactions were performed in 96-well plates using 20 ng of template DNA, 1 μL of each paired indexed primer set (10 μM), 25 μL of NEBNext High-Fidelity 2X PCR Master Mix (New England BioLabs) in a total reaction volume of 50 uL in 96-well plates. PCR cycling condition used was 2 min at 95 °C; 30 cycles of 20 s at 95 °C, 15 s at 55 °C, 5 min at 72 °C; followed by 10 min at 72 °C. Random samples were evaluated for successful amplification on a 1% agarose gel at 100 V for 30 min. PCR products were cleaned using Agencourt AMPure XP system (Beckman Coulter) and quantified with a Spectramax Quant Accuclear Nano dsDNA Assay kit (Molecular Devices). A library pool was constructed by combining equal amounts of each PCR product, which was subsequently quality checked (Agilent High Sensitivity DNA kit with Agilent Bioanalyzer) and quantified (KAPA HiFi HotStart ReadyMix; Kapa Biosystems). Illumina MiSeq with MiSeq Reagent kit v3 was used for sequencing.
2.6. Sequencing Data Analysis
Sequences were de-multiplexed and trimmed (using MiSeq Reporter v2.2) to eliminate barcodes and primers, after which contigs were generated using Mothur v1.36.1 (Schloss et al., 2009). Contigs having a length of 252–253 bp, no ambiguous bases and homopolymers shorter than 8 bp were kept and further de-replicated with USEARCH v9.0.2132 (Edgar, 2013). Unique sequences except singletons were clustered at 97% sequence homology and chimeras were filtered by the UPARSE-OTU algorithm. The depths of the resulting OTU table ranged from 5,074 to 200,084 with a median of 73,366. The Ribosomal Database Project (RDP) Classifier v2.12 (Q. Wang et al., 2007) was used to assign the taxonomies of OTU representative sequences with a 50% confidence threshold.
The OTU table was rarefied to 11,680 sequences per sample, and then used for microbiota α diversity (Shannon index and OTU richness) and β diversity analyses with Qiime 1.9.1 (Caporaso et al., 2010). The phylogenetic tree was generated from OTU representative sequences as previously described (Zhang et al., 2017). Principle Coordinate Analyses (PCoA) were performed on weighted UniFrac distances between bacterial communities (Lozupone & Knight, 2005). The differential clustering of bacterial communities was evaluated by ADONIS tests using the vegan R package v2.4–1 (Oksanen et al., 2015).
Relative abundances of bacterial phyla and genera calculated from non-rarefied OTU table were used to discover features altered by MSE supplementation. For animals on the same diet, features that were present in less than 50% of samples and features with average relative abundances lower than 0.02% in both the treatment and control groups were filtered out. For each time point, data matrices were permutated 10,000 times, and P values represent fraction of times that permuted differences assessed by Welch’s t test were greater than or equal to real differences. P values from multiple testing were adjusted (q values) using the Benjamini-Hochberg false discovery rate (FDR) with a significance level of 0.05 (Benjamini & Hochberg, 1995).
2.7. Total bacterial load analysis by qPCR targeting 16S rRNA gene
Genomic DNA of fecal samples were extracted using the DNeasy PowerSoil Kit (QIAGEN). DNA quality and quantity of the samples were determined using the NanoDrop 1000 (NanoDrop Technologies) and the DNA was diluted to 2.5 ng/μL for qPCR analysis. Universal primers (Integrated DNA Technologies) for 16S rRNA genes were used for analysis: U341F: 5’-CCTACGGGRSGCAGCAG-3’ and U515R: 5’-TACCGCGGCKGCTGRCAC-3’ (Baker et al., 2003; Y. Wang & Qian, 2009). Genomic DNA was extracted from Akkermansia muciniphila (ATCC BAA-835) to generate the standard curve for quantifying the total bacterial and archaeal 16S rRNA gene counts, which was then normalized to the amount of fecal sample extracted (referred to as ‘bacterial load’ since the relative abundance of Archaea is normally lower than 1% in mice). The following information was used for calculation: 1 ng of A.muciniphila genomic DNA corresponds to 3.48 × 105 genome copies, i.e. 1.044 × 106 16S rRNA gene copies.
2.8. Statistical analysis
All statistical analyses were performed using GraphPad Prism 7.03, unless otherwise specified. Data was analyzed for outliers using the robust regression followed by outlier identification (ROUT) method and normality was evaluated using the D’Agostino & Pearson normality test. Subsequently, analyses were performed using repeated measures ANOVA followed by Tukey’s post hoc test or Kruskal-Wallis test followed by Dunn’s post hoc test based on the normality of the data set. P < 0.05 was considered significant.
3. RESULTS
3.1. MSE alleviated VHFD-induced adiposity
Supplementation with MSE in the VHFD group resulted in lower body weights compared to the corresponding control over the course of the study, while animals in the LFD group showed no difference in body weight between the treatment and control (Fig 2A). A significant decrease in body weight was observed within the first week of treatment for animals on the VHFD+MSE diet compared with its control, which corresponded with a short-term reduction in food intake (Fig 2B). After the first week, mice on the VHFD+MSE diet maintained lower body weights than its control even though food intake normalized to similar levels between the two groups (Fig 2B). The average weekly food intake was 2.8±0.3, 2.6±0.3, 2.8±0.2 and 2.8±0.3 mg/d per mouse on the VHFD, VHFD+MSE, LFD and LFD+MSE diets, respectively, corresponding to an average treatment dose of 161±19 and 335±23 mg MIC-1/kg bw/d for mice on the VHFD+MSE and LFD+MSE diets, respectively. Furthermore, from cage-side observations, there were no toxic effects in any of the treatment groups during the course of the study.
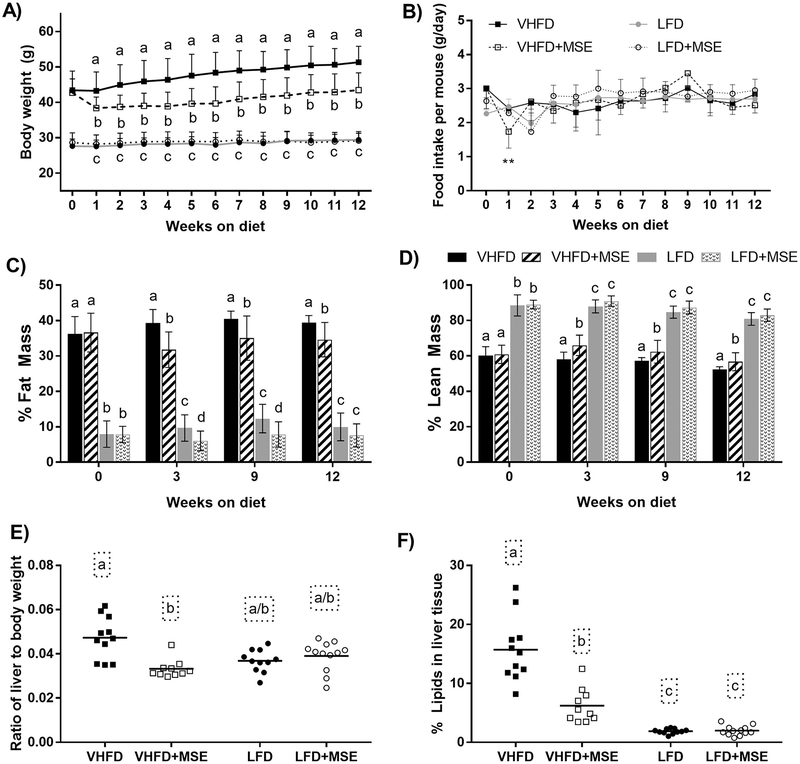
Figure 2: Changes in body weight and body composition in mice on a VHFD or LFD with or without MSE supplementation for 12 weeks.
A) Body weight gain, B) Weekly food intake per mouse per day, C) Fat mass composition as a percentage of body weight, and D) Lean mass composition as a percentage of body weight. E) Ratio of liver to body weight and F) % lipids in liver. For panels A-D, data are mean ± SD and for panel E-F, data represents the mean (line) and each data point corresponds to a biological sample (n = 11/12). Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc tests or Kruskal-Wallis tests followed by Dunn’s (multiple comparisons) tests based on the distribution of the data. Statistical significance is indicated by different letters at P < 0.05.
MSE supplementation resulted in reduced adiposity in mice (Fig 2C–D). The fat mass in both VHFD and LFD groups supplemented with MSE was significantly decreased compared to the corresponding control groups from week 3 to 12 (Fig 2C), while the lean mass was significantly increased only in the VHFD+MSE-fed group (Fig 2D). Reduced adiposity was also observed in the liver; liver weights were significantly lower due to reduced fat accumulation in VHFD+MSE-fed mice, with no significant changes observed in LFD-fed mice (Fig 2E–F). In the VHFD groups, the liver weights of the treatment mice were 23% lower than the control mice and similar to those of the LFD control mice (Fig 2E). Lipid content in the liver was decreased by 60% in mice on VHFD+MSE compared to the VHFD control (Fig 2F). Additionally, the VHFD+MSE livers were noticeably a darker red color than the control, which looked paler (photos not shown). No differences were observed in the liver weights or their lipid content within the LFD group.
3.2. MSE improved glucose tolerance
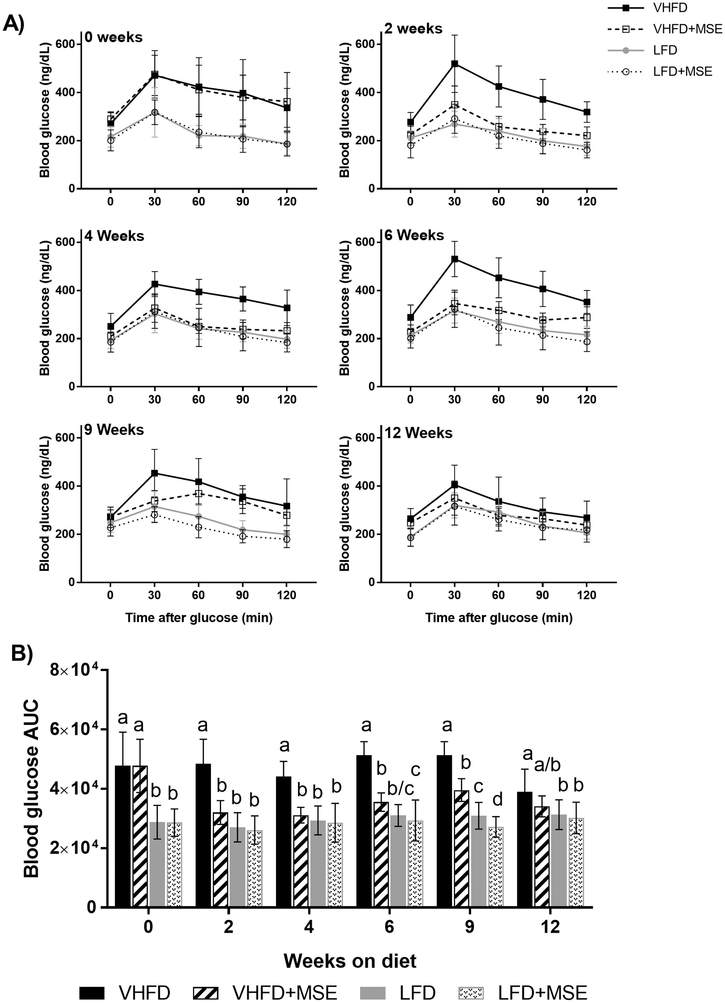
Treatment with MSE in the VHFD groups significantly improved glucose tolerance within the first 2 weeks of treatment and throughout the study, while no significant differences were observed in mice in the LFD groups, except at 9 weeks (Fig 3A–B). In the VHFD groups, MSE-treated mice had reduced blood glucose area under the curve (AUC) compared to the control mice during the 12 weeks on the diet (Fig 3B). In the LFD groups, the only reduction in blood glucose AUC was observed in MSE-treated mice at 9 weeks, compared to the control group (Fig 3B). The blood glucose response of mice on the VHFD+MSE were similar to that of the LFD control except at 9 weeks (Fig 3A), despite the difference in weight of animals on the VHFD compared to the LFD control.
Figure 3: Effects of MSE on the glucose tolerance of mice on a VHFD and LFD with or without MSE supplementation for 12 weeks.
A) Blood glucose levels during OGTT at 0, 2, 4, 6, 9 and 12 weeks and B) blood glucose AUC. Data are mean ± SD (n = 11/12). Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc tests or Kruskal-Wallis tests followed by Dunn’s (multiple comparisons) tests based on the distribution of the data. Statistical significance is indicated by different letters at P < 0.05.
3.3. MSE altered anti-inflammatory and antioxidant gene expression
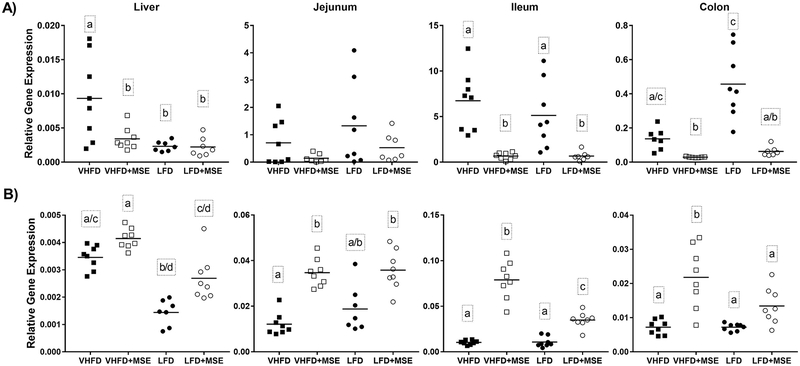
Supplementation with MSE reduced inflammatory gene expression of iNOS (Fig 4A) and upregulated antioxidant gene expression of NQO1 (Fig 4B) in both the VHFD and LFD groups. In the VHFD groups, iNOS expression was significantly decreased in the liver (2.7-fold), ileum (10.1-fold) and colon (4.7-fold) of MSE-treated animals, and was non-significantly lower in the jejunum (5.0-fold) (Fig 4A). Gene expression of the other inflammatory markers analyzed (IL-1β, IL-6 and TNF-α) was non-significantly reduced in most tissues by MSE treatment compared to VHFD control animals (Supplementary Fig 1). NQO-1 gene expression was significantly upregulated by MSE treatment in the jejunum (2.9-fold), ileum (7.6-fold), and colon (3.0-fold) compared to VHFD control mice. Non-significant upregulation of NQO-1 was also observed in the liver (Fig 4B).
Figure 4: Effects of MSE on inflammatory and antioxidant gene expression in mice on a VHFD and LFD with or without MSE supplementation for 12 weeks.
Gene expression of (A) iNOS A) and (B) NQO-1 in liver, jejunum, ileum and colon tissue of C57BL/6J mice on a VHFD and LFD with or without MSE supplementation for 12 weeks. Data represent the mean (line) and each data point corresponds to a biological sample (n=8). Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc tests or Kruskal-Wallis tests followed by Dunn’s (multiple comparisons) tests based on the distribution of the data. Statistical significance is indicated by different letters at P < 0.05.
Among the LFD groups, a significant decrease in iNOS expression was observed in the ileum (7.7-fold) and colon (7.3-fold), and a non-significant increase was observed in the liver or jejunum (Fig 4A). Gene expression of the other inflammatory markers analyzed (IL-1β, IL-6 and TNF-α) was non-significantly reduced in most tissues by MSE treatment compared to LFD control animals (Supplementary Fig 1). Significant upregulation of NQO-1 gene expression by MSE treatment was observed in the ileum (3.2-fold), and colon (1.9-fold) compared to the control, however it was not statistically significant in the liver (1.9-fold) and jejunum (1.9-fold) (Fig 4B).
3.4. MSE modulated the gut microbial community
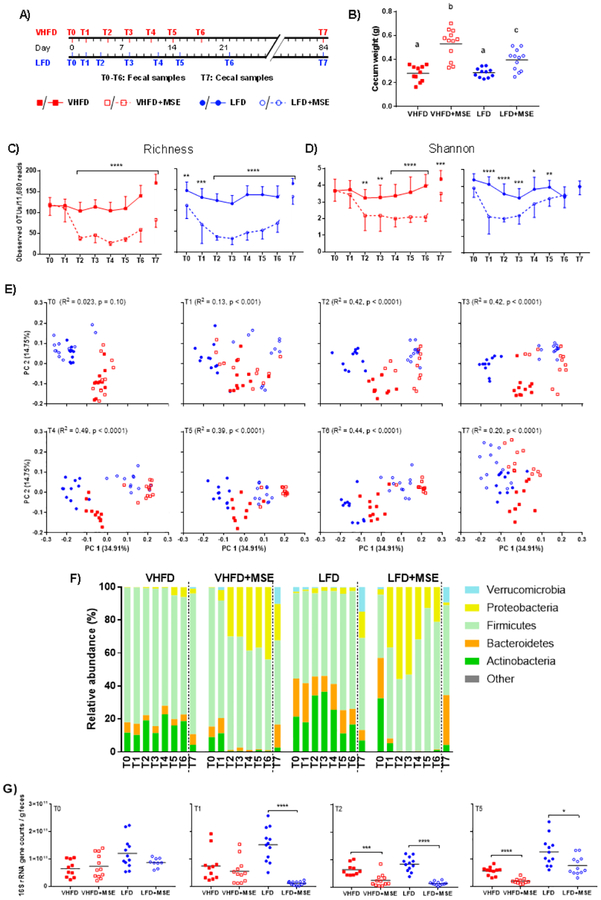
16S rRNA gene sequencing was performed on fecal samples collected on days 0 (T0), 2 (T1), 4/5 (T2), 8 (T3), 11/12 (T4), 14/15 (T5), 18/22 (T6) and cecal samples collected at the endpoint (week 12, T7) (Fig 5A). MSE-treated mice on both VHFD and LFD diets exhibited significantly enlarged ceca, especially those on the VHFD+MSE diet, however no difference was observed between the VHFD and LFD control groups (Fig 5B). MSE supplementation induced strong reduction of bacterial α diversity, represented by numbers of observed OTUs (richness; Fig 5C) and Shannon index (Fig 5D). The bacterial α diversity of LFD-fed mice showed a quicker response to MSE treatment than that of VHFD-fed mice, decreasing significantly after 2 days of treatment (T1), reaching the lowest point on day 4–8 (T2-T3), and starting to recover thereafter. This trend was especially evident for the Shannon index, which takes both the microbial richness and evenness into account. In the VHFD+MSE-fed group, bacterial α diversity became significantly lower than the VHFD control group on day 5 (T2), and remained lower until the endpoint of the study (Fig 5C–D). Principle coordinate analysis (PCoA) based on weighted UniFrac distance, showed clear clustering by diet base (R2 = 0.51, P < 0.0001, ADONIS test) at the baseline (T0; Fig 5E). Two days into the treatment period (T1), bacterial communities were distinguishable by MSE supplementation (R2 = 0.13, P < 0.001), and the differentiation became larger, explaining around 40% of the variance (ADONIS tests, P < 0.0001), in the following 2–3 weeks (T2-T6). In line with the response-recovery process revealed by bacterial α diversity, the separation of MSE-treated groups from control groups on the PCoA plots became less significant at the end of the study (ADONIS test, R2 = 0.20, P < 0.0001).
Figure 5: Gut microbiome changes in mice on a VHFD and LFD with or without MSE supplementation for 12 weeks.
A) Schematic figure showing sampling schedule for microbiota analyses. B) The weight of entire cecal tissue with content. One-way ANOVA followed by Tukey’s post hoc test was performed, where different letters represent statistical significance (P < 0.05). C) OTU richness and D) Shannon index as indexes for microbiota α diversity. Multiple t tests were performed within each diet group between the treatment and corresponding control. E) Principle coordinate analysis (PCoA) on weighted UniFrac distances. ADONIS tests were performed to assess differential clustering caused by MSE supplementation and diet base. Listed R2 values and P values represent the percentages of variation explained by MSE supplementation and corresponding significance. F) Relative abundance of the five dominant bacterial phyla. The rest low-abundance phyla were combined into the ‘Other’ category. Statistical results are shown in Supplementary Table 1. G) Bacterial+archaeal load determined by qPCR from feces at baseline (T0), T1, T2 and T5. In panels C-E, analyses were performed on 11,680 sequences per sample, while non-rarefied data was used in panel F. For panels B and G, data represent the mean (line) and each data point corresponds to a biological sample, for panels C-D, data are mean ± SD (n = 11/12). Asterisks represent significant differences between the treatment and control groups (, *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001).
At the phylum level, there was a prominently lower relative abundance of Actinobacteria and Bacteroidetes and higher abundance of Proteobacteria as a result of MSE supplementation regardless of diet base (Fig 5F and Supplementary Table 2). Proteobacteria increased to a maximum relative abundance of 43.99 ± 11.63% (T6) for VHFD+MSE-fed mice and 55.89 ± 13.85% (T2) for LFD+MSE-fed mice. The bacterial compositional changes at the phylum level showed similar patterns as observed for the bacterial α and β diversity, i.e., quick response-recovery process in LFD+MSE-fed mice while slower response-recovery in VHFD+MSE-fed mice. Both VHFD and LFD groups treated with MSE partially regained their gut microbiome as indicated by the cecal community structure at the phylum level at the endpoint (12 weeks).
At the genus level, MSE supplementation induced a broad range effect, with changes observed in the relative abundance of 95.85 ± 9.35% and 99.01 ± 6.03% of classified genera for VHFD groups and LFD groups, respectively (Supplementary Fig 2). A number of dominant genera (with average relative abundances greater than 15% either in the VHFD+MSE/LFD+MSE groups or the control groups) were dramatically altered, showing more than 30-fold differences in relative abundances between the MSE-treated groups and corresponding control groups. Some of the most notable changes in genera were observed at T2 (day 4/5) and these included: Olsenella (relative abundance, VHFD 16.96±14.36% vs. VHFD+MSE 0.27±0.56%) and Bifidobacterium (LFD 32.62±15.03% vs. LFD+MSE 0.24±0.19%), within the phylum Actinobacteria, were lower in MSE-treated groups. Enterobacter (VHFD 0.46±0.34% vs. VHFD+MSE 25.34±26.43%) and Eschrichia/Shigella (LFD 0.94±1.27% vs. LFD+MSE 28.27±22.92%), within the phylum Proteobacteria, were higher in MSE-treated groups. Allobaculum (LFD 15.93±13.11% vs. LFD+MSE 0.13±0.11%) was lower, while Enterococcus (VHFD 0.47±0.45% vs. VHFD+MSE 24.24±15.06%) was higher, both within the phylum Firmicutes, due to MSE supplementation. Regardless of diet base, MSE supplementation profoundly affected the relative abundances of different bacterial genera.
The total bacterial load in fecal samples at T0, T1, T2 and T5 was analyzed to determine the changes in the quantity of bacteria in the gut in response to the treatment (Fig 5G). These time points correlate with the initial changes in the bacterial community (T1 and T2) and the first glucose tolerance test performed (T5). At baseline (T0), there was no difference in the bacterial load between the treatment groups and the corresponding controls. Similar to the pattern revealed by 16S rRNA gene sequencing, the LFD group showed a rapid response to MSE treatment, with a significant reduction in bacterial load at T1, T2 and T5, while the bacterial load in the VHFD treatment group was significantly reduced at T2 and T5.
4. DISCUSSION
Moringa has been used for decades for the treatment of various inflammatory related chronic illnesses including diabetes. A limited number of studies have shown the benefits of poorly characterized moringa leaf extracts in models of hyperglycemia and diabetes (Omodanisi et al., 2017; Tuorkey, 2016). The suggested phytochemicals in moringa associated with these effects include the phenolic compounds (quercetin and chlorogenic acid), and the ITCs, which are well-known for their anti-inflammatory and antioxidant properties (Mbikay, 2012; Waterman et al., 2015). Two studies have shown that moringa seeds exhibited anti-hyperglycemic and anti-diabetic effects in alloxan-induced and streptozotocin-induced diabetes in rats, respectively (Ajibola et al., 2014; Al-Malki & El Rabey, 2015). Unlike moringa leaves, moringa seeds contain high levels of glucosinolate-1, which can be converted into MIC-1, and very little, if any, phenolic compounds (Amaglo et al., 2010; Jaja-Chimedza et al., 2017). This current study has shown that a well-characterized moringa seed extract rich in MIC-1 exerted anti-hyperglycemic properties and that this activity is likely associated with MIC-1.
Two potential mechanisms by which MSE mitigates the inflammatory response and improves metabolic health are activation of Nrf2 and its downstream targets and/or modulation of the gut microbiota. The Nrf2 stress response is thought to be an important regulator in oxidative-stress and chronic inflammatory conditions (Singh et al., 2010). Under normal conditions, Nrf2 is sequestered in the cytosol by Kelch-like ECH-associated protein (Keap1) and targeted for proteosomal degradation, while under stressed conditions, Nrf2 dissociates from Keap1 and translocates to the nucleus, where it interacts with the antioxidant response element (ARE) to induce the transcription of its target genes (Singh et al., 2010). Cruciferous ITCs (especially sulforaphane) are known inducers of Nrf2, interacting with the Keap1 subunit to allow Nrf2 to dissociate from Keap1 (which sequesters Nrf2 and tags it for proteosomal degradation), and translocate to the nucleus for transcription of its target phase II detoxifying genes (Hu et al., 2006; Prawan et al., 2008). We have previously shown that MIC-1 has anti-inflammatory and antioxidant properties in RAW 264.7 murine macrophages when stimulated with LPS, significantly downregulating the gene expression of iNOS and other inflammatory markers as well as upregulating the gene expression of NQO1 and other antioxidant markers (Jaja-Chimedza et al., 2017). MIC-1 has been shown to be a potent inducer of NQO-1 in intestinal tissue and cell lines, with activity comparable to or higher than sulforaphane (Dinkova-Kostova et al., 2004). Another study has shown that MIC-1 (also called moringin) significantly increases the nuclear expression of Nrf2 in LPS-stimulated macrophages (Rajan et al., 2016), supporting the direct regulation of Nrf2 by MIC-1. In this study, MSE significantly regulated the gene expression of iNOS and NQO1 in vivo, which are associated with chronic inflammation, oxidative stress, and obesity-induced insulin resistance in mice (Fujimoto et al., 2005; Munday et al., 2010; Shinozaki et al., 2011), supporting the role of MIC-1 as the primary bioactive component of MSE and the mechanism by which MSE improved the glucose tolerance and overall metabolic health of the mice.
Another plausible mechanism of MIC-1 action is based on the antibiotic properties of this compound, making it an effective modulator of the gut microbiota, which can influence the host metabolism through regulating energy balance, glucose metabolism, lipid metabolism and inflammation (Cani et al., 2012; David et al., 2014). Adverse changes to the intestinal integrity in response to the HFD can be partially negated by antibiotic treatment, resulting in the recovery of the epithelial integrity, improved glucose and insulin tolerance, reduced adiposity, inflammation and oxidative stress (Cani, Delzenne, et al., 2008; Carvalho et al., 2012; Membrez et al., 2008). Metagenomic analysis showed that antibiotic treatment of the HFD-fed mice dramatically reduced total bacterial abundance and increased the relative ratio of Proteobacteria to Bacteroidetes, Firmicutes and Verrucomicrobia. These changes were accompanied by reduced LPS levels and improved intestinal barrier. Accumulating evidence thus suggests that antimicrobial compounds can reduce the production of gut-derived inflammatory signals and improve gut and metabolic health (Carvalho et al., 2012).
Our data showed that MSE-supplementation significantly altered the gut microbiota of both VHFD and LFD-fed mice by reducing the total bacterial load and α-diversity and modifying the overall bacterial community. Enlarged cecum, observed in MSE-treated mice, normally reflects disturbed cecal fermentation caused by reduced bacterial load and/or complexity, as seen in germ-free and antibiotic-treated mice (Thompson & Trexler, 1971). 16S rRNA gene sequencing showed that MSE supplementation affected at least 95% of the classified genera, significantly reducing Actinobacteria and Bacteroidetes spp., with variable response in Firmicutes spp. Even though MSE supplementation resulted in an increased relative abundance of Gammaproteobacteria (Enterobacter and Escherichia/Shigella), which are known producers of pro-inflammatory LPS (Zhao, 2013), we still observed improved glucose tolerance and reduced adiposity, inflammation and oxidative stress. We suggest that the reduction in total bacterial load, across all taxa, translates to lower levels of circulating LPS.
Moringa seeds have been traditionally used for water purification due to their antimicrobial properties. Among the identified antimicrobial compounds is MIC-1, showing activity against a number of Gram-positive bacteria including Staphylococcus aureus, Staphylococcus epidermis, Bacillus subtilis and Mycobacterium phlei, while being less active against the Gram-negative bacteria, Escherichia coli and Serratia marcescens (Eilert et al., 1981; Padla et al., 2012). MSE showed a strong antibiotic effect by reducing the total bacteria in the gut of the mice and, which was likely caused by MIC-1, which comprises 47% of MSE.
In summary, MSE significantly improved the metabolic health in diet-induced obese and insulin resistant mice, manifested in greater glucose tolerance and lower adiposity, inflammation and oxidative stress. This effect is likely driven by MIC-1 activation of the transcription factor Nrf2, a key regulator of the phase II detoxifying pathways. In addition, MSE, most likely via antibacterial effects of MIC-1, suppresses gut microbiota, possibly reducing metabolic endotoxemia and improving overall metabolic health. These data validate the traditional use of moringa in the treatment of diabetes and provide further insight into understanding its mechanism of action.
Supplementary Material
Table 1: Composition of rodent diets.
Table 2: Statistical analysis of the relative abundance at phylum level between MSE-treated groups and controls.
Supplementary Figure 1: Anti-inflammatory gene expression of IL-1β, IL-6 and TNF-α in the liver, jejunum, ileum and colon of C57BL/6J mice supplemented with MSE on both a VHFD and LFD for 12 weeks. Statistical analyses were performed using one-way ANOVA followed by Tukey’s post hoc test. No statistical significance was observed between the groups at p < 0.05.
Supplementary Figure 2: Dietary MSE altered bacterial composition at genus level. Heatmaps showing the average relative abundances of bacterial genera differing between MSE-supplemented groups and control groups. For fecal samples at T0-T6 and cecal samples at T7, statistical comparison of the two groups within the same diet base was done by 10,000 times of permutation; p values represent fraction of times that permuted differences assessed by Welch’s t-test were greater than or equal to real differences, and were adjusted by false discovery rate (FDR) correction (*q < 0.05, **q < 0.01, ***q < 0.001). Alterations that were common in both VHFD and LFD conditions were indicated by bold characters; genera that were more abundant in the MSE-supplemented groups are indicated in red and the opposite in blue; others with changes in both directions between VHFD and LFD groups or between different time points within groups of the same diet are indicated in black.
Highlights.
Isothiocyanate-rich moringa seed extract improves metabolic health in obese mice.
Moringa seed extract modulates antioxidant and inflammatory gene expression.
Moringa seed extract drastically modulates the gut microbiota.
Two potential mechanisms for the anti-diabetic effects of moringa.
Acknowledgments:
The authors would like to thank Hetalben Kalariya, Samvitha Cherravuru, Dr. Gang Pan and Irina Tmenova for their technical assistance. AJC, LZ and IR take full responsibility for the contents of the manuscript.
Funding: This publication was supported by the National Center For Complementary & Integrative Health and the Office of Dietary Supplements of the National Institutes of Health under Award Number P50AT002776 and 5T32AT004094, which fund the Botanical Research Center of Pennington Biomedical Research Center and the Department of Plant Biology in the School of Environmental and Biological Sciences (SEBS) of Rutgers University, and by the New Jersey Agricultural Experiment Station at Rutgers University (AJ, BLG, PK and IR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This project/work used genomics core facilities that are supported in part by COBRE (NIH8 1P30GM118430–01) and NORC (NIH 2P30DK072476) center grants from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: IR has a financial interest in Nutrasorb LLC and is an inventor on the patent application of MSE.
Data deposition: DNA sequences encoding bacterial 16S rRNA V4 region reported in this paper have been deposited in the Sequence Read Archive (SRA) under the accession number SRP127415.
References
- Ajibola M, Eunice O, & Stephanie IN (2014). Effects of Aqueous Extract of Moringa oleifera Seeds on Alloxan Induced Hyperglycemia. Basic Sciences of Medicine, 3(3), 37–42. [Google Scholar]
- Al-Malki AL, & El Rabey HA (2015). The Antidiabetic Effect of Low Doses of Moringa oleifera Lam. Seeds on Streptozotocin Induced Diabetes and Diabetic Nephropathy in Male Rats. BioMed Research International, 2015, 381040. doi: 10.1155/2015/381040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaglo NK, Bennett RN, Lo Curto RB, Rosa EAS, Lo Turco V, Giuffrida A, Curto AL, Crea F, & Timpo GM (2010). Profiling selected phytochemicals and nutrients in different tissues of the multipurpose tree Moringa oleifera L., grown in Ghana. Food Chemistry, 122(4), 1047–1054. doi: 10.1016/j.foodchem.2010.03.073 [DOI] [Google Scholar]
- Anwar F, Latif S, Ashraf M, & Gilani AH (2007). Moringa oleifera: a food plant with multiple medicinal uses. Phytother Res, 21(1), 17–25. doi: 10.1002/ptr.2023 [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, & Cowan DA (2003). Review and re-analysis of domain-specific 16S primers. J Microbiol Methods, 55(3), 541–555. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Brunelli D, Tavecchio M, Falcioni C, Frapolli R, Erba E, Iori R, Rollin P, Barillari J, Manzotti C, Morazzoni P, & D’Incalci M (2010). The isothiocyanate produced from glucomoringin inhibits NF-kB and reduces myeloma growth in nude mice in vivo. Biochem Pharmacol, 79(8), 1141–1148. doi: 10.1016/j.bcp.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, & Burcelin R (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes, 56(7), 1761–1772. doi: 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, & Burcelin R (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes, 57(6), 1470–1481. doi: 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- Cani PD, & Delzenne NM (2011). The gut microbiome as therapeutic target. Pharmacol Ther, 130(2), 202–212. doi: 10.1016/j.pharmthera.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Cani PD, Delzenne NM, Amar J, & Burcelin R (2008). Role of gut microflora in the development of obesity and insulin resistance following high-fat diet feeding. Pathol Biol (Paris), 56(5), 305–309. doi: 10.1016/j.patbio.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Cani PD, Osto M, Geurts L, & Everard A (2012). Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes, 3(4), 279–288. doi: 10.4161/gmic.19625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, & Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods, 7(5), 335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier A (2017). Metabolic Syndrome and Oxidative Stress: A Complex Relationship. Antioxid Redox Signal, 26(9), 429–431. doi: 10.1089/ars.2016.6929 [DOI] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JBC, & Saad MJA (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia, 55(10), 2823–2834. doi: 10.1007/s00125-012-2648-4 [DOI] [PubMed] [Google Scholar]
- Cequier-Sanchez E, Rodriguez C, Ravelo AG, & Zarate R (2008). Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J Agric Food Chem, 56(12), 4297–4303. doi: 10.1021/jf073471e [DOI] [PubMed] [Google Scholar]
- Chartoumpekis DV, & Kensler TW (2013). New player on an old field; the keap1/Nrf2 pathway as a target for treatment of type 2 diabetes and metabolic syndrome. Curr Diabetes Rev, 9(2), 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheenpracha S, Park EJ, Yoshida WY, Barit C, Wall M, Pezzuto JM, & Chang LC (2010). Potential anti-inflammatory phenolic glycosides from the medicinal plant Moringa oleifera fruits. Bioorg Med Chem, 18(17), 6598–6602. doi: 10.1016/j.bmc.2010.03.057 [DOI] [PubMed] [Google Scholar]
- Das A, & Mukhopadhyay S (2011). The evil axis of obesity, inflammation and type-2 diabetes. Endocr Metab Immune Disord Drug Targets, 11(1), 23–31. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, & Turnbaugh PJ (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505(7484), 559–563. doi: 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, & Lund PK (2011). Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr Opin Clin Nutr Metab Care, 14(4), 328–333. doi: 10.1097/MCO.0b013e3283478727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Fahey JW, & Talalay P (2004). Chemical structures of inducers of nicotinamide quinone oxidoreductase 1 (NQO1). Methods Enzymol, 382, 423–448. doi: 10.1016/s0076-6879(04)82023-8 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods, 10(10), 996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Eilert U, Wolters B, & Nahrstedt A (1981). The antibiotic principle of seeds of Moringa oleifera and Moringa stenopetala. Planta Med, 42(1), 55–61. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, & Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem, 226(1), 497–509. [PubMed] [Google Scholar]
- Fujimoto M, Shimizu N, Kunii K, Martyn JA, Ueki K, & Kaneki M (2005). A role for iNOS in fasting hyperglycemia and impaired insulin signaling in the liver of obese diabetic mice. Diabetes, 54(5), 1340–1348. [DOI] [PubMed] [Google Scholar]
- Galuppo M, Nicola G, Iori R, Dell #039 Utri P, Bramanti P, & Mazzon E (2013). Antibacterial Activity of Glucomoringin Bioactivated with Myrosinase against Two Important Pathogens Affecting the Health of Long-Term Patients in Hospitals. Molecules, 18(11), 14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, & Kong AN (2006). Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett, 243(2), 170–192. doi: 10.1016/j.canlet.2005.11.050 [DOI] [PubMed] [Google Scholar]
- Jaja-Chimedza A, Graf BL, Simmler C, Kim Y, Kuhn P, Pauli GF, & Raskin I (2017). Biochemical characterization and anti-inflammatory properties of an isothiocyanate-enriched moringa (Moringa oleifera) seed extract. PLoS ONE, 12(8), e0182658. doi: 10.1371/journal.pone.0182658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, & Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol, 79(17), 5112–5120. doi: 10.1128/aem.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol, 1(6), a001651. doi: 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone A, Spada A, Battezzati A, Schiraldi A, Aristil J, & Bertoli S (2015). Cultivation, genetic, ethnopharmacology, phytochemistry and pharmacology of Moringa oleifera leaves: An overview. International journal of molecular sciences, 16(6), 12791–12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, & Knight R (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol, 71(12), 8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbikay M (2012). Therapeutic potential of Moringa oleifera leaves in chronic hyperglycemia and dyslipidemia: a review. Frontiers in pharmacology, 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Mace K, & Chou CJ (2008). Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. Faseb j, 22(7), 2416–2426. doi: 10.1096/fj.07-102723 [DOI] [PubMed] [Google Scholar]
- Munday R, Zhang Y, Paonessa JD, Munday CM, Wilkins AL, & Babu J (2010). Synthesis, biological evaluation, and structure-activity relationships of dithiolethiones as inducers of cytoprotective phase 2 enzymes. J Med Chem, 53 (12), 4761–4767. doi: 10.1021/jm100425v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, & Wagner H (2015). Vegan: community ecology package. R package vegan, vers. 2.2–1.
- Olefsky JM, & Glass CK (2010). Macrophages, inflammation, and insulin resistance. Annu Rev Physiol, 72, 219–246. doi: 10.1146/annurev-physiol-021909-135846 [DOI] [PubMed] [Google Scholar]
- Omodanisi EI, Aboua YG, & Oguntibeju OO (2017). Assessment of the Anti-Hyperglycaemic, Anti-Inflammatory and Antioxidant Activities of the Methanol Extract of Moringa Oleifera in Diabetes-Induced Nephrotoxic Male Wistar Rats. Molecules, 22(4). doi: 10.3390/molecules22040439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padla EP, Solis LT, Levida RM, Shen CC, & Ragasa CY (2012). Antimicrobial isothiocyanates from the seeds of Moringa oleifera Lam. Z Naturforsch C, 67(11–12), 557–564. [DOI] [PubMed] [Google Scholar]
- Prawan A, Keum YS, Khor TO, Yu S, Nair S, Li W, Hu L, & Kong AN (2008). Structural influence of isothiocyanates on the antioxidant response element (ARE)-mediated heme oxygenase-1 (HO-1) expression. Pharm Res, 25(4), 836–844. doi: 10.1007/s11095-007-9370-9 [DOI] [PubMed] [Google Scholar]
- Rajan TS, Giacoppo S, Iori R, De Nicola GR, Grassi G, Pollastro F, Bramanti P, & Mazzon E (2016). Anti-inflammatory and antioxidant effects of a combination of cannabidiol and moringin in LPS-stimulated macrophages. Fitoterapia, 112, 104–115. doi: 10.1016/j.fitote.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Rim Jeon S, Ha Lee K, Ha Shin D, Sang Kwon S, & Sung Hwang J (2014). Synergistic antimicrobial efficacy of mesoporous ZnO loaded with 4-(alpha-L-rhamnosyloxy)-benzyl isothiocyanate isolated from the Moringa oleifera seed. J Gen Appl Microbiol, 60(6), 251–255. doi: 10.2323/jgam.60.251 [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, & Weber CF (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol, 75(23), 7537–7541. doi: 10.1128/aem.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, & Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc, 3(6), 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shinozaki S, Choi CS, Shimizu N, Yamada M, Kim M, Zhang T, Shiota G, Dong HH, Kim YB, & Kaneki M (2011). Liver-specific inducible nitric-oxide synthase expression is sufficient to cause hepatic insulin resistance and mild hyperglycemia in mice. J Biol Chem, 286(40), 34959–34975. doi: 10.1074/jbc.M110.187666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Vrishni S, Singh BK, Rahman I, & Kakkar P (2010). Nrf2-ARE stress response mechanism: a control point in oxidative stress-mediated dysfunctions and chronic inflammatory diseases. Free Radic Res, 44(11), 1267–1288. doi: 10.3109/10715762.2010.507670 [DOI] [PubMed] [Google Scholar]
- Thompson GR, & Trexler PC (1971). Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut, 12(3), 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, & Mithen R (2009). Glucosinolates, isothiocyanates and human health. Phytochemistry Reviews, 8(1), 269–282. doi: 10.1007/s11101-008-9103-7 [DOI] [Google Scholar]
- Tumer TB, Rojas-Silva P, Poulev A, Raskin I, & Waterman C (2015). Direct and indirect antioxidant activity of polyphenol- and isothiocyanate-enriched fractions from Moringa oleifera. J Agric Food Chem, 63(5), 1505–1513. doi: 10.1021/jf505014n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorkey MJ (2016). Effects of Moringa oleifera aqueous leaf extract in alloxan induced diabetic mice. Interv Med Appl Sci, 8(3), 109–117. doi: 10.1556/1646.8.2016.3.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. doi: 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, & Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol, 73(16), 5261–5267. doi: 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, & Qian PY (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE, 4(10), e7401. doi: 10.1371/journal.pone.0007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardyn JD, Ponsford AH, & Sanderson CM (2015). Dissecting molecular cross-talk between Nrf2 and NF-kappaB response pathways. Biochem Soc Trans, 43(4), 621–626. doi: 10.1042/bst20150014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman C, Cheng DM, Rojas-Silva P, Poulev A, Dreifus J, Lila MA, & Raskin I (2014). Stable, water extractable isothiocyanates from Moringa oleifera leaves attenuate inflammation in vitro. Phytochemistry, 103, 114–122. doi: 10.1016/j.phytochem.2014.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman C, Rojas-Silva P, Tumer TB, Kuhn P, Richard AJ, Wicks S, Stephens JM, Wang Z, Mynatt R, Cefalu W, & Raskin I (2015). Isothiocyanate-rich Moringa oleifera extract reduces weight gain, insulin resistance, and hepatic gluconeogenesis in mice. Mol Nutr Food Res, 59(6), 1013–1024. doi: 10.1002/mnfr.201400679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PW, D’Agostino RB, Parise H, Sullivan L, & Meigs JB (2005). Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation, 112(20), 3066–3072. doi: 10.1161/circulationaha.105.539528 [DOI] [PubMed] [Google Scholar]
- Zhang L, Bahl MI, Roager HM, Fonvig CE, Hellgren LI, Frandsen HL, Pedersen O, Holm JC, Hansen T, & Licht TR (2017). Environmental spread of microbes impacts the development of metabolic phenotypes in mice transplanted with microbial communities from humans. Isme j, 11(3), 676–690. doi: 10.1038/ismej.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L (2013). The gut microbiota and obesity: from correlation to causality. Nature Reviews Microbiology, 11, 639. doi: 10.1038/nrmicro3089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: Composition of rodent diets.
Table 2: Statistical analysis of the relative abundance at phylum level between MSE-treated groups and controls.
Supplementary Figure 1: Anti-inflammatory gene expression of IL-1β, IL-6 and TNF-α in the liver, jejunum, ileum and colon of C57BL/6J mice supplemented with MSE on both a VHFD and LFD for 12 weeks. Statistical analyses were performed using one-way ANOVA followed by Tukey’s post hoc test. No statistical significance was observed between the groups at p < 0.05.
Supplementary Figure 2: Dietary MSE altered bacterial composition at genus level. Heatmaps showing the average relative abundances of bacterial genera differing between MSE-supplemented groups and control groups. For fecal samples at T0-T6 and cecal samples at T7, statistical comparison of the two groups within the same diet base was done by 10,000 times of permutation; p values represent fraction of times that permuted differences assessed by Welch’s t-test were greater than or equal to real differences, and were adjusted by false discovery rate (FDR) correction (*q < 0.05, **q < 0.01, ***q < 0.001). Alterations that were common in both VHFD and LFD conditions were indicated by bold characters; genera that were more abundant in the MSE-supplemented groups are indicated in red and the opposite in blue; others with changes in both directions between VHFD and LFD groups or between different time points within groups of the same diet are indicated in black.