Abstract
Next generation sequencing has uncovered a trove of short noncoding RNAs (e.g microRNAs) and long noncoding RNAs (lncRNAs) that act as molecular rheostats in the control of diverse homeostatic processes. Meanwhile, the tsunamic emergence of clustered regularly interspaced short palindromic repeats (CRISPR) editing has transformed our influence over all DNA-carrying entities, heralding global CRISPRization. This is evident in biomedical research where the ease and low-cost of CRISPR editing has made it the preferred method of manipulating the mouse genome, facilitating rapid discovery of genome function in an in vivo context. Here, CRISPR genome editing components are updated for elucidating lncRNA function in mice. Various strategies are highlighted for understanding lncRNA function in intergenic sequence space, as host genes that harbor microRNAs or other genes, and as natural antisense, overlapping or intronic genes. Also discussed is CRISPR editing of mice carrying human lncRNAs as well as the editing of competing endogenous RNAs. The information described herein should assist labs in the rigorous design of experiments that interrogate lncRNA function in mice where complex disease processes can be modeled thus accelerating translational discovery.
Keywords: CRISPR, long noncoding RNA, mouse, genetics, genome editing
Nearly 50 years ago, Susumu Ohno (1928-2000) correctly estimated the size of our haploid genome, but incorrectly suggested there being “so much ‘junk’ DNA in our genome” [1]. Due in large measure to efforts of the ENCODE Consortium [2], we now appreciate that much of our genome, beyond protein-coding genes, is functional with tens of thousands of noncoding genes and millions of transcription factor binding sites (TFBS). Among noncoding genes, the class of long noncoding RNAs (lncRNAs), defined broadly as processed transcripts >200 nucleotides in length with no translated open reading frame, bear striking resemblance to protein-coding genes inasmuch as they are RNA polymerase II-dependent, multi-exonic and spliced, 5’ capped, and frequently polyadenylated [3]. LncRNAs encompass numerous subclasses of noncoding RNA genes, distinguished primarily by genomic position relative to other gene loci (below). The first lncRNAs (XIST and H19) were identified in the early 1990s [4, 5], but very few would be reported over the next 20 years. Current estimates of the number of human lncRNAs vary considerably but a recent update of LNCipedia (https://lncipedia.org/) reveals nearly 50,000, a number that more than doubles all protein-coding genes [6], Unlike microRNAs, with anticipated functions related to mRNA degradation and/or translational repression, lncRNAs have diverse, unpredictable functions in the cell ranging from nuclear gene transcription and architecture to cytoplasmic roles in translation, signaling, cellular trafficking, and mitochondrial homeostasis [7, 8]. Importantly, of the 50,000 human lncRNAs listed in LNCipedia, only 3% (1,555) have some functional annotation [6]. Further, much of the sequence variation associated with cardiovascular disease occurs in noncoding sequence space where lncRNAs reside [9, 10]. These facts highlight the urgent need to define the function of lncRNAs in vivo, particularly those expressed in cells of the normal and pathological vessel wall [11-14].
As detailed in the reviews of this special issue of Vascular Pharmacology, a growing number of vascular lncRNAs have been discovered since 2010 (Figure 1). Initial studies to elucidate vascular lncRNA function are most often done in cell culture models; however, definitive proof for the function of these and other soon-to-be discovered vascular lncRNAs will require genetic experiments in animal models where an lncRNA is deleted, mutated, or ectopically expressed [15, 16]. This practical review serves to update advances in CRISPR genome editing of the mouse and the opportunities and challenges of harnessing this game-changing technology to elucidate lncRNA function in an in vivo context.
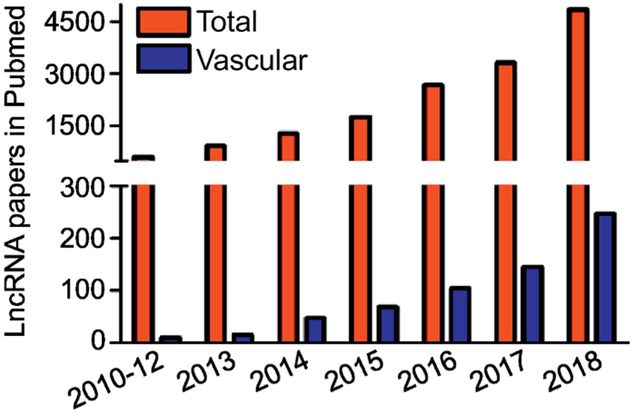
Figure 1.
Pubmed lncRNA publications since 2010. Shown are the total and vascular biology-related publications on lncRNAs in Pubmed beginning in 2010. A steady rise in vascular lncRNA publications is evident beginning in 2013.
CRISPR Components
Three components comprise the CRISPR system: (1) a programmable guide RNA that escorts (2) the Class 2, Type II-A Cas9 endonuclease for double-strand breakage and, when directed, (3) precision-mediated repair through a single-stranded or double-stranded DNA template carrying sequence substitutions or small to moderately-sized inserts such as a polyadenylation signal sequence. Two-component CRISPR editing (Cas9 and guide RNA alone) is used to disrupt protein-coding reading frames (not discussed here), to modulate expression of a gene via deactivated or ‘dead’ Cas9 (dCas9) conjugated to either a transactivator or transrepressor [17], or to generate small or very large deletions [18, 19]. Three-component CRISPR editing (Cas9, guide RNA, and repair template) is used to introduce precision edits such as a single base substitution or the introduction of foreign sequences such as loxP sites for the generation of a conditional knockout. For more historical information about the basic biology of CRISPR components and applications in mouse genome editing, the reader is referred to several previous reviews [20-23].
Updated CRISPR Editing in Mice
Innovations in Guide RNA
CRISPR-Cas9 genome editing in the mouse was first reported in 2013 [24-29] and has now all but supplanted traditional methods of modifying the mouse genome. Early CRISPR studies injected in vitro transcribed and purified guide RNA into the mouse zygote. The in-house cloning, purification, and activity testing of guide RNAs was time-and labor-intensive, often taking weeks to complete. More recent developments have enhanced the speed and efficiency of CRISPR-Cas9 genome editing in mice by improving the preparation and activity of the guide RNA. For example, it is no longer necessary to clone and purify the guide RNA. Instead, many companies provide synthetic guide RNA either as a single-guide RNA as originally reported [30] or as independent CRISPR RNA and trans-activating CRISPR RNA, ready for injection into the mouse zygote (e.g., https://www.synthego.com/). Importantly, synthetic guide RNA lacks a 5’ triphosphate which has been shown to elicit innate immunity with massive induction of interferon-dependent genes in both rodent and human cells [31, 32], though this is unlikely to present a problem in the mouse zygote. Defining optimal guide RNAs is made simple with on-line programs such as “Breaking-Cas” (http://bioinfogp.cnb.csic.es/tools/breakingcas) [33] and CRISPOR (http://crispor.tefor.net/), which combines different measures of specificity as well as an informative summary of guide RNA off-targeting [34]. Note that the original design tool from Feng Zhang’s lab (crispr.mit.edu) [35] is no longer available. Recently, additional insights have been gained in optimal guide RNA design with the finding that 3’ terminal GC-rich sequences are inefficient in positioning Cas9 at targeted sites in the genome [36].
An important consideration in guide RNA design is unintended editing or offtargeting. In contrast to editing cultured cells where off-targeting events can be permanent, off-target edits in mice can be segregated from the on-target edit through breeding so long as the unintended edit is not in linkage disequilibrium with the intended edit. In this context, the off-target summary in CRISPOR [34] provides notation as to the potential for linkage disequilibrium. Although a previous report indicated pervasive off-targeting in the mouse [37], this work was subsequently retracted based on independent labs showing flaws in design and interpretation of findings [38-42]. The latter reports are in agreement with several studies showing little off-target CRISPR editing in the mouse [43-45]. The fact that a low-scoring guide RNA (with 100s of predicted off-targets) can induce many off-targets in the mouse [46] underscores the importance of carefully selecting an optimal guide RNA for mouse genome editing. A guide RNA score of 66 or higher was considered optimal to minimize off-targeting in mice [44]. Even still, investigators should evaluate whether predicted unintended edits in linkage disequilibrium with the intended edit exist and, if so, interrogate any sequence changes through PCR and Sanger sequencing of founder mice. Of course, on-target sequence fidelity must also be evaluated by Sanger sequencing since insertions and deletions, beyond the desired outcome, are possible. A recent, unbiased approach for assessing off-targeting in vivo has been reported with very high efficiency [46].
Innovations in Cas9 Endonucleases
A number of innovations have been reported for Cas9 endonucleases. For example, most mouse cores now inject purified Cas9 protein with the synthetic guide RNA as a ribonucleoprotein (RNP) complex [47]. Since the vast majority of guide RNAs successfully target Cas9 endonuclease to the desired sequence for editing, complex activity assays such as the T7E1 assay or split reporter assay [48] are no longer used in some mouse facilities. On the other hand, a simple in vitro digestion should be done to confirm proper sequence targeting of the Cas9 endonuclease (e.g., https://www.synthego.com/). RNPs mediate rapid (one hour) genome editing in cultured cells [49], and elegant imaging studies from the labs of Jennifer Doudna and Robert Tjian showed that the RNP surveils the genome and finds its target in minutes [50]. This is relevant because a major hurdle to overcome in any type of germline genome editing is the phenomenon of mosaicism [51, 52]. The rapid cleavage of zygotic DNA with RNPs minimizes mosaicism and enhances germline transmission of CRISPR edits in mice [47]. In contrast, the time necessary for Cas9 mRNA translation and nuclear import may exceed the time to zygotic division, thus favoring mosaicism. Importantly, germline transmission of CRISPR edits in the mouse is highly favored though on rare occasion we have seen the absence of a transmitted CRISPR-edited allele to the first filial generation; PCR analysis of founder male sperm can avoid protracted breeding efforts that fail to reveal passage of the edited allele through the germline.
Another innovation has been the genetic engineering of Cas9 variants that display reduced off-targeting [53-56]. Recently, the so-called high fidelity Cas9 was shown to be superior to previous recombinant Cas9 variants and is readily available commercially, though it has yet to be tested in mouse zygotes [57]. Smaller Cas9 endonucleases from other species of bacteria exist with longer guide RNA and PAM sequences that likely confer even greater specificity than the more popular Streptococcus pyogenes Cas9 variants [58]. Moreover, other classes of Cas endonuclease have distinct cutting properties and PAM recognition sequences to expand the CRISPR armamentarium for genome editing in the mouse [59-62]. Transiently acting Cas9 is an additional development that reduces mosaicism and attending toxicity associated with Cas9, a subject that has largely been overlooked in the editing community, particularly with respect to somatic editing [63, 64].
Base editing involves yet another innovation of Cas9 where the catalytic endonuclease domains are mutated and base modifying enzymes, cytosine or adenine deaminases, are fused to Cas9. The resultant recombinant dCas9 can directly edit a single base in the genome without introducing a double-strand break [65]. Base editing has enormous potential for precision editing of clinically-relevant variants in noncoding RNAs and other noncoding sequences such as regulatory elements. There has been some reporting of base editing in mouse embryos [66-68], but most mouse labs continue to use wild-type Cas9 protein for germline editing, likely because of the limited 4-5 base pair window for base conversion with so-called ‘bystander editing’ [65].
Innovations in Donor Templates
Innovations in donor templates for three-component CRISPR have also evolved. Previously, there was a limitation of 200 nucleotides for the synthesis of single-strand oligonucleotides (ssODNs), the preferred donor template in three-component CRISPR. Thus, insertions of many polyadenylation signal sequences for transcriptional arrest of an lncRNA using ssODNs were not possible. Today, there are commercial sources of long ssODNs that are synthesized up to 2,000 nucleotide in length (e.g., Megamers, https://www.idtdna.com). These long ssODNs can also be designed to carry loxP sites flanking the intervening sequence to be removed upon Cre-mediated excision in a process known as efficient additions with ssDNA inserts CRISPR [69]. If longer donor templates are needed (e.g., knocking in a human lncRNA into the mouse), then targeted integration with linearized dsDNA-CRISPR (Tild-CRISPR) may be considered, where up to 6-kb of sequence has been successfully integrated in mouse embryos [70].
Innovations in Delivery of CRISPR Components to Mouse Embryos
Finally, there are a number of new methods that simplify the delivery of CRISPR components into mouse zygotes and/or enhance editing efficiency. Electroporation of mouse embryos has been widely adopted in mouse cores. This approach circumvents the technically challenging task of microinjecting zygotes while improving targeting efficiency [71-73]. CRISPR with long ssODNs inducing conditional knockout alleles utilizes electroporation in zygotes to generate floxed mice [74]. Electroporation of embryos has also been extended to the intact pregnant female in a process known as genome-editing via oviductal nucleic acids delivery [75]. This method further democratizes CRISPR editing such that most any lab can perform simple noncoding sequence deletions in mice. Three-component CRISPR editing is less efficient than two-component editing in the mouse [22]. However, a recent method of injecting mouse embryos at the two-cell stage, where the G2 phase of the cell cycle favoring three-component CRISPR editing outcomes is extended and chromatin is more accessible, has increased efficiencies of three-component CRISPR knockins more than 10-fold [76].
To summarize, several innovations in CRISPR components and delivery mechanisms have simplified and democratized this revolutionary technology in the mouse zygote. It is now possible, from conception of the experiment to genotyping founder mice, to obtain CRISPRized mice in as few as 5-6 weeks (Figure 2). Moreover, the cost of generating such mice has fallen to as little as two-thousand dollars at some academic centers. The continued refinement of CRISPR editing in the mouse coincides with a growing interest in lncRNAs in vascular biology (Figure 1). As more lncRNAs are discovered, especially in mouse tissue and cell screens, there will be a pressing need to ascertain function in the complex milieu of a living animal where disease modeling can be done and the phenotyping is more extensive over simple cell culture models.
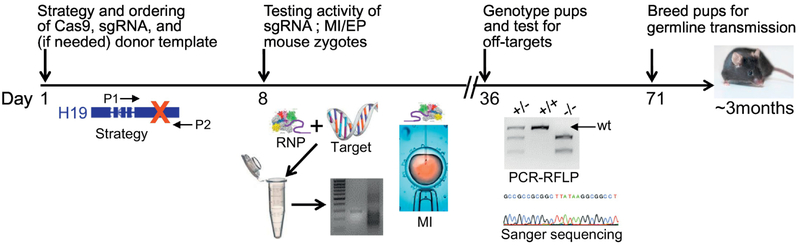
Figure 2. Timeline for generating CRISPRized mice.
The continued refinement in CRISPR components (e.g., store-bought ribonucleoprotein complexes [RNP] of Cas9 and guide RNA) as well as methods of delivery (microinjection, MI versus electroporation, EP) allow for the generation of genetically-modified mice in as little as 5 weeks. Critical steps include the strategy in design with PCR primers (P1, P2) for genotyping founder mice by a PCR restriction fragment length polymorphism (RFLP)-like method and testing for offtargets by Sanger sequencing. Note that germline transmission of CRISPR edited genes typically can occur within 3 months from conception of the project.
CRISPR Editing LncRNAs in Mice
The reviews in this special issue of Vascular Pharmacology extend and complement earlier reviews of functional noncoding genes in vascular biology [11-14] as well as general strategies for the discovery and functional assessment of lncRNAs [15]. Initial lncRNA studies are necessarily done in cultured model systems where full transcript annotation is done by 5’ and 3’ RACE. These experiments provide much-needed information such as the presence of additional lncRNA variants arising from alternative splicing, transcription initiation, or polyadenylation. Expression level profiling and the definition of transcriptional control of the lncRNA would be informative for later in vivo editing experiments. Moreover, cellular localization studies using quantitative single transcript RNA-FISH are essential in formulating hypotheses that test the function of lncRNAs. Loss and gain-of-function experiments in cultured vascular cells provide some insight into lncRNA function, but are of limited utility as they do not address lncRNA function in the complex milieu of an animal that can be subjected to an array of pathophysiological stressors. Several lncRNAs have been genetically inactivated in mice and in zebrafish, with almost all knockouts compatible with embryonic development and viability, consistent with the idea that lncRNAs fine-tune regulatory processes such as local gene expression [77-83]. Although several studies have interrogated the biology of lncRNAs under pathophysiological conditions related to the vessel wall, none have used the CRISPR editing system in the mouse zygote [84-89]. Below, we consider several strategies for germline transmitted CRISPR editing to elucidate lncRNA function in the mouse based on the genomic position of the lncRNA. We begin with the simplest scenario and work up to the more complex, highlighting strengths and weaknesses of each CRISPR editing approach.
CRISPRing Intergenic LncRNAs
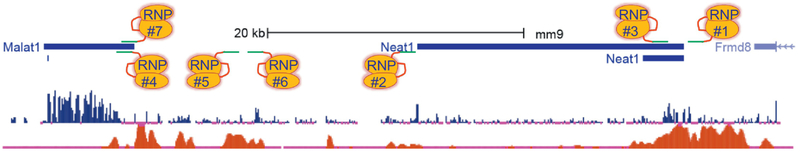
We define long intergenic noncoding RNAs (lincRNAs) as those lncRNAs residing between two other gene loci where all three gene loci are independently transcribed. Accordingly, an lncRNA whose 5’ end is found closely juxtaposed to a neighboring 5’ locus may not be a bonafide lincRNA as it would likely be co-regulated by shared promoter elements. There are thousands of lincRNAs and many show sequence conservation and chromosomal synteny between mouse and human [90, 91]. LincRNAs are the easiest class of lncRNA to target with CRISPR in the mouse and are amenable to all methods of CRISPR editing. Consider Neat1, an lincRNA shown recently to promote VSMC phenotypic modulation [88]. This lincRNA is transcribed ∼20-kb upstream of another lincRNA (Malat1), shown to have function in both EC [92] and VSMC [93], and ∼6kb downstream of a protein-coding gene (Frmd8) (Figure 3). One could delete the entire 21-kb Neat1 gene using two RNPs as shown (RNP1 and RNP2, Figure 3). Indeed, a similarly-sized deletion was done for the lincRNA, Rian, the first lncRNA to undergo CRISPR editing in the mouse [94]. Another lincRNA, Gene model 26878 (Gm26878), was deleted in the mouse and found to result in neonatal lethality [95]. More recently, a 6.7-kb deletion of Norad revealed a premature aging phenotype in mice [82]. Genotyping such lincRNA deletions is relatively straight-forward with primers located at least 100-200 nucleotides outside the RNPs, though larger deletions or inversions have been reported requiring careful assessment of founder mice [96]. One obvious concern with such a deletion approach is the inadvertent excision of smaller transcription units or regulatory elements that control distal gene expression. Though the latter concern could be addressed by assessing chromatin marks within and around the lncRNA as well as RNA-seq experiments in a specific vascular cell type, accurate interpretation is complicated by the presence of deleted sequences across all cells of the mouse with potential cell non-autonomous effects. On the other hand, the locus of an lincRNA could be floxed with the insertion of two loxP sequences on the same allele to allow for vascular cell-restricted loss of the lincRNA, using appropriate Cre driver mice [97, 98]. The original method of floxing mice with CRISPR calls for Cas9, two guide RNAs, and 2 ssODNs and while the method was reported to have moderately high efficiency [27], this notion has recently been challenged (https://www.biorxiv.org/content/early/2018/09/01/393231). RNA-seq experiments would still need to be done to determine whether a distal-acting regulatory region was removed. If there is only one promoter driving lincRNA expression, it should be informative to either make a deletion of the promoter and a portion of first exon using two RNPs or insert loxP sites around the promoter and first exon for vascular cell-restricted inactivation as has been done for some protein-coding genes [99] (RNP1 and RNP3, Figure 3). However, in the case of Neat1, we see a potential problem with each of these approaches; loss of a shorter, more abundantly expressed transcript located at the 5’ end of the gene (Figure 3). Prior to CRISPR, differentiating the effects of these two variants of Neat1 was problematic due to their identical sequences at the 5’ end. Recently, however, the function of the two Neat1 variants was elucidated through clever positioning of RNPs and insertion of transgenes that could differentially arrest transcription of each isoform [100]. This example underscores the critical importance of fully annotating lincRNA transcripts prior to any CRISPR editing.
Figure 3. CRISPR editing strategies for lincRNAs.
Screenshot of UCSC Genome Browser tracks are shown with (from top to bottom) the protein-coding and lincRNA genes, vertebrate conservation track (highly conserved sequences indicated by blue peaks), and chromatin signature. The chromatin signature here and in Figures 4-5 represents H3K27Ac in 8 week old mouse hearts. In this figure and below, RNPs are depicted as orange ovals (wildtype Cas9 protein) and red (invariant scaffold RNA) and green (variable, user defined protospacer sequence) lines that constitute the guide RNA. RNPs are numbered here and below to simplify discussion of their genomic location. Genomic scale bar (in kilobases, kb) and the species and genome assembly number (mm, mus musculus and assembly 9) are indicated here and below. Please note, the strategies shown here and below are examples only and are not meant to indicate actual targeting of these specific lincRNAs, some of which (e.g., Malat1 and Neat1) have already been targeted in the mouse. See text for more details.
There is at least one more consideration with deleting an lincRNA and that relates to generating microdeletions (less than 100 nucleotides) within the lincRNA itself. Unlike protein-coding genes, where well-defined functional domains are observed, lincRNA sequences rarely offer insight into functional motifs that can be easily excised with one or two RNPs. However, biochemical studies that define contact points between an lincRNA and RNA, DNA, or protein may reveal functional sequences [101, 102] which could be deleted with RNPs in close proximity to the motif. Note, however, that such an approach could disrupt the overall structure of the lincRNA, thus imparting new macromolecular associations and potential non-physiological activities. In vitro studies that narrow the binding sequence could result in a more subtle edit of a functional motif within an lincRNA.
Assuming there is only one lincRNA transcript type, as suggested in the well-studied Malat1 lincRNA (Figure 3), what other options beyond deleting sequences are available to the investigator? One idea would be to insert a polyadenylation signal sequence just downstream of the transcription start site to arrest transcription of the lincRNA (RNP4 in Figure 3). A similar strategy was used in cells to show that despite >90% suppression of the Lockd lincRNA, there was no effect on expression of the neighboring Cdkn1b gene suggesting that transcription of Lockd is dispensable for cis-acting transcriptional events [103]. On the other hand, deleting the 25-kb Lockd lincRNA resulted in 70% inhibition in Cdkn1b transcription supporting loss of a critical enhancer and TFBS functioning independently of Lockd transcription to effect Cdkn1b expression [103]. A contrary example exists with the Upperhand lincRNA, located upstream of Hand2, an essential cardiac transcription factor. Here, transcriptional arrest of Upperhand with a polyadenylation signal sequence abolished expression of Hand2 whereas knockdown of Upperhand had no effect [104]. The disparate findings between Lockd and Upperhand highlight the unpredictable nature of lincRNAs, and the regulatory elements in and around the locus, in controlling local gene expression. More importantly, they demonstrate the need for combinatorial experiments in vitro that can inform the best in vivo strategy to follow for elucidating lincRNA in the mouse. The length of polyadenylation signal sequences are generally >100 nucleotides requiring a dsDNA repair template for insertion into the genome [103-105]; however, a shorter polyadenylation signal of 49 nucleotides can easily be synthesized as an ssODN and was used to arrest lncRNA transcription in vitro [105]. Moreover, the Charme lincRNA was recently inactivated in mice by inserting a short polyadenylation signal followed by strong transcriptional termination sequences in an ssODN [106]. An even smaller polyadenylation signal sequence has yet to be tested in arresting lincRNA transcription [107].
Another method to study loss-of-function of an lincRNA in vivo is through CRISPR editing of an enhancer or critical TFBS. Several studies have used CRISPR-Cas9 to delete essential enhancers associated with protein-coding genes in mice [108-111], but there is virtually no such information with respect to the regulation of lincRNAs. A recent study of lincRNA-Cox2 (aka Ptgs2os) used a variety of CRISPR approaches to suggest the presence of an enhancer RNA upstream of the lincRNA [112]. The use of chromatin signatures (e.g., H3K27ac) and sequence conservation can guide investigators to define potential enhancers (RNP5 and RNP6, Figure 3). Enhancers are often hundreds of bases in length and comprise multiple TFBS, the smallest regulatory unit in the genome. Thus, genotyping deletion of an enhancer with primers flanking the predicted double-strand break is straight-forward as long as a larger than expected deletion does not occur.
For finer resolution edits, it should be possible to mutate a single TFBS using three-component CRISPR (RNP6, Figure 3). Historically, this has been a rather daunting task with only one study ever reported using conventional knockout technology to modify a TFBS without leaving a genomic scar or cavity [113]. The first regulatory element edit using CRISPR in the mouse involved a three nucleotide substitution within a TFBS in the first intron of Cnn1; mice homozygous for this edit showed a virtual knockout of this smooth muscle-restricted gene [114]. Genotyping such subtle edits requires either an RFLP assay or PCR primers that discriminate the wildtype versus mutant allele [114]. A subsequent study, using paired TALENs surrounding a GATA1 site in the Alas2 locus, revealed a sharp reduction of Alas2 expression and embryonic lethality in mice [115]. The results of editing TFBS controlling Cnn1 and Alas2 are somewhat surprising given that gene transcription is thought to involve multiple, often redundant, regulatory elements. It will be interesting to determine if additional examples exist in other protein-coding genes as well as lincRNAs and, if so, whether conserved single nucleotide variants are present in human populations that can be modeled in mice for further functional analysis.
An advantage of targeting a single enhancer or, better, a single TFBS, is the subtle nature of the edit and the ability to override loss in target gene expression with, for example, CRISPR activation (CRISPRa) [17]. The latter studies would likely require in vitro culturing of vascular cells from CRISPRized mice since CRISPRa is a transient phenomenon that is not amenable to the aforementioned types of CRISPR editing in the mouse zygote given that no permanent editing of DNA occurs. Recently, CRISPRa was used to reverse obesity in a haploinsufficient mouse [116].
A final method of targeting an lincRNA in vivo could be to directly repress transcription using CRISPR interference (CRISPRi), which originally was reported as a single-guide RNA-mediated method of targeting the non-template strand (that is the plus or Watson strand of DNA) with dCas9 to sterically block transcription of a target gene [117]. Subsequent work showed greater repression of target genes by joining dCas9 with the Krüppel-associated box (KRAB) domain found in an array of zinc finger transcriptional repressors [118]. Although CRISPRi has already been used to interrogate lncRNA function in cultured cells [119, 120], and shown to be superior to other methods of gene silencing (e.g., RNAi) [121], its application in the mouse for analyzing lincRNA function has yet to be reported. It should be possible to inducibly activate CRISPRi in a vascular cell-restricted manner to repress an lincRNA in vivo. Note that the guide RNA (RNP7, Figure 3), which would need to be delivered to the vessel wall via viral-mediated transfer, is targeted to the proximal promoter region within a window of −50 to +300 bp from the TSS; guide RNAs outside this interval exhibit distance-dependent loss in effective gene repression [120]. This fact, again, emphasizes the need for careful in vitro characterization of the 5’ end of an lincRNA prior to studies in the mouse.
Because lincRNAs, as defined here, are well-separated from neighboring gene loci, each method of CRISPR editing may be used to elucidate loss-of-function phenotypes in the mouse. However, as is the case with protein-coding genes, variations in phenotype may be observed depending on the strategy of inactivating the lincRNA [78, 104]. Strain (or sub-strain due to genetic drift), age, gender, and the nature of a stressor are other important variables to consider in the interpretation of lincRNA loss-of-function phenotypes in mice.
CRISPRing Host LncRNAs
Some lncRNAs are a host gene for other lncRNAs or microRNAs. For example, Carmn, first reported as a conserved super-enhancer associated lncRNA [122], is a host gene for miR143/145, a known mediator of the VSMC differentiated phenotype [123-125] (Figure 4). Moreover, Carmn overlaps at its 3’ end with Bvht, a convergently transcribed mouse-restricted lncRNA shown initially to be involved in cardiac muscle specification [126], but apparently dispensable for normal mouse development [78]. Several CRISPR strategies are available to elucidate the role of Carmn in the mouse. The key here and in other host lncRNAs is to differentiate between the host lncRNA and gene loci within it. Accordingly, and in contrast to an lincRNA (Figure 3), deletion of the entire Carmn locus with or without loxP sites would be contraindicated given that two other gene loci would be deleted or disrupted (RNP1 and RNP2, Figure 4). Note, however, that deletion of the 5’ promoter region and first exon of Carmn (RNP1 and RNP3, Figure 4) may be informative so long as the miR143/145 cluster is not dependent on the Carmn promoter for expression. Previous work has shown the presence of an upstream, conserved enhancer region harboring an SRF-binding CArG box that controls miR143/145 expression in the context of reporter mice [124, 125]. Whether this CArG box is necessary for miR143/145 expression in its native genomic milieu, as shown for Cnn1 [114], can easily be addressed using three-component CRISPR editing (RNP4, Figure 4). The presence of an H3K27Ac chromatin signature in this region highlights the importance of utilizing existing ENCODE data to gain insight into potentially important regulatory regions of a host lncRNA or its internal gene loci. If the host lncRNA is transcriptionally uncoupled from the internal gene(s) within, it should be possible to target a critical upstream promoter element (RNP1, Figure 4), insert a polyadenylation signal within the first exon (RNP5, Figure 4) or target the promoter region with CRISPRi (RNP1, Figure 4). The latter, however, is technically challenging for reasons outlined above.
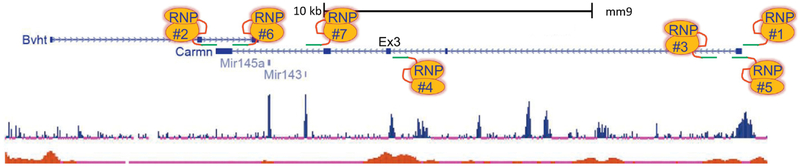
Figure 4. CRISPR editing strategies for a host lncRNA.
Screenshot of the multi-exonic Carmn host lncRNA. Blue squares/rectangles are exons separated by introns depicted as a line with arrows in the direction of transcription. Note the presence of highly conserved miR143/145 cluster and an overlapping lncRNA (Bvht). RNP#4 overlays a conserved sequence upstream of exon 3 (Ex3) of Carmn that includes a consensus CArG box. Based on previous mouse studies [114], mutation of this CArG box would be expected to abolish miR143/145 expression in VSMC of the mouse. See text for more details.
If internal gene loci are co-transcribed and processed with the host lncRNA, what CRISPR strategy could be used to elucidate the function of the host lncRNA apart from the internal gene(s)? If the host lncRNA is multi-exonic, as is Carmn, one approach could be to make moderate deletions of a terminal exon (RNP2 and RNP6, Figure 4) or internal exons (RNP4 and RNP7, Figure 4). It would be essential not to disrupt splicing of the host lncRNA (or in the case here, the 3’ overlapping Bvht locus) or excision of a critical regulatory element. RNA-seq experiments could address the latter. Note that in the present example, excision of internal Carmn exons (RNP4 and RNP7, Figure 4) would be contraindicated since a potentially important CArG box for miR143/145 expression would be excised. The best case scenario for any loss-of-function analysis of an lncRNA would be the in vitro elucidation of a critical motif that could be point mutated, much like what has been done for other lncRNAs [101, 102]. The recent categorization of lncRNAs based on short, 8-10 nucleotide sequences called k-mers, which appear to predict protein binding and cellular localization of lncRNAs, offers some computational guidance to functionality and subtle, three-component CRISPR editing [127].
CRISPRing Intronic LncRNAs
LncRNAs may be found within introns of coding genes and noncoding genes in either the sense or antisense orientation. Sense intronic lncRNAs have recently been included in the class of stable intronic sequence RNAs and can arise through splicing-dependent and splicing-independent processes [128]. They generally are co-transcribed from the host gene; thus, targeting sense lncRNA transcription with RNPs that insert a polyadenylation signal, enable CRISPRi, or disrupt a proximal regulatory sequence (RNP1, Figure 5) may be ineffective approaches to mediate loss-of-function. On the other hand, each of these approaches could be utilized with the less frequent antisense intronic lncRNAs or sense intronic lncRNAs that are independently transcribed from the host gene. It is likely that elucidating the function of most sense intronic lncRNA will involve deleting most or all of the lncRNA either with or without loxP sites (RNP2 and RNP3, Figure 5). For example, the muscle-restricted Syisl sense intronic lncRNA was deleted with two RNPs resulting in poor expansion of satellite cells during skeletal muscle repair in the mouse [129]. Importantly, despite its close juxtaposition to a splice donor in the host (Synpo2) gene, there was little change in expression of Synpo2 upon deletion of Syisl, consistent with the independent transcription of Syisl [129]. Since a binding site for MYOD1 was identified in the proximal promoter region of Syisl [129], it may prove informative and less aggressive to make a subtle mutation in the MYOD1-binding E-box to inactivate Syisl. There is precedent for effectively generating loss-of-function mice with subtle mutations in a single TFBS [113-115]. The nuclear intronic sense lncRNA, Panct1, was floxed and shown to have a critical role in the maintenance of pluripotency in mouse embryonic stem cells [130]. Interestingly, a short sequence motif was discovered in Panct1 through chromatin isolation by RNA purification and this mutant Panct1 failed to rescue the loss in stem cell pluripotency observed when the endogenous Panct1 gene was deleted [130]. The latter result demonstrates a rare example of a functional lncRNA motif and its candidacy for subtle, three-component CRISPR editing in the mouse. Thus, we can see that depending on the nature of the intronic lncRNA and insights gained through in vitro biochemical and promoter studies, most any approach to generating a loss-of-function mouse with CRISPR editing could work.
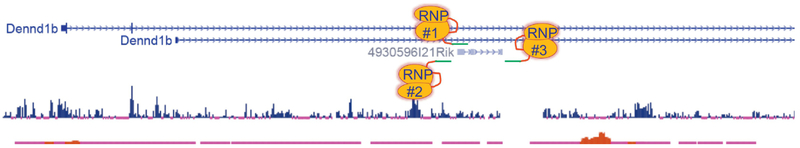
Figure 5. CRISPR editing strategies for an intronic lncRNA.
Screenshot of an lncRNA (4930596121Rik) within a protein-coding gene (Dennd1b). Note that the lncRNA is transcribed in the same (sense) direction as Dennd1b. See text for more details.
CRISPRing Natural Antisense or Overlapping LncRNAs
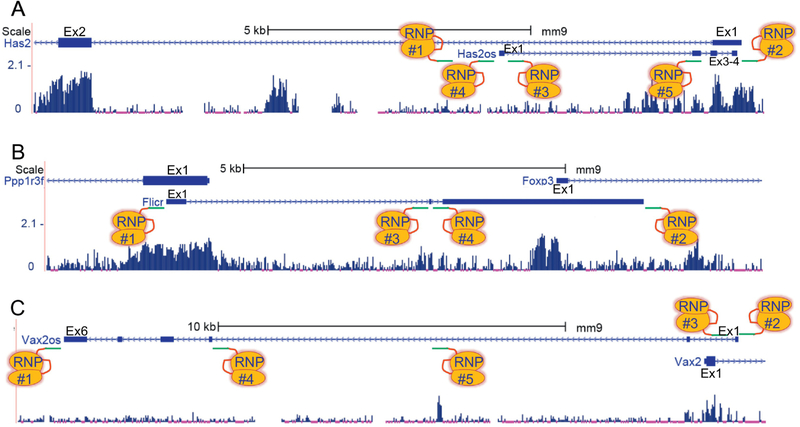
Thousands of annotated lncRNAs are found in close proximity to other gene loci either as natural antisense transcripts (NATs) or 5’ or 3’ overlapping lncRNAs that do not have exons intersecting opposingly transcribed exons. Several vascular cell NATs have been defined [13], including the very first vascular-restricted lncRNA discovered called Tie1-AS [131]. In contrast to the aforementioned lincRNAs and intronic lncRNAs, those of the NAT and overlapping classes are less easily targeted in the mouse with CRISPR editing. For example, the conserved NAT Has2os (os for opposite strand), which is involved with matrix remodeling in VSMC [132], has two exons (Ex3-4) that overlap the opposingly transcribed exon (Ex1) of the protein-coding gene, Has2 (Figure 6A). Deleting the entire Has2os locus may disrupt expression of Has2 (RNP1 and RNP2, Figure 6A). However, since exon 1 of Has2os is several kilobases away from exon 1 of Has2, a microdeletion of the promoter/exon1 of Has2os is attractive, especially given the absence of highly conserved sequences that would be deleted (RNP3 and RNP4, Figure 6A).
Figure 6. CRISPR editing strategies for natural antisense transcript (NAT) lncRNAs and overlapping lncRNAs.
(A) Screenshot of the NAT lncRNA, Has2os (os signifies opposite strand). This lncRNA is transcribed in the antisense orientation to the proteincoding gene, Has2. Note the much higher conservation of Has2 versus the Has2os lncRNA. Exons here and below are labeled as Ex1, Ex2, etc. (B) Screenshot of the Flicr lncRNA that overlaps two flanking protein-coding genes. (C) Screenshot of a 5’ overlapping lncRNA (Vax2os) whose exons do not intersect exons of the antisense transcribed protein-coding gene, Vax2. See text for more details.
Alternatively, targeting exon 1 of Has2os for polyadenylation signal sequence insertion (RNP3, Figure 6A) or the promoter region with either CRISPRi or a subtle edit in a Has2os-specific regulatory element with three-component CRISPR (RNP4, Figure 6A), would likely confer Has2os restricted loss-of-function (Figure 6A). Importantly, when a first exon of a NAT lncRNA falls within a few hundred bases of the overlapping exon 1, then CRISPRi or edits to the promoter region of the lncRNA may impact the overlapping gene. However, insertion of a polyadenylation signal sequence in an lncRNA exon in close juxtaposition to an overlapping exon would unlikely impact overlapping transcription [133]. Thus, three-component CRISPR-mediated insertion of a polyadenylation signal should be widely applicable across different lncRNA classes. Of course, defining a structural or regulatory motif in a single exon of a NAT lncRNA, as has been described in other lncRNA genes [101, 102, 129], would allow for subtle editing of an exon in three-component CRISPR (RNP5, Figure 6A), so long as the edit does not influence an overlapping proteincoding exon. In this context, an intriguing synonymous SNP found in an exon of the EGFR locus was found to affect a NAT lncRNA, conferring heightened sensitivity to cancer therapy in humans [134]. This unexpected finding should alert vascular biologists to variations in lncRNA sequences that may have hidden, clinically-relevant functions worthy of dissection in the mouse. The lncRNASNP2 database (http://bioinfo.life.hust.edu.cn) contains a trove of lncRNA sequence variants that, if found in conserved sequences of an orthologous mouse lncRNA, could be modeled in the mouse with three-component CRISPR editing as reported for a vascular-associated coding SNP [135]. Of note, polymorphisms in the promoter region of the NEAT1 lncRNA result in reduced expression and protein binding making these potentially viable substitutions to study in the mouse [136].
A more difficult scenario exists with the NAT lncRNA, Flicr [80]. Here, we see Flicr exons overlapping exons in two flanking protein-coding genes (Figure 6B). Deleting the Flicr locus in its entirety is a bad strategy as it would likely result in a triple knockout of the lncRNA and the two overlapping protein-coding genes (RNP1 and RNP2, Figure 6B). Targeting the first exon or promoter region of the annotated Flicr locus would impact the overlapping and oppositely transcribed Ppp1r3f coding sequence (RNP1, Figure 6B). Thus, the Flicr gene was targeted with either a single RNP that disrupted splicing at exon 2 (RNP3, Figure 6B) or a pair of RNPs that deleted much of exon 2 (RNP3 and RNP4, Figure 6B). Remarkably, a phenotype associated with reduced Foxp3 expression in regulatory T cells was discovered with reduced incidence of experimentally-induced diabetes [80].
As with Flicr, a 5’ overlapping lncRNA could be challenging to target with CRISPR editing. For example, Vax2os is a multi-exonic lncRNA whose complete deletion would impact the expression of the Vax2 protein-coding gene (RNP1 and RNP2, Figure 6C). However, as with Has2os above, a microdeletion of the promoter/exon 1 of Vax2os represents a more desirable strategy since it would less likely impact Vax2 expression (RNP2 and RNP3, Figure 6C). Alternative approaches to targeting Vax2os would be inserting a polyadenylation signal in exon 1 (RNP2, Figure 6C), making an internal deletion of exons (e.g., RNP1 and RNP4, Figure 6C) or, as with Flicr, targeting specific splice sites to generate a truncated lncRNA (e.g., RNP4, Figure 6C). In general, targeting regions of high sequence conservation may be ill-advised since such sequences could represent small transcription units or regulatory elements controlling distal gene expression (RNP5, Figure 6C). It should be noted that targeting the promoter or first exon of 3’ overlapping lncRNAs may be less intrusive to the 3’ end of the overlapping gene, whether proteincoding or noncoding though disruption of annotated microRNA binding sites in the 3’ untranslated region should be avoided (not shown). As with any of the aforementioned strategies of CRISPR editing an lncRNA in mice, basic experimentation in cultured vascular cells should be done to gain some insight into how the lncRNA should be edited in vivo [15].
CRISPRing Human LncRNAs by BAC Editing in Mice
In contrast to protein-coding genes and microRNAs, lncRNAs generally lack sequence similarity across species. While there are clear experimental advantages to studying mouse-specific lncRNAs, their relevance to human biology may not be so readily apparent (e.g., Bvht). On the other hand, the study of human-specific lncRNAs is complicated by the limited experimental tools to unravel function in vivo. Here, we highlight a human vascular lncRNA called SENCR, which functions to mediate or maintain EC and VSMC differentiation [137-139]. SENCR is a multi-exonic 5’ overlapping lncRNA that coincides with the opposingly transcribed FLI1 transcription factor (Figure 7). One could perform CRISPR editing of such an lncRNA in an established human EC line (e.g., EA.hy926 cells); however, these cells are aneuploid making three-component CRISPR editing of a regulatory element or the insertion of loxP sites or a polyadenylation signal an arduous task. Accordingly, investigators working with immortalized human cell lines should define by karyotyping the number of chromosomes harboring the lncRNA. CRISPR editing human embryonic stem cells or inducible pluripotent stem cells represents a suitable alternative, but experimentation is rather limited.
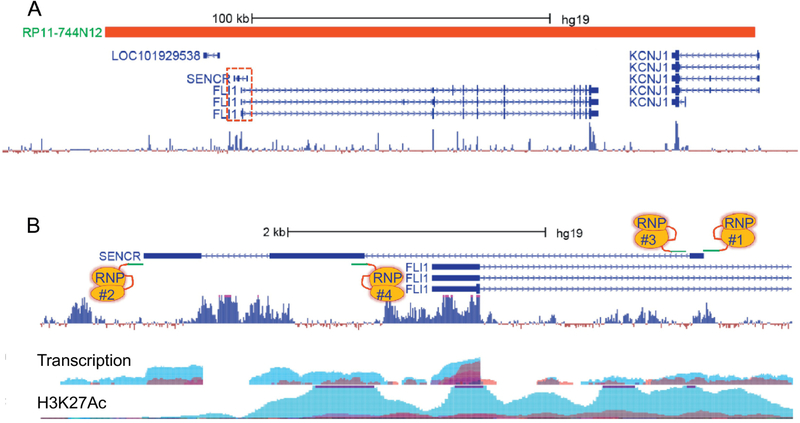
Figure 7. BAC editing strategies for a human lncRNA integrated in the mouse.
(A) Screenshot showing a BAC (RP11-744N12) harboring the human-specific SENCR lncRNA that overlaps the 5’ end of the FLI1 transcription factor. Note that this screenshot is from human genome (hg), assembly 19. (B) Magnified region (stippled red box in panel A) showing the three exons of SENCR in relation to FLI1. The tracks below SENCR/FLI1 represent ENCODE data relating to transcription and H3K27Ac. The teal colored peaks indicate transcription and H3K27Ac in HUVEC. Note the distinction in transcription of each of the exons of SENCR (teal associated HUVEC) versus FLI1 (multi-colored exon 1, indicating transcription in multiple cell types). Note scale bars in panel A versus panel B. See text for more details.
On the other hand, humanizing mice with bacterial artificial chromosomes (BACs) that carry human genes and all neighboring regulatory elements has been successful in pinpointing enhancers and regulatory elements (or variations therein) effecting gene expression in vivo [140-143]. CRISPR has been used to replace a region of the mouse genome with a BAC carrying orthologous human sequences [144]. The latter approach, however, carries the risk of deleting a critical mouse-specific regulatory element that results in embryonic lethality [145]. Consequently, we recently used piggyBAC transposon-mediated BAC transgenesis to integrate a BAC carrying SENCR in the mouse [139, 146]. SENCR was expressed on the surface endothelium of the mouse aorta and, more importantly, was shown to be most abundant in regions of the aorta exhibiting laminar shear stress, a finding observed in cultured human endothelial cells [139]. Of course, BACs often carry additional transcription units. In the case of an lncRNA-mRNA gene pair, such as SENCR-FLI1, this could be advantageous in elucidating any local cis-acting effects on gene transcription. Moreover, it should be possible to inactivate the endogenous Fli1 gene and study the biology of human FLI1 ± SENCR in the mouse. The overarching assumption in humanized mouse models is that the human gene(s) faithfully recapitulates expression and function in the mouse, which may not always be the case.
How might a human lncRNA be edited in a humanized mouse within the context of a BAC? Any of the aforementioned strategies is possible depending on the type of lncRNA. However, it is necessary to first validate expression of the human genes and delineate the boundaries of the BAC. In addition, it is imperative to define founder mice with a single copy of the BAC using digital droplet PCR or Southern blotting before commencing with BAC editing; accurate BAC editing will be very difficult with multiple copies of the BAC. Further, BAC editing is best done in hemizygous mice where only one chromosome carries the BAC-containing lncRNA. Similar to other lncRNAs in close apposition to a protein-coding or noncoding gene, BAC editing of SENCR would not involve the deletion of the entire SENCR locus as there would be simultaneous deletion of the FLI1 promoter and first exon, effectively creating a double knockout (RNP1 and RNP2, Figure 7B). Similarly, removal of terminal exons in SENCR could disrupt proximal promoter elements controlling FLI1 expression (RNP2 and RNP4, Figure 7B). A reasonable starting point would be to either create a microdeletion of the first exon (RNP1 and RNP3, Figure 7B) or insert a short polyadenylation signal in the first exon of SENCR (RNP1, Figure 7B). Note that insertion of a polyadenylation signal in the sense strand of exon 1 of SENCR would not be expected to impinge on transcription of FLI1 [133]. If key upstream regulatory sites controlling SENCR were discovered, a more subtle CRISPR edit could be performed as previously reported for a protein-coding gene [114]. The same could be said for a functional sequence motif within SENCR identified through computational and biochemical means. Any deletions or insertions, however small, would necessitate RNA-seq experiments to rule out unintended consequences on the general transcriptome [147]. We recommend performing RNA-seq experiments, even with regulatory SNP edits, to rule out potentially confounding changes in gene expression following CRISPR editing in the mouse.
CRISPRing Competing Endogenous (Sponge) RNAs
Regarding the competing endogenous RNA (ceRNA) concept, which posits that an lncRNA “sponges” a specific pool of microRNA to derepress target protein expression, much of the hyperbole stems from an unusual circular RNA (CDR1as) that has 73 binding sites for miR-7 and is resistant to miR-7-mediated deadenylation and degradation [148]. Hundreds of papers have proposed lncRNAs functioning as competing endogenous RNAs. However, since most lncRNAs are of low abundance, the vast majority of these may not be of much biological significance in vivo due to asymmetry in expression levels of the lncRNA (lower/less stable) versus the microRNA and target mRNA (higher/more stable). Moreover, there is a notable lack of rigor in formally proving a bonafide sponge RNA [149]. For example, demonstration of a ceRNA would require colocalization studies of the lncRNA and microRNA as shown originally for CDR1as [148] and CRISPR-mediated deletion of the putative ceRNA with subsequent measures of microRNA target proteins [150]. The penultimate experiment would be to CRISPR edit the microRNA binding element (MRE) within the lncRNA and assess microRNA targets; however, this has yet to be done for any lncRNA. There have been efforts to target MREs in protein-coding genes of Drosophila, zebrafish, and mice [151, 152] and at least one MRE within a risk allele for coronary artery disease exists that could undergo CRISPR editing in the mouse [153]. A website has been created (miR-CRISPR) to facilitate CRISPR targeting of putative MREs in lncRNAs [151]. If several MREs in an lncRNA are suspected to function as a sponge, the multiplex nature of CRISPR allows for targeting of each MRE simultaneously [154, 155]. This may require longer dsDNA templates [70] carrying the MRE mutations; however, if the MREs are in close proximity to one another (within 20-30 bases) then an ssODN approach might work [156]. Alternatively, multiplex base editing could be done as reported for targeting two TFBS located 9-kb apart in the mouse [68]. The advantage of the latter would be the absence of double-strand breaks that could delete intervening sequences between two MREs. On the other hand, base editing generally alters only one base and that may not be sufficient to functionally knockout an MRE. We strongly recommend against continued reporting of so-called sponge lncRNAs in the absence of more rigorous analysis.
Summary and Perspective
Mammalian genomes are pervasively transcribed with an expanding class of lncRNAs [157]. In addition, there are thousands of other long noncoding transcripts such as pseudogenes, circular RNAs, and enhancer RNAs, each of which is amenable to approaches of gene editing outlined here. Before embarking on any CRISPR editing project involving a long noncoding RNA in the mouse, its genomic landscape must be thoroughly interrogated with the latest version of the UCSC Genome Browser [158]. Full length transcripts must be defined to ensure all transcribed sequence is delineated. Moreover, cellular localization, transcriptional control in reporter assays, and knockdown experiments are necessary to gain insight into the regulation and function of a new lncRNA [15]. A recent report suggests most lncRNAs may not be targetable with CRISPR editing [159]; here, however, we propose a number of strategies that should allow most any lncRNA to be targeted in the mouse, although advantages and disadvantages exist for each based on the position of the lncRNA. These approaches capitalize on new innovations in CRISPR-Cas9 editing of the mouse, including in vivo BAC-editing of a human lncRNA. When contemplating CRISPR editing in the mouse, ‘less is more’; therefore, the more subtle the edit (e.g., microdeletions or single/multiple base substitutions), the less chance of confounding effects stemming from loss of a proteincoding gene or structural/regulatory element. It should also be mentioned that previously annotated lncRNAs have turned out to be protein-coding genes [160] and some proteincoding genes have noncoding functions [161, 162]. Three-component CRISPR editing of AUG (or CUG) codons could distinguish between a coding versus noncoding function.
In this review, we have promoted the broadened use of CRISPR to edit lncRNAs in the mouse. The time and expense in generating a mouse over a cell line has diminished considerably and there is much more flexibility in generating a mouse line, where diploid vascular cells carrying edited lncRNAs can be readily cultured, than an aneuploid cell line carrying indelible edits. We do recognize, however, there is great power in CRISPR editing human inducible pluripotent stem (iPS) cells where human disease modeling in the mouse is feasible [163]. Indeed, recent CRISPR editing of the cardiovascular risk haplotype in the ANRIL (CDKN2B-AS1) lncRNA revealed intriguing pathological phenotypes when edited human iPS cells were differentiated to a VSMC phenotype [164]. An atherosclerosis risk allele exists in another lncRNA that awaits formal proof of function using CRISPR in iPS cells [165]. Of course, all biomedical research using CRISPR editing has the ultimate goal of effecting meaningful measures to improve the health of human beings. The recent revelation of reckless germline genome editing of the CCR5 gene in twin Chinese girls named “LuLu” and “Nana” [166] must galvanize scientists and the global community to proceed with this powerful technology in an ethically, socially, and scientifically responsible manner [167].
Acknowledgments
Mouse CRISPR work is supported by grants from the National Institutes of Health (HL132574, HL138987, HL136224 to JMM and HL122686 and HL139794 to XL) and an American Heart Association Post-doctoral Fellowship Award (17POST3360938) to QL. We extend our sincere thanks to Lin Gan and Xiaoling Xie of the University of Rochester Mouse Genome Editing Resource for their expertise in generating CRISPR edited mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ohno S, So much "junk" DNA in our genome, Brookhaven Symp Biol, 23 (1972) 366–370. [PubMed] [Google Scholar]
- [2].Encode Project Consortium, An integrated encyclopedia of DNA elements in the human genome, Nature, 489 (2012) 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression, Genome Res, 22 (2012) 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. , A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome, Nature, 349 (1991) 38–44. [DOI] [PubMed] [Google Scholar]
- [5].Brannan CI, Dees EC, Ingram RS, Tilghman SM, The product of the H19 gene may function as an RNA, Mol Cell Biol, 10 (1990) 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, et al. , LNCipedia 5: towards a reference set of human long non-coding RNAs, Nucleic Acids Res, 47 (2019) D135–D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kopp F, Mendell JT, Functional classification and experimental dissection of long noncoding RNAs, Cell, 172 (2018) 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M, Cytoplasmic functions of long noncoding RNAs, Wiley interdisciplinary reviews RNA, 9 (2018) e1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D, LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases, Nucleic Acids Res, 47 (2019) D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, et al. , Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease, Nat Genet, 49 (2017) 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leung A, Natarajan R, Noncoding RNAs in vascular disease, Curr Opin Cardiol, 29 (2014) 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Uchida S, Dimmeler S, Long noncoding RNAs in cardiovascular diseases, Circ Res, 116 (2015) 737–50. [DOI] [PubMed] [Google Scholar]
- [13].Miano JM, Long X, The short and long of noncoding sequences in the control of vascular cell phenotypes, Cell Mol Life Sci, 72 (2015) 3457–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deng L, Bradshaw AC, Baker AH, Role of noncoding RNA in vascular remodelling, Curr Opin Lipidol, 27 (2016) 439–48. [DOI] [PubMed] [Google Scholar]
- [15].Freedman JE, Miano JM, Challenges and opportunities in linking long noncoding RNAs to cardiovascular, lung, and blood diseases, Arterioscler Thromb Vasc Biol, 37 (2016) 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haemmig S, Feinberg MW, Targeting lncRNAs in cardiovascular disease: options and expeditions, Circ Res, 120 (2017) 620–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].La Russa MF, Qi LS, The new state of the art: Cas9 for gene activation and repression, Mol Cell Biol, 35 (2015) 3800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fujii W, Kawasaki K, Sugiura K, Naito K, Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease, Nucleic Acids Res, 41 (2013) e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Adikusuma F, Williams N, Grutzner F, Hughes J, Thomas P, Targeted deletion of an entire chromosome using CRISPR/Cas9, Mol Ther, 25 (2017) 1736–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Harms DW, Quadros RM, Seruggia D, Ohtsuka M, Takahashi G, Montoliu L, et al. Mouse genome editing using the CRISPR/Cas system. Curr Protoc Hum Genet 2014; 83: 15.7.1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Singh P, Schimenti JC, Bolcun-Filas E, A mouse geneticist's practical guide to CRISPR applications, Genetics, 199 (2015) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miano JM, Zhu QM, Lowenstein CJ, A CRISPR path to engineering new genetic mouse models for cardiovascular research, Arterioscler Thromb Vasc Biol, 36 (2016) 1058–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Birling MC, Herault Y, Pavlovic G, Modeling human disease in rodents by CRISPR/Cas9 genome editing, Mamm Genome, 28 (2017) 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M, Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA, Sci Rep, 3 (2013) 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering, Cell, 153 (2013) 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, et al. , Correction of a genetic disease in mouse via use of CRISPR-Cas9, Cell Stem Cell, 13 (2013) 659–62. [DOI] [PubMed] [Google Scholar]
- [27].Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering, Cell, 154 (2013) 1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. , Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity, Cell, 154 (2013) 1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. , Generation of gene-modified mice via Cas9/RNA-mediated gene targeting, Cell Res, 23 (2013) 720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 337 (2012) 816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wienert B, Shin J, Zelin E, Pestal K, Corn JE, In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway, PLoS Biol, 16 (2018) e2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim S, Koo T, Jee HG, Cho HY, Lee G, Lim DG, et al. , CRISPR RNAs trigger innate immune responses in human cells, Genome Res, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Oliveros JC, Franch M, Tabas-Madrid D, San-Leon D, Montoliu L, Cubas P, et al. , Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes, Nucleic Acids Res, 44 (2016) W267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, et al. , Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR, Genome Biol, 17 (2016) 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. , DNA targeting specificity of RNA-guided Cas9 nucleases, Nat Biotechnol, 31 (2013) 827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Graf R, Li X, Chu VT, Rajewsky K, sgRNA sequence motifs blocking efficient CRISPR/Cas9-mediated gene editing, Cell Reports, 26 (2019) 1098–1103 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB, Unexpected mutations after CRISPR-Cas9 editing in vivo, Nat Methods, 14 (2017) 547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nutter LMJ, Heaney JD, Lloyd KCK, Murray SA, Seavitt JR, Skarnes WC, et al. , Response to "Unexpected mutations after CRISPR-Cas9 editing in vivo", Nat Methods, 15 (2018) 235–236. [DOI] [PubMed] [Google Scholar]
- [39].Wilson CJ, Fennell T, Bothmer A, Maeder ML, Reyon D, Cotta-Ramusino C, et al. , Response to "Unexpected mutations after CRISPR-Cas9 editing in vivo", Nat Methods, 15 (2018) 236–237. [DOI] [PubMed] [Google Scholar]
- [40].Lareau CA, Clement K, Hsu JY, Pattanayak V, Joung JK, Aryee MJ, et al. , Response to "Unexpected mutations after CRISPR-Cas9 editing in vivo", Nat Methods, 15 (2018) 238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lescarbeau RM, Murray B, Barnes TM, Bermingham N, Response to "Unexpected mutations after CRISPR-Cas9 editing in vivo", Nat Methods, 15 (2018) 237. [DOI] [PubMed] [Google Scholar]
- [42].Kim ST, Park J, Kim D, Kim K, Bae S, Schlesner M, et al. , Response to "Unexpected mutations after CRISPR-Cas9 editing in vivo", Nat Methods, 15 (2018) 239–240. [DOI] [PubMed] [Google Scholar]
- [43].Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, et al. , Off-target mutations are rare in Cas9-modified mice, Nat Methods, 12 (2015) 479. [DOI] [PubMed] [Google Scholar]
- [44].Anderson KR, Haeussler M, Watanabe C, Janakiraman V, Lund J, Modrusan Z, et al. , CRISPR off-target analysis in genetically engineered rats and mice, Nat Methods, 15 (2018) 512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Iyer V, Boroviak K, Thomas M, Doe B, Riva L, Ryder E, et al. , No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice, PLoS Genet, 14 (2018) e1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, et al. , In vivo CRISPR editing with no detectable genome-wide off-target mutations, Nature, 561 (2018) 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, et al. , Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice, Genome Biol, 16 (2015) 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sentmanat MF, Peters ST, Florian CP, Connelly JP, Pruett-Miller SM, A survey of validation strategies for CRISPR-Cas9 editing, Sci Rep, 8 (2018) 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kim S, Kim D, Cho SW, Kim J, Kim JS, Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins, Genome Res, 24 (2014) 1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, et al. , Dynamics of CRISPR-Cas9 genome interrogation in living cells, Science, 350 (2015) 823–6. [DOI] [PubMed] [Google Scholar]
- [51].Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, et al. , Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes, Dev Biol, 393 (2014) 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Oliver D, Yuan S, McSwiggin H, Yan W, Pervasive genotypic mosaicism in founder mice derived from genome editing through pronuclear injection, PLoS One, 10 (2015) e0129457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F, Rationally engineered Cas9 nucleases with improved specificity, Science, 351 (2016) 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. , High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects, Nature, 529 (2016) 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. , Enhanced proofreading governs CRISPR-Cas9 targeting accuracy, Nature, 550 (2017) 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. , Evolved Cas9 variants with broad PAM compatibility and high DNA specificity, Nature, 556 (2018) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. , A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells, Nat Med, 24 (2018) 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Amrani N, Gao XD, Liu P, Edraki A, Mir A, Ibraheim R, et al. , NmeCas9 is an intrinsically high-fidelity genome-editing platform, Genome Biol, 19 (2018) 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. , Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system, Cell, 163 (2015) 759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, et al. , CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice, Science advances, 3 (2017) e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, et al. , Engineering of CRISPR-Cas12b for human genome editing, Nature communications, 10 (2019) 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, et al. , CasX enzymes comprise a distinct family of RNA-guided genome editors, Nature, 566 (2019) 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yang S, Li S, Li XJ, Shortening the half-life of Cas9 maintains its gene editing ability and reduces neuronal toxicity, Cell Rep, 25 (2018) 2653–2659 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Petris G, Casini A, Montagna C, Lorenzin F, Prandi D, Romanel A, et al. , Hit and go CAS9 delivered through a lentiviral based self-limiting circuit, Nature communications, 8 (2017) 15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rees HA, Liu DR, Base editing: precision chemistry on the genome and transcriptome of living cells, Nature reviews Genetics, 19 (2018) 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, et al. , Highly efficient RNA-guided base editing in mouse embryos, Nat Biotechnol, 35 (2017) 435–437. [DOI] [PubMed] [Google Scholar]
- [67].Liang P, Sun H, Sun Y, Zhang X, Xie X, Zhang J, et al. , Effective gene editing by high-fidelity base editor 2 in mouse zygotes, Protein & cell, 8 (2017) 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lee HK, Willi M, Smith HE, Miller SM, Liu DR, Liu C, et al. , Simultaneous targeting of linked loci in mouse embryos using base editing, Sci Rep, 9 (2019) 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, et al. , Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins, Genome Biol, 18 (2017) 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yao X, Zhang M, Wang X, Ying W, Hu X, Dai P, et al. , Tild-CRISPR allows for efficient and precise gene knockin in mouse and human cells, Dev Cell, 45 (2018) 526–536 e5. [DOI] [PubMed] [Google Scholar]
- [71].Teixeira M, Py BF, Bosc C, Laubreton D, Moutin MJ, Marvel J, et al. , Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing, Sci Rep, 8 (2018) 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kaneko T, Mashimo T, Simple genome editing of rodent intact embryos by electroporation, PLoS One, 10 (2015) e0142755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, et al. , Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease, Genetics, 200 (2015) 423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Miyasaka Y, Uno Y, Yoshimi K, Kunihiro Y, Yoshimura T, Tanaka T, et al. , CLICK: one-step generation of conditional knockout mice, BMC Genomics, 19 (2018) 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ohtsuka M, Sato M, Miura H, Takabayashi S, Matsuyama M, Koyano T, et al. , i-GONAD: a robust method for in situ germline genome engineering using CRISPR nucleases, Genome Biol, 19 (2018) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gu B, Posfai E, Rossant J, Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos, Nat Biotechnol, 36 (2018) 632–637. [DOI] [PubMed] [Google Scholar]
- [77].Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. , Multiple knockout mouse models reveal lincRNAs are required for life and brain development, eLife, 2 (2013) e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Han X, Luo S, Peng G, Lu JY, Cui G, Liu L, et al. , Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development, J Mol Cell Biol, 10 (2018) 175–178. [DOI] [PubMed] [Google Scholar]
- [79].Goudarzi M, Berg K, Pieper LM, Schier AF, Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility, eLife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C, Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity, Proc Natl Acad Sci U S A, 114 (2017) E3472–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Perry RB, Hezroni H, Goldrich MJ, Ulitsky I, Regulation of neuroregeneration by long noncoding RNAs, Mol Cell, 72 (2018) 553–567 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kopp F, Elguindy MM, Yalvac ME, Zhang H, Chen B, Gillett FA, et al. , PUMILIO hyperactivity drives premature aging of Norad-deficient mice, eLife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hansmeier NR, Widdershooven PJM, Khani S, Kornfeld JW, Rapid generation of long noncoding RNA knockout mice using CRISPR/Cas9 technology, Non-coding RNA, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, et al. , LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity, Circulation, 130 (2014) 1452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, et al. , Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction, Cell Death Dis, 7 (2016) e2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, et al. , Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA, Nat Med, 24 (2018) 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, et al. , H19 induces abdominal aortic aneurysm development and progression, Circulation, 138 (2018) 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ahmed ASI, Dong K, Liu J, Wen T, Yu L, Xu F, et al. , Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells, Proc Natl Acad Sci U S A, 115 (2018) E8660–E8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cremer S, Michalik KM, Fischer A, Pfisterer L, Jae N, Winter C, et al. , Hematopoietic deficiency of the long non-coding RNA MALAT1 promotes atherosclerosis and plaque inflammation, Circulation, (2018). [DOI] [PubMed] [Google Scholar]
- [90].Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. , Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals, Nature, 458 (2009) 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. , Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev, 25 (2011) 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, et al. , Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth, Circ Res, 114 (2014) 1389–97. [DOI] [PubMed] [Google Scholar]
- [93].Yu CK, Xu T, Assoian RK, Rader DJ, Mining the stiffness-sensitive transcriptome in human vascular smooth muscle cells identifies long noncoding RNA stiffness regulators, Arterioscler Thromb Vasc Biol, 38 (2018) 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, et al. , Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9, RNA Biol, 11 (2014) 829–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Szafranski P, Karolak JA, Lanza D, Gajecka M, Heaney J, Stankiewicz P, CRISPR/Cas9-mediated deletion of lncRNA Gm26878 in the distant Foxf1 enhancer region, Mamm Genome, 28 (2017) 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kosicki M, Tomberg K, Bradley A, Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements, Nat Biotechnol, 36 (2018) 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Payne S, De Val S, Neal A, Endothelial-specific Cre mouse models, Arterioscler Thromb Vasc Biol, 38 (2018) 2550–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chakraborty R, Saddouk FZ, Carrao AC, Krause DS, Greif DM, Martin KA, Promoters to study vascular smooth muscle: mistaken identity?, Arterioscler Thromb Vasc Biol, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Miano JM, Ramanan N, Georger MA, de Mesy-Bentley KL, Emerson RL, Balza RO Jr., et al. , Restricted inactivation of serum response factor to the cardiovascular system, Proc Natl Acad Sci U S A, 101 (2004) 17132–17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li R, Harvey AR, Hodgetts SI, Fox AH, Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles, RNA, 23 (2017) 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang B, Gunawardane L, Niazi F, Jahanbani F, Chen X, Valadkhan S, A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA, Mol Cell Biol, 34 (2014) 2318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, et al. , A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage, Mol Cell, 64 (2016) 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, et al. , Unlinking an lncRNA from its associated cis element, Mol Cell, 62 (2016) 104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN, Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development, Nature, 539 (2016) 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. , Local regulation of gene expression by lncRNA promoters, transcription and splicing, Nature, 539 (2016) 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ballarino M, Cipriano A, Tita R, Santini T, Desideri F, Morlando M, et al. , Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice, EMBO J, 37 (2018) pii: e99697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].McFarland TJ, Zhang Y, Atchaneeyaskul LO, Francis P, Stout JT, Appukuttan B, Evaluation of a novel short polyadenylation signal as an alternative to the SV40 polyadenylation signal, Plasmid, 56 (2006) 62–7. [DOI] [PubMed] [Google Scholar]
- [108].Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. , BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis, Nature, 527 (2015) 192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hu J, Verzi MP, Robinson AS, Tang PL, Hua LL, Xu SM, et al. , Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway, Development, 142 (2015) 2775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Seruggia D, Fernandez A, Cantero M, Pelczar P, Montoliu L, Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis, Nucleic Acids Res, 43 (2015) 4855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Metser G, Shin HY, Wang C, Yoo KH, Oh S, Villarino AV, et al. , An autoregulatory enhancer controls mammary-specific STAT5 functions, Nucleic Acids Res, 44 (2016) 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Elling R, Robinson EK, Shapleigh B, Liapis SC, Covarrubias S, Katzman S, et al. , Genetic models reveal cis and trans immune-regulatory activities for lincRNA-Cox2, Cell Rep, 25 (2018) 1511–1524 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, et al. , Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning, Development, 125 (1998) 1025–36. [DOI] [PubMed] [Google Scholar]
- [114].Han Y, Slivano OJ, Christie CK, Cheng AW, Miano JM, CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice--brief report, Arterioscler Thromb Vasc Biol, 35 (2015) 312–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhang Y, Zhang J, An W, Wan Y, Ma S, Yin J, et al. , Intron 1 GATA site enhances ALAS2 expression indispensably during erythroid differentiation, Nucleic Acids Res, 45 (2017) 657–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, et al. , CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency, Science, 363 (2019) pii: eaau0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. , Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression, Cell, 152 (2013) 1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. , CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes, Cell, 154 (2013) 442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. , CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells, Science, 355 (2017) pii: aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. , Genome-scale CRISPR-mediated control of gene repression and activation, Cell, 159 (2014) 647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Stojic L, Lun ATL, Mangei J, Mascalchi P, Quarantotti V, Barr AR, et al. , Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis, Nucleic Acids Res, 46 (2018) 5950–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, et al. , CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis, J Mol Cell Cardiol, 89 (2015) 98–112. [DOI] [PubMed] [Google Scholar]
- [123].Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, et al. , MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation, Circ Res, 105 (2009) 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. , miR-145 and miR-143 regulate smooth muscle cell fate and plasticity, Nature, 460 (2009) 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. , MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury, Genes Dev, 23 (2009) 2166–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. , Braveheart, a long noncoding RNA required for cardiovascular lineage commitment, Cell, 152 (2013) 570–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kirk JM, Kim SO, Inoue K, Smola MJ, Lee DM, Schertzer MD, et al. , Functional classification of long non-coding RNAs by k-mer content, Nat Genet, 50 (2018) 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chan SN, Pek JW, Stable intronic sequence RNAs (sisRNAs): an expanding universe, Trends Biochem Sci, 44 (2019) 258–272. [DOI] [PubMed] [Google Scholar]
- [129].Jin JJ, Lv W, Xia P, Xu ZY, Zheng AD, Wang XJ, et al. , Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2, Proc Natl Acad Sci U S A, 115 (2018) E9802–E9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chakraborty D, Paszkowski-Rogacz M, Berger N, Ding L, Mircetic J, Fu J, et al. , lncRNA Panct1 maintains mouse embryonic stem cell identity by regulating TOBF1 recruitment to Oct-Sox sequences in early G1, Cell Rep, 21 (2017) 3012–3021. [DOI] [PubMed] [Google Scholar]
- [131].Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. , A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo, Blood, 115 (2010) 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M, et al. , Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation, J Biol Chem, 289 (2014) 28816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Liu Y, Han X, Yuan J, Geng T, Chen S, Hu X, et al. , Biallelic insertion of a transcriptional terminator via the CRISPR/Cas9 system efficiently silences expression of protein-coding and non-coding RNA genes, J Biol Chem, 292 (2017) 5624–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, et al. , Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma, Nat Med, 23 (2017) 1167–1175. [DOI] [PubMed] [Google Scholar]
- [135].Zhu QM, Ko KA, Ture S, Mastrangelo MA, Chen MH, Johnson AD, et al. , Novel thrombotic function of a human SNP in STXBP5 revealed by CRISPR/Cas9 gene editing in mice, Arterioscler Thromb Vasc Biol, 37 (2017) 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Rheinbay E, Parasuraman P, Grimsby J, Tiao G, Engreitz JM, Kim J, et al. , Recurrent and functional regulatory mutations in breast cancer, Nature, 547 (2017) 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, et al. , Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA, Arterioscler Thromb Vasc Biol, 34 (2014) 1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, et al. , A role for the long non-coding RNA SENCR in commitment and function of endothelial cells, Mol Ther, 24 (2016) 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, et al. , SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4, Proc Natl Acad Sci U S A, 116 (2019) 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wang X, Raghavan A, Peters DT, Pashos EE, Rader DJ, Musunuru K, Interrogation of the atherosclerosis-associated SORT1 (Sortilin 1) locus with primary human hepatocytes, induced pluripotent stem cell-hepatocytes, and locus-humanized mice, Arterioscler Thromb Vasc Biol, 38 (2018) 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Sokhi UK, Liber MP, Frye L, Park S, Kang K, Pannellini T, et al. , Dissection and function of autoimmunity-associated TNFAIP3 (A20) gene enhancers in humanized mouse models, Nature communications, 9 (2018) 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Giraldo P, Montoliu L, Size matters: use of YACs, BACs, and PACs in transgenic animals, Transgenic Res, 10 (2001) 83–103. [DOI] [PubMed] [Google Scholar]
- [143].Long X, Miano JM, Remote control of gene expression, J Biol Chem, 282 (2007) 15941–15945. [DOI] [PubMed] [Google Scholar]
- [144].Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T, ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes, Nature communications, 7 (2016) 10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Long X, Slivano OJ, Cowan SL, Georger MA, Lee TH, Miano JM, Smooth muscle calponin: an unconventional CArG-dependent gene that antagonizes neointimal formation, Arterioscler Thromb Vasc Biol, 31 (2011) 2172–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Jung CJ, Menoret S, Brusselle L, Tesson L, Usal C, Chenouard V, et al. , Comparative analysis of piggyBac, CRISPR/Cas9 and TALEN mediated BAC transgenesis in the zygote for the generation of humanized SIRPA rats, Sci Rep, 6 (2016) 31455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, Schulze C, et al. , Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression, FASEB J, 24 (2010) 1714–24. [DOI] [PubMed] [Google Scholar]
- [148].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. , Natural RNA circles function as efficient microRNA sponges, Nature, 495 (2013) 384–8. [DOI] [PubMed] [Google Scholar]
- [149].Ulitsky I, Interactions between short and long noncoding RNAs, FEBS Lett, 592 (2018) 2874–2883. [DOI] [PubMed] [Google Scholar]
- [150].Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. , Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function, Science, 357 (2017) pii: eaam8526. [DOI] [PubMed] [Google Scholar]
- [151].Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, McGowan S, et al. , Understanding functional miRNA-target interactions in vivo by site-specific genome engineering, Nature communications, 5 (2014) 4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Mildner A, Chapnik E, Varol D, Aychek T, Lampl N, Rivkin N, et al. , MicroRNA-142 controls thymocyte proliferation, Eur J Immunol, 47 (2017) 1142–1152. [DOI] [PubMed] [Google Scholar]
- [153].Miller CL, Haas U, Diaz R, Leeper NJ, Kundu RK, Patlolla B, et al. , Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation, PLoS Genet, 10 (2014) e1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. , Multiplex genome engineering using CRISPR/Cas systems, Science, 339 (2013) 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, et al. , Genome-wide inactivation of porcine endogenous retroviruses (PERVs), Science, 350 (2015) 1101–4. [DOI] [PubMed] [Google Scholar]
- [156].Halim D, Wilson MP, Oliver D, Brosens E, Verheij JB, Han Y, et al. , Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice, Proc Natl Acad Sci U S A, 114 (2017) E2739–E2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, et al. , The reality of pervasive transcription, PLoS Biol, 9 (2011) e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, et al. , The UCSC Genome Browser database: 2019 update, Nucleic Acids Res, 47 (2019) D853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S, Challenges of CRISPR/Cas9 applications for long non-coding RNA genes, Nucleic Acids Res, 45 (2017) e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Makarewich CA, Olson EN, Mining for micropeptides, Trends Cell Biol, 27 (2017) 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gilot D, Migault M, Bachelot L, Journe F, Rogiers A, Donnou-Fournet E, et al. , A non-coding function of TYRP1 mRNA promotes melanoma growth, Nat Cell Biol, 19 (2017) 1348–1357. [DOI] [PubMed] [Google Scholar]
- [162].Kumari P, Sampath K, cncRNAs: Bi-functional RNAs with protein coding and noncoding functions, Semin Cell Dev Biol, 47–48 (2015) 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Soldner F, Jaenisch R, Stem cells, genome editing, and the path to translational medicine, Cell, 175 (2018) 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lo Sardo V, Chubukov P, Ferguson W, Kumar A, Teng EL, Duran M, et al. , Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing, Cell, 175 (2018) 1796–1810 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang DD, Wang WT, Xiong J, Xie XM, Cui SS, Zhao ZG, et al. , Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NFkappaB pathway in human monocytes, Sci Rep, 7 (2017) 46204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Lovell-Badge R, CRISPR babies: a view from the centre of the storm, Development, 146 (2019) pii: dev175778. [DOI] [PubMed] [Google Scholar]
- [167].Coller BS, Ethics of human genome editing, Annu Rev Med, 70 (2019) 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]