Summary
Recent work of Besseling and Bringmann (2016) identified a molecular intervention for C. elegans in which premature segregation of maternal and paternal chromosomes in the fertilized oocyte can produce viable animals exhibiting a non-Mendelian inheritance pattern. Overexpression in embryos of a single protein regulating chromosome segregation (GPR-1) provides a germline derived clonally from a single parental gamete. We present a collection of strains and cytological assays to consistently generate and track non-Mendelian inheritance. These tools allow reproducible and high-frequency (~80%) production of non-Mendelian inheritance, the facile and simultaneous homozygosis for all nuclear chromosomes in a single generation, the precise exchange of nuclear and mitochondrial genomes between strains, and the assessments of non-canonical mitosis events. We show the utility of these strains by demonstrating a rapid assessment of cell lineage requirements (AB versus P1) for a set of genes (lin-2, lin-3, lin-12, and lin-31) with roles in C. elegans vulval development.
eTOC blurb:
Non-Mendelian inheritance in C. elegans results in chimeric animals with cells containing only maternal or paternal nuclear DNA. Artiles et al. develop a toolkit of strains with a high frequency of non-Mendelian segregation and monitor inheritance by fluorescence, thereby enabling experiments requiring detection of non-canonical inheritance events across large populations.
Graphical Abstract
Introduction
Prior to the ‘modern’ era of genetics (Mendel, 1866), a variety of intuitive expectations of inheritance mechanisms predominated, ranging from mixing models in which offspring carried a uniform mixture of maternal and paternal determinants to mosaic models in which some tissues were maternally-derived in character and others derived paternally (e.g., Charaka, “The Charaka Samhita”, ~200 BCE, Chapter 3, citing Atreya). Indeed, some such mechanisms have a place in biological systems, with the simple “Mendelian” (and diploid) rules relevant to only a subset of organisms and situations (e.g. Wollman et al., 1956; Fujiwara et al., 1997; de la Filia et al., 2015; Matsuura, 2017; Blay et al., 2018; Ramachandran and McDaniel, 2018).
In an example of strong interest to the biomedical community, human inheritance is (i) governed in an overwhelming majority of cases by Mendelian rules, and (ii) is subject to rare exceptions where components of an individual are clonally derived from duplication of a haploid genome from either a maternal or paternal source (Robinson, 2000; Kotzot, 2008; Conlin et al., 2010; Winberg et al., 2010; Darcy et al., 2015; Bens et al., 2017). A descriptor that has been used for the latter (rare) form of inheritance is “mosaic genome-wide uniparental isodisomy”. “Mosaic” refers to the fact that different tissues in the individual have different genetic compositions, in this case with some tissues carrying chromosomes only from a single parent and others carrying chromosomes from the other parent or both parents. “Genome-wide” refers to such a DNA composition observed simultaneously for all nuclear chromosomes. “Uniparental isodisomy” refers to a diploid situation in which both alleles at any locus derive from a single copy in the parent (so the individual cells are functionally homozygous at all loci that have not been subject to mutagenesis in this generation). Numerous studies in the area of genomic imprinting have shown that chromosomal derivation from maternal or paternal gametes can be essential for specific phenotypic characteristics and viability (Kermicle, 1970; Adalsteinsson and Ferguson-Smith, 2014). In one human example, an individual who obtains both of their copies of chromosome 15 from one parent will manifest neurodevelopmental syndromes (Angelman or Praeder-Willi) arising from differences in imprinting (Girardot et al., 2013). Full uniparental inheritance in humans is thought to lead to inviability (Girardot et al., 2013; Adalsteinsson and Ferguson-Smith, 2014), although the possibility that compensating processes might mitigate this has been discussed. Furthermore, it appears that requirements for parental derivation are not universal amongst tissue types, with a very small number of reported cases where humans exhibit genomewide uniparental disomy in a limited subset of tissues (e.g. Kotzot, 2008; Conlin et al., 2010; Winberg et al., 2010; Darcy et al., 2015; Bens et al., 2017).
By contrast to well-characterized vertebrate examples where biparental inheritance is critical, certain invertebrate model systems show no evidence for a biparental inheritance requirement. For both C. elegans and D. melanogaster, inheritance of individual chromosomes or of the entire genome from a single parent (and hence inheritance through a single gamete type) appears compatible with normal development and function (Fuyama, 1986; Haack and Hodgkin, 1991; Komma and Endow, 1995), although both species have been shown to exhibit capabilities for imprinting of loci and chromosomes (Golic et al., 1998; Maggert and Golic, 2002; Sha and Fire, 2005; Bean et al., 2004). The lack of an imprinting requirement in the two major invertebrate models raises the possibility of directed engineering of inheritance in which some or all cells derive their entire genomes from a single parent.
Numerous distinct mechanisms have been observed to produce uniparental inheritance of one or more chromosomes, including parthenogenesis (activation of oocytes with no sperm contribution; (e.g. Ramachandran and McDaniel, 2018), early mitotic events (e.g. Komma and Endow, 1995), and later events in which an entire parental genome is silenced and lost to future generations (e.g. Sánchez, 2014). All may operate in certain “normal” circumstances in specific organisms and in specific rare or pathological circumstances in human biology.
In addition to studies of normal (and rare/pathological) inheritance, interventions in experimental model systems that yield non-canonical inheritance patterns are of considerable use (i) for understanding different contributions of parental genomes in development, (ii) for generating and propagating strains with highly-defined genetic backgrounds, and (iii) as tools for understanding the nature and regulation of chromosome segregation. Of additional utility are interventions in which uniparental inheritance can be engineered following experimental intervention. Such interventions have been proposed as tools for the production of functionally mosaic embryos and for facilitating construction of strains with broad homozygosity (Eicher and Hoppe, 1973; Anderegg and Markert, 1986; Komma and Endow, 1995; Seymour et al., 2012). Despite the evident promise of such approaches, technical hurdles in their application have prevented widespread use in routine genetics.
Recently, Besseling and Bringmann (2016) demonstrated that mosaic uniparental inheritance could be achieved by overexpressing the conserved microtubule force regulator GPR-1 in stages leading to the first cleavage in the C. elegans embryo. As diagrammed in Figure 1, this intervention leads to the disruption of the process by which maternal and paternal chromosomes are mixed and segregated in the pre-cleavage C. elegans embryo, and hence to the production of embryos and animals in which the maternal and paternal genetic contributions are partitioned exclusively rather than equally at the first division between the first two blastomeres in the C. elegans embryo (AB and P1). Germ cells (sperm and oocytes) are derived exclusively from the P1 lineage, with the result that the generation following a non-Mendelian segregation event contains nuclear genomic DNA derived from individual chromosomes of only one parent in the original cross (not shown in the diagram). This feature can be used to create isogenic animals in just two generations with possible applications for generating strains or mutagenized strains with broad homozygosity.
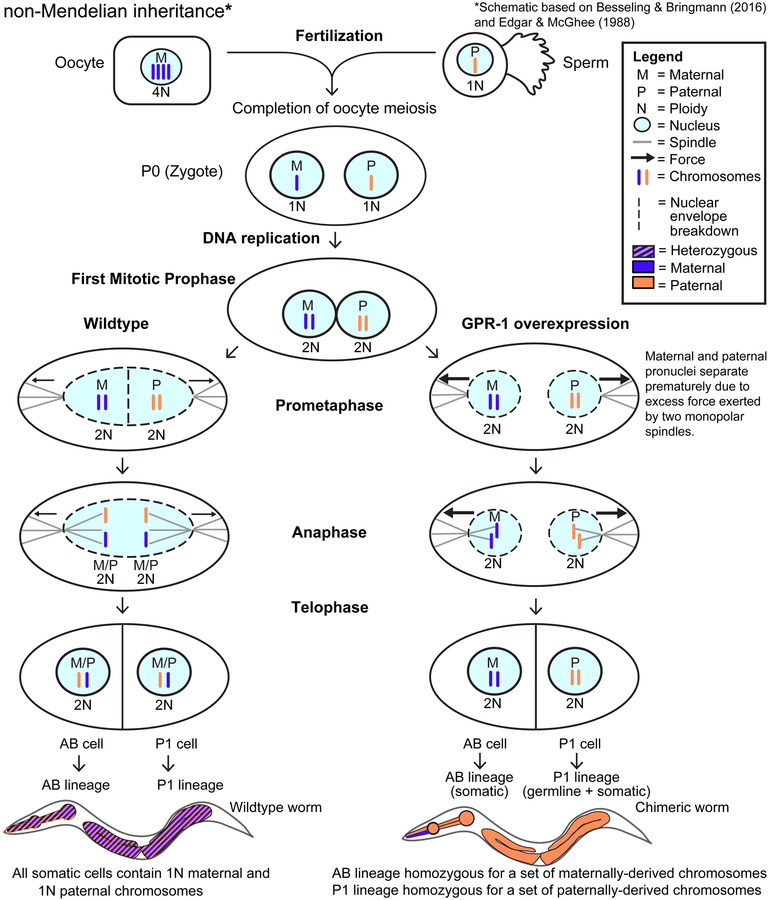
Figure 1. Overexpression of the microtubule force regulator GPR-1 induces non-Mendelian inheritance.
Diagram of zygotic division in wildtype (Edgar and McGhee, 1988) and GPR-1 overexpressing (OE) cells (Besseling and Bringmann, 2016). In wildtype, fertilization combines the oocyte and sperm to form the zygote (P0 cell). As meiosis completes, 3N DNA content is lost from the maternal pro-nucleus by polar body extrusion. The prophase zygote contains two pronuclei, containing 1N paternal (P, orange) or 1N maternal (M, blue) chromosomes, respectively. During S phase, chromosomes are duplicated, resulting in 2N DNA content in each pronucleus in the form of sister chromatids. In wildtype zygotes, the duplicated chromosomes are arranged on the metaphase plate. During anaphase, sister chromatids are separated resulting in one copy of each parental chromosome being inherited by each daughter cell. This zygotic division yields the two founder cells of the P1 and AB lineages. As expected from Mendelian inheritance, every somatic cell in wildtype animals contains a full complement of paternal (1N) and maternal (1N) chromosomes. In contrast, in the GPR-1 overexpression strain additional force is exerted on pronuclei which results in premature separation during prometaphase. This results in maternal and paternal chromosomes remaining at opposite poles thus preventing proper chromatid segregation. GPR-1(OE) worms that have undergone abnormal chromosome partitioning become chimeric, with a P1 lineage that is homozygous for paternal chromosomes and an AB lineage homozygous for maternal chromosomes. Because the P1 lineage gives rise to the germline, all oocytes and sperm from a GPR-1(OE) animal contain identical, paternally-derived chromosomes.
Besseling and Bringmann’s demonstration of rewired first mitosis in response to GPR-1 overexpression raises many exciting possibilities for strain engineering, while also leaving opportunities for improvements that might enable broad usage of this approach for developmental and physiological studies. The original GPR-1 over-expression transgene was prone to silencing (Besseling and Bringmann, 2016) (H. Bringmann, personal communication) making GPR-1 overexpression difficult to maintain. Also, the initial study used a very precise transgene expression pattern that, while highly specific, required in-depth microscopic analysis to assess whether non-Mendelian inheritance had occurred in any individual worm. Here we generate and characterize a toolkit of GPR-1 over-expressing strains with the following features: (i) consistent over-expression of GPR-1, (ii) reproducible high-penetrance of non-Mendelian segregation events (iii) direct, low-magnification visual assessment of Mendelian vs. non-Mendelian inheritance in individual F1 cross progeny, (iv) facile strain maintenance, and (v) incorporation of markers for quick identification of P1/AB mosaics. Working with these strains, we provide examples of several distinct capabilities: the facile production of genomewide homozygous (‘uniparental isodisomic’) animals, the ability to functionally distinguish between genes acting in the two C. elegans founder lineages (AB and P1), the production of strains carrying mitochondrial and nuclear genomes from different parental strains, and the ability to readily detect non-canonical inheritance events among large populations of animals. Cenik et al., in the accompanying paper, further demonstrate the use of the methodology in the investigation of cell non-autonomous interactions during C. elegans development [ref].
Results and discussion
Non-Mendelian inheritance in stable lines overexpressing GPR-1.
To generate strains with stable overexpression of GPR-1, we assembled a silencing-resistant GFP::GPR-1 transgene using principles from Frøkjaer-Jensen et al. (2016) (Figure 2A). The construct was designed with the following features: (i) A strong female-germline-specific RNA-Pol II promoter from the mex-5 gene (Merritt et al., 2008). (ii) A GFP-GPR-1 coding region optimized for high expression (Redemann et al., 2011) and with at least three mismatches relative to known piRNAs (Batista et al., 2008) to prevent spurious transgene silencing (Bagijn et al., 2012; Lee et al., 2012). (iii) To produce strong and robust expression, the synthetic gene contains multiple introns and a 3’ UTR rich in Periodic An/Tn Clusters (PATCs), a prevalent non-coding DNA feature in C. elegans which reduces transgene silencing in the animal’s germline (Fire et al., 2006; Frøkjaer-Jensen et al., 2016). We generated several independent strains with the transgene stably integrated as a single copy in the genome by Mos1 transposition (Frøkjaer-Jensen et al., 2014).
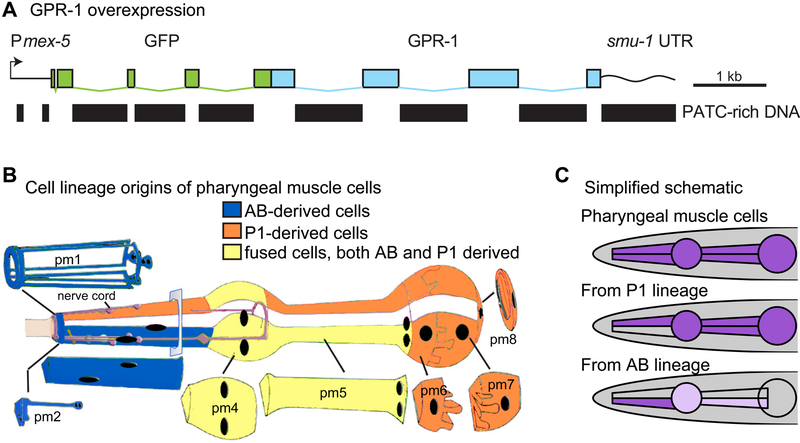
Figure 2. Developing an easily tractable system for non-Mendelian chromosomal inheritance by stable GPR-1 overexpression and pharyngeal muscle fluorescence.
(A) A silencing-resistant GPR-1 transgene, expressed under a germline-specific promoter (Pmex-5), was tagged with GFP at the N-terminus. PATC-rich DNA segments, which can counteract germline silencing, were incorporated into the transgene and are indicated as black bars (Frøkjaer-Jensen et al., 2016). A PATC-rich 3’ UTR from smu-1 was also included. (B) Lineages of pharyngeal muscle (PM) cells, adapted from Wormbook (Mango, 2007). PM1, PM2 muscle rings are derived entirely from the AB cell lineage (blue). Muscle ring PM3 consists of three lobes, two derived from the AB-lineage (blue) and one from the P1-lineage (orange). PM4 and PM5 consist of multinucleate cells resulting from the fusion of both AB and P1-derived cells (yellow). PM6 and PM7 consist entirely of P1-derived cells (orange). (C) Simplified schematic of fluorescently expressing AB, P1, and fused cells as viewed under a dissection microscope. Note that the fluorescence intensity in fused cells depends on the proportion of nuclei from the AB versus P1 lineage. For example, in the AB-derived pattern, expression is limited to 1/6 nuclei in PM4 and 2/6 nuclei in PM5, resulting in partial expression and dim fluorescence. The fainter fluorescence in these compartments can be difficult to visualize. Also, PM1 is not indicated in the schematic as it is hard to observe at low magnification fluorescence microscopy.
We first sought to set up a simple cross that could readily identify cases of mosaic uniparental inheritance. By using a genetic marker active in a tissue derived nearly-exclusively from one of the two founder cells, we can identify such events by direct inspection of the phenotype. As an initial assessment of potential non-canonical zygotic division, we used a simple cross in which males heterozygous for a temperature-sensitive loss-of-function allele of the body wall muscle myosin gene unc-54 (referred to here as unc-54(lf)/+ males) were crossed to hermaphrodites with no movement defect (unc-54(+) hermaphrodites) (Table 1). The recessive character of the unc-54(lf) mutation ensures that progeny obtained from a standard cross between unc-54(lf) heterozygote males with a wild type C. elegans hermaphrodite would all be expected to be movement competent (with both unc-54(lf)/+ and +/+ cross progeny showing full motility) (Brenner, 1974; Epstein et al., 1974). Crosses in which a noncanonical zygotic division event occurs in the zygote, by contrast, could be expected to yield atypical F1 progeny with the two founder blastomeres P1 and AB each deriving from a single maternal or parental haplotype. The unc-54 gene functions in the body wall musculature of the animal, a tissue derived almost completely (94 of 95 cells) from the P1 blastomere (Sulston et al., 1983; Epstein et al., 1974). F1 progeny in which the P1 blastomere genome derives from a single unc-54(lf) sperm would be expected to exhibit a near-complete unc-54(lf) phenotype. While a standard C. elegans cross produces a 1:1 mixture of male (XO) and hermaphrodite (XX) progeny, we note that noncanonical Unc progeny from the above crosses are expected to be exclusively hermaphrodite. This initially non-intuitive gender restriction derives from a consideration of the two sperm categories produced by the parental C. elegans XO males. Such male-derived sperm bear either a single X chromosome or no X chromosome. X-bearing sperm are expected to produce (after the standard genome duplication) a fully XX P1 lineage in the noncanonical early mitosis, while sperm bearing no X chromosome are expected to produce nullo-X P1 lineages. Based on the many essential genes on the X chromosome, the latter progeny class is not viable (Besseling and Bringmann). Thus, only XX animals (hermaphrodites) would be expected amongst viable uncoordinated F1 cross progeny. Given these genetic expectations, the occurrence (and hermaphrodite specificity) of a recessive Unc-54 phenotype in the first generation following a cross of candidate hermaphrodites with unc-54(lf)/+ males is a highly suggestive indicator of potential non-canonical mitosis events.
Table 1. Phenotypic classes of cross progeny expected from mating GPR-1(OE) hermaphrodites with unc-54(lf)/+ males.
The genotypes of the AB and P1 cell lineages, sex chromosome, and expected phenotypes are described. See also STAR Methods.
| F1 cross progeny classes | Genotype of AB lineage | Genotype of P1 lineage (including 94/95 wall musculature cells) | Sex Chromosomes | Phenotype (sex) | Phenotype (movement) |
|---|---|---|---|---|---|
| Mendelian | unc-54(lf)/+ | unc-54(lf)/+ | XO | male | wildtype |
| Mendelian | unc-54(lf)/+ | unc-54(lf)/+ | XX | hermaphrodite | wildtype |
| Mendelian | + | + | XO | male | wildtype |
| Mendelian | + | + | XX | hermaphrodite | wildtype |
| Non-Mendelian | + | unc-54(lf) | XX | hermaphrodite | Unc |
| Non-Mendelian | + | + | XX | hermaphrodite | wildtype |
| Non-Mendelian | unc-54(lf) | + | XX | hermaphrodite | wildtype |
When two candidate GPR-1 overexpression strains were crossed to unc-54(lf)/+ males, we observed phenotypically Unc worms predicted to result from non-canonical zygotic division. As expected, such animals were uniformly hermaphrodites. Self fertilization of Unc F1 progeny resulted in populations of unc-54(lf) homozygotes, as expected given the derivation of the germline from the P1 blastomere. Silencing of GPR-1 overexpression strains was also evident in such crosses: when using a hermaphrodite population with silenced GFP fluorescence from the GFP::GPR-1 transgene, we observed no Unc cross progeny. We thus hypothesized that it might be possible to use the GFP::GPR-1 signal as a surrogate to screen for strains with sustained GPR-1 overexpression. Additional candidate strains were obtained (PD1593, PD1594, PD1595) in which GFP germline fluorescence persisted through multiple generations of passaging at a variety of temperatures (16°C, 20°C, 25°C). One of the most stable strains (PD1594) was chosen for further characterization. PD1594 fertility was characterized by monitoring 36 individual L4 worms, of which 33/36 (92%) had viable progeny. Whole genome sequencing revealed that the transgene in PD1594 is located near the left tip of chromosome III at position 680195 with respect to the WBcel235/ce11 genome assembly.
Use of pharyngeal fluorescence markers to track non-canonical zygotic division.
Besseling and Bringmann (2016) used a broadly expressed fluorescent marker to identify mosaic animals. While enabling detection of potential mosaics through microscopic analysis, the generally-expressed markers are of limited value in studies requiring screening or selection of moderate to large numbers of animals. We thus sought alternate fluorescent markers that could be incorporated in either parent in the non-canonical cross and could be used under a low-magnification fluorescence dissecting microscope to identify cross progeny with non-canonical inheritance. As a marker of particular utility, we chose GFP and mCherry constructs expressed in pharyngeal muscles. The C. elegans pharynx consists of eight pharyngeal muscle (PM) rings and is chimeric with respect to the AB and P1 cell lineages. Differential chromosomal derivation of AB and P1 genomes can thus be identified by differential fluorescence in the AB and P1-derived subpopulations of the pharynx. The lineal derivation of the pharyngeal musculature is positionally distinctive, with posterior muscle rings (PM6, PM7, and PM8) deriving completely from the P1 lineage, while portions of the anterior (PM1, PM2, and a lobe of PM3) are completely AB-derived (Figure 2B) (Sulston et al., 1983). PM4 and PM5 consist of multinucleate cells resulting from the fusion of both AB and P1-derived cells. A simplified schematic of pharyngeal cell lineages in Figure 2C shows how the cell subpopulations can create distinct patterns. The myo-2 promoter drives expression throughout the pharyngeal muscle (PM) cells (Miller et al., 1986; Okkema and Fire, 1994), making it an ideal tool to generate easily discernable patterns to visually indicate animals with a wild-type or aberrant zygotic division.
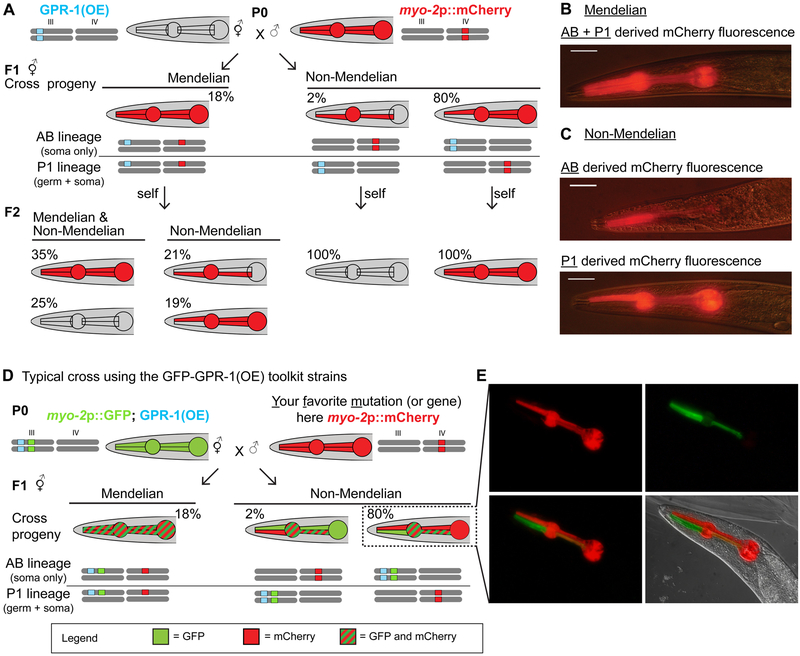
To visualize pharyngeal mosaicism, males containing an integrated myo-2p::mCherry dominantly visible transgene (VS21) were crossed to GPR-1 overexpressing (GPR-1(OE)) hermaphrodites (PD1594) (Figure 3A). Cross progeny exhibited the three expected pharyngeal fluorescence patterns: 1) uniform mCherry throughout the pharynx, indicative of a wildtype zygotic division (18% ± 5%, mean ± SEM) of hermaphrodites, 2) mCherry expression restricted to the AB-lineage, indicative of a non-Mendelian zygotic division resulting in a maternally derived P1 lineage (2% ± 1%) of hermaphrodites), and 3) mCherry expression restricted to P1-derived cells, indicative of a non-Mendelian zygotic division with a paternally derived P1 lineage (80% ± 6%) of hermaphrodites) (Figure 3A–C and Table 2). Sperm entry into the zygote (Goldstein and Hird, 1996) determines a cell’s posterior axis and the biased segregation of paternal DNA into the posterior P1 lineage is therefore expected by simple relative positioning. These results indicate AB/P1 mosaicism rates comparable to the highest rates observed by Besseling and Bringmann (2016). We assayed fertility of non-Mendelian classes of progeny and found that both P1-patterned hermaphrodites (33/34 (97%)) and AB-patterned hermaphrodites (6/6 (100%)) were highly fertile. As expected, male progeny were almost exclusively of the canonical mitosis pattern (mCherry throughout the pharynx). We did, however, identify occasional (<1%) progeny exhibiting partial male morphology and a P1 pharyngeal fluorescence pattern. These seemingly intersex animals neither mated nor were self-fertile and could conceivably have resulted from mitotic X chromosome loss in a single blastomere (Hunter and Wood, 1992). Single F1 hermaphrodites of all three pharyngeal fluorescence patterns were allowed to self-fertilize, and patterns of the resulting F2 were observed. P1 or AB-patterned F1 hermaphrodites yielded progeny with homogeneous pharyngeal patterns, consistent with an isogenic F1 germline. Uniformly fluorescent “Mendelian” F1 yielded a high proportion of P1 and AB patterned F2 animals indicating that a single maternal copy of the GPR-1 over-expressing chromosome is sufficient to induce non-Mendelian inheritance.
Figure 3. Pharyngeal myo-2p::mCherry patterns in GPR-1(OE) crosses.
(A) We crossed GPR-1 overexpressing hermaphrodites (GPR-1(OE), PD1594) to males with pharyngeal mCherry (myo-2p::mCherry, VS21) and characterized resulting classes of hermaphroditic F1 cross-progeny by fluorescence. Chromosome schematics indicate classic Mendelian segregation with maternal and paternal DNA “mixing” (left) and non-Mendelian segregation with no DNA mixing (right). Chromosomes are indicated by grey bars and genotype is indicated by grey (wildtype) or colored (blue = GPR-1(OE), red = myo-2p::Cherry) boxes. F1 segregation (see Table 2A: Mendelian = 18% (± 5%), Non-Mendelian (Maternal DNA-> P1 lineage) = 2% (± 1%), and Non-Mendelian (Paternal DNA-> P1 lineage) = 80% (± 6%), mean ± SEM (N = 4 crosses)). F2 self-progeny of four Mendelian heterozygous F1s were also classified and quantified (Table 2B: full pharynx 35% ± 4%, P1 lineage only 19% ± 2%, AB lineage only 21% ± 2%, no fluorescence 25% ± 3%). (B-C) Photomicrographs of mCherry expression in the pharynx of F1 hermaphrodite cross progeny: (B) full pharyngeal expression (C) Top: AB-derived pharyngeal expression. We note that faint mCherry expression in PM4 and PM5 is expected, as only 3 of 12 nuclei are derived from AB lineage. Bottom: P1-derived pharyngeal expression. (D) A typical cross using a GPR-1(OE) toolkit strain to analyze expression of Your Favorite Mutation/gene (YFM) in chimeric worms exemplified here by using myo-2p::mCherry as YFM. We crossed pharyngeal GFP marked GPR-1 overexpressing hermaphrodites (myo-2p::GFP; GPR-1(OE), PD2220) with myo-2p::mCherry homozygous males (YFM). Left: Hermaphroditic “Mendelian” cross progeny uniformly expressing both GFP and mCherry throughout their pharynxes (18%). Right: Hermaphroditic “non-Mendelian” cross progeny, which have presumably undergone an abnormal zygotic division resulting in maternally derived chromosomes in the AB cell lineage and paternally derived chromosomes in the P1 cell lineage (80%), expressing GFP in all AB-derived pharyngeal tissue. Half of these chimeric animals also expressed paternally inherited mCherry exclusively in their P1-derived pharyngeal tissue. Expected % of hermaphroditic cross progeny of each class are shown. (E) Fluorescence microscopy showing the pharyngeal pattern of F1 non-Mendelian cross progeny from (D). Top. mCherry is expressed throughout the P1-derived pharyngeal cells. GFP is expressed throughout the AB-derived pharyngeal cells. Bottom. Merged fluorescent images and DIC image overlay.
Table 2A. Frequencies of distinct mosaic cross progeny classes resulting from maternal GFP-GPR-1 expression.
F1 cross progeny of VS21 (myo-2p::mCherry::unc-54 3 ‘UTR) males crossed with PD1594 (mex-5p::gfp::gpr-1::smu-1 3’UTR) hermaphrodites were examined for pharyngeal fluorescence expression patterns. The number and percentage of F1 hermaphrodites that displayed each pattern is described.
| # | N (%) | N (%) | N (%) | N |
|---|---|---|---|---|
| 1 | 57 (80.3%) | 2 (2.8%) | 12 (16.9%) | 71 |
| 2 | 52 (88.1%) | 1 (1.7%) | 6 (10.2%) | 59 |
| 3 | 29 (64.4%) | 1 (2.2%) | 15 (33.3%) | 45 |
| 4 | 51 (87.9%) | 0 (0.0%) | 7 (12.0%) | 58 |
| All hermaphrodite cross progeny | 189 (80.2%) | 4 (1.7%) | 40 (18.1%) | 233 |
Construction of a collection of fluorescently marked toolkit strains
We sought to generate a collection of fluorescently marked strains that can be used for non-Mendelian crosses. Constructing marked strains with the GPR-1 overexpression loci entails an added complication that the majority of the progeny from such strains (the ~80% from non-canonical mitosis) carry uniparental germlines not useful for strain construction. Fortunately, the modest fraction of progeny deriving from Mendelian segregation provides a means to obtain the desired doubles, with these animals conveniently distinguishable through the uniform fluorescence throughout-the-pharynx. A listing of toolkit strains is presented in the Key Resources Table, with a schematic for such strain constructions presented in Figure 3A–C and a cross exemplifying the use of a toolkit strain illustrated in Figure 3D–E and Figure S1.
Mosaic analysis of vulval development using GPR-1(OE) toolkit strains
By providing an efficient means to produce and identify AB/P1 genetic mosaics, the fluorescently marked GPR-1(OE) strains should facilitate a variety of site-of-action assessments that would otherwise be both challenging and labor intensive. As a test case for such analysis, we have assessed AB versus P1 lineage requirements for a set of genes with roles in patterning the C. elegans vulva (Figure 4, Figure S1, and Table S1). The C. elegans vulva forms during late larval development from a set of epithelial precursor cells (the AB-derived vulval precursor cells [“VPCs”]), with the pattern reflecting a combination of extrinsic signals from the P1 derived anchor cell (“AC”) and interactions amongst VPCs. Numerous genetic contributions to this process have been defined from mosaic analysis or inferred from expression patterns, phenotypes, and epistasis experiments (Schmid and Hajnal, 2015). As a result, many of these genes have been classified as acting either in the AB or P1 lineage in vulval development.
Figure 4. Analysis of vulval development mutants.
(A) Vulval development relies in part on communication between the Anchor Cell (AC) and the Vulval Precursor Cells (VPCs) via EGF-like and Notch-like signaling pathways. (B-D) Cell lineage diagrams illustrating how (B) VPCs and the AC/VU precursors are derived from the two lineages (AB and P1) resulting from division of the zygote. (C) Cell genotypes in AB/P1 mosaics with a mutation restricted to the P1 lineage will include mutant AC/VU precursors and wild-type VPCs and (D) conversely, cell genotypes in AB/P1 mosaics with a mutation restricted to the AB lineage will include wild-type AC/VU precursors and mutant VPCs (E) A summary of phenotypes observed when the listed vulval development mutation was present in either the P1 lineage (including AC/VU precursors) or the AB lineage (including VPCs). Gene, allele used, previously inferred/expected role, expected lineage of action, and phenotype observation (aggregate number of worms exhibiting stated phenotype/number examined @23°C and corresponding percentage) is listed. Additional data and details, including the crosses used to generate these chimeric worms, are described in Figure S1 and Table S1.
We chose to examine four vulval development genes to distinguish roles in AB versus P1 by mosaic analysis. lin-3 encodes an EGF (epidermal growth factor)-like ligand that is secreted by the anchor cell, inducing vulval lineages in nearby VPCs (Hill and Sternberg, 1992). lin-2 encodes a component thought to act at the level of signal reception in the VPCs (Hoskins et al., 1996). lin-31 encodes an HNF/Forkhead transcription factor regulating specification in the VPC nuclei (Miller et al., 1993). lin-12 encodes a notch-like transmembrane receptor acting in both in P1 and AB lineages (required for specification of the anchor cell in the P1 lineage and for lateral inhibition of “primary” vulval fates in the AB-derived VPC cells) (Greenwald et al., 1983; Seydoux and Greenwald, 1989; Sternberg and Horvitz, 1989a; Yochem et al., 1988). AB/P1 mosaics for all four genes were readily produced, each using a single genetic cross, including both reciprocal classes in most cases (Table S1A).
For the three genes thought to contribute to vulval development from a single lineage (lin-3, lin-2, and lin-31), all results were consistent with the expected lineage-of-action (AB for lin-2 and lin-31, P1 for lin-3) (Table S1B). Allowing a slightly more complex challenge for the GPR-1(OE) methodology, we used two gain-of-function alleles of lin-12 expected to have different phenotypic effects on vulval development depending on their presence in AB versus P1 lineage. From previous work (Greenwald and Seydoux, 1990; Sternberg and Horvitz, 1989b), a lin-12(gf) allele can act in the P1 lineage to block anchor cell specification, leaving a vulval primordium with no inductive signal (equivalent to lin-3), while the presence of a lin-12(gf) signal reception only in the AB-derived VPCs can produce duplicated vulval structures (Multivulval or “Muv” phenotype; particularly evident with allele n952). We observed the expected phenotypes in both P1-limited and AB-limited lin-12(gf) mosaics for both alleles, with the expected higher incidence of Muv phenotypes with the n952 allele (Table S1B).
While these results do not replace the many careful localization, epistasis, lineage, and mosaic analyses carried out on the well-studied vulval system, they do provide a strong confirmation that the GPR-1(OE) toolkit can reliably dissect genetic contributions from the two founder lineages in C. elegans mosaics, thus contributing substantially to the toolkit available for studies of localized gene function in this model organism.
Generation of chimeric animals for analysis of male-specific phenotypes:
As previously noted, male-specific phenotypes cannot be readily investigated using GPR-1 overexpression due to the heterogametic nature of the C. elegans male gender. With whole genome duplication just before the first mitosis, the two constituent lineages can either be (XX) or (nullo-X) but not easily the single X (XO) configuration leading to male development. Fortunately, there are several means to obtain animals with male characteristics with simple diploid and homogametic character (Hodgkin, 2002; Hodgkin and Brenner, 1977). An unusual loss of function allele of the tra-2 gene, tra-2(q276), provides a good example of such a configuration. Fully diploid XX homozygotes for this allele show a remarkably complete gender transformation, with male phenotypes and behaviors and the ability to sire viable progeny in crosses (Chen et al., 2003). To apply this for production of functionally mosaic males, we needed to construct strains in which both AB and P1 lineages would be XX tra-2(q276) homozygotes but where these two lineages could carry different genetic markers. Such strains were constructed by using a balancer chromosome (mnC1) to maintain stocks as hermaphrodites and by including fluorescent markers that allow definitive identification of chimeric trans-males. Several crosses (diagramed in Figure S2) confirmed that these strains could indeed be used to produce such chimeric males. As yet we have not had success in generating cross progeny from these males, although this may reflect a burden of genetic markers in the strains rather than any fundamental characteristic of mating. Despite the genetic complexity, we expect that the ability to produce mosaic male-phenotype animals using GPR-1 overexpression and tra-2(q276) gender transformation will provide a valuable tool for investigating both lineage-specific contributions to male morphology and behavior and parent-of-origin effects on chromosome structure and function.
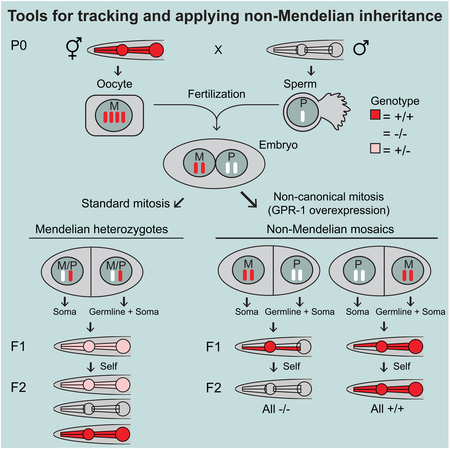
Nuclear/mitochondria genome exchange and one-step isogenesis through non-canonical genetics
Non-Mendelian inheritance patterns should allow novel, and potentially useful, genetic manipulations. For example, matings with GPR-1(OE) hermaphrodites should allow rapid creation of nuclear-genome-wide isogenic strains since the genetic makeup of a non-Mendelian segregation event is derived from a single copy of each chromosome. In C. elegans, and potentially other genetically tractable organisms where a similar inheritance could be reproducibly forced, rapid homozygosing would facilitate genetic screens. Similarly, cytoplasmic factors such as the genetic material in mitochondria are almost exclusively contributed by the oocyte (Tsang and Lemire, 2002; Feng et al., 2001). Therefore, the progeny of a male crossed with GPR-1(OE) hermaphrodites will receive only maternal mitochondria and other cytoplasmic factors. Such experiments could, for example, test models of trans-generational inheritance mediated by small RNAs (Rechavi and Lev, 2017) or speciation events driven by mitochondrial and nuclear incompatibilities (Jhuang et al., 2017). To test the feasibility of such experiments, and further confirm the functionality of the GPR-1(OE) strains, we sought to create a set of strains that each contain a nuclear genome entirely derived from the male parent, and a mitochondrial genome entirely derived from the maternal parent. We chose to utilize a wild strain (PX174) that has diverged sufficiently from the wildtype lab reference strain (N2) to allow a definitive assessment of chromosome-wide derivation throughout the genome. PX174 males were crossed to two of the GPR-1(OE) toolkit strains, PD2117 and PD2118 (Figure 5A), and AB-fluorescence-patterned F1 progeny from each cross were singled and allowed to self to create a line. The genomes of four resulting strains (two from each cross) were subjected to shotgun sequencing followed by SNP calling. Comparison between parental and progeny DNA showed that all four strains contained mitochondrial SNPs exclusively matching the maternal mitochondrial genome, whereas the nuclear genomes were derived exclusively from the paternal genome (Figure 5B). These experiments confirm both the maternal derivation of mtDNA for C. elegans as in other organisms (e.g. Reich and Luck, 1966; Hutchison Iii et al., 1974; Feng et al., 2001; Tsang and Lemire, 2002) and the ability to exchange mitochondrial and nuclear genomes using the GPR-1 overexpression strains as an intermediate facilitator.
Figure 5. Nuclear/mitochondria genome exchange and one-step isogenesis through non-canonical genetics.
(A) Experimental schematic. We generated nuclear/mitochondrial genome hybrid strains by crossing fluorescently marked GPR-1(OE) strains (either PD2217 or PD2218) with a wild strain (PX174) containing many polymorphisms relative to the laboratory N2 strain. Non-Mendelian F1 progeny were selected based on AB-specific expression of a maternally derived pharyngeal fluorescence reporter (either myo-2::GFP or myo-2::mCherry). Hybrid strains were generated by picking self-progeny. Hybrid strains (PD2231, PD2232, PD2233, PD2234), predicted to have a nuclear genome entirely derived from the male parent and a mitochondrial genome entirely derived from the maternal parent were sequenced along with the parent strains (PX174, PD2217, PD2218). Note that sperm-derived paternal mitochondria, which are rapidly eliminated by autophagy after fertilization (Sato and Sato, 2011), were omitted from the schematic for clarity. (B) SNPs unique to strain PX174 (relative to N2 reference strain VC2010) were determined using a Kmer-based approach. For each parental and hybrid strain, the number of sequencing reads matching the paternal (orange) and maternal (blue) base at each SNP position along the chromosome are plotted.
Limitation of prevalent mosaic uniparental isodisomy under laboratory conditions to engineered populations.
Fascinated by the ability of C. elegans to (i) proceed through development following an aberrant first cleavage, and (ii) to produce a viable population from such events, we wondered whether this process might be a natural means for animals of this genus to navigate challenges such as a low local population in maintaining haplotypes and populations. In particular, situations where numerous related strains might cohabit an environment in a shared niche but remain incompatible for “standard” Mendelian (crossing-based) reproduction might be less deleterious to the species and genus (see Schwander and Oldroyd, 2016 for additional discussion) if there were an option (albeit potentially rare) for the kind of noncanonical embryonic behavior enabled by the GPR-1 strain. Numerous previous genetic studies (Brenner, 1974; Hodgkin et al., 1979) have not been suggestive of such noncanonical behavior as a common event, but it remained possible that events were missed in the many C. elegans genetic studies that have been published. Regardless of the plausibility of any evolutionary argument, it seemed important to assess the fidelity of chromosome mixing after fertilization.
We used the GFP reporter markers described above to carry out a medium scale search for “spontaneous” aberrant first-mitosis events such as those observed above. We examined 2094 cross progeny from myo-2p::mCherry males mated with wild type worms at 25°C, screening for worms with an abnormal pharyngeal pattern reminiscent of GPR-1(OE) induced abnormalities. No abnormal pharyngeal patterns were found, suggesting that viable progeny resulting from such events are rare (if present at all). We note that such biological alterations might be under biological control and it will be of interest to determine (through mutagenesis or by varying environmental conditions) whether conditions exist where a mosaic first cleavage is used by the organism at detectable frequencies.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew Z. Fire (afire@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans
C. elegans animals were grown on nematode growth media (NGM) plates seeded with OP50 bacterial cultures as described (Brenner, 1974). Transgenic strains were maintained and passaged using standard techniques. Derivation of transgenic GPR-1(OE) lines was initially at 25°C; lines have been maintained at temperatures between 16°C and 23°C. Crosses were carried out at 23°C, with noncanonical inheritance confirmed at 23°C and 25°C.
METHOD DETAILS
Generating GPR-1 overexpression strains
We generated a silencing-resistant GPR-1 transgene, tagged with GFP at the N-terminus, using step-wise assembly with codon optimization (Redemann et al., 2011), gene synthesis (IDT), Gibson assembly (Gibson et al., 2009), Golden gate assembly (Engler et al., 2009), and Gateway assembly (Invitrogen). A manuscript with a detailed description of the assembly method is in preparation (Froekjaer-Jensen et al.).
In brief, we optimized the coding region of GPR-1 used by (Besseling and Bringmann, 2016) for high expression with a web-based tool from Redemann et al. (2011) (https://worm.mpicbg.de/codons/cgi-bin/optimize.py) with the parameter: COI = 1, three synthetic introns, and no RNAi resistance. BsaI and BsmBI sites were removed from the optimized coding sequence for compatibility with Golden Gate cloning. piRNA binding sites with less than three mismatches were removed by synonymous base pair substitutions. The codon-optimized gene was ordered in three fragments (gCFJ92, gCFJ93, gCFJ94; sequence files in File S1) by gene synthesis (IDT) as “G-blocks” compatible with blunt cloning. The gene fragments were blunt end cloned into a shuttle vector and sequence verified by Sanger sequencing. Each gene fragment contained a synthetic intron into which we inserted approximately 900 base pairs of PATC-rich DNA using Golden Gate assembly (Engler et al., 2009) with a BsaI restriction enzyme (NEB). The three PATC-rich GPR-1 fragments were combined with a PATC-rich (3 × 750bp) codon-optimized GFP fragment by a second round of Golden Gate assembly (Engler et al., 2009) using BsmBI (NEB) to make a full-length GFP::GPR-1 gene in a three-fragment Gateway compatible vector (Invitrogen). The final construct (pCFJ2522; Genbank annotated sequence in File S1) was generated by Gateway assembly of a germline specific promoter mex-5 (Zeiser et al., 2011), GFP::GPR-1, and a PATC-rich 3’UTR from smu-1 (Fire et al., 2006) into a miniMos transposon vector backbone containing an unc-119 positive selection marker and a peel-1 negative selection marker (Frøkjaer-Jensen et al., 2014).
The pCFJ2522 plasmid was injected at 10 ng/ul into unc-119 animals derived from PS6038 together with a plasmid expressing Mos1 transposase, red fluorescent markers, and stuffer DNA from a 1 kb ladder to generate single-copy transposon insertions (Frøkjaer-Jensen et al., 2014). We isolated animals with a transposon insertion based on homozygous rescue of the unc-119 phenotype, absence of the red co-injection markers, and no lethality from the peel-1 toxin after heat shock. From 16 independent insertions, we identified 5 strains with stable GFP expression at 25°C. Transgene silencing in the germline is common (Kelly et al., 1997), either acute or gradually. Silencing is especially common at lower temperatures (15°C to 20°C) compared to 25°C (Strome et al., 2001). We therefore propagated three strains (PD1593, PD1594, PD1595) at 16°C, 20°C and 25°C for several generations; no silencing was observed based on GFP fluorescence over three generations. The miniMos insertion sites were mapped using inverse PCR as previously described (Frøkjaer-Jensen et al., 2014) and locations are noted in the Key Resources Table. We picked one strain, PD1594, for further characterization. The PD1594 insertion site was also mapped by whole-genome sequencing to Chr. III:680195 as described below.
Fertility Assays
Fertility was assayed by determining the percentage of individual L3–L4 staged worms that eventually gave rise to viable progeny.
Imaging and Scoring of fluorescent markers
Pharyngeal and body wall fluorescence was scored using a fluorescence dissecting microscope. A compound microscope with a 40x oil objective was used for scoring germline fluorescence of GFP::GPR-1(OE) expressing strains and for all imaging.
Validation of GPR-1(OE) strains
To test for non-Mendelian zygotic division, GPR-1(OE) candidate strains (non-Unc worms containing a GFP+ germline) were crossed to unc-54(unc-54(e1301)::gfp::TAA::NSUTR)/+ males derived from mating PD2882 with PD1074 males (Table 1). Strains capable of yielding phenotypically Unc F1 were identified as GPR-1 overexpressors.
As an additional test for non-Mendelian zygotic division GPR-1(OE) candidate strains were crossed to a set of strains expressing fluorescent markers in either the pharynx (VS21, CGC18, CGC38) or both the pharynx and body wall muscle (EG4887). Strains capable of yielding F1 with pharyngeal fluorescence restricted to either the AB or P1 lineage were identified as GPR-1 overexpressors.
Fluorescently marked GPR-1(OE) toolkit strains
GPR-1 overexpressing hermaphrodites (PD1594) were mated with males containing either mCherry or GFP pharyngeal markers (VS21, CGC18, CGC38). F1 hermaphrodites with homogeneous fluorescence throughout the pharynx (indicating that they were Mendelian heterozygotes) were singled and allowed to self-fertilize. The resulting F2 animals exhibited both canonical and non-canonical pharyngeal fluorescence. The high proportion of P1 and AB patterned F2 animals indicates that a single maternal copy of the GPR-1 over-expressing chromosome is sufficient to induce non-Mendelian inheritance. F2 animals were again self-fertilized and the resulting pharyngeal patterns were observed. The progeny of P1-patterned F2 animals consistently gave rise to F3 progeny homozygous for the fluorescent pharyngeal marker, and these populations were assayed for GFP::GPR-1(OE) homozygosity by fluorescence microscopy to identify double homozygous strains (PD2217, PD2218, PD2219, PD2220). In contrast, AB-patterned F2 progeny gave rise to F3 animals lacking the fluorescent pharyngeal marker (data not shown). PD2219 was subsequently mated with males containing a myo-2::mCherry, myo-3::mCherry transgene expressed in nuclei (EG4887), and double or triple homozygous populations were selected in a similar manner (PD2224, PD2227). Additional GPR-1(OE) strains (PD1593, PD1595) were similarly mated with mCherry or GFP pharyngeal markers (VS21, CGC18) to generate strains with alternate GPR-1(OE) transgene locations (PD2238, PD2239, PD2240, PD2242). This collection of fluorescently marked toolkit strains are listed in the Key Resources Table. The genotype of the unc-119 locus was determined by PCR amplification using primers AF-KLA-396 and AF-KLA-397 and subsequent Sanger sequencing using AF-KLA-398 by MCLAB.
Tracking non-Mendelian inheritance
To demonstrate fluorescent tracking of mosaics using a toolkit strain, an mCherry/+ male (derived from mating N2 males with VS21) was crossed with a GPR-1(OE) toolkit strain marked with GFP pharyngeal fluorescence (PD2218). This resulted in a large population of chimeric, non-Mendelian F1 cross progeny which were readily identifiable by their AB-derived pharyngeal GFP pattern. Half of these also contained myo-2:mCherry expressed in the P1-lineage. In these dual labeled chimeric animals, P1-derived pharyngeal tissue expresses GFP and contains exclusively paternally derived chromosomes, whereas AB-derived pharyngeal tissue expresses mCherry and contains exclusively maternally derived chromosomes.
Construction of a lin-31(e1417); GPR-1(OE), myo-2p::GFP strain
To allow identification of cross progeny, we utilized the myo-2p::mCherry marker in VS21 (hjSi20 [myo-2p::mCherry::unc-54 3’ UTR] IV). VS21 males were crossed with MT301 hermaphrodites and the resulting myo-2p::mCherry/+, lin-31(n301)/+ males were mated with PD2218 (ccTi1594[mex-5p::gfp::gpr-1::smu-1 3’ UTR, cbr-unc-119(+)] umnIs7 [myo-2p::GFP + NeoR] III). A single F1 hermaphrodite exhibiting a homogeneous pharyngeal mCherry pattern (indicative of cross progeny resulting from Mendelian chromosomal segregation) was allowed to self-fertilize. F2 Progeny exhibiting GFP+ in the P1-derived pharyngeal cells, but lacking mCherry, were singled out and allowed to self-fertilize. The resulting populations were monitored for Muv phenotype and germline GFP expression. Two phenotypically mCherry-, pharyngeal GFP+, germline GFP+, Muv strains were identified (PD2279, PD2280).
Facile construction of a triple mutant strain using transient GPR-1 overexpression
To build a lin-2(e1309); him-3(e1147), myo-2p::mCherry strain, we took advantage of homozygosis induced by GPR-1(OE). Heterozygous males for all three markers were mated with a GPR-1(OE) strain and non-Mendelian cross progeny identified, generating a population of chimeric F1, each with a genome-wide homozygous germline. To do this, VS21 males were crossed with PD4769 (lin-2(e1309) X; him-3(e1147) IV) hermaphrodites and the resulting lin-2(e1309)/+ X; him-3(e1147)/myo-2p::mCherry IV males were mated with PD2218 hermaphrodites. Individual progeny (mCherry+, GFP-) in the P1-derived pharyngeal cells were singled out and allowed to self. The resulting homozygous mCherry+ populations were screened for Him, Vul and germline GFP+ phenotypes, yielding PD2278 (lin-2(e1309) X; him-3(e1147), hjSi20 [myo-2p::mCherry::unc-54 3’UTR] IV). We note that this strategy is particularly useful in generating homozygous animals with dominant visible markers such as fluorescence.
Analysis of lin mutations in Chimeric worms.
Homozygous males carrying a vulval development mutation (strains PD4769, PD2278, MT2021(lin-12(n952) III; him-5(e1467)V) or MT1329(lin-12(n302) III; him-5(e1467) V)) were mated with pharyngeal mCherry (PD2217) or GFP (PD2218) marked GPR-1(OE) hermaphrodites at 23°C as described in Table S1A. Non-Mendelian cross progeny were identified by their pharyngeal fluorescence patterns and were singled out onto fresh plates. Maternally inherited fluorescence marked which cell lineage (AB or P1) had inherited maternal chromosomes and indicated that the other lineage was homozygous for the paternally inherited vulval development mutation (see Figure S1). 9 to13 animals were scored for the percentage of Muv (multivulval), Egl (egg-laying defective), inviable eggs laid, and overall fertility by monitoring for 7 days. We note that in some cases we also utilized a myo-2p::mCherry paternal marker, as it greatly eased simultaneous detection of cross progeny and non-Mendelian pharyngeal patterns.
In cases where it was difficult to obtain a homozygous male containing the vulval mutation (MT301(lin-31(n301) II) and CB1417(lin-3(e1417) IV)), hermaphrodites were first mated with VS21 or N2 (Fire Lab N2 [wild type] clonally derived from Brenner’s original N2 strain (Brenner, 1974)) males respectively. The resulting heterozygotes were mated with PD2217 or PD2218 hermaphrodites, and chimeric progeny were selected and analyzed as described above. In these cases, the inherited genotype was inferred by scoring the phenotype of the F2 progeny, which reflected that the homozygous germline-derived from the paternal (or in one case maternal) parent. A reciprocal cross was also performed in order to look at a higher fraction of chimeric animals containing homozygous lin-31(n301) in the VPCs (AB lineage). VS21 males were mated with PD2279 or PD2280 (lin-2(e1309) X; him-3(e1147), hjSi20 [myo-2p::mCherry::unc-54 3’UTR] IV) hermaphrodites, and the resulting chimeric animals were selected and analyzed as described above.
Construction of toolkit balancer strains
Heterozygous males containing one copy of a chromosomal inversion were generated by crossing N2 males to FX30123 (tmC24), PD7119 (mIn1), CGC43(mnC1), CGC48 (mnC1), and were then crossed to the GPR-1(OE) toolkit strains, in various combinations. Heterozygous F1 Mendelian cross progeny were selected based on a full pharyngeal fluorescence pattern and were allowed to self-fertilize. Single F2 progeny containing fluorescence only in the P1-lineage derived pharyngeal cells were allowed to self-fertilize. F3 populations which scored positive for pharyngeal fluorescence and GFP expression in the germline were selected as double (or in some case triple) mutant strains. Any heterozygote strain (including any balancer-containing heterozygote) carrying the GPR-1(OE) transgene will be intrinsically more difficult to maintain than the original non-GPR-1(OE) counterpart, since maintenance will require heterozygote progeny while a majority of progeny exhibit genome-wide homozygosity.
We note that we were unable to generate either an mIn1; GPR-1(OE) or mnC1; GPR-1(OE) double homozygous strain. This appeared to reflect combined viability effects of the different markers and GPR-1(OE) and not a genetic linkage, since three GPR-1(OE) loci on different chromosomes presented similar challenges.
Construction of tra-2(q276); GPR-1(OE) strains
In order to generate chimeric male worms, we utilized a unique tra-2(q276) allele for which XX animals develop as mating-competent males. We note considerable precedent in using transformed XX males for specific genetic tests, despite the complexity of producing such strains (e.g. (Hodgkin et al., 1979), who used a similar strategy [described in their paper in detail, albeit in small print] to check for defects in male spermatogenesis in chromosome segregation mutants. Two heterozygous tra-2(q276)/mnC1 [myo-2p::GFP] II; GPR-1(OE) III strains, one containing an additional myo-2p::mCherry marker, (PD2275 and PD2286) were constructed as follows:
An intermediate strain, PD2246, was made by mating JK987 (tra-2(q276)/mnC1 [dpy-10(e128) unc-52(e444)] II) XX males with CGC43 (unc-4(e120)/mnC1 [dpy-10(e128) unc-52(e444) umnIs32 [myo-2p::GFP + NeoR] II) hermaphrodites, and selecting GFP+ worms with wild type movement that yielded populations capable of producing XX males lacking pharyngeal GFP. The resulting strain was saved as PD2246 (tra-2(q276)/mnC1 [dpy-10(e128) unc-52(e444) umnIs32] umnIs32 [myo-2p::GFP + NeoR, II: 11755713] II).
A second intermediate strain, PD2252, was constructed by first mating VS21 (hjSi20 [myo-2p::mCherry::unc-54 3’UTR]) males with CGC43 hermaphrodites, and subsequently mating the resulting mnC1/+, myo-2p::mCherry/+ males with PD2246 hermaphrodites. Individual mCherry+, GFP+ progeny, also capable of producing XX males lacking pharyngeal GFP, were identified. A population grown from one such individual was saved as PD2252 (tra-2(q276)/mnC1 [dpy-10(e128) unc-52(e444) umnIs32] umnIs32 [myo-2p::GFP + NeoR, II: 11755713] II).
Finally, PD2286 and PD2275 were constructed as follows: PD2252 XX males were mated with PD1594 (ccTi1594[Pmex-5::gfp::gpr-1::smu-1 3’ UTR, cbr-unc-119(+), III:680195*] unc-119(ed3) III.) hermaphrodites. A heterozygous tra-2(q276)/+ II; GPR-1(OE)/+ III; myo-2p::mCherry/+ IV F1 resulting from Mendelian chromosomal inheritance was identified by its homogeneous full pharyngeal mCherry pattern and was allowed to self fertilize. F2 XX males exhibiting mCherry+ pharynxes (which could be either Mendelian or non-Mendelian F1 progeny) were mated with PD2246 (tra-2(q276)/mnC1) hermaphrodites. Resulting hermaphrodite cross progeny with homogeneous GFP+, mCherry+ pharynxes were singled and allowed to self-fertilize. Populations were monitored for chimeric pharynxes indicative of GPR-1(OE). Further selection yielded a homozygous GPR-1(OE) population that was verified by uniform germline GFP expression. Homogeneous mCherry+ and mCherry-animals were identified by fluorescence microscopy and saved as mCherry positive strain PD2275 (tra-2(q276)/mnC1 [dpy-10(e128) unc-52(e444) umnIs32] umnIs32 [myo-2p::GFP + NeoR, II: 11755713] II; ccTi1594[mex-5p::gfp::gpr-1::smu-1 3’ UTR, cbr-unc-119(+), III:680195*], unc-119(ed3)? III; hjSi20 [myo-2p::mCherry::unc-54 3’UTR] IV) and mCherry negative strain PD2286 (tra-2(q276)/mnC1 [dpy-10(e128) unc-52(e444) umnIs32] umnIs32 [myo-2p::GFP + NeoR, II: 11755713] II; ccTi1594[mex-5p::gfp::gpr-1::smu-1 3’ UTR, cbr-unc-119(+), III:680195*], unc-119(ed3)? III). We note that PD 2275 and PD2286 may carry the unc-119(ed3) mutation.
* Note that chromosome location is given with respect to the WBcel11 assembly.
Generation of and tracking of chimeric males
PD2275 males were mated with PD2286 hermaphrodites exhibiting homogeneous GFP+ pharyngeal patterns indicative of a Mendelian heterozygote. Resulting cross progeny (diagrammed in Figure S2) fell into six classes (all of which were homozygous for GPR-1(OE)):
Mendelian
1. tra-2(q276) II, myo-2p::mCherry/+ XX males
2. tra-2(q276)/mnC1 [myo-2p::GFP] II; myo-2p::mCherry/+ IV hermaphrodites
Common non-Mendelian chimeras resulting from P1 inheritance of paternal chromosomes
3. tra-2(q276) in the AB lineage, tra-2(q276) II; myo-2p::mCherry IV in the P1 lineage, XX males
4. mnC1 [myo-2p::GFP] in the AB lineage, tra-2(q276) II; myo-2p::mCherry IV in the P1 lineage
Rare non-Mendelian chimeras resulting from P1 inheritance of maternal chromosomes
5. tra-2(q276) II and myo-2p::mCherry in the AB lineage and tra-2(q276) in the P1 lineage
6. tra-2(q276) II and myo-2p::mCherry in the AB lineage and mnC1[myo-2p::GFP] in the P1 lineage
Generation of Mitochondrial hybrid strains
PX174 males were crossed to PD2217 and PD2218. Two AB-fluorescence-patterned F1 progeny from each cross were singled and allowed to self-fertilize to create strains: (PD2231, PD2232) and (PD2233, PD2234) respectively.
DNA extraction, Sequencing and Analysis
C. elegans DNA from mixed developmental stages was treated essentially as was described by Johnson et al. (2006) for their purification of nucleosome cores. Following resuspension of the initial ethanol precipitate, DNA sequencing libraries were made using a standard Nextera tagmentation kit (FC-121–1030) and subsequently sequenced on a MiSeq Genome Analyzer (Illumina, Inc.). Reads were mapped to the C. elegans genome (WBcel235) using BWA (version 0.6.1-r104). Reads were filtered to require a mapping score of 20. The genomic position of the GPR-1(OE) MiniMos insertion in PD1594 was determined by finding chimeric reads containing a sequence spanning the junction of the transgene and the genome.
Nuclear and mitochondrial genome derivation
A subset of high-confidence single-nucleotide-polymorphisms (SNPs) was first identified from a listing of k-mer counts with k=25 for reference sequence datasets for PX174 (Thompson et al., 2013) and N2 (from our independent Illumina dataset) as follows: First, all possible 25-mers were assigned to groups of four based on identical sequences in positions 1–12 and 14–25 and different sequences in the central position (position 13). Second, positions to be considered as candidates for representative high-confidence SNPs were defined as cases where (i) Only one of the k-mer group matches the reference C. elegans (N2) genome (version ws220) and this k-mer is in a single copy in the reference genome, (ii) this k-mer sequence was the predominant member k-mer for the relevant group in the experimental N2 dataset, and (iii) a different k-mer from the group was predominant in the experimental PX174 dataset. Predominance was defined based on characteristics of the individual datasets with a simple arithmetic test.
For predominance of the expected canonical base in the N2 dataset, we applied (N2_Reference_Counts_At_Base_n) > 6* (N2_NonReference_Counts_At_Base_n) + 6
For predominance of a non-canonical base in the PX174 dataset, we used (PX174_NonReference_Counts_At_Base_n) > 5* (N2_Reference_Counts_At_Base_n) + 6
The constant value 6 added to the left side of each inequality ensures that a minimum number of reads is present before calling an assignment definitive. The multiplier (minimum ratio once 6 instances have been tallied) is 6 for comparing a single “reference” allele to three alternative alleles, and is lowered to 5 when three “alternative” alleles are compared to a single reference allele (the latter adjustment to account for the possibility of a low level of random sequence-error events). Filtering was then applied to the set of potential SNPs to remove a small number that appeared to have much larger or smaller numbers of reads than expected in the N2 or PX174 datasets (we removed any k-mer with a representation that was 5-fold above or below the mean for the relevant chromosome). For graphical representation, experimental Illumina reads for the strains shown in Figure 4 were parsed into k-mers and counts for canonical N2 (blue), PX174 variant (orange), and other sequences (labeled green; rarely observed). Vertical lines at each SNP position for each strain are colored by the numbers of canonical and non-canonical reads at the SNP position. For positions with less than 10 identifiable k-mer matches at any position, the total height of the vertical line was scaled down proportionally, for sites with at least 10 reads, a single line height is used for display. The predominance of N2 derivation in the nuclear chromosomes in the hybrid lines, and of PX174 derivation in the mitochondrial DNA is evident in examining the plots both in aggregate and detail. A small number of exceptional reads (and in a few cases, loci) are observed in which the sequences appear to derive more equally or from the unexpected parent. Manual inspection for a number of these cases revealed to be representative of (very rare) back-mutation events or sequencing errors.
QUANTIFICATION AND STATISTICAL ANALYSIS
Values listed in the manuscript are average values ± standard error of the mean. No statistical tests were performed.
DATA AND SOFTWARE AVAILABILITY
Raw data been deposited in the SRA (Short Read Archive), BioProject ID PRJNA472811.
ADDITIONAL RESOURCES
Not applicable
Supplementary Material
File S1. DNA sequences, Related to STAR Methods.
A Crosses used to generate chimeric worms containing a lin mutation in either the AB or P1 cell lineage. To generate mosaic animals, males carrying one of the following mutations: lin-2(e1309), lin-3(e1417), lin-12(n952), lin-12(n302), lin-31(n301), were mated with fluorescently marked GPR-1(OE) hermaphrodites. The strain name, genotype, and relevant characteristics are listed for each parent in crosses 1–15. Crosses 1 and 6 were used to generate heterozygous males carrying a lin mutation. Crosses 2–5 and 7–15 were used to generate chimeric animals for phenotypic analysis.
B Phenotypes of chimeric F1 worms containing a lin mutation in either the AB or P1 cell lineage, and their parents. F1 cross progeny (cross-listed in first column) exhibiting non-Mendelian pharyngeal fluorescence patterns were singled out onto fresh plates and scored for the percentage of Muv (multivulval), Egl (egg-laying defective), Sterile (defined here as any worm that failed to give rise to a viable population of progeny for reasons including failure to produce eggs and production and laying of inviable eggs), and overall fertility. Single worms from parental strains (listed in first column) were assayed similarly for comparison. The relevant genotype of the AC/VU precursors (P1 lineage) and the VPCs (AB lineage) was inferred from the fluorescence pattern and listed in column 2 and 3 respectively (see Figure S1 for a diagram of the crossing strategy)
Table 2B. Frequencies of distinct mosaic self progeny classes resulting from maternal GFP-GPR-1 expression.
F2 self progeny from single F1 containing mCherry expression restricted to the P1 cell lineage were examined for pharyngeal fluorescence expression patterns. The number and percentage of F2 hermaphrodites that displayed each pattern is described.
| # | N (%) | N (%) | N (%) | N (%) | N |
|---|---|---|---|---|---|
| 1 | 15 (18.5%) | 16 (19.8%) | 32 (39.5%) | 18 (22.2%) | 81 |
| 2 | 22 (22.5%) | 20 (20.4%) | 24 (24.5%) | 32 (32.7%) | 98 |
| 3 | 25 (23.4%) | 18 (16.8%) | 39 (36.5%) | 25 (23.4%) | 107 |
| 4 | 11 (13.8%) | 20 (25.0%) | 32 (40.0%) | 17 (21.3%) | 80 |
| All hermaphrodite self progeny | 73 (19.5%) | 74 (20.5%) | 127 (35.1%) | 92 (24.9%) | 366 |
Highlights:
Non-Mendelian inheritance can be stably maintained using optimized GPR-1 transgenes.
Fluorescently marked toolkit strains enable precise tracking of inheritance mode.
Marking allows derivation of sperm-derived clonal populations from any fertile male.
Toolkit enables precise tests for lineage-of-action and non-nuclear inheritance.
Acknowledgments
We thank Henrik Bringmann for communications on the nature of noncanonical inheritance, Elif Sarinay Cenik and Sedona Murphy for observations on the gpr-1(oe) strains and their genetic interactions, Josh Arribere for the gfp-tagged unc-54 allele, Lamia Wahba for pointing out biological aspects of noncanonical inheritance, Loren Hansen for guidance in SNP calling, Tim Schedl, Michael Ailion, Maria Sallee, and Anne Villeneuve for helpful discussions, Tomoko Tabuchi and Susan Strome for communicating results prior to publication, and Elif Sarinay Cenik, Nimit Jain, Massa Shoura, and Ryan Bell for their critical reading of the manuscript. This work was supported by grant NIGMS-R01-GM37706/GM130366 (to AF). Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Adalsteinsson BT, and Ferguson-Smith AC (2014). Epigenetic control of the genome-lessons from genomic imprinting. Genes (Basel) 5, 635–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderegg C, and Markert CL (1986). Successful rescue of microsurgically produced homozygous uniparental mouse embryos via production of aggregation chimeras. Proc. Natl. Acad. Sci. U.S.A. 83, 6509–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. (2008). PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean CJ, Schaner CE, and Kelly WG (2004). Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet 36, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bens S, Luedeke M, Richter T, Graf M, Kolarova J, Barbi G, Lato K, Barth TF, and Siebert R (2017). Mosaic genome-wide maternal isodiploidy: an extreme form of imprinting disorder presenting as prenatal diagnostic challenge. Clin Epigenetics 9, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseling J, and Bringmann H (2016). Engineered non-Mendelian inheritance of entire parental genomes in C. elegans. Nat Biotech 34, 982–986. [DOI] [PubMed] [Google Scholar]
- Blay C, Planes S, and Ky C-L (2018). Crossing phenotype heritability and candidate gene expression in grafted black-lipped pearl oyster Pinctada margaritifera, an animal chimera. J. Hered [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spence AM, and Schachter H (2003). Isolation of null alleles of the Caenorhabditis elegans gly-12, gly-13 and gly-14 genes, all of which encode UDP-GlcNAc: alpha-3-D-mannoside beta1,2-N-acetylglucosaminyltransferase I activity. Biochimie 85, 391–401. [DOI] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, and Spinner NB (2010). Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet 19, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcy D, Atwal PS, Angell C, Gadi I, and Wallerstein R (2015). Mosaic paternal genome-wide uniparental isodisomy with down syndrome. Am. J. Med. Genet. A 167A, 2463–2469. [DOI] [PubMed] [Google Scholar]
- Dejima K, Hori S, Iwata S, Suehiro Y, Yoshina S, Motohashi T, and Mitani S (2018). An Aneuploidy-Free and Structurally Defined Balancer Chromosome Toolkit for Caenorhabditis elegans. Cell Rep 22, 232–241. [DOI] [PubMed] [Google Scholar]
- Edgar LG, and McGhee JD (1988). DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53, 589–599. [DOI] [PubMed] [Google Scholar]
- Eicher EM, and Hoppe PC (1973). Use of chimeras to transmit lethal genes in the mouse and to demonstrate allelism of the two X-linked male lethal genes jp and msd. J. Exp. Zool 183, 181–184. [DOI] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, and Marillonnet S (2009). Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS ONE 4, e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Waterston RH, and Brenner S (1974). A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. Journal of Molecular Biology 90, 291–300. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussière F, and Hekimi S (2001). Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644. [DOI] [PubMed] [Google Scholar]
- de la Filia AG, Bain SA, and Ross L (2015). Haplodiploidy and the reproductive ecology of Arthropods. Current Opinion in Insect Science 9, 36–43. [DOI] [PubMed] [Google Scholar]
- Fire A, Alcazar R, and Tan F (2006). Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics 173, 1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Ailion M, and Lockery SR (2008). Ammonium-acetate is sensed by gustatory and olfactory neurons in Caenorhabditis elegans. PLoS ONE 3, e2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Ailion M, and Jorgensen EM (2012). Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, and Jorgensen EM (2014). Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Meth 11, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Jain N, Hansen L, Davis MW, Li Y, Zhao D, Rebora K, Millet JRM, Liu X, Kim SK, et al. (2016). An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 166, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara A, Abe S, Yamaha E, Yamazaki F, and Yoshida MC (1997). Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma 106, 44–52. [DOI] [PubMed] [Google Scholar]
- Fuyama Y (1986). Genetics of Parthenogenesis in DROSOPHILA MELANOGASTER. II. Characterization of a Gynogenetically Reproducing Strain. Genetics 114, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Girardot M, Feil R, and Llères D (2013). Epigenetic deregulation of genomic imprinting in humans: causal mechanisms and clinical implications. Epigenomics 5, 715–728. [DOI] [PubMed] [Google Scholar]
- Goldstein B, and Hird SN (1996). Specification of the anteroposterior axis in Caenorhabditis elegans. Development 122, 1467–1474. [DOI] [PubMed] [Google Scholar]
- Golic KG, Golic MM, and Pimpinelli S (1998). Imprinted control of gene activity in Drosophila. Curr. Biol 8, 1273–1276. [DOI] [PubMed] [Google Scholar]
- Greenwald I, and Seydoux G (1990). Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346, 197–199. [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, and Horvitz HR (1983). The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34, 435–444. [DOI] [PubMed] [Google Scholar]
- Haack H, and Hodgkin J (1991). Tests for parental imprinting in the nematode Caenorhabditis elegans. Mol. Gen. Genet 228, 482–485. [DOI] [PubMed] [Google Scholar]
- Hill RJ, and Sternberg PW (1992). The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358, 470–476. [DOI] [PubMed] [Google Scholar]
- Hodgkin J (2002). Exploring the envelope. Systematic alteration in the sex-determination system of the nematode caenorhabditis elegans. Genetics 162, 767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, and Brenner S (1977). Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86, 275–287. [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, and Brenner S (1979). Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics 91, 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R, Hajnal AF, Harp SA, and Kim SK (1996). The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development 122, 97–111. [DOI] [PubMed] [Google Scholar]
- Hunter CP, and Wood WB (1992). Evidence from mosaic analysis of the masculinizing gene her-1 for cell interactions in C. elegans sex determination. Nature 355, 551–555. [DOI] [PubMed] [Google Scholar]
- Hutchison Iii C. a., Newbold JE, Potter SS, and Edgell MH (1974). Maternal inheritance of mammalian mitochondrial DNA. Nature 251, 536–538. [DOI] [PubMed] [Google Scholar]
- Jhuang H-Y, Lee H-Y, and Leu J-Y (2017). Mitochondrial-nuclear co-evolution leads to hybrid incompatibility through pentatricopeptide repeat proteins. EMBO Rep. 18, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Tan FJ, McCullough HL, Riordan DP, and Fire AZ (2006). Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 16, 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, and Fire A (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL (1970). Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komma DJ, and Endow SA (1995). Haploidy and androgenesis in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 92, 11884–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzot D (2008). Complex and segmental uniparental disomy updated. J. Med. Genet 45, 545–556. [DOI] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D Jr, and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, and Golic KG (2002). The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics 162, 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S (2007). The C. elegans pharynx: a model for organogenesis WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K (2017). Evolution of the asexual queen succession system and its underlying mechanisms in termites. J. Exp. Biol 220, 63–72. [DOI] [PubMed] [Google Scholar]
- Mendel G (1866). Versuche über Plflanzenhybriden. Verhandlungen Des Naturforschenden Vereines in Brünn, Bd. IV Für Das Jahr 1865 3–47. [Google Scholar]
- Merritt C, Rasoloson D, Ko D, and Seydoux G (2008). 3’ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol 18, 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Stockdale FE, and Karn J (1986). Immunological identification of the genes encoding the four myosin heavy chain isoforms of Caenorhabditis elegans. PNAS 83, 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LM, Gallegos ME, Morisseau BA, and Kim SK (1993). lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev. 7, 933–947. [DOI] [PubMed] [Google Scholar]
- Okkema PG, and Fire A (1994). The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development 120, 2175–2186. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, and McDaniel C (2018). Parthenogenesis in birds: A review. Reproduction. [DOI] [PubMed] [Google Scholar]
- Rechavi O, and Lev I (2017). Principles of Transgenerational Small RNA Inheritance in Caenorhabditis elegans. Curr. Biol 27, R720–R730. [DOI] [PubMed] [Google Scholar]
- Redemann S, Schloissnig S, Ernst S, Pozniakowsky A, Ayloo S, Hyman AA, and Bringmann H (2011). Codon adaptation-based control of protein expression in C. elegans. Nat Meth 8, 250–252. [DOI] [PubMed] [Google Scholar]
- Reich E, and Luck DJ (1966). Replication and inheritance of mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 55, 1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP (2000). Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 22, 452–459. [DOI] [PubMed] [Google Scholar]
- Sánchez L (2014). Sex-determining mechanisms in insects based on imprinting and elimination of chromosomes. Sex Dev 8, 83–103. [DOI] [PubMed] [Google Scholar]
- Sato M, and Sato K (2011). Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Schmid T, and Hajnal A (2015). Signal transduction during C. elegans vulval development: a NeverEnding story. Curr. Opin. Genet. Dev 32, 1–9. [DOI] [PubMed] [Google Scholar]
- Schwander T, and Oldroyd BP (2016). Androgenesis: where males hijack eggs to clone themselves. Philos. Trans. R. Soc. Lond., B, Biol. Sci 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, and Greenwald I (1989). Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell 57, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Seymour DK, Filiault DL, Henry IM, Monson-Miller J, Ravi M, Pang A, Comai L, Chan SWL, and Maloof JN (2012). Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc. Natl. Acad. Sci. U.S.A. 109, 4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha K, and Fire A (2005). Imprinting capacity of gamete lineages in Caenorhabditis elegans. Genetics 170, 1633–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, and Horvitz HR (1989a). The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell 58, 679–693. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, and Horvitz HR (1989b). The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell 58, 679–693. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, and Saxton W (2001). Spindle Dynamics and the Role of γ-Tubulin in EarlyCaenorhabditis elegans Embryos. Mol. Biol. Cell 12, 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, and Lemire BD (2002). Stable heteroplasmy but differential inheritance of a large mitochondrial DNA deletion in nematodes. Biochem. Cell Biol. 80, 645–654. [DOI] [PubMed] [Google Scholar]
- Winberg J, Gustavsson P, Lagerstedt-Robinson K, Blennow E, Lundin J, Iwarsson E, Nordenström A, Anderlid B-M, Bondeson M-L, Nordenskjöld A, et al. (2010). Chimerism resulting from parthenogenetic activation and dispermic fertilization. Am. J. Med. Genet. A 152A, 2277–2286. [DOI] [PubMed] [Google Scholar]
- Wollman EL, Jacob F, and Hayes W (1956). Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb. Symp. Quant. Biol 21, 141–162. [DOI] [PubMed] [Google Scholar]
- Yochem J, Weston K, and Greenwald I (1988). The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 335, 547–550. [DOI] [PubMed] [Google Scholar]
- Zeiser E, Frøkjaer-Jensen C, Jorgensen E, and Ahringer J (2011). MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS ONE 6, e20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Fire AZ (2010). Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Developmental Biology 344, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. DNA sequences, Related to STAR Methods.
A Crosses used to generate chimeric worms containing a lin mutation in either the AB or P1 cell lineage. To generate mosaic animals, males carrying one of the following mutations: lin-2(e1309), lin-3(e1417), lin-12(n952), lin-12(n302), lin-31(n301), were mated with fluorescently marked GPR-1(OE) hermaphrodites. The strain name, genotype, and relevant characteristics are listed for each parent in crosses 1–15. Crosses 1 and 6 were used to generate heterozygous males carrying a lin mutation. Crosses 2–5 and 7–15 were used to generate chimeric animals for phenotypic analysis.
B Phenotypes of chimeric F1 worms containing a lin mutation in either the AB or P1 cell lineage, and their parents. F1 cross progeny (cross-listed in first column) exhibiting non-Mendelian pharyngeal fluorescence patterns were singled out onto fresh plates and scored for the percentage of Muv (multivulval), Egl (egg-laying defective), Sterile (defined here as any worm that failed to give rise to a viable population of progeny for reasons including failure to produce eggs and production and laying of inviable eggs), and overall fertility. Single worms from parental strains (listed in first column) were assayed similarly for comparison. The relevant genotype of the AC/VU precursors (P1 lineage) and the VPCs (AB lineage) was inferred from the fluorescence pattern and listed in column 2 and 3 respectively (see Figure S1 for a diagram of the crossing strategy)
Data Availability Statement
Raw data been deposited in the SRA (Short Read Archive), BioProject ID PRJNA472811.