Abstract
Chronic kidney disease (CKD) is prevalent across the world, and kidney function is well defined by an estimated glomerular filtration rate (eGFR). The progression of kidney disease can be predicted if the future eGFR can be accurately estimated using predictive analytics. In this study, we developed and validated a prediction model of eGFR by data extracted from a regional health system. This dataset includes demographic, clinical and laboratory information from primary care clinics. The model was built using Random Forest regression and evaluated using Goodness-of-fit statistics and discrimination metrics. After data preprocessing, the patient cohort for model development and validation contained 61740 patients. The final model included eGFR, age, gender, body mass index (BMI), obesity, hypertension, and diabetes, which achieved a mean coefficient of determination of 0.95. The estimated eGFRs were used to classify patients into CKD stages with high macro-averaged and micro-averaged metrics. In conclusion, a model using real-world electronic medical records (EMR) data can accurately predict future kidney functions and provide clinical decision support.
INTRODUCTION
The increasing incidence of chronic kidney disease (CKD) in the United States and around the world lays an enormous burden on healthcare [1, 2]. By December 2015, there were 703,243 prevalent patients with End Stage Renal Disease (ESRD), with the unadjusted incident rate of 378 per million population [3]. In 2017, there were approximately 500,000 patients on different dialysis modalities (91% are on hemodialysis), 20,000 received transplants [3]. Treatments that are effective in patients with advanced CKD also increase health care costs and lead to adverse effects [4]. Thus it is essential to identify earlier stage CKD and prevent its progression to ESRD [5]. However, the biggest challenge is that most people do not have any signs or symptoms in the early stages of CKD and go undetected until an advanced stage [6].
Early identification and targeted intervention of CKD have attracted considerable attention from clinicians and researchers since both have the potential to reduce the number of patients progressing to ESRD and lower the mortality rate related to CKD and associated healthcare costs [7]. With the growing availability of Electronic Medication Record (EMR) data, various computational predictive models for disease progression have been developed to facilitate the decision-making process of health care providers [4, 8, 9]. Choi et al. have classified disease progression models into two categories based on the extent of targeted diseases: models focusing on a specific disease and those focusing on a broader range of conditions [10]. Among those disease-specific progression models, some are validating specific hypotheses of disease progression based on experts’ knowledge [4, 11, 12], while others are driven by application of advanced statistical methods [13–15]. Approaches that can be generalized to model the progression of multiple diseases have been proposed, where statistical methods and machine learning techniques are widely used [16–18]. For kidney disease, different models have been developed in predicting CKD stages to ESRD over time and in predict variations of GFR in patients [7, 9, 19–22].
The application of statistical models and machine learning techniques have been rapidly-growing in estimating health and disease outcomes [23]. Cerqueira et al. developed a model using the Cox proportional hazard regression in predicting the risks that pre-dialysis pediatric patients progress to ESRD from CKD [9]. Decruyenaere et al. compared the performances of machine learning methods with logistic regression in predicting the occurrence of delayed renal graft in renal transplant patients [24]. Their results showed that linear support vector machine outperformed logistic regression in sensitivity. Kumar compared six machine learning classifiers (Random Forest, Sequential Minimal Optimization, NaiveBayes, Radial Basis Function, Multilayer Perceptron Classifier, and SimpleLogistic) in CKD classification and identified that Random forest outperformed the other classifiers [25].
Estimated glomerular filtration rates (eGFRs) have been implemented in primary care to assist the early detection and staging of CKD [26, 27]. The eGFR formula [28] is
| (Equation 1) |
Where eGFR (estimated glomerular filtration rate) = mL/min/1.73 m2; SCr (standardized serum creatinine) = mg/dL, κ = 0.7 (females) or 0.9 (males), α = −0.329 (females) or −0.411 (males), min = indicates the minimum of SCr/κ or 1, max = indicates the maximum of SCr/κ or 1, and age = years. Although routine reporting eGFR had positive effects in clinical practice, including prevention of CKD progression and reduction of CKD related complications, there are still concerns in its negative effects caused by over-diagnosis [26]. Other than reporting eGFR, studies have begun using an alternative measurement, such as eGFR decline derived from eGFR to evaluate and predict CKD progression [29, 30]. Studies have investigated the association between eGFR change and ESRD risk and mortality risk respectively, where age and gender factors have been taken into account [29, 31, 32]. Higher eGFR decline levels were proved to be associated with greater hazard ratios of ESRD in several clinical trials [33–35]. However, a smaller percentage of eGFR change, which is a reflection of the shortterm treatment effect of kidney disease, is underexamined [29].
The future renal function of a CKD patient can be predicted if their GFR variations can be predicted since GFR is the best test in measuring the level of kidney function [3, 7, 36]. Consequently, the time to reach GFR thresholds corresponding to stages of CKD can be anticipated. An integrated intelligent fuzzy expert system has been used in predicting future GFR based on selected clinical variables and demonstrated reliable accuracy [7]. However, there is still a lack of efficient methods for predicting the individual level timeframe of CKD progression [37]. Specifically, Random Forest Regression, featured with a reduction in overfitting and less variance, has not been used to predict the progression of renal function yet. This study predicted future eGFR values using Random Forest regression based on real-world EMR data representing the general population in the upper Midwest. The main aim of this study is to propose an efficient and reliable clinical tool that allows us to identify at an earlier stage and preemptively suggest the preventive strategies that can attenuate the development of this challenging disease in patients that reside in our agricultural communities.
METHODS
Data acquisition
The dataset used in this study comes from real-world clinical data. We built up a cohort consisting of 120,495 patients aged from 20 to 80 in Sioux Falls, SD, region that receiving primary care from Sanford Health. By consulting with the nephrologist, we pulled out data elements influencing GFR variations for this cohort from the comprehensive Sanford EMR database for years 2009–2017. None of the identifiable information was extracted to protect patients’ privacy. We are focusing on the progression of CKD, so only the “clinical” encounter data was included. Those data elements contain patients’ eGFR records for years 2009–2017, the ICD-10 codes [38] for CKD, Hypertension, Diabetes, and Obesity, and their demographic information comprising Age, Gender, Race. A detailed description of the data elements is given in Table 1.
Table 1.
Predictor and covariate data type breakdown.
| Feature | Data elements |
|---|---|
| Predictor | |
| eGFR | All clinical encounter eGFR data with testing dates were pulled out for each patient |
| Covariates | |
| Age | Continuous |
| Gender | Categorical |
| Race/Ethnicity | Categorical |
| BMI | Continuous |
| Hypertension | Flagged for each patient (ICD-10: I10, I11, I12, I13, I15, I16) |
| Diabetes | Flagged for each patient (ICD-10: E08, E09, E10, E11, E13) |
| Obesity | Flagged for each patient (ICD-10: E66.9) |
Data pre-processing
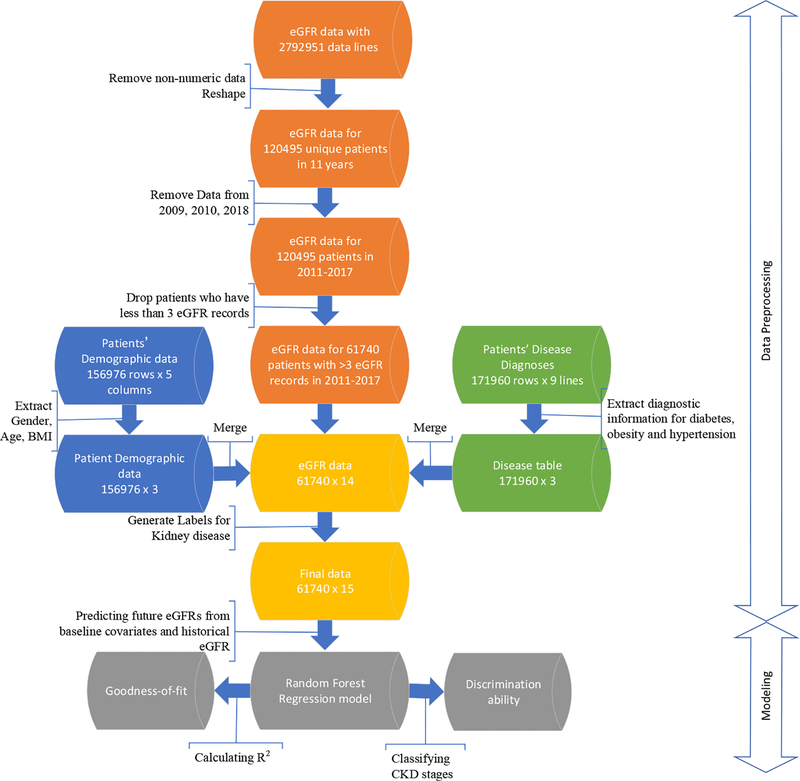
The extracted data were formatted into three separate tables: (1) eGFR table with rows representing patients and columns containing eGFR for multiple years; (2) Demographic table consisting of demographic information; and (3) Disease table composed of diagnosis status of hypertension, diabetes, and obesity. The processing of these data tables is illustrated in Figure 1 and described below.
Figure 1:
Workflow of the data preprocessing, including initial eGFR data, demographic and disease information, and data merging and filtering. This process resulted in 61740 samples with 15 variables each.
The eGFR table has 120,495 unique patients and 10 columns, each of which representing eGFR records in years 2009–2018. First, the non-numeric eGFR records (e.g. “>90”)) were considered as missing data and marked as “NA.” For patients with more than one eGFR values in a specific year, the median of these values was calculated and kept for that year in the table. The following table shows the numbers and percentages of missing records for each column.
As shown in Table 2, more than 95% eGFR records are missing in 2009 and 2010, so data from these two years were omitted. Since the data in 2018 was not complete when the data was extracted, we also excluded the records in this year. Patient lines were removed from the data if they have no more than three available records from 2011 to 2017. The final eGFR table has 61740 unique patients and 7 years eGFR data for each patient with at least three eGFR values.
Next, the different CKD stages were determined by eGFR values in the physical laboratory. Therefore, the CKD stages true labels were created using eGFR. The minimum eGFR value in each of the years between 2011 and 2017 was evaluated first, and then the CKD stages labels were produced based on the following equation:
Table 2.
Percentages of missing eGFR records for years from 2009 to 2018.
| Year | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 |
|---|---|---|---|---|---|---|---|---|---|---|
| Missing Data |
1204 91 |
1148 02 |
8119 0 |
7056 5 |
6891 8 |
6207 9 |
6113 4 |
6078 1 |
6058 7 |
8265 4 |
| Missing Data Percent age |
99.99 % |
95.28 % |
67.38 % |
58.56 % |
57.20 % |
51.52 % |
50.74 % |
50.44 % |
50.28 % |
68.60 % |
| (Equation 2) |
The true labels were also merged into eGFR matrix based on their index (patient ID).
The current eGFR matrix includes 61740 unique patients, and each patient has 7 years eGFR values from 2011 to 2017 and labels for the CKD stage from 1 to 5. The final data table was created by merging the eGFR table with the demographic table and the disease table by matching their patient IDs.
Data summary
A total of 61740 patients with different CKD stages were included in the data analysis. Summary statistics of the final data table were given in Table 3. The demographic characteristics and the disease diagnoses represented the patients’ status at the time the data was extracted from the EMR system. Mean and standard deviation were calculated for continuous variables, and count and proportion were given for discrete variables. Of those 61740 patients, there are only 6 patients have CKD-1 which indicated that the data lines for patients without CKD or with CKD-1 were removed in the data preprocessing steps. 78.7% and 19% of the cohort have CKD-2 and CKD-3 respectively, and patients with CKD-4 and CKD-5 were only 2.36%. We ignored the group of CKD-1 in the following description of this cohort since there were too few patients to draw a conclusion from. The mean age was 57 (SD=12.77) for the whole cohort with variations among various CKD groups. There were more female patients than male patients in CKD 1–4 while there were more CKD-5 cases in male patients. The majority of this cohort were white patients (99.9%). The rates of hypertension and diabetes increased as the kidney disease was getting worse. The obesity rate was relatively consistent among those CKD groups.
Table 3.
Descriptive statistics of the patient cohort with a breakdown of counts and percentages falling within each stage of chronic kidney disease.
| Total | CKD-1 | CKD-2 | CKD-3 | CKD-4 | CKD-5 | |
|---|---|---|---|---|---|---|
| Number | 61740 | 6 (<0.01%) |
48568 (78.7%) |
11769 (19%) |
909 (1.47%) |
488 (0.79%) |
| Age (Years) | 57.27 ±12.77 |
50.66 ±6.8 |
55.34 ±12.5 |
64.62 ±11 |
64.44 ±12.6 |
58.38 ±13.1 |
| Gender, M | 26843 (43.5%) |
4 (66.7%) |
21708 (44.7%) |
4466 (37.9%) |
409 (45%) |
256 (52.4%) |
| Gender, F | 34897 (56.5%) |
2 (33.3%) |
26860 (55.3%) |
7303 (62.1%) |
500 (55%) |
232 (47.6%) |
| Race, white | 61685 (99.9%) |
6 (100%) |
48535 (99.9%) |
11751 (99.8%) |
906 (99.7%) |
487 (99.8%) |
| Race, non-white | 55 (0.01%) |
0 (0%) |
33 (0.01%) |
18 (0.2%) |
3 (0.3%) |
1 (0.02%) |
| Hypertension | 32086 (52%) |
4 (66.7%) |
22675 (46.7%) |
8241 (70%) |
742 (81.6%) |
424 (86.9%) |
| Diabetes | 12763 (20.7%) |
0 (0%) |
8232 (16.9%) |
3744 (31.8%) |
485 (53.4%) |
302 (61.9%) |
| Obesity | 12654 (20.5%) |
1 (16.7%) |
9754 (20.1%) |
2583 (21.9%) |
217 (23.9%) |
99 (20.3%) |
Overall this patient cohort represented the patient population with kidney damages instead of representing the population from primary care; thus we used it to develop a predictive model in estimating the patient’s future eGFR value.
Construction of Random Forest Regression model
The longitudinal design of this study enables the estimation the future eGFR value from the past eGFR values adjusted by clinical covariates. We selected Random Forest regression as the primary model because of its efficiency and accuracy to predict 1 year, 2 years and 3 years eGFRs from the historical eGFR records between years 2011 −2014.
Baseline covariates and predictors:
The variables included in the analysis were baseline eGFR, age, gender, ethnicity, body mass index (BMI), hypertension, diabetes, obesity.
Outcome:
eGFR values in the year 2015, 2016, and 2017 were considered as the outcome variable. This is based on the consensus that GFR is the best measure of kidney function [36].
Model development:
the inputs of this model are the attributes of the ith patient denoted by a vector Xi = (xi1,…, xin) which includes eGFR values from multiple years and other covariates listed in Table 1. The output is the future eGFR for the ith patient denoted by Gij where j indicating a future year.
In the computational experiment, we used the processed dataset with 61740 unique patients. For building the model in predicting eGFR of 2015, the patient must have recorded eGFR in 2015, and at least two recorded eGFR between 2011 to 2014. Similar requirements were used in predicting eGFR of 2016 and 2017. Other years’ eGFR values were imputed and filled by the median eGFR value of each patient. All models were built using scikit-learn package [39]. The parameters of Random Forest Regressor were determined using the grid-search method. Only two parameters, number of estimators and maximum number of features, were tuned because they can determine numbers of trees in forest and how the tree will split and grow. We also randomly split the dataset and repeat the training process five times with different sets to avoid over-fitting for our models.
Assessment of model performance
Goodness-of-fit
The model fit of the proposed Random Forest Regression was measured using the coefficient of determination R2 to show how well the fitted eGFR value approximates the real eGFR value. R2 is a measure used to represent the percent of variation explained,i.e., the proportion of variance in the dependent variable that can directly be attributed to variance in the independent variables. An R2 of 1 would indicate all changes we see in the dependent variable are caused by changing our independent variables, whereas an R2 of 0 means no such direct impact. We also checked the residual plot since randomly distributed residuals indicate the model fits the data well.
Discrimination
The estimated eGFR values were used to classify patients into different CKD stages based on Equation 1. Both micro-average and macro-average were generated to illustrate the classification accuracy of the Random Forest model.
RESULTS
eGFR prediction with Random Forest regression
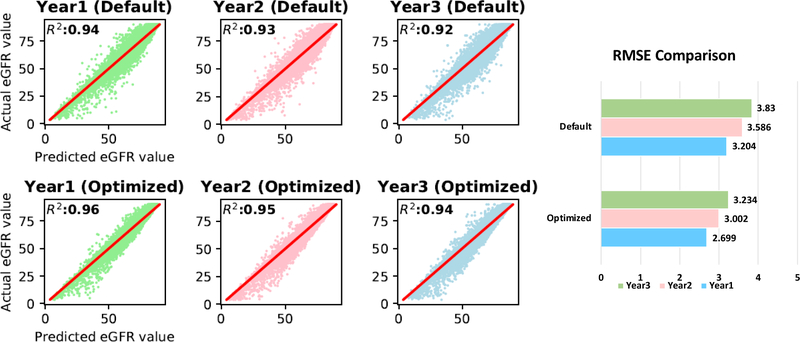
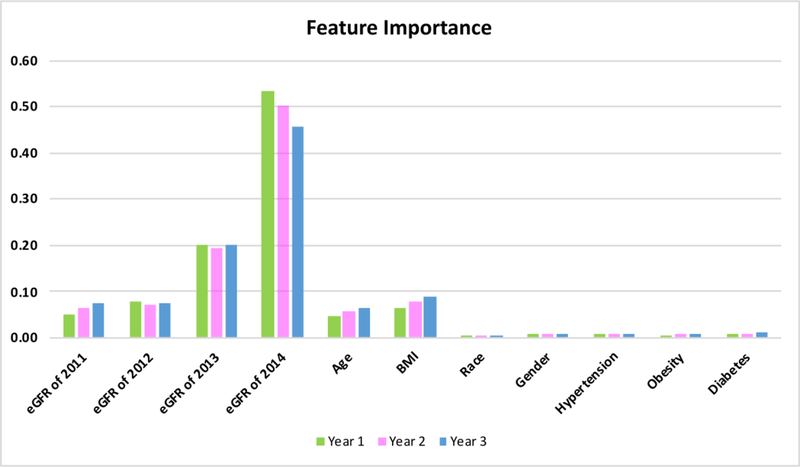
In Random Forest regression analysis, the predicting accuracy was enhanced by optimizing the values of hyperparameters, where the default values and the optimized values of the hyperparameters were shown in Table 4. The predicted versus observed eGFR values in years 1–3 were plotted for both the default and optimized hyperparameters in Figure 2. The R2 was increased from default to optimized hyperparameters in each of the three years. The Root of Mean Squared Error (RMSE) in Figure 2 illustrated that the optimized hyperparameters provided a more accurate prediction that the default values. It is also worse noticing that the prediction accuracy decreased over time. With the optimal parameters, we further examined the importance of the features included in the analysis whose results were given in Figure 3. It is not surprising that previous eGFR records played essential roles than other features since eGFR is decreasing continuously over time. Although the information of age and BMI are considered in estimating GFR using the eGFR formula [28], predictions based solely on the previous eGFR are not sufficient. Age and BMI, as illustrated in Figure 3, still contribute to 4.7%~9% to the future three years of eGFR respectively (Supplement Table S1). All the other features, including Race, Gender, Obesity, Hypertension, and Diabetes, accounted for a total of 2.7%~3.9% of the variances (Supplement Table S1).
Table 4:
Hyperparameters used in the Random Forest Regression for the default and optimized models.
| Default | Optimized | |
|---|---|---|
| # of trees | 10 | 100 |
| Max depth | None | None |
| Max sample split | 2 | 2 |
| Min samples leaf | 1 | 1 |
| Max features | 11 | 8 |
| Bootstrap | True | True |
Figure 2:
Goodness of fit based on R2of the Random Forest Regression model in predicting eGFR in year 1 to year 3 for the default and optimized models. RMSE comparison for each year is also provided for the default and optimized models.
Figure 3:
Feature importance in predicting eGFR values in years 1–3 using optimized parameter values in Random Forest Regression. eGFR values from the prior year are the most impactful feature on the future prediction, as expected. Age and BMI also contribute significantly to the future prediction, while other demographic and disease information was much less impactful.
Performances of the predicted eGFR in disease classification
The predicted CKD stage labels were generated from the estimated eGFR values using Equation 1. Then the confusion matrix of a tabular summary of the actual CKD status versus the predicted CKD status was created for each of the future three years and was shown in Table S2 of the supplementary file. The confusion matrices were further assessed by both per class metrics and metrics for multi-class classification to demonstrate the classification accuracy of the Random Forest prediction model. In per class metrics, Precision, Recall, and F1 were taken into account to assess the performance respecting each class [40]. Macro-averaged and micro-averaged metrics were adopted to evaluate the multi-way classification. In Table 5, we showcased these metrics for all three years. In Year 1, CKD-2 and CKD-3 were accurately classified using the estimated eGFR with all three metrics greater than 0.9, while the classification accuracy of CKD-4 and CKD-5 decreased to around 0.8. In Year 2 and 3, the per class metrics demonstrated similar patterns as in Year 1. The multi-class classifications performed well in all three years. Besides, there was no apparent temporal pattern of the prediction accuracy.
Table 5:
Evaluation metrics of CKD classifications based on predicted eGFR. Precision, Recall, and F1-score for each class for each year was used to show performance. Additionally, macro- and micro-averaged metrics were used for each year to show overall performance, regardless of the categorization. CKD-1 was left out due to no methods predicting a CKD-1 result for any year for any patients.
| Per-class accuracy | Macro-averaged metrics | Micro-averaged metrics |
||||||
|---|---|---|---|---|---|---|---|---|
| Precisio n |
Reca ll |
F1 | Macro precision |
Macro recall |
MacroF 1 |
|||
| Year 1 | CKD- 2 |
0.980 | 1.000 | 0.99 0 |
||||
| CKD- 3 |
0.983 | 0.918 | 0.95 0 |
|||||
| CKD- 4 |
0.901 | 0.787 | 0.84 0 |
|||||
| CKD- 5 |
1.000 | 0.801 | 0.88 9 |
|||||
| Year 2 | CKD- 2 |
0.978 | 1.000 | 0.98 9 |
||||
| CKD- 3 |
0.983 | 0.923 | 0.95 2 |
|||||
| CKD- 4 |
0.913 | 0.770 | 0.83 5 |
|||||
| CKD- 5 |
1.000 | 0.835 | 0.91 0 |
|||||
| Year 3 | CKD- 2 |
0.980 | 1.000 | 0.99 0 |
||||
| CKD- 3 |
0.985 | 0.933 | 0.95 9 |
|||||
| CKD- 4 |
0.892 | 0.814 | 0.85 1 |
|||||
| CKD- 5 |
1.000 | 0.782 | 0.87 8 |
|||||
DISCUSSION
In this study, we proposed a model in predicting future eGFR values, which is based on Random Forest regression that can efficiently learn from the real world EMR data and accurately predict future patient outcomes. We validated this model on an EMR dataset extracted from a health system located in the Great Plains. The computational experiment achieved an average R2 of 0.95 over three years with small variation. And an 88% Macro Recall and a 96% Macro Precision by averaging over three years were obtained by dividing patients into different CKD stages using estimated eGFRs. Besides, we identified the crucial features that contribute to the variation of future eGFRs, which include recent eGFR records, Age and BMI. Therefore, our proposed predictive model of eGFR has excellent potential to be developed into a clinical decision support tool to assist doctors in providing preventive advice to patients.
One of the limitations of this work is that only patients with numeric eGFR records were included, which exclude those patients without CKD symptoms in the study period. However, those excluded patients can serve as a control group whose clinical information can be incorporated into the predictive model to adjust the parameter estimations. Also, the current study only contained historical eGFRs, demographic characteristics, and relevant disease diagnoses. Studies have shown that an individual’s genetic and phenotypic characteristics both affect their risk in developing kidney disease, including genetic mutations, a family history, gender, ethnicity, age, obesity, socioeconomic status, smoking, nephrotoxins, acute kidney injury, diabetes mellitus, and hypertension [41]. Thus we are planning to address those issues in future studies to improve the practicability of the predictive model of eGFR in support of patient care.
Supplementary Material
HIGHLIGHTS.
Future kidney function can be predicted from Electronic Medical Records
Historical estimated glomerular filtration rate, demographic features, and relative disease contribute to the prediction of kidney function
Predicted estimated glomerular filtration rate accurately classify patients into chronic kidney disease stages
FUNDING
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 5P20GM121341.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Coresh J, et al. , Prevalence of chronic kidney disease in the United States. JAMA, 2007. 298(17): p. 2038–47. [DOI] [PubMed] [Google Scholar]
- 2.Webster AC, et al. , Chronic kidney disease. The Lancet, 2017. 389(10075): p. 1238–1252. [DOI] [PubMed] [Google Scholar]
- 3.System USRD, USRDS annual data report: Epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. 2017. [Google Scholar]
- 4.Tangri N, et al. , A predictive model for progression of chronic kidney disease to kidney failure. Jama, 2011. 305(15): p. 1553–1559. [DOI] [PubMed] [Google Scholar]
- 5.Locatelli F, Vecchio LD, and Pozzoni P, The importance of early detection of chronic kidney disease. Nephrology Dialysis Transplantation, 2002. 17(11): p. 2–7. [DOI] [PubMed] [Google Scholar]
- 6.Prevention C f.D.C.a. Age-adjusted prevalence of CKD Stages 1–4 by Gender 1999–2012. [cited 2016 December 6]; Available from: https://nccd.cdc.gov.
- 7.Norouzi J, et al. , Predicting renal failure progression in chronic kidney disease using integrated intelligent fuzzy expert system. Computational and mathematical methods in medicine, 2016. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taal M and Brenner B, Predicting initiation and progression of chronic kidney disease: developing renal risk scores. Kidney international, 2006. 70(10): p. 1694–1705. [DOI] [PubMed] [Google Scholar]
- 9.Cerqueira DC, et al. , A predictive model of progression of CKD to ESRD in a predialysis pediatric interdisciplinary program. Clinical Journal of the American Society of Nephrology, 2014: p. CJN. 06630613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi E, et al. Doctor ai: Predicting clinical events via recurrent neural networks. in Machine Learning for Healthcare Conference 2016. [PMC free article] [PubMed] [Google Scholar]
- 11.De Winter W, et al. , A mechanism-based disease progression model for comparison of longterm effects of pioglitazone, metformin and gliclazide on disease processes underlying type 2 diabetes mellitus. Journal of pharmacokinetics and pharmacodynamics, 2006. 33(3): p. 313–343. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, et al. , Disease progression meta-analysis model in Alzheimer’s disease. Alzheimer’s & Dementia, 2010. 6(1): p. 39–53. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y-Y, et al. Longitudinal modeling of glaucoma progression using 2-dimensional continuoustime hidden markov model in International Conference on Medical Image Computing and Computer-Assisted Intervention. 2013. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson CH, et al. , Multistate Markov models for disease progression with classification error. Journal of the Royal Statistical Society: Series D (The Statistician), 2003. 52(2): p. 193–209. [Google Scholar]
- 15.Zhou J, et al. Modeling disease progression via fused sparse group lasso. in Proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining 2012. ACM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Sontag D, and Wang F. Unsupervised learning of disease progression models. in Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining 2014. ACM. [Google Scholar]
- 17.Choi E, et al. Constructing disease network and temporal progression model via context-sensitive hawkes process. in Data Mining (ICDM), 2015 IEEE International Conference on 2015. IEEE. [Google Scholar]
- 18.Ranganath R, et al. The Survival Filter: Joint Survival Analysis with a Latent Time Series. in UAI. 2015. [Google Scholar]
- 19.Obrador GT, et al. , Establishing the global kidney disease prevention network (KDPN): a position statement from the National Kidney Foundation. American Journal of Kidney Diseases, 2011. 57(3): p. 361–370. [DOI] [PubMed] [Google Scholar]
- 20.Rucci P, et al. , A clinical stratification tool for chronic kidney disease progression rate based on classification tree analysis. Nephrology Dialysis Transplantation, 2013. 29(3): p. 603–610. [DOI] [PubMed] [Google Scholar]
- 21.Stevens LA, et al. , Assessing kidney function—measured and estimated glomerular filtration rate. New England Journal of Medicine, 2006. 354(23): p. 2473–2483. [DOI] [PubMed] [Google Scholar]
- 22.Gaspari F, et al. , Performance of different prediction equations for estimating renal function in kidney transplantation. American Journal of Transplantation, 2004. 4(11): p. 1826–1835. [DOI] [PubMed] [Google Scholar]
- 23.Wainberg M, et al. , Deep learning in biomedicine. Nature biotechnology, 2018. 36(9): p. 829. [DOI] [PubMed] [Google Scholar]
- 24.Decruyenaere A, et al. , Prediction of delayed graft function after kidney transplantation: comparison between logistic regression and machine learning methods. BMC medical informatics and decision making, 2015. 15(1): p. 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar M, Prediction of chronic kidney disease using random forest machine learning algorithm. International Journal of Computer Science and Mobile Computing, 2016. 5(2): p. 24–33. [Google Scholar]
- 26.Levin A and Stevens PE, Early detection of CKD: the benefits, limitations and effects on prognosis. Nature Reviews Nephrology, 2011. 7(8): p. 446. [DOI] [PubMed] [Google Scholar]
- 27.Levey AS, Becker C, and Inker LA, Glomerular Filtration Rate and Albuminuria for Detection and Staging of Acute and Chronic Kidney Disease in Adults: A Systematic Review. Jama, 2015. 313(8): p. 837–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham AG, et al. , Longitudinal formulas to estimate GFR in children with CKD. Clinical Journal of the American Society of Nephrology, 2009. 4(11): p. 1724–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coresh J, et al. , Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. Jama, 2014. 311(24): p. 2518–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins RM, et al. , GFR decline and mortality risk among patients with chronic kidney disease. Clinical Journal of the American Society of Nephrology, 2011: p. CJN. 00470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallan SI, et al. , Age and association of kidney measures with mortality and end-stage renal disease. Jama, 2012. 308(22): p. 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitsch D, et al. , Associations of estimated glomerular filtration rate and albuminuria with mortality and renal failure by sex: a meta-analysis. Bmj, 2013. 346: p. f324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heerspink HJL, et al. , Estimated GFR decline as a surrogate end point for kidney failure: a post hoc analysis from the Reduction of End Points in Non-Insulin-Dependent Diabetes With the Angiotensin II Antagonist Losartan (RENAAL) study and Irbesartan Diabetic Nephropathy Trial (IDNT). American Journal of Kidney Diseases, 2014. 63(2): p. 244–250. [DOI] [PubMed] [Google Scholar]
- 34.Inker LA, et al. , GFR decline as an alternative end point to kidney failure in clinical trials: a meta-analysis of treatment effects from 37 randomized trials. American Journal of Kidney Diseases, 2014. 64(6): p. 848–859. [DOI] [PubMed] [Google Scholar]
- 35.Greene T, Teng C, and Ying J, Validity and statistical power of alternative eGFR-based endpoints: A report from an NKF FDA Workshop. J Am Soc Nephrol, 2013. 24: p. 151A. [Google Scholar]
- 36.Baumgarten M and Gehr T, Chronic kidney disease: detection and evaluation. American family physician, 2011. 84(10): p. 1138. [PubMed] [Google Scholar]
- 37.Yadollahpour A, Applications of expert systems in management of chronic kidney disease: a review of predicting techniques. Oriental Journal of Computer Science and Technology, 2014. 7(2): p. 306–315. [Google Scholar]
- 38.Organization, W.H., ICD-10: International statistical classification of diseases and health-related problems. Geneva: WHO, 1992. [Google Scholar]
- 39.Pedregosa F, et al. , Scikit-learn: Machine learning in Python. Journal of machine learning research, 2011. 12(Oct): p. 2825–2830. [Google Scholar]
- 40.Du XQ, et al. , DeepPPI: boosting prediction of protein-protein interactions with deep neural networks. Journal of Chemical Information and Modeling, 2017. 57(6): p. 1499–1510. [DOI] [PubMed] [Google Scholar]
- 41.Kazancioglu R, Risk factors for chronic kidney disease: an update. Kidney international supplements, 2013. 3(4): p. 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.