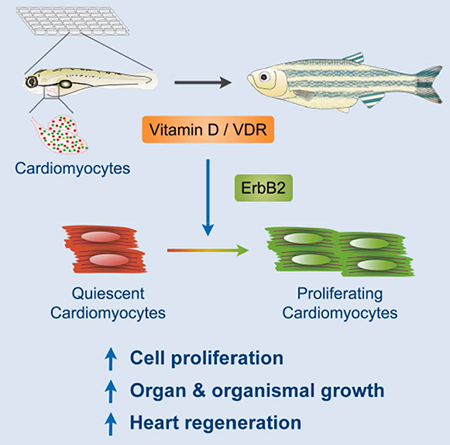
SUMMARY
Attaining proper organ size during development and regeneration hinges on activity of mitogenic factors. Here, we performed a large-scale chemical screen in embryonic zebrafish to identify cardiomyocyte mitogens. Although commonly considered antiproliferative, vitamin D analogues like alfacalcidol had rapid, potent mitogenic effects on embryonic and adult cardiomyocytes in vivo. Moreover, pharmacologic or genetic manipulation of vitamin D signaling controlled proliferation in multiple adult cell types and dictated growth rates in embryonic and juvenile zebrafish. Tissue-specific modulation of vitamin D receptor (VDR) signaling had organ-restricted effects, with cardiac VDR activation causing cardiomegaly. Alfacalcidol enhanced the regenerative response of injured zebrafish hearts, whereas VDR blockade inhibited regeneration. Alfacalcidol activated cardiac expression of genes associated with ErbB2 signaling, while ErbB2 inhibition blunted its effects on cell proliferation. Our findings identify vitamin D as mitogenic for cardiomyocytes and other cell types in zebrafish and indicate a mechanism to regulate organ size and regeneration.
Graphical Abstract
eTOC Blurb
By chemical screening, Han et al. find that the nutrient vitamin D promotes cardiomyocyte proliferation during tissue growth, homeostasis, and injury-induced regeneration in zebrafish, requiring intact ErbB2 signaling for its effects. They also that vitamin D has broad and potent mitogenic effects on a variety of cell types and stages.
INTRODUCTION
Adult mammalian cardiomyocytes (CMs) can renew at a limited rate (Bergmann et al., 2009; Bergmann et al., 2015), yet there is minimal regeneration of lost CMs after myocardial infarction (MI). Adult zebrafish and neonatal mice or swine can regenerate heart muscle lost to severe trauma, through dedifferentiation and proliferation of spared cardiomyocytes (CMs) (Jopling et al., 2010; Kikuchi et al., 2010; Porrello et al., 2011; Poss et al., 2002; Ye et al., 2018; Zhu et al., 2018). Recent reports indicate that forced expression of Cyclins and/or CDKs can activate spared CMs to re-enter cell cycle and improve heart function after injury (Hassink et al., 2008; Mohamed et al., 2018). Additional genetic factors have been implicated in promoting CM proliferation in various contexts, including Neuregulin1/ErbB2 (Bersell et al., 2009; D’Uva et al., 2015; Gemberling et al., 2015), and YAP/TAZ transcription factors, which are normally restrained by Hippo in CMs (Heallen et al., 2013; von Gise et al., 2012; Xin et al., 2013). Discovery of new influences that regulate CM proliferation can illuminate how and why heart regeneration occurs, and how to trigger cardiogenesis effectively after MI.
Chemical screening is a powerful technique to discover biological regulators. The zebrafish has been widely used for high-throughput chemical screening, owing to its small size, transparency, high fecundity and rapid development. To monitor CM proliferation in live zebrafish embryos, we recently generated fluorescent ubiquitin-based cell cycle indicator (FUCCI) dual transgenes under control of the zebrafish cmlc2 promoter (cmlc2:FUCCI) (Choi et al., 2013). In this system, Venus-hGeminin fusion protein accumulates at S/G2/M phases in CMs and degrades quickly afterwards, whereas mCherry-zCdt1 fusion protein is degraded at S/G2/M and accumulates during G1/G0 (Sakaue-Sawano et al., 2008). Thus, by direct visual inspection in live zebrafish, one can readily track all or most proliferating CMs and the effects of experimental manipulations on cardiogenesis.
Here, we employed the FUCCI detection system in a large-scale chemical screen for compounds that impact CM proliferation in zebrafish embryos. Unexpectedly, we found that vitamin D analogues, commonly considered to be anti-proliferative influences in mammalian cells, are potent cardiogenic factors in both embryonic and adult zebrafish. We show that increased vitamin D signaling enhances cell proliferation in many cell types and contexts, including cardiac regeneration. Manipulation of vitamin D signaling in growing zebrafish has major effects on animal size, and locally dysregulating the pathway in specific cell types can reduce or increase cardiac size. Transcriptome profiling and pharmacological perturbation implicated ErbB2 signaling as a key growth network regulated by vitamin D. Our results reveal vitamin D signaling as a profound mitogenic influence in a vertebrate model system.
RESULTS
Chemical Screening Identifies Vitamin D Analogues as Activators of Embryonic Cardiomyocyte Proliferation
To discover compounds that regulate CM proliferation in zebrafish, we screened the Prestwick Chemical Library for effects on the number of Venus-hGeminin+; mCherry-zCdt1− (FUCCI+) CMs in cmlc2:FUCCI embryos after a 24-hour treatment starting at 3 days post fertilization (dpf; Figure 1A). We found that 104 and 26 compounds from a total of 1200 tested drugs increased or decreased the number of FUCCI+ CMs, respectively (Figure S1A). Modulator classes include neurotransmission, steroid hormone effectors, antimicrobial influences, and ion channel regulators (Figure S1B; Table S1). Two out of the 18 hormone effectors, alfacalcidol (Alfa) and calcipotriene, are vitamin D analogues.
Figure 1. Vitamin D Analogues Promote Cardiomyocyte Proliferation in Zebrafish.
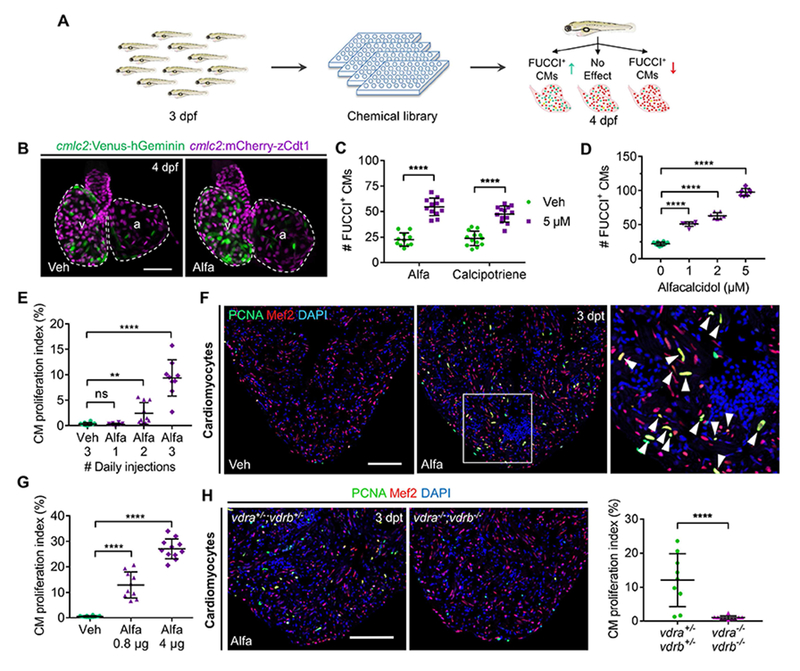
(A) Cartoon schematic of in vivo chemical screen for regulators of CM proliferation using cmlc2:FUCCI zebrafish embryos.
(B) Maximum intensity projection (MIP) images of dissected cmlc2:FUCCI hearts. Veh, vehicle; v, ventricle; a, atrium. Scale bar, 50 μm.
(C) Quantification of FUCCI+ CMs after 24-h treatment. n = 11-13.
(D) Quantification of FUCCI+ CMs after 24-h treatment. n = 7-10.
(E) Quantification of adult ventricular CM proliferation indices. n = 9-10.
(F) Representative images of PCNA/Mef2 staining quantified in (E). Boxed area is enlarged on the right. Arrowheads indicate PCNA+ CMs. Scale bar, 100 μm.
(G) Ventricular CM proliferation indices of vehicle- or Alfa-injected adult zebrafish. n = 10-12.
(H) Representative images and quantification of ventricular PCNA/Mef2 staining. n = 9-10. Scale bar, 100 μm.
Vitamin D, widely studied for over 80 years, is a steroid pro-hormone that can be hydroxylated sequentially in the liver and kidney to yield the active hormone, ercalcitriol (D2) or calcitriol (D3), in vertebrates (Bikle, 2014). Alfa is a vitamin D analogue that can be hydroxylated to form calcitriol directly in the liver (Ringe and Schacht, 2007). The active hormones ercalcitriol and calcitriol can bind to vitamin D receptors (VDRs) and regulate downstream gene expression in target cells (Bikle, 2014). Notably, many studies have reported vitamin D analogues to have anti-proliferative effects in mammalian cells (Ma et al., 2016; Samuel and Sitrin, 2008), including cultured CMs (Hlaing et al., 2014; Nibbelink et al., 2007; O’Connell et al., 1997). In our screen, treatment with Alfa elevated the number of FUCCI+ CMs in live embryonic zebrafish by 141%, the largest increase among library compounds. Similarly, calcipotriene also increased FUCCI+ CMs by 100% (Figures 1B and 1C). In addition, both Alfa and calcitriol had dose-dependent effects on the number of FUCCI+ CMs (Figures 1D and S1C). By contrast, treatment with the VDR-specific inhibitor PS121912 (Sidhu et al., 2014) caused a 39% reduction in the number of FUCCI+ CMs (Figure S1C). These findings indicate that vitamin D is a potent activator of CM proliferation in embryonic zebrafish.
Vitamin D Signaling Stimulates Cell Cycle Re-entry in Adult Cardiomyocytes and Other Cell Types
Adult zebrafish hearts are similar to adult mammalian hearts in that the majority of CMs are quiescent under normal conditions (Wills et al., 2008). To determine the potency of vitamin D in mature cardiac tissue, we injected Alfa intraperitoneally in adult zebrafish. One day after Alfa injection, CMs remained quiescent, with no significant difference in the percentage of CMs positive for the proliferation marker PCNA compared to vehicle-injected fish (Figure 1E). After two daily Alfa injections, the CM proliferation index increased moderately in 4 of 10 animals (Figure 1E). By contrast, the CM proliferation indices of all animals rose in a dose-dependent manner after 3 daily Alfa injections, to an average index of 27% at the highest dose, a 50.9-fold increase over vehicle-injected controls (an average index of 0.52%) (Figures 1F and 1G). We found no evidence of enhanced apoptosis upon Alfa treatment that might indirectly lead to CM proliferation in adult hearts (data not shown).
To interrogate broader effects of vitamin D, we next asked whether Alfa injections increased indicators of proliferation in several additional adult zebrafish cell types. We found that Alfa boosted proliferation indices of non-muscular epicardial and endocardial cells of the heart by 26.6-fold and 4.7-fold, respectively (Figures S1D and S1E). In addition, Alfa enhanced the proliferation of adult liver cells, including hepatocytes by 9.0-fold and biliary duct cells by 4.0-fold (Figures S1F and S1G). Alfa also stimulated proliferation of dermal osteoblasts and basal epithelial cells in fins, corneal epithelial cells, and retinal cells (Figure S1H-J). Retinal cells in the ciliary marginal zone (CMZ), a dense region of progenitor cells, were more responsive to Alfa than mature cells (Figure S1H-J). Interestingly, a recent study reported that vitamin D signaling can promote hematopoietic progenitor cell proliferation in zebrafish (Cortes et al., 2016). Together these findings indicate that vitamin D signaling is mitogenic for a wide spectrum of adult zebrafish cells.
To determine how cell cycle progression is affected, we imaged tcf21:FUCCI cardiac tissue explants treated with vehicle or calcitriol and cultured ex vivo for long time periods (Figure S1K; Video S1) (Cao et al., 2017). Compared to vehicle controls, the number of tcf21-expressing epicardial cells bypassing the G1 checkpoint and entering S phase (characterized by a red (false-colored as magenta) to green conversion; Figure S1L) was increased by 79% on average over a 69-hour period during continual calcitriol treatment (Figure S1N). The number of cells traversing the G2 checkpoint to M phase (characterized by breakdown of nuclear membrane and subsequent dispersal of the green signal; Figure S1M) was also increased, but only by 14% (Figure S1O). These data suggest that the principal effect of vitamin D treatment is surpassing the G1 checkpoint to initiate the cell cycle.
Vitamin D Signaling Rapidly Accelerates Organismal Growth
Our results indicated that many zebrafish tissues respond to vitamin D treatment. To visualize vitamin D signaling activity in live zebrafish, we knocked in a monomeric EGFP-polyA (mGFP) cassette at the cyp24a1 start codon. cyp24a1 encodes a mitochondrial cytochrome P450 enzyme that catabolizes calcitriol to inactivate vitamin D signaling and is a direct downstream target gene of vitamin D signaling (Bikle, 2014). cyp24a1mGFP embryos displayed conspicuous mGFP fluorescence in cardiac ventricles (Figure S2), indicating active vitamin D signaling during heart development. When incubated with Alfa, cyp24a1-driven mGFP expression was increased sharply throughout embryonic zebrafish tissues with the greatest enhancement in liver (Figure S2), validating this line as a reporter of embryonic vitamin D signaling levels.
To examine how vitamin D analogues affect organismal growth, we first tested if they increased embryonic cell proliferation and size. Indeed, Alfa treatments longer than 9 hours elevated the number of cells positive for the mitotic marker phospho-Histone H3 (Figure 2A) throughout the trunks of 3 dpf embryos. Strikingly, we found that the total lengths of Alfa- or calcipotriene-treated embryos were increased by ~3% after treatment from 3 to 4 dpf over vehicle controls, representing a 43% (Alfa) or 30% (calcipotriene) faster growth rate during the 24-hour treatment (Figure 2B). To test if vitamin D had effects on post-metamorphic growth, we treated 5 weeks-post-fertilization (wpf) juvenile zebrafish with Alfa for 15 or 30 days. We found that Alfa-treated animals were 10% longer after 30 days of treatment, having grown substantially (33%) more than vehicle-treated clutchmates (Figures 2C and 2D). These data demonstrate that exogenous vitamin D is sufficient to accelerate zebrafish growth and boost animal size.
Figure 2. Vitamin D Signaling Enhances Organismal Size.
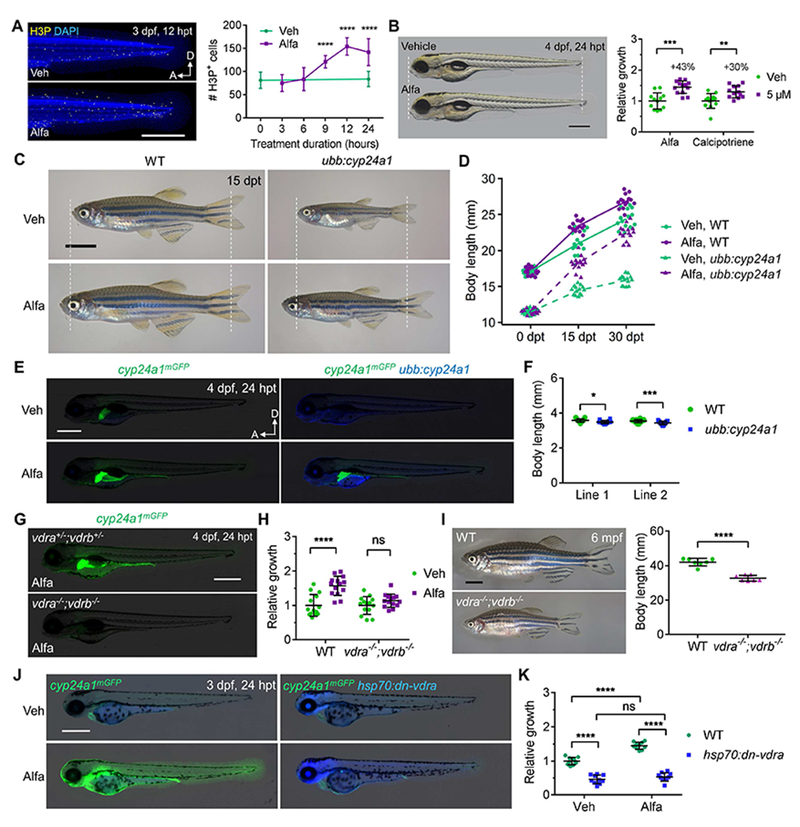
(A) MIP images and quantification of H3P staining of 3 dpf embryos. n = 11-13. Scale bar, 500 μm.
(B) Example and quantification of 4 dpf zebrafish growth. Scale bar, 500 μm. n = 12-14.
(C) Growth and rescue of 2 mpf juvenile WT and ubb:cyp24a1 zebrafish. Scale bar, 4 mm.
(D) Quantification of zebrafish length before and after treatment. n = 14-15.
(E) mGFP and TagBFP expression in 4 dpf embryos treated with vehicle or 10 nM Alfa. Scale bar, 500 μm.
(F) Quantified body length of two independent ubb:cyp24a1 lines and WT siblings at 4 dpf. n=14-23.
(G) cyp24a1mGFP expression in 1 μM Alfa-treated embryos.
(H) Quantification of relative growth of 4 dpf embryos treated with vehicle or Alfa for 24 hours. n = 13.
(I) Representative images and body length quantification of 6 mpf zebrafish. Scale bar, 5 mm. n = 7.
(J) mGFP and TagBFP expression of 3 dpf embryos after a single heat-shock and 24-h drug treatment. Scale bar, 500 μm.
(K) Quantified relative growth of 4 dpf embryos after a heat shock and 24-h drug treatment. n = 10.
See also Figure S2.
To examine how the size-enhancing effects of Alfa reflect endogenous roles, we disabled vitamin D signaling by multiple different ways. To deplete endogenous vitamin D metabolites, we generated transgenic ubb:cyp24a1 zebrafish that constitutively express cyp24a1 via the ubiquitin promoter (Mosimann et al., 2011) (Figure 2E). Total body lengths in two independent lines of ubb:cyp24a1 animals were ~3% shorter than those of wild-type siblings at 4 dpf (Figure 2F), and 33% shorter at 5 wpf (line 2; Figure 2D). A low (10 nM) dose of Alfa was sufficient to rescue cyp24a1mGFP expression and growth rate in ubb:cyp24a1 animals (Figures 2C-2E), indicating that the transgene product had negligible toxicity and could be saturated by substrate. To genetically disable endogenous VDRs, we generated zebrafish containing mutations in vdra and vdrb using CRISPR/Cas9-based gene editing (Varshney et al., 2015). Knockdown of zebrafish vdra and vdrb using morpholino oligonucleotides was reported to cause pericardial edema (Kwon, 2016), although we did not observe this phenotype in vdra−/−; vdrb−/− mutants. Nevertheless, homozygous mutations in both vdr genes abolished cyp24a1mGFP expression and rapid embryonic growth in response to Alfa treatment, indicating a genetic requirement for VDRs in this response (Figures 2G and 2H). Additionally, vdra−/−; vdrb−/− mutant zebrafish were 22% smaller in length than wild-type siblings at 6 months post fertilization (mpf) (Figure 2I). They also demonstrated no increase in the proliferative response of adult CMs after 3 daily Alfa injections, indicating a genetic requirement for VDRs in the mitogenic effects of Alfa (Figure 1H). To block VDR signaling in a conditional manner, we generated transgenic lines enabling heat shock (hsp70 promoter)-inducible expression of a dominant negative vdra (dn-vdra) cassette throughout the animal. Heat-shock treatment at 3 dpf inhibited growth in hsp70:dn-vdra embryos by 54% versus controls over the following 24 hours, and it eliminated the growth response to Alfa (Figures 2J and 2K). Collectively, these findings identify vitamin D signaling as an endogenous pathway that controls organ and organismal size in zebrafish.
Vitamin D Regulates Organ Size through Tissue-Intrinsic Signaling
As a circulating nutrient, vitamin D could influence cell proliferation directly by VDR signaling in the organs itself, and/or through a relay mechanism that relies on VDR signaling in other organs. To distinguish such mechanisms in CMs, we generated transgenic lines that express dn-vdra or constitutively active vdra (ca-vdra) via the cmlc2 promoter. When crossed to cyp24a1mGFP animals, cmlc2:dn-vdra zebrafish displayed little or no cardiac mGFP fluorescence and unchanged fluorescence elsewhere (Figures 3A and S3A), indicating specific inhibition of cardiac VDR signaling. By contrast, cmlc2:ca-vdra; cyp24a1mGFP animals had sharply elevated mGFP fluorescence in both cardiac chambers and unchanged mGFP fluorescence elsewhere (Figures 3B and S3B), indicating targeted enhancement of cardiac VDR signaling. To assess effects of these transgenes on CM proliferation, we examined them in the cmlc2:FUCCI or cmlc2:H2A-EGFP (labeling all CM nuclei) backgrounds. cmlc2:dn-vdra reduced the average number of FUCCI+ CMs by 63% at 4 dpf, yielding hearts with 31% fewer total CMs on average, whereas cmlc2:ca-vdra increased the number of FUCCI+ CMs by 171% and the total number of CMs by 74% (Figures 3C-3D and S3C-S3D). Notably, juvenile cmlc2:ca-vdra animals displayed overt cardiomegaly, with massive hearts dwarfing those of clutchmates by 1 mpf (Figures 3E-3F). Ventricular walls were dysmorphic and thickened (Figure S3E), and we cannot rule out the possibility of direct or indirect contributions through cell hypertrophy during these events. By contrast, assessment of heart-to-body size ratios in juvenile cmlc2:dn-vdra animals versus WT siblings indicated a 27% smaller ratio during VDR inhibition (Figure S3F). Thus, cardiac-intrinsic changes in VDR signaling are sufficient to regulate CM proliferation and heart size.
Figure 3. Tissue-Intrinsic Effects of Vitamin D Signaling and Impact on Heart Regeneration.
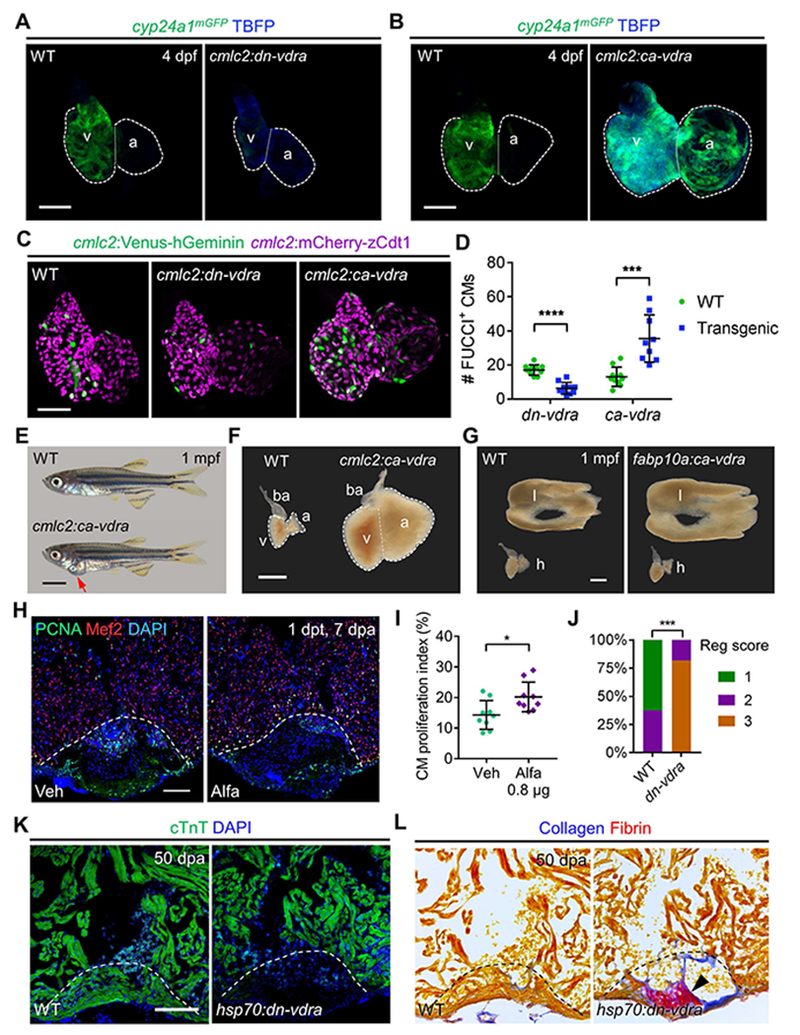
(A) MIP images of cyp24a1mGFP expression in dissected hearts. Scale bar, 50 μm.
(B) MIP images of cyp24a1mGFP expression in dissected hearts. Scale bar, 50 μm.
(C) MIP images of dissected cmlc2:FUCCI hearts in indicated transgenic backgrounds. Scale bar, 50 μm.
(D) Quantification of FUCCI+ CMs in (C). n=9-10.
(E) Examples of 1 mpf cmlc2:ca-vdra and WT zebrafish. Red arrow indicates gross pericardial enlargement caused by cardiomegaly in cmlc2:ca-vdra zebrafish. Scale bar, 2 mm.
(F) Darkfield images of dissected 1 mpf hearts. a, atrium; ba, bulbus arteriosus; v, ventricle. Scale bar, 500 μm.
(G) Darkfield images of dissected livers (l) and hearts (h) from 1 mpf zebrafish. Scale bar, 500 μm.
(H) Examples of Mef2/PCNA staining of 7 dpa hearts after a single injection. Dashed lines represent amputation planes. Scale bar, 100 μm.
(I) Quantification of CM proliferation indices in (H). n = 9.
(J) Quantified heart regeneration scores of WT or hsp70:dn-vdra(dn-vdra) zebrafish given daily heat-shocks from 6 to 50 dpa. Regeneration scores (Reg scores) indicate vigorous regeneration (1), partial regeneration (2) or blocked regeneration (3), as shown in Figure S3N. n = 8-11. ***p < 0.001 (Two-tailed Fisher’s exact test).
(K) Representative images of cTnT staining at 50 dpa, after daily heat-shock from 6 dpa. Scale bar, 100 μm.
(L) Representative images of AFOG stains to indicate Fibrin (red) and Collagen (blue) within the same hearts in (K). Arrowhead indicates scar tissue.
See also Figure S3.
To supplement these experiments, we examined whether modulation of VDR signaling in a distant organ, the liver, influenced cardiac cell proliferation or size. Among all embryonic tissues, we noted that liver displayed the sharpest increase in cyp24a1mGFP-directed fluorescence in response to Alfa treatment (Figure S2). New transgenic lines expressing dn-vdra or ca-vdra driven by fabp10a regulatory sequences in hepatocytes directionally controlled hepatic cyp24a1 reporter fluorescence in zebrafish (Figure S3G). In these animals, embryonic hearts had normal numbers of FUCCI+ CMs and were of a similar size as those of clutchmates (Figures S3H and S3I). Interestingly, whereas the hearts of juvenile fabp10a:ca-vdra zebrafish appeared normal, their livers were grossly enlarged (Figure 3G). Taken together, our data indicate that vitamin D dictates CM proliferation and controls heart size in growing zebrafish through intrinsic signaling, and that this principal can extend to other organs.
Vitamin D Signaling Regulates Heart Regeneration
Zebrafish possess a wide range of tissues with an elevated capacity to regenerate after major injury. In uninjured hearts, sparse cyp24a1mGFP fluorescence was detectable in CMs and other cardiac cells. This fluorescence became more prominent in ventricles injured by partial resection or induced CM ablation (Figures S3J and S3K) although cardiac cyp24a1mGFP expression patterns and levels varied among animals. To test whether vitamin D treatment augments proliferation of CMs during heart regeneration, we partially resected adult zebrafish ventricles and injected Alfa at 6 days post resection (dpa), one day before harvesting tissues at 7 dpa. By contrast with CMs of uninjured animals that have little or no response to a single Alfa injection (Figure 1E), a single treatment was sufficient to increase CM proliferation indices by 41% in regenerating hearts (Figures 3H and 3I). This result is consistent with the notion that vitamin D is more potent in CMs that are sensitized by injury or primed for regeneration programs. VDR inhibition by heat shock-induced dn-vdra transgene expression from 6-7 dpa had an opposite effect, reducing CM proliferation by 69% (Figure S3L). Moreover, sustained expression of dn-vdra after 6 dpa blocked heart regeneration, with ventricles retaining large scars in the injury sites at 50 dpa (Figures 3J-3L and S3N). Thus, vitamin D signaling is essential for normal heart regeneration.
Stimulation of heart regeneration in mammals by secreted factors may be more complex than in zebrafish, as mammalian adult CMs have a lower proliferative capacity and/or more hurdles to division like polyploidy (Gonzalez-Rosa et al., 2018; Patterson et al., 2017). Vitamin D signaling has been reported to protect mouse CMs from pathological hypertrophy by regulation of calcium handling (Chen et al., 2011; Choudhury et al., 2014; Ford et al., 2018), but we have identified no clear evidence from published literature for hyperplastic effects in mammalian hearts. To examine potential effects on cultured mouse P7 CMs, we treated with different concentrations of calcitriol and examined the expression of the proliferation marker Ki67 after 3 days. The frequency of Ki67+ CMs was increased in a dose-dependent manner (Figure S3M), suggesting that vitamin D supplementation is sufficient to induce mouse CM cycling in vitro.
Mitogenic Effects of Vitamin D Require ErbB2 Signaling
Vitamin D has been reported to regulate calcium homeostasis, vasculogenesis, and fatty acid oxidation in embryonic zebrafish (Lin et al., 2012; Merrigan and Kennedy, 2017; Peng et al., 2017), and VDR transcriptional targets are reported to be species- and context-dependent (Campbell and Trump, 2017). To identify downstream genes and pathways underlying vitamin D-induced cell proliferation, we sequenced cardiac transcriptomes of zebrafish injected with vehicle or Alfa. Among 14,962 recovered genes, 767 showed >2-fold higher levels with Alfa treatment, whereas levels of 495 genes were reduced by 50% or more (Table S2A). Functional clustering analysis indicated that levels of many genes involved in cell cycle regulation and DNA repair were increased, including cyclins (A2/B1/B2/B3/D2/E1/E2/F/J), cdks (1/2/6), e2fs (1/3/4/7/8) and mcms (2/3/4/5/6/7/8/9/10) (Figure 4A; Table S2B), consistent with the observed effects on cell proliferation and G1 checkpoint release. In addition, genes involved in metabolic pathways like steroid, nucleotide, and amino acid biosynthesis and/or degradation were enriched upon Alfa injection (Table S2B), although we detected no common metabolite level changes induced by Alfa among adult hearts, livers, and whole embryos by metabolomic analysis (Figure S4A; Table S3).
Figure 4. ErbB2 Blockade Inhibits Vitamin D-Induced Cardiomyocyte Proliferation.
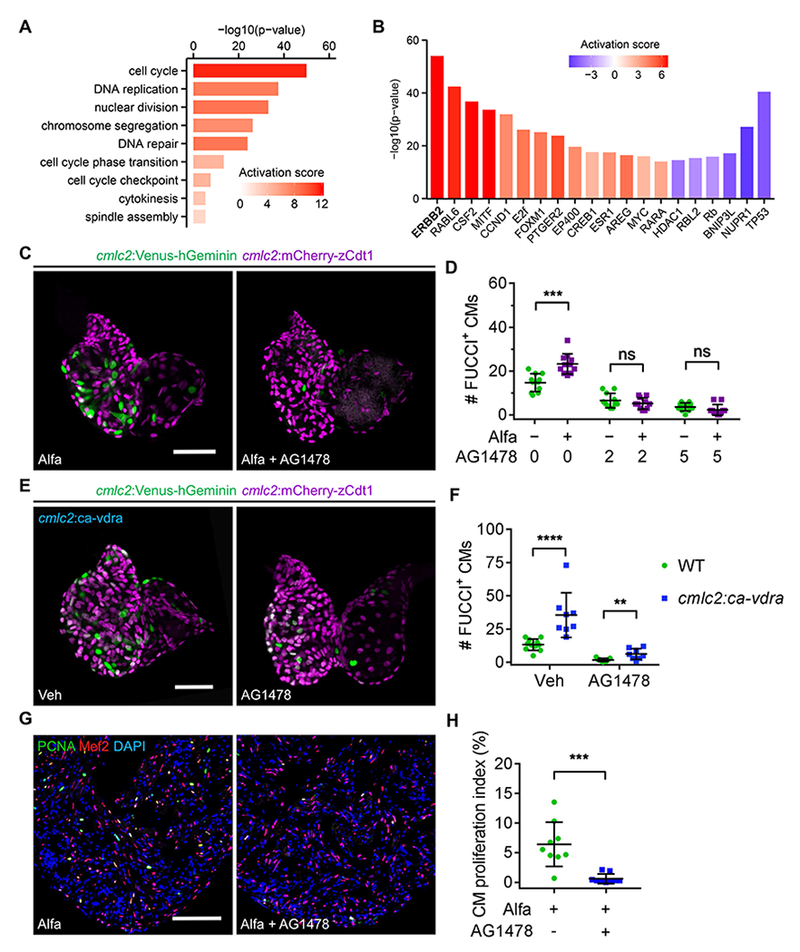
(A) Selected functional cluster enrichment of differentially expressed genes. The activation score infers the activation score of the corresponding process.
(B) Selected common upstream regulators of differentially expressed genes in adult hearts after three Alfa injections. The p-value measures the statistical significance of the overlap between differentially expressed genes and the genes under predicted control of a regulator, and the Activation score infers the activation state and score of a regulator, either activating (positive) or inhibiting (negative).
(C) MIP images of dissected 4 dpf cmlc2:FUCCI hearts after 24-h treatment. Scale bar, 50 μm.
(D) Quantification of FUCCI+ CMs of 4 dpf cmlc2:FUCCI embryos after 24-h treatment. n = 11.
(E) MIP images of dissected 4 dpf hearts after 24-h treatment. Scale bar, 50 μm.
(F) Quantification of FUCCI+ CMs of 4 dpf embryos after 24-h treatment. n= 8-11. (G) PCNA/Mef2 staining of ventricles from Alfa-injected zebrafish treated with vehicle or AG1478 for 24 h starting at 2 h after the last Alfa injection. Scale bar, 100 μm.
(H) Quantification of CM proliferation indices of experiments in (H). n = 9. ***p < 0.001. See also Figures S4, Table S2 and S3.
To identify common upstream regulators of differentially expressed genes, we analyzed RNA-Seq datasets using Ingenuity Pathway Analysis (IPA) software (Kramer et al., 2014). From this analysis, the EGF receptor family member ErbB2 emerged as the signaling effector with the highest enrichment and activation scores (Figure 4B; Table S2C); 135 ErbB2-associated genes were differentially expressed (Figure S4B; Table S2D). Notably, ErbB2 and one of its activating ligands Nrg1 have been functionally implicated as CM mitogens in zebrafish and mice (Bersell et al., 2009; D’Uva et al., 2015; Gemberling et al., 2015), with Nrg1 overexpression able to boost CM proliferation in the absence of injury in adult zebrafish (Gemberling et al., 2015). Also, daily calcitriol administration to rats over a period of 6 weeks was associated with modest cardiac increases in Nrg1 and ErbB2 proteins (Dang et al., 2016). We performed quantitative PCR for 19 ligand, receptor, and target genes, finding 9 of which had at least 2-fold increases in hearts of Alfa-treated animals (Figure S4C). To test if ErbB2 signaling is required for the effects of vitamin D signaling, we treated zebrafish embryos or adults with Alfa and the commonly used ErbB2 inhibitor, AG1478. AG1478 reduced CM proliferation in cmlc2:FUCCI embryos, and it abolished Alfa stimulation of CM proliferation (Figures 4C and 4D). Moreover, AG1478 blocked the increase in CM proliferation caused by cardiac expression of a ca-vdra transgene (Figures 4E and 4F). In addition, AG1478 inhibited Alfa-induced osteoblast proliferation (Figure S4D) in adults and Alfa-induced growth in zebrafish embryos (Figures S4E and S4F). Notably, AG1478 incubation reduced the CM proliferation index in Alfa-injected adult zebrafish to 0.6%, comparable to the index of vehicle-injected zebrafish and ~94% lower than that of animals treated with Alfa alone in these experiments (Figures 4G and 4H). Taken together, these data indicate that the mitogenic and growth-promoting effects of vitamin D on zebrafish CMs and other cell types require ErbB2 signaling.
DISCUSSION
Modulating VDR activity selectively in zebrafish cardiomyocytes or hepatocytes is sufficient to control heart or liver size, respectively. This finding indicates that vitamin D effects do not require signaling in distant tissues, though it does not rule out possible influences of systemic signaling. It is also possible that local relays involving secreted mitogenic factors or cell-cell interactions within these tissues are activated by vitamin D signaling, rather than a formal cell-autonomous effect. Because vitamin D is a circulating factor, the effects we report make it a candidate signal to synchronize growth among different tissues in growing zebrafish – that is, to achieve proportional growth. This synchrony was maintained in whole-animal manipulation of vitamin D signaling, but cardiac or hepatic growth no longer tracked with animal growth when VDR activity was selectively manipulated. How vitamin D modulates intracellular ErbB2 signaling requires further investigation; one possible mechanism is by sensitizing cells to activation of this pathway. Thousands of VDR binding elements exist throughout vertebrate genomes (Carlberg and Seuter, 2009), and systematic methods to identify VDR interactomes in different developmental contexts should be useful to illuminate key mechanisms.
The majority of studies assessing proliferation of mammalian cells in response to vitamin D have reported inhibitory effects, most notably in tumor cells (Ma et al., 2016; Samuel and Sitrin, 2008). Previous studies of mammalian CMs have suggested that VDR signaling regulates CM differentiation and hypertrophy and represses CM cycling (Hlaing et al., 2014; Nibbelink et al., 2007; O’Connell et al., 1997), whereas our data indicate that elevated vitamin D concentrations can increase cycling. The effects of vitamin D on cell proliferation are likely context- and/or concentration-dependent, and are affected by species’ distinct VDR target gene repertoires and collaborating regulatory sequences.
Vitamin D deficiency has been associated with various cardiovascular diseases, and VDR signaling has been reported to protect ischemic myocardium through anti-inflammatory, anti-fibrotic, and anti-apoptotic mechanisms (Bae et al., 2013; Chen et al., 2011; Simpson et al., 2007; Yao et al., 2015). Vitamin D and its analogues have been widely used to prevent and/or treat many diseases, including rickets, osteoporosis, osteomalacia, hypocalcemia, hypertension, hypercholesterolemia, hyperparathyroidism diabetes, and psoriasis. Yet, excessive vitamin D can lead to hypercalcemia and vascular calcification (Pilz et al., 2016; Tebben et al., 2016), challenging the use of systemic vitamin D administration to enhance tissue regeneration. Our results indicate that modulation of vitamin D signaling or sensitivity, e.g. targeted or tailored mechanisms to increase VDR activation in specific cell types without impacting physiological calcium handling, can be incorporated into potential methods to boost regeneration of the injured heart or other tissues.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kenneth Poss (kenneth.poss@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish and Mice
Wild-type or transgenic male and female zebrafish of the outbred Ekkwill and Ekkwill/AB strains were used in this study. Four to 12-month-old zebrafish were used for adult experiments, and ages of embryonic and juvenile zebrafish were indicated in the text. Water temperature was maintained at 28°C for embryos up to 5 dpf and 27°C afterwards. After transfer to system racks at 5 dpf, all fish were fed with artemia and age-dependent dry diets, i.e., Larval AP100 Z1 to Z3 (Zeigler) for 5–14 dpf, GEMMA Micro 75 for 2–4 wpf, GEMMA Micro 150 for 4–5 wpf, GEMMA Micro 300 for 5 wpf to 3 mpf and a mixture of GEMMA Micro 500 and Gemma Wean Diamond 0.5 after 3 mpf. The vitamin D3 levels in the diet are 2800 IU/kg for GEMMA Micro and 2400 IU/kg for Gemma Wean Diamond. All transgenic strains were analyzed as hemizygotes unless otherwise indicated. Partial ventricular resection surgeries were performed as previously described (Poss et al., 2002). Briefly, animals were anaesthetized and placed ventral side up in a prepared sponge bed. Straight iridectomy scissors were used to make a small incision through the ventral body and pericardial sac, and curved iridectomy scissors were used to remove approximately 20% of the cardiac ventricle. To genetically ablate ~50% of cardiomyocytes, zebrafish with cmlc2:CreERpd10; bactin2:loxp-mCherry-STOP-loxp-DTApd36 transgenes were treated with 0.4 μM tamoxifen for 18 h and hearts were collected 7 days later (Wang et al., 2011). For embryonic heat shock experiments, animals maintained at 28°C were incubated at 38°C for 30 minutes and then washed with egg water at room temperature. For adult heat shock experiments, animals maintained at 28°C were heat shocked daily at 37 to 38°C for 30 minutes as described (Lee et al., 2005), starting at 6 days post-surgery until tissues were harvested. All zebrafish procedures were performed in accordance with animal use guidelines at Duke University. Neonatal day 7 (P7) P7 ICR mice were used to isolate and culture primary cardiac cells. Animal procedures were approved by Animal Care and Use Committees of Duke University and the Weizmann Institute of Science.
METHOD DETAILS
Generation of cyp24a1mGFP zebrafish
To insert an mGFP-polyA cassette into the cyp24a1 locus, we designed a TALEN pair (left arm: 5’-GACAACTTTTACGCACG-3’, right arm: 5’-GCGCTCTCATCTTGAG-3’) targeting the start codon region of cyp24a1 gene. After validating the efficiency of the TALEN nuclease, we introduced the TALEN sequences and a common right arm sequence (5’-GTATACTACTGCGGCTAT-3’) into a donor vector that contains a mGFP-polyA reporter cassette, a ubiquitous ef1α:mCherry-polyA reporter cassette, and two FRT sequences flanking the vector sequences (Figure S1B). Then, the three TALEN nuclease mRNAs and donor vector were co-injected into one-cell stage embryos. The vector-free stable mGFP+mCherry− transgenic embryos were screened under a stereofluorescence microscope and further confirmed by PCR and Sanger sequencing.
Generation of ubb:cyp24a1 zebrafish
The full-length cyp24a1 coding sequences were amplified from 2 dpf zebrafish cDNA using the following primers: forward 5’-GAA TTT GTT TAC AGG GAT CCA CCG GTG CCA CCA TGA GAG CGC ACT TGC A-3’ and reverse 5’-GAA GTT TGT AGC GCC GCT TCC GGA GCG TGG AAC AAA AGC CA-3’. 3.8 kb ubb promoter sequences (Mosimann et al., 2011) were amplified from adult genomic DNA using the following primers: forward 5’-CCA CCT CGA GGC TGT GAC ATG GTG TT-3’ and reverse 5’-GAC TAC CGG TGG ATC CCT GTA AAC AAA TTC AAA GTA AGA TTA GC-3’. Then, the two sequences were subcloned sequentially into a P2A-TagBFP vector. Purified plasmid was linearized with I-SceI meganuclease and injected into one-cell stage embryos. Stable transgenic embryos were screened under a stereofluorescence microscope.
Generation of hsp70:dn-vdra and hsp70:dn-vdrb zebrafish
Full-length coding sequences of vdra were amplified from zebrafish embryonic cDNA and subcloned into an mRNA in vitro transcription vector using the following primers: vdra-forward, 5’-CTC GAA TTC ACC GGT CAC CAT GCT TAC GGA AAA TAG TGC C-3’; vdra-reverse, 5’-GGT ACT AGT CGG CCG CTA GGA CAC CTC ACT CC-3’. Human E420A-equivalent dominant negative mutations (Malloy et al., 2011) were introduced using the following primers: vdra-mut, 5’-CGC ACT AGT CGG CCG CTA GGA CAC CTC ACT CCC GAA CAC AGC CAG CACCAG-3’. Wild-type and dominant-negative vdr gene cassettes were transcribed, and mRNA was injected into one-cell stage cyp24a1mGFP embryos to test their effects on cyp24a1 expression. The hsp70 promoter was PCR amplified from an hsp70 vector (Han et al., 2011) using the following primers: hsp70-forward, 5’-CTT CGT CTC AGT ACC CTC GAG TCA GGG GTG TCG CTT GGT T-3’; hsp70-reverse, 5’-GCT CGT CTC AGG ATC CGA TTG ATT TCA AGA AAC TGC AA-3’. Then, the hsp70 promoter fragment and in-frame dn-vdra sequences were amplified and subcloned into the P2A-TagBFP vector. Stable transgenic lines were generated using I-SceI-mediated transgenesis and screened under a stereofluorescence microscope following heat shock.
Generation of cmlc2:dn-vdra and cmlc2:ca-vdra zebrafish
E420A-equivalent dn-vdra sequences were excised from the hsp70:dn-vdra construct and subcloned into a 5 kb cmlc2 promoter construct using AgeI and NotI. Stable transgenic lines were generated using I-SceI-mediated transgenesis and screened under a stereofluorescence microscope. To generate the constitutively active vdra construct, the FF version of dual-VP16 sequences (Asakawa et al., 2008) was synthesized from Integrated DNA Technologies and fused to 3’ end of wild-type vdra sequences, referred to as ca-vdra. Then, a 900 bp cmlc2 promoter fragment (Huang et al., 2003) and ca-vdra-P2A-TagBFP2 sequences were subcloned into a Tol2/I-SceI construct using Golden Gate cloning (Engler et al., 2008). Stable transgenic lines were generated using Tol2-mediated transgenesis and screened under a stereofluorescence microscope.
Generation of krtt1c19e:H2A-mCherry zebrafish
The 4 kb krtt1c19e promoter sequence (Lee et al., 2014) was amplified using the primers 5’-CCG CGG CAA CAA CAA TCC ACC TCA AGA GT-3’ to add a SacII site to the 5’ end and 5’-GGA TCC GAT GGT GGT TGG TGT CTT ACT CTG-3’ to add a BamHI site to the 3’ end, then digested with those restriction enzymes and subcloned into the pSKS-1 vector. H2A-mCherry-polyA sequence was amplified using primers 5’-ATC GAT GCC GCC ACC ATG GCA GGT GGA AAA GCA GG-3’ to add a ClaI site to the 5’ end and 5’-CTC GAG GAT ACA TTG ATG AGT TTG GAC AAA CC-3’ to add a XhoI site the the 3’ end, then subcloned into the krtt1c19e promoter-containing vector. The resulting plasmid was purified and injected into one-cell stage embryos.
Generation of fabp10a:dn-vdra and fabp10a:ca-vdra zebrafish
dn-vdra-2A-TagBFP2 and ca-vdra-2A-TagBFP2 sequences were subcloned into middle entry vectors and subjected to Gateway cloning with a p5E-fabp10a entry vector (a kind gift from Drs. Brian Link and John Rawls) and a Tol2/I-SceI destination vector to generate final fabp10a:dn-vdra and fabp10a:ca-vdra vectors. Stable transgenic lines were generated using Tol2-mediated transgenesis and screened under a stereofluorescence microscope.
Generation of vdra and vdrb mutant zebrafish
vdra and vdrb knockout alleles were generated using CRISPR/Cas9 techniques. Two guide RNAs targeting sequences encoding DNA binding domains of Vdra and Vdrb proteins were designed using CHOPCHOP website (Montague et al., 2014), synthesized as described (Varshney et al., 2015) and co-injected with Cas9 protein (PNA Bio) to generate deletional vdra and vdrb mutants lacking functional DNA binding domain. Target sequences of guide RNAs are 5’-GGCGTTGAAGTGGAATCCGGTGG-3’ and 5’- GATCATCAACTCTCTGGTGGAGG-3’ for vdra, 5’-GAACTCATCTGGCACTTGAGTGG-3’ and 5’-GAAGCGTGAAAAGTCAGAGTAGG-3’ for vdrb (underlined trinucleotides indicate PAM sequences). Injected embryos were raised to adulthood and screened as sperm samples and/or F1 embryos using the following primers: vdra-fwd: 5’-TCTCTCAGCCAATCAGGAGTG-3’, vdra-rev: 5’-TAATAGATCAGAGCAGCCGAC-3’; vdrb-fwd: 5’-CTTCAGGTAAACGTGCACACA-3’, vdrb-rev: 5’-TACCGGCAGAGAAAAGGTCAT-3’.
Chemical screening and drug treatment
cmlc2:FUCCI zebrafish were crossed with wild-type AB strain animals to produce hemizygous embryos. Five 3 dpf embryos were placed into each well of 48-well plates containing 0.5 mL fish water for initial screening using Prestwick Chemical Library (http://www.prestwickchemical.com/libraries-screening-lib-pcl.html). Then, embryos were treated with 5 μM compounds for 24 hours before examination using a stereofluorescence microscope. Embryos visually scored with more or fewer Venus-positive CMs were fixed with 4% paraformaldehyde (PFA) and analyzed by confocal microscopy.
For vitamin D agonist or antagonist treatment, alfacalcidol (S1468, Selleck Chemicals) and calcitriol (S1466, Selleck Chemicals) were dissolved in DMSO (for embryos) or ethanol (for injection into adults) to make 10 mM stock solutions. Final concentrations were 5 μM and 1 μM, respectively, unless otherwise indicated for embryo treatment. For adult injections, drugs were diluted with ethanol and water to make 200 μM injection-ready solutions in 10% ethanol. Then, 3 daily 10 μL solution were injected intraperitoneally into adult zebrafish unless otherwise indicated. For long-term treatment and rescue experiments, juvenile zebrafish were incubated in aquarium water with Vehicle or 10 nM alfacalcidol and fed with artemia twice every day. Fish water was changed every other day to maintain quality. PS121912 was a kind gift from Dr. Alexander Arnold (Sidhu et al., 2014). AG-1478 (S2728, Selleck Chemicals) were purchased and dissolved in DMSO to make 10 mM stock solutions.
Epicardial explants culture and imaging
tcf21:FUCCI hearts were collected, cut into large pieces, and cultured using Fibronectin-coated dishes as described (Cao et al., 2017). Culture media were replaced with fresh media 3 days after culture and drugs were added 1–2 hours before live imaging the next day. Confocal images were captured once every 5 minutes for 3 days using a Zeiss 710 inverted microscope.
Mouse cardiomyocyte cultures
Primary cardiac cells were isolated as described (Bassat et al., 2017). Hearts were harvested from neonatal day 7 (P7) ICR mice, and cardiac cells were isolated using a neonatal dissociation kit (gentleMACS, Miltenyi Biotec) according to the manufacturer’s instructions. Cardiac cells were cultured in gelatin-coated (0.02%, G1393, Sigma-Aldrich) 96 wells plates, using “rich” media (DMEM/F12 medium supplemented with l-glutamine, Na-pyruvate, non-essential amino acids, penicillin/streptomycin, 5% horse serum and 10% fetal bovine serum (FBS)) at 37°C and 5% CO2, allowing them to adhere. 2 days post seeding, the medium was replaced with serum-free medium containing vehicle or 0.1 μM, 1 μM or 10 μM calcitriol for 72 hours. Cells were then fixed in 4% PFA and stained for cardiomyocyte marker (cTnT) and proliferation marker (Ki67).
Histology and imaging
Tissues were fixed with 4% PFA, and 10 μm cryosections were used in this study. Immunofluorescence was performed as described previously (Kikuchi et al., 2011). Briefly, slides were washed in PBT (Phosphate-buffered saline + 0.1% Tween-20), blocked in NCS-PBT (PBT with 10% heat-inactivated calf serum and 1% DMSO) and 2% horse serum, and then incubated in NCS-PBT with primary antibodies at 37°C for 3 h or 4°C overnight. After washes in PBT, slides were incubated with fluorescently labeled secondary antibodies at 37°C for one hour, washed in PBT, and mounted. Staining with anti-PCNA antibodies required citrate buffer (10 mM citric acid, 0.05% Tween-20, pH 6.0) treatment, involving heating to 98°C, boiling for 25 min, and cooling for 20 min before PBT washes and blocking. For EdU incorporation, zebrafish were injected intraperitoneally with 10 μL of 10 mM EdU (A10055, Sigma) 6 hours (for fin epithelial cells) or 24 hours (for other tissues) before tissue collection. EdU was detected through a click reaction as described previously using fluorescent azide (Alexa Fluor 488 or 647, Thermo Fisher) (Salic and Mitchison, 2008). Primary antibodies used in this study were: anti-Mef2 (sc-313, Santa Cruz Biotechnology), anti-PCNA (P8825, Sigma), anti-phospho-Histone H3 (9701S, 9706S, Cell Signaling Technology), anti-Hnf4a (ab201460, Abcam), anti-p63 (CM163A, Biocare Medical), anti-cTnT (MS-295-PABX, Thermo Fisher Scientific; ab33589, Abcam), anti-Ki67 (275R, Cell Marque), anti-Ki-67 (41–5698-80, Thermo Fisher Scientific). Secondary antibodies (Invitrogen, 1:200–1:500) used in this study were highly cross-absorbed Alexa Fluor 488/546/594 goat anti-rabbit or antimouse antibodies. All confocal images were acquired with Zeiss LSM 700 or Zeiss LSM 880 microscopes unless otherwise stated. Acid fuchsin orange g (AFOG) stains to visualize fibrin and collagen was performed and imaged as described (Poss et al., 2002). Briefly, sections were hydrated, placed in Bouin’s solution at 60°C, rinsed in water followed by 5 minutes in 1% phosphomolbdic acid and water again, then stained in an AFOG solution of 1 g analine blue, 2 g orange g, and 3 g acid fuchsin in 200 ml water, pH 1.09 for 5 minutes. Slides were then rinsed in water, dehydrated, cleared and mounted, and imaged using a Leica DM6000 microscope. Whole-mounted tissues and embryos were imaged with a Zeiss Axio Zoom microscope.
RNA sequencing and quantitative PCR
Adult zebrafish were injected with vehicle or Alfa every 24 hours over 3 days, and euthanized with tricaine for heart collection 24 hours after the last injection. Total RNA from 10 pooled hearts of two biological replicates were extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instruction. Total RNA were treated with DNaseI to remove genomic DNA, purified with Quick-RNA MiniPrep kit (Zymo Research), and then submitted for library preparation and sequences using an Illumina HiSeq2000 at the Duke Center for Genomic and Computational Biology. Reads were mapped to the zebrafish genome (GRCz10) using Tophat2 (Kim et al., 2013), counted with featureCounts (Liao et al., 2014), and differentially expressed genes were analyzed using edgeR (Robinson et al., 2010). Functional clustering analysis was performed using the DAVID Bioinformatics Resources website (Huang da et al., 2009). The zebrafish gene IDs were mapped to human homologous gene IDs to perform pathway and upstream regulator analysis using QIAGEN Ingenuity Pathway Analysis software (Kramer et al., 2014). For qPCR, purified total RNAs were reverse transcribed with Superscript III Reverse Transcriptase (Thermo Scientific) and qPCRs were performed using the Roche Light Cycler 480, Roche UPL probes, and LightCycler 480 Probes Master. All experiments were performed using biological duplicates and technical triplicates. Primer efficiencies were tested and selected for at least 1.80. The relative expression levels were calculated with real efficiencies.
Metabolomic analysis
For embryos, 3 dpf embryos treated with vehicle or 5 μM Alfa for 24 hours were deyolked (11 per sample) and stored at −80°C. For hearts and livers, adult zebrafish were injected with vehicle or Alfa daily for 3 days and dissected 24 hours after the last injection before collecting hearts (4 per sample) and livers (2 per sample). All tissues were analyzed with 5 biological replicates and submitted for metabolite preparation and analysis to Duke University Proteomics and Metabolomics Core Facility. Each sample was sonicated, homogenized and processed using the AbsoluteIDQ p180 kit (Biocrates Innsbruck, Austria) according to manufacturer’s protocol to prepare metabolites. Finally, metabolites were diluted and subjected to UPLC analysis (amino acids and biogenic amines) or flow injection analysis (acylcarnitines, sphingolipids, and glycerophospholipids).
QUANTIFICATION AND STATISTICAL ANALYSIS
Clutchmates were randomized into different treatment groups for each experiment. No animal or sample was excluded from the analysis unless the animal died during the procedure or deformed/destroyed due to fixation or dissection. All experiments were performed with at least two biological replicates, and at least 10 samples were used for each experiment unless otherwise indicated. All statistical values are displayed as Mean ± Standard Deviation. Statistics tests were calculated using two-tailed Student’s t-tests when normality test was passed or Mann-Whitney Rank Sum test otherwise. ns, not significant, *p < 0.05, *** p < 0.001, **** p < 0.0001.
DATA AND SOFTWARE AVAILABILITY
All sequencing data supporting the findings in this study were deposited to the GEO database under the accession number GSE112826. All other relevant data are available from the corresponding author on request.
Supplementary Material
Table S1. List of compounds showing positive or negative effects on cardiomyocyte proliferation. Related to Figure 1.
Table S2. Analysis of transcriptome datasets of adult hearts after vehicle or alfacalcidol injection. Related to Figure 4.
(A) Differential gene analysis results of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Effects of alfacalcidol on expression of individual genes are shown as log(fold-change) and log (counts per million). Genes are ranked by ascending p-values.
(B) Functional clustering analysis results of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Gene ontology categories and terms are listed in rows ranked by ascending FDR.
(C) Ingenuity pathway analysis of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Common upstream regulators are listed in Column A, ranked by ascending p-value.
(D) List of 135 genes predicted to be regulated by ErbB2 signaling based on Ingenuity pathway analysis. Column information and values are the same as listed in Table S2A.
Table S3. Metabolomic analysis of adult heart and liver after three daily vehicle or alfacalcidol injections, and of 4 dpf zebrafish embryos treated with alfacalcidol or vehicle for 24 hours. Column values are described in Row 1. Related to Figure 4.
Video S1. Time lapse video showing the proliferation and migration of epicardial cells ex vivo using tcf21:FUCCI heart explants, treated with vehicle (left) or 1 μM calcitriol (right). Related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-phospho-Histone H3 (Ser10) | Cell Signaling Technology | Cat#9701; RRID:AB_331535 |
| Mouse polyclonal anti-phospho-Histone H3 (Ser10) | Cell Signaling Technology | Cat#9706; RRID:AB_331748 |
| Rabbit polyclonal anti-Mef-2 | Santa Cruz Biotechnology | Cat#sc-313; RRID:AB_631920 |
| Mouse monoclonal anti-PCNA | Sigma-Aldrich | Cat#P8825; RRID:AB_477413 |
| Rabbit monoclonal anti-HNF-4-alpha | Abcam | Cat#ab201460 |
| Mouse monoclonal anti-Troponin T | Lab Vision | Cat#MS-295-PABX; RRID:AB_61810 |
| Mouse monoclonal anti-cardiac Troponin T | Abcam | Cat#ab33589; RRID:AB_727035 |
| Mouse monoclonal anti-Ki-67 | Cell Marque Corp | Cat#275R; RRID:AB_1158031 |
| Rat monoclonal anti-Ki-67, eFlour 570 conjugated | Thermo Fisher Scientific | Cat#41-5698-80; RRID:AB_11219874 |
| Mouse monoclonal anti-p63 | Biocare Medical | Cat#CM163A; RRID:AB_10582730 |
| Alexa Fluor 488 Goat anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-11034; RRID:AB_2576217 |
| Alexa Fluor 488 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A-11001; RRID:AB_2534069 |
| Alexa Fluor 546 Goat anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-11035; RRID:AB_143051 |
| Alexa Fluor 546 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A-11030; RRID:AB_144695 |
| Alexa Fluor 594 Goat anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-11037; RRID:AB_2534095 |
| Alexa Fluor 594 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A-11032; RRID:AB_141672 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Metronidazole | Sigma-Aldrich | Cat#M1547 |
| Fibronectin | Thermo Fisher Scientific | Cat#33010018 |
| 5-ethynyl-2’-deoxyuridine | Thermo Fisher Scientific | Cat#A10044 |
| AG-1478 | Selleck Chemicals | Cat#S2728 |
| Alfacalcidol | Selleck Chemicals | Cat#S1468 |
| Calcitriol | Selleck Chemicals | Cat#S1466 |
| DAPI | Thermo Fisher Scientific | Cat#D3571 |
| Alexa Fluor 488 Azide | Thermo Fisher Scientific | Cat#A10266 |
| Alexa Fluor 594 Azide | Thermo Fisher Scientific | Cat#A10270 |
| Alexa Fluor 488 Click-iT Plus TUNEL Assay | Thermo Fisher Scientific | Cat#C10617 |
| Prestwick Chemical Library | Prestwick Chemical | N/A |
| PS121912 | University of Wisconsin-Milwaukee | N/A |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: Tg(cmlc2:mCherry-zCdt1)pd57 | (Choi et al., 2013) | pd57 |
| Zebrafish: Tg(cmlc2:Venus-hGeminin)pd58 | (Choi et al., 2013) | pd58 |
| Zebrafish: Tg(cmlc2:H2A-EGFP)pd115 | (Foglia et al., 2016) | pd115 |
| Zebrafish: Tg(tcf21:mAG-zGeminin)pd254 | (Cao et al., 2017) | pd254 |
| Zebrafish: Tg(tcf21:zCdt1-mKO2)pd255 | (Cao et al., 2017) | pd255 |
| Zebrafish: Tg(kdrl:H2B-EGFP)mu122 | (Kochhan et al., 2013) | mu122 |
| Zebrafish: Tg(bactin2:loxP-mCherry-STOP-loxP-DTA176)pd36 | (Wang et al., 2011) | pd36 |
| Zebrafish: Tg(Tp1:H2B-mCherry) | (Ninov et al., 2012) | N/A |
| Zebrafish: Tg(fabp10a:nls-mCherry) | (Choi et al., 2014) | N/A |
| Zebrafish: Tg(krtt1c19e:H2A-m Cherry)pd309 | This paper | pd309 |
| Zebrafish: Tg(osx:mCherry-zCdt1)pd270 | Unpublished | pd270 |
| Zebrafish: Tg(osx:Venus-hGeminin)pd271 | Unpublished | pd271 |
| Zebrafish: cyp24a1mGFP | This paper | pd272 |
| Zebrafish: Tg(ubb:cyp24a1) pd273 | This paper | pd273 |
| Zebrafish: Tg(ubb:cyp24a1)pd274 | This paper | pd274 |
| Zebrafish: Tg(hsp70:dn-vdra)pd276 | This paper | pd276 |
| Zebrafish: Tg(cmlc2:dn-vdra)pd301 | This paper | pd301 |
| Zebrafish: Tg(cmlc2:ca-vdra)pd302 | This paper | pd302 |
| Zebrafish: Tg(fabp10a:dn-vdra)pd303 | This paper | pd303 |
| Zebrafish: Tg(fabp10a:ca-vdra)pd304 | This paper | pd304 |
| Zebrafish: vdrapd307 | This paper | pd307 |
| Zebrafish: vdrbpd308 | This paper | pd308 |
| Software and Algorithms | ||
| Tophat2 | (Kim et al., 2013) | Version 2.1.1 |
| featureCounts | (Liao et al., 2014) | Version 1.5.3 |
| edgeR | (Robinson et al., 2010) | Version 3.20.9 |
| DAVID Bioinformatics Resources | (Huang da et al., 2009) | Version 6.8 |
| Ingenuity Pathway Analysis | Qiagen, (Kramer et al., 2014) | 830011 |
HIGHLIGHTS.
Vitamin D activates cardiomyocyte proliferation in several contexts in zebrafish
Vitamin D controls growth rate and cycling in a wide range of adult cells
Tissue-specific modulation of vitamin D activity controls cardiac size
ErbB2 inhibition blunts vitamin D-induced cell proliferation
ACKNOWLEDGMENTS
We thank Duke Zebrafish Core Facilities staff for zebrafish care; Duke Computing Cluster for computing support; Duke University Proteomics and Metabolomics Core Facility for metabolomics analysis; A. Arnold for PS121912; J. Rawls and B. Link for DNA constructs; M. Bagnat, M. Goll and D. Shin for zebrafish lines; R. Karra and N. Bursac for discussions; and J. Kang for comments on the manuscript. This work was supported by predoctoral fellowships from American Heart Association (AHA) to A.C. and National Science Foundation to V.A.T. and B.D.C.; postdoctoral fellowships from AHA to J.C., from Swiss National Science Foundation to V.C., and from National Institutes of Health (NIH) to W.Y.C.; grants from NIH (R01 HL081674, R01 GM074057, R01 HL131319), March of Dimes, and AHA to K.D.P., and a grant from Fondation Leducq to K.D.P. and E.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, and Kawakami K (2008). Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A 105, 1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Singh SS, Yu H, Lee JY, Cho BR, and Kang PM (2013). Vitamin D signaling pathway plays an important role in the development of heart failure after myocardial infarction. J Appl Physiol (1985) 114, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. (2009). Evidence for cardiomyocyte renewal in humans. Science 324, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, et al. (2015). Dynamics of Cell Generation and Turnover in the Human Heart. Cell 161, 1566–1575. [DOI] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, and Kuhn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. [DOI] [PubMed] [Google Scholar]
- Bikle DD (2014). Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol 21, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MJ, and Trump DL (2017). Vitamin D Receptor Signaling and Cancer. Endocrinol Metab Clin North Am 46, 1009–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wang J, Jackman CP, Cox AH, Trembley MA, Balowski JJ, Cox BD, De Simone A, Dickson AL, Di Talia S, et al. (2017). Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev Cell 42, 600–615 e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg C, and Seuter S (2009). A genomic perspective on vitamin D signaling. Anticancer Res 29, 3485–3493. [PubMed] [Google Scholar]
- Chen S, Law CS, Grigsby CL, Olsen K, Hong TT, Zhang Y, Yeghiazarians Y, and Gardner DG (2011). Cardiomyocyte-specific deletion of the vitamin D receptor gene results in cardiac hypertrophy. Circulation 124, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi TY, Ninov N, Stainier DY, and Shin D (2014). Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Gemberling M, Wang J, Holdway JE, Shen MC, Karlstrom RO, and Poss KD (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Bae S, Ke Q, Lee JY, Singh SS, St-Arnaud R, Monte FD, and Kang PM (2014). Abnormal calcium handling and exaggerated cardiac dysfunction in mice with defective vitamin d signaling. PLoS One 9, e108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Chen MJ, Stachura DL, Liu SY, Kwan W, Wright F, Vo LT, Theodore LN, Esain V, Frost IM, et al. (2016). Developmental Vitamin D Availability Impacts Hematopoietic Stem Cell Production. Cell Rep 17, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Uva G, Aharonov A, Lauriola M, Kain D, Yahalom-Ronen Y, Carvalho S, Weisinger K, Bassat E, Rajchman D, Yifa O, et al. (2015). ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol 17, 627–638. [DOI] [PubMed] [Google Scholar]
- Dang R, Guo Y, Zhu Y, Yang R, Cai H, and Jiang P (2016). Chronic administration of calcitriol enhanced neuregulin-1/ErbB signaling in rat myocardium. Pharmazie 71, 192–195. [PubMed] [Google Scholar]
- Engler C, Kandzia R, and Marillonnet S (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglia MJ, Cao J, Tornini VA, and Poss KD (2016). Multicolor mapping of the cardiomyocyte proliferation dynamics that construct the atrium. Development 143, 1688–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford K, Latic N, Slavic S, Zeitz U, Dolezal M, Andrukhov O, Erben RG, and Andrukhova O (2018). Lack of vitamin D signalling per se does not aggravate cardiac functional impairment induced by myocardial infarction in mice. PLoS One 13, e0204803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Karra R, Dickson AL, and Poss KD (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, and Burns CG (2018). Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell 44, 433–446 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Mu Y, Li X, Xu P, Tong J, Liu Z, Ma T, Zeng G, Yang S, Du J, et al. (2011). Grhl2 deficiency impairs otic development and hearing ability in a zebrafish model of the progressive dominant hearing loss DFNA28. Hum Mol Genet 20, 3213–3226. [DOI] [PubMed] [Google Scholar]
- Hassink RJ, Pasumarthi KB, Nakajima H, Rubart M, Soonpaa MH, de la Riviere AB, Doevendans PA, and Field LJ (2008). Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc Res 78, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, and Martin JF (2013). Hippo signaling impedes adult heart regeneration. Development 140, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlaing SM, Garcia LA, Contreras JR, Norris KC, Ferrini MG, and Artaza JN (2014). 1,25-Vitamin D3 promotes cardiac differentiation through modulation of the WNT signaling pathway. J Mol Endocrinol 53, 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, and Tsai HJ (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn 228, 30–40. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, and Izpisua Belmonte JC (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, and Poss KD (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, and Poss KD (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhan E, Lenard A, Ellertsdottir E, Herwig L, Affolter M, Belting HG, and Siekmann AF (2013). Blood flow changes coincide with cellular rearrangements during blood vessel pruning in zebrafish embryos. PLoS One 8, e75060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., and Tugendreich S (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ (2016). Vitamin D receptor signaling is required for heart development in zebrafish embryo. Biochem Biophys Res Commun 470, 575–578. [DOI] [PubMed] [Google Scholar]
- Lee RT, Asharani PV, and Carney TJ (2014). Basal keratinocytes contribute to all strata of the adult zebrafish epidermis. PLoS One 9, e84858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, and Poss KD (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173–5183. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin CH, Su CH, Tseng DY, Ding FC, and Hwang PP (2012). Action of vitamin D and the receptor, VDRa, in calcium handling in zebrafish (Danio rerio). PLoS One 7, e45650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Johnson CS, and Trump DL (2016). Mechanistic Insights of Vitamin D Anticancer Effects. Vitam Horm 100, 395–431. [DOI] [PubMed] [Google Scholar]
- Malloy PJ, Zhou Y, Wang J, Hiort O, and Feldman D (2011). Hereditary vitamin D-resistant rickets (HVDRR) owing to a heterozygous mutation in the vitamin D receptor. J Bone Miner Res 26, 2710–2718. [DOI] [PubMed] [Google Scholar]
- Merrigan SL, and Kennedy BN (2017). Vitamin D receptor agonists regulate ocular developmental angiogenesis and modulate expression of dre-miR-21 and VEGF. Br J Pharmacol 174, 2636–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S, and Srivastava D (2018). Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 173, 104–116 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42, W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, and Zon LI (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbelink KA, Tishkoff DX, Hershey SD, Rahman A, and Simpson RU (2007). 1,25(OH)2-vitamin D3 actions on cell proliferation, size, gene expression, and receptor localization, in the HL-1 cardiac myocyte. J Steroid Biochem Mol Biol 103, 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninov N, Borius M, and Stainier DY (2012). Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139, 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell TD, Berry JE, Jarvis AK, Somerman MJ, and Simpson RU (1997). 1,25-Dihydroxyvitamin D3 regulation of cardiac myocyte proliferation and hypertrophy. Am J Physiol 272, H1751–1758. [DOI] [PubMed] [Google Scholar]
- Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, et al. (2017). Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 49, 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Shang G, Wang W, Chen X, Lou Q, Zhai G, Li D, Du Z, Ye Y, Jin X, et al. (2017). Fatty Acid Oxidation in Zebrafish Adipose Tissue Is Promoted by 1alpha,25(OH)2D3. Cell Rep 19, 1444–1455. [DOI] [PubMed] [Google Scholar]
- Pilz S, Verheyen N, Grubler MR, Tomaschitz A, and Marz W (2016). Vitamin D and cardiovascular disease prevention. Nat Rev Cardiol 13, 404–417. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, and Sadek HA (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, and Keating MT (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- Ringe JD, and Schacht E (2007). Improving the outcome of established therapies for osteoporosis by adding the active D-hormone analog alfacalcidol. Rheumatol Int 28, 103–111. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Salic A, and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel S, and Sitrin MD (2008). Vitamin D’s role in cell proliferation and differentiation. Nutr Rev 66, S116–124. [DOI] [PubMed] [Google Scholar]
- Sidhu PS, Nassif N, McCallum MM, Teske K, Feleke B, Yuan NY, Nandhikonda P, Cook JM, Singh RK, Bikle DD, et al. (2014). Development of novel Vitamin D Receptor-Coactivator Inhibitors. ACS Med Chem Lett 5, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RU, Hershey SH, and Nibbelink KA (2007). Characterization of heart size and blood pressure in the vitamin D receptor knockout mouse. J Steroid Biochem Mol Biol 103, 521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebben PJ, Singh RJ, and Kumar R (2016). Vitamin D-Mediated Hypercalcemia: Mechanisms, Diagnosis, and Treatment. Endocr Rev 37, 521–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, et al. (2015). High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 25, 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A 109, 2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Holdway JE, Major RJ, and Poss KD (2008). Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135, 183–192. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A 110, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Ying X, Zhao Y, Yuan A, He Q, Tong H, Ding S, Liu J, Peng X, Gao E, et al. (2015). Vitamin D receptor activation protects against myocardial reperfusion injury through inhibition of apoptosis and modulation of autophagy. Antioxid Redox Signal 22, 633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, D’Agostino G, Loo SJ, Wang CX, Su LP, Tan SH, Tee GZ, Pua CJ, Pena EM, Cheng RB, et al. (2018). Early Regenerative Capacity in the Porcine Heart. Circulation 10.1161/CIRCULATIONAHA.117.031542. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhang E, Zhao M, Chong Z, Fan C, Tang Y, Hunter JD, Borovjagin AV, Walcott GP, Chen JY, et al. (2018). Regenerative Potential of Neonatal Porcine Hearts. Circulation 10.1161/CIRCULATIONAHA.118.034886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of compounds showing positive or negative effects on cardiomyocyte proliferation. Related to Figure 1.
Table S2. Analysis of transcriptome datasets of adult hearts after vehicle or alfacalcidol injection. Related to Figure 4.
(A) Differential gene analysis results of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Effects of alfacalcidol on expression of individual genes are shown as log(fold-change) and log (counts per million). Genes are ranked by ascending p-values.
(B) Functional clustering analysis results of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Gene ontology categories and terms are listed in rows ranked by ascending FDR.
(C) Ingenuity pathway analysis of transcriptomes of hearts from zebrafish treated with vehicle, versus hearts of zebrafish treated with alfacalcidol. Common upstream regulators are listed in Column A, ranked by ascending p-value.
(D) List of 135 genes predicted to be regulated by ErbB2 signaling based on Ingenuity pathway analysis. Column information and values are the same as listed in Table S2A.
Table S3. Metabolomic analysis of adult heart and liver after three daily vehicle or alfacalcidol injections, and of 4 dpf zebrafish embryos treated with alfacalcidol or vehicle for 24 hours. Column values are described in Row 1. Related to Figure 4.
Video S1. Time lapse video showing the proliferation and migration of epicardial cells ex vivo using tcf21:FUCCI heart explants, treated with vehicle (left) or 1 μM calcitriol (right). Related to Figure 1.
Data Availability Statement
All sequencing data supporting the findings in this study were deposited to the GEO database under the accession number GSE112826. All other relevant data are available from the corresponding author on request.


