Abstract
Klebsiella species cause a wide range of diseases including pneumonia, urinary tract infections (UTIs), bloodstream infections and sepsis. These infections are particularly a problem among neonates, elderly and immunocompromised individuals. Klebsiella is also responsible for a significant number of community-acquired infections. A defining feature of these infections is their morbidity and mortality, and the Klebsiella strains associated with them are considered hypervirulent. The increasing isolation of multidrug-resistant strains has significantly narrowed, or in some settings completely removed, the therapeutic options for the treatment of Klebsiella infections. Not surprisingly, this pathogen has then been singled out as an ‘urgent threat to human health’ by several organisations. This review summarises the tremendous progress that has been made to uncover the sophisticated immune evasion strategies of K. pneumoniae. The co-evolution of Klebsiella in response to the challenge of an activated immune has made Klebsiella a formidable pathogen exploiting stealth strategies and actively suppressing innate immune defences to overcome host responses to survive in the tissues. A better understanding of Klebsiella immune evasion strategies in the context of the host–pathogen interactions is pivotal to develop new therapeutics, which can be based on antagonising the anti-immune strategies of this pathogen.
Keywords: Klebsiella, innate immunity, virulence
The human pathogen Klebsiella pneumoniae: a master puppeteer of manipulation of our body to counteract our defences.
INTRODUCTION
Klebsiella pneumoniae was first described by Carl Friedlander in 1882 as a bacterium isolated from the lungs of patients who had died from pneumonia (Friedlander 1882). Klebsiella species are ubiquitously found in nature including water, soil and animals, and they can colonise medical devices and the healthcare environment (Podschun and Ullmann 1998; Podschun et al.2001). Klebsiella species are considered opportunistic pathogens colonising mucosal surfaces without causing pathology; however, from mucosae Klebsiella may disseminate to other tissues causing life-threatening infections including pneumonia, UTIs, bloodstream infections and sepsis (Paczosa and Mecsas 2016). K. pneumoniae infections are particularly a problem among neonates, elderly and immunocompromised individuals within the healthcare setting (Magill et al.2014). This organism is also responsible for a significant number of community-acquired infections worldwide (Ko et al.2002). Defining features of these infections are the ability to metastatically spread and their significant morbidity and mortality (Paczosa and Mecsas 2016). Klebsiella strains associated with these infections are regarded as hypervirulent, and recent epidemiological studies indicate that these strains share specific genetic characteristics (Holt et al.2015).
K. pneumoniae is gaining attention due to the rise in the number of infections and the increasing number of strains resistant to antibiotics. More than a third of the K. pneumoniae isolates reported to the European Centre for Disease Prevention and Control were resistant to at least one antimicrobial group, the combined resistance to fluoroquinolones, third-generation cephalosporins and aminoglycosides being the most common resistance phenotype (European Centre for Disease Prevention and Control Antimicrobial resistance (EARS-Net) 2018). Furthermore, Klebsiella species are a known reservoir for antibiotic-resistant genes, which can spread to other Gram-negative bacteria. In fact, many of the antibiotic-resistant genes now commonly found in multidrug-resistant organisms were firstly described in Klebsiella. Very few therapeutic options are left for patients infected with multidrug-resistant K. pneumoniae with additional resistance to carbapenems, and are often limited to combination therapy and to colistin. Alarmingly, recent studies have recognised that several K. pneumoniae virulent and multidrug-resistant clones have access to a mobile pool of virulence and antimicrobial resistance genes (Holt et al.2015; Lam et al.2018; MC Lam et al.2018), making then possible the emergence of a multidrug, hypervirulent K. pneumoniae clone capable of causing untreatable infections in healthy individuals. Unfortunately, there are already reports describing the isolation of such strains (Zhang et al.2015, 2016; Gu et al.2018; Yao et al.2018). Despite its clinical relevance, our understanding of K. pneumoniae pathogenesis contains considerable gaps, thereby making a compelling case to better understand its infection biology to design new strategies to treat Klebsiella infections.
Recent excellent reviews have covered the epidemiology of Klebsiella-triggered infections, the mechanisms of resistance to antibiotics and the description of some of the virulence factors of this pathogen (Paczosa and Mecsas 2016; Navon-Venezia, Kondratyeva and Carattoli 2017; Martin and Bachman 2018). This review focuses on the complex interaction between Klebsiella species and the innate immune system, and summarises our understanding of Klebsiella anti-immune strategies. Although central to the infection biology of multidrug-resistant pathogens such as Klebsiella, the repertoire of their adaptations to the human immune system are generally overlooked. However, the co-evolution of these bacteria in response to the challenge of an activated immune system has made them formidable pathogens. As will be apparent in this review, Klebsiella can no longer be considered only as a stealth pathogen. Klebsiella has developed an array of systems that ‘surgically strike’ key regulators and effectors of the host immune system, placing this pathogen as a master puppeteer controlling several anti-immune evasion systems to overcome host responses to survive in the tissues.
INNATE IMMUNE DEFENCES AGAINST BACTERIAL INFECTIONS
Infection can be viewed as a consequence of specific interactions between pathogens and the host, involving the early interaction with the innate system, which includes mechanical, chemical and cellular barriers. Mucociliary clearance is one of the first mechanical defences faced by any pathogen in the respiratory tract. Pathogens may be trapped in a blanket of mucus which covers the airways and is constantly propelled by cilia from the distal to proximal lung airways. The flow of urine in conjunction with its low pH prevents colonisation of the genitourinary tract, whereas peristalsis and the mucus lining of the gastrointestinal tract limit the attachment of bacteria to the gut epithelium. The presence of digestive enzymes, bile and the acid pH in the stomach further prevents pathogen colonisation of the gastrointestinal tract.
Once pathogens overcome these mechanical barriers, they face the challenge of chemical defences, chiefly the complement system, collectins and antimicrobial peptides. In the classical pathway of activation of the complement cascade, C1q recognises pathogen- or damage-associated patterns (such as IgG, IgM or CRP) on foreign or apoptotic cells, inducing the formation of the C3 convertase (C4b2b) (Holers 2014). In the lectin pathway, mannose-binding lectins and ficolins bind to carbohydrates leading to the activation of C4b2b, which subsequently activates C3 in its active fragments C3a and C3b (Holers 2014). The deposition of the latter on surfaces leads to the binding of factor B and conversion into the alternative pathway C3 convertase (C3bBb), which cleaves more C3 into C3b, thereby amplifying the complement response (Holers 2014). Opsonisation by C1q, C3b and its degradation products induces phagocytosis via a panel of complement receptors (Ricklin et al.2016). In addition, complement factors such as C3a and C5a are powerful chemoattractants guiding neutrophils, macrophages and monocytes to the sites of infection (Ricklin et al.2016). Complement also shapes inflammatory responses activated via pathogen recognition receptors (PRRs), and even dictates the differentiation of T cells, thereby acting as player and mediator in immune surveillance (Ricklin et al.2010).
Collectins are a family of proteins that include mannan-binding lectin (mannose-binding protein) and lung surfactant proteins (SPs), SP-A to D (Holmskov, Thiel and Jensenius 2003). These proteins share a common structure made of a C-terminal-located C-type lectin domain which is attached to a collagen-like region via an alpha-helical coiled-coil neck region (Holmskov, Thiel and Jensenius 2003). Collectins bind surface carbohydrates in pathogens leading to the opsonisation, agglutination or killing of the pathogen. Interestingly, lung SPs have also immunomodulatory roles by governing phagocytosis and controlling inflammation (Sano and Kuroki 2005; Kuroki, Takahashi and Nishitani 2007). Additional defences against bacterial infections include antimicrobial peptides and proteins, and cathelicidins produced by epithelial cells, neutrophils and macrophages in response to infection. The levels of these antimicrobials in the site of infection may reach hundreds of micrograms creating a harsh environment. Defensins and lysozyme have potent antibacterial activity against Gram-positive and -negative bacteria. The antibacterial action is based on electrostatic interaction with the anionic bacterial surface leading to membrane perturbations. LL-37/hCAP-18, the only cathelicidin found in humans, is also antimicrobial. Interestingly, defensins and cathelicidins have additional multiple roles influencing diverse processes such as cell proliferation and migration, immune modulation, wound healing, angiogenesis and the release of cytokines (Ganz 2003; Bowdish, Davidson and Hancock 2006).
Upon infection, the host activates a sophisticated program dedicated to clear the pathogen by activation of germ-line encoded receptors referred to as pathogen recognition receptors (PRRs). Data support the notion that there is a common host response associated to infection, the so-called ‘alarm signal’, mainly controlled by PRRs (Jenner and Young 2005; Lachmandas et al.2016; Li et al.2016; Martinez et al.2017). Elements of this antimicrobial programme are antimicrobial molecules, cytokines, chemokines and IFNs. Early production of type I IFN is required to limit initial viral replication. However, type I IFN-dependent responses can no longer be considered virus specific since a body of mainly recent data indicates that type I IFNs are also produced in response to bacteria. However, depending on the bacterial infection, type I IFNs exert seemingly opposing functions (Boxx and Cheng 2016; Kovarik et al.2016).
The most extensively studied mammalian (human and mouse) PRRs belong to the ‘Toll-like’ receptors (TLRs), the nucleotide-binding and oligomerisation domain-like receptors (NLRs) and the retinoic acid inducible gene I (RIG-I)-like receptor (RLR) families (Takeuchi and Akira 2010). Activation of all these PRRs converges on the activation of mitogen-activated protein kinases (MAPKs), and a limited set of transcriptional factors, mainly IRF3, IRF7 and NF-κB. These factors act cooperatively to activate the transcription of genes. Several members of the NLR protein family, NLRP1, NLRP3, NLRC4, may assemble into a multiprotein platform, known as inflammasome, to induce caspase-1 activation (Latz, Xiao and Stutz 2013; Guo, Callaway and Ting 2015). This protease is responsible for the cytosolic processing of pro-IL-1β and pro-IL-18 and for the secretion of their mature active forms. IL-1β and IL-18 exert crucial roles orchestrating immune responses to control infections. Activation of caspase-1 also triggers a form of cell death called ‘pyroptosis’. The role of pyroptosis as a bona fide cell-autonomous defence mechanism is still poorly understood, although recent evidence indicates that pyroptosis triggers pore-induced intracellular traps that capture bacteria and lead to their clearance by efferocytosis (Miao et al.2010; Jorgensen et al.2016). Other inflammatory caspases, caspase-11 in mouse and caspases4/5 in humans, detect cytosolic lipopolysaccharide (LPS) and trigger the activation of the so-called non-canonical inflammasome to produce IL1β and induce cell death (Hagar et al.2013; Shi et al.2014).
Several PRRs detect RNA (Schlee and Hartmann 2016). TLR3 and TLR7 detect double-stranded RNA in the endosome, whereas TLR7 and TLR8 sense single-stranded RNA. The helicases RIG-I and melanoma differentiation associated gene 5 (MDA5) also detect double-stranded RNA in the cytosol. Stimulation of these receptors results in the production of type I IFN, as well as the expression of IFN-stimulated genes. There is still limited knowledge on the possible contribution of these RNA-sensing receptors to bacterial defence.
Only in the past years, the molecular basis of cytosolic DNA sensing by the innate immune system has begun to be revealed (Paludan and Bowie 2013). Several DNA sensors were identified, and a new family of DNA sensors called AIM2-like receptors formed by, at least, IFI16, AIM2 and p202 (a negative regulator of AIM2), which are all PYHIN proteins, has been proposed. STING is the central adaptor for cytosolic DNA sensing directing TBK1 to activate IRF3 for DNA sensing pathways. The role of STING during bacterial diseases is controversial, ranging from protective to detrimental effects for the host (Marinho et al.2017).
MODELS TO ASSESS KLEBSIELLA SPECIES INFECTION BIOLOGY
Several research models have been implemented to assess Klebsiella infection biology (Table 1). However, the vast majority of evidence on the interplay between Klebsiella species and the immune system has been obtained by infecting rodents, chiefly mice. Inbred mouse strains allow the study of genetically identical cohorts, whereas the development of methods for the creation of transgenic, knock-out and knock-in mice has provided powerful tools to investigate Klebsiella infection biology. Outcomes of infection vary with the mouse strain used, infection with 102 CFUs leads to bacteremia and death within 72 h in CD-I, CBA/J and BALB/c mice while C57BL/6 mice are more resistant (Mehrad and Standiford 1999). The mouse model has been extensively used to investigate two clinical manifestations of Klebsiella infections: pneumonia and sepsis. Studies of Klebsiella-induced pneumonia in animal models date back to 1947 with induction of pneumonia in rats with intratracheal instillation of K. pneumoniae (Sale, Smith and Wood 1947; Sale and Wood 1947). The mouse pneumonia model recapitulates key features of Klebsiella-induced pneumonia in humans, namely the massive inflammation characterised by an influx of polymorphonuclear neutrophils, and oedema. The results obtained with this model have uncovered receptors and molecules implicated in host defence against the pathogen. Moreover, the mouse model has been useful to provide mechanistic evidence on why some health factors such as alcohol abuse, obesity, poor glycaemic control in diabetic patients and respiratory viral infection are associated with increased susceptibility to Klebsiella infections (Mancuso et al.2002; Happel et al.2006; Ballinger and Standiford 2010).
Table 1.
Models to assess K. pneumoniae infection biology.
| Infection model | Useful to assess | Advantages | Limitations |
|---|---|---|---|
| Mus musculus | Recapitulates many clinical aspects of Klebsiella infections. | Well-established model. Available knock-out and knock-in animals. | Costs. Differences between mice and human immune system. |
| Dyctiostelium discoideium | Recapitulates interactions with phagocytic cells. | Easy to handle. Available genetic tools, and bank of mutant strains. | Growth conditions (temperature and growth medium to assess virulence). |
| Drosophila melanogaster | Early interactions with ancient antibacterial mechanisms. | Easy to handle. Available genetic tools, and bank of mutant strains. | Growth conditions. Not clear how to translate findings in this model to model human disease. |
| Caenorhabditis elegans | Early interactions between a pathogen and a host. | Easy to handle. Available genetic tools, and bank of mutant strains. | Growth conditions. Not clear how to translate findings in this model to model human disease. |
| G. mellonella | Recapitulates interactions with innate immune system (effectors and phagocytes). | Easy to handle. Growth conditions (temperature). Good correlation with the mouse model in terms of assessing virulence. | Lack of genetic tools, and bank of mutant strains. |
| Danio rerio | Interactions with a complex immune system | Possibility of easily imaging infection. Available genetic tools, and bank of mutant strains. | Need for costly infrastructure. Not clear how to translate findings in this model to model human disease. |
In the mouse model, Klebsiella-induced pneumonia is achieved either via intratracheal/endobronchial instillation or via intranasal inoculation, each of which has its limitations. Intratracheal/endobronchial instillation delivers the inoculum to the lower respiratory track bypassing the host barriers of the upper airways but modelling oropharyngeal aspiration. The intratracheal method of infection often results in a higher ratio of infecting organisms to local defences at the site of infection than those achieved with other inoculation routes. This results in an exuberant inflammatory response and tissue destruction already few hours after infection. We suggest that the intratracheal/endobronchial inoculation route should be the one choice to investigate the biology of Klebsiella-induced lung injury. However, to gain insights into Klebsiella-triggered respiratory infections, we favour the intranasal inoculation route because it captures the interaction between the pathogen and the defences of upper and lower respiratory track. Furthermore, it is a simple method of infection. Its major limitation is the variable deposition of microorganisms in the lungs which may lead to significant differences between infected mice.
Intratracheal, intraperitoneal and intravenous routes of infection are used to model Klebsiella-triggered sepsis, whereas intraperitoneal and orogastric routes of infection are used to investigate the virulence of Klebsiella strains inducing pyogenic liver abscess (Siu et al.2012). This syndrome was anecdotally reported in Taiwan in the 1980s, although now it seems to be spreading to countries outside Asia. Clinical evidence suggests that healthy adults carry the virulent strains in their intestines, and liver abscess occur when bacteria translocate across the intestinal epithelium (Siu et al.2012). Experiments done in mice provide initial evidence supporting this notion (Tu et al.2009); however, it should be noted that this infection model only demonstrates the ability of these strains to spread from the gut to other organs. Of note, there are no specific Klebsiella genetic features associated with these infections, suggesting that perhaps host factors might play a critical role in the outcome of this Klebsiella-triggered pyogenic liver abscess. Currently, there is no well-developed model to investigate the gut colonisation by Klebsiella species. Recently, Krapp and colleagues (Krapp et al.2017) have developed a subcutaneous model of infection to model Klebsiella-induced skin and soft tissue infections. These are rare clinical manifestations also associated with hypervirulent strains.
Although the mouse model has proven useful to illuminate K. pneumoniae infection biology, it is important to acknowledge its limitations. There exist significant differences between mice and humans in immune system development, activation and response to infection (Mizgerd and Skerrett 2008). For example, circulating neutrophil counts are lower in mice than in humans (Haley 2003), and mouse neutrophils lack defensins (Eisenhauer and Lehrer 1992). There are no murine homologs of several chemokines including IL8, although mice express other chemoattractant cytokines (Strieter et al.1996). Species differences also exist in the receptors sensing infections. There are 10 known functional TLRs in humans and 12 in mice (Takeuchi and Akira 2010); TLRs 1–9 are conserved in both species, although the tissue expression and transcriptional regulation also differ (Rehli 2002). There is general conservation between mouse and human TLR-controlled signalling pathways; however, there are notable differences in the use of signalling adaptors (Sun et al.2016). In addition, the ligand specificities and affinities of TLRs also differ in humans and mice. For example, human TLR4 exquisitely discriminates between lipid A structures, whereas murine TLR4 does not and, as result, there are differences in the inflammatory responses induced by lipid As with different structures (Hajjar et al.2002; Montminy et al.2006). These discrepancies and others (Mizgerd and Skerrett 2008) should be carefully considered in interpreting experiments to translate the findings to human disease.
More recently, other non-mammalian infection models have been tested to investigate Klebsiella pathogenesis to circumvent ethical and costs issues associated with animal research, and to potentially facilitate large-scale analysis of virulence. The social amoeba Dictyostelium discoideum is a phagocytic cell that can be used to screen potential roles of phagocytic immune cells such as neutrophils and macrophages (Dunn et al.2018). There is evidence demonstrating that D. discoideum is a valuable system for studying how pathogens evade fundamental processes of phagocytic cells (Dunn et al.2018). Benghezal and colleagues (Benghezal et al.2006) carried out a two-dimensional virulence array to identify D. discoideum genes implicated in host defence against Klebsiella, and Klebsiella genes require to survive in the attenuated D. discoideum host. phg1 and kil1, the D. discoideum genes identified as essential to kill intracellular Klebsiella, have homologues in mammalian cells, although their potential contribution to the physiology of phagocytic cells has not been studied yet. The screen of bacterial mutants revealed that the surface polysaccharides expressed by Klebsiella, the capsule polysaccharide (CPS) and the LPS play a crucial role in the interaction with D. discoideum (Benghezal et al.2006). Additional studies have shown that, in addition to the CPS and the LPS O-polysaccharide and core, the outer membrane proteins (OMP) OmpA and OmpK36, and the lipid A decorations with aminoarabinose and palmitate are also necessary to avoid predation by D. discoideum (March et al.2013). Interestingly, these factors are also required to limit phagocytosis by mouse alveolar macrophages (March et al.2013), suggesting that K. pneumoniae exploits the same factors to interact with social amoeba and macrophages. Dictyostelium discoideum has also proven to be useful to model the interaction between Klebsiella and human neutrophils (Pan et al.2011), where CPS and LPS O-polysaccharide being also the important factors governing the interaction of Klebsiella with neutrophils. Further reinforcing the importance of Klebsiella surface polysaccharide on the interaction with phagocytes, D. discoideum specifically senses Klebsiella CPS to activate a predation programme (Lima et al.2014).
The nematode Caenorhabditis elegans and Drosopila melanogaster have also been used to identify host pathways implicated in host defence against Klebsiella. In C. elegans, PI3K-AKT/mTOR and the MAPK p38 are required for host protection against K. pneumonia (Kamaladevi and Balamurugan 2015, 2017), whereas Phg1, important in the Klebsiella-D. discoideum interplay, is also essential to resist K. pneumoniae infection by D. melanogaster (Benghezal et al.2006). Whether these pathways play any role in mammalian defence against K. pneumoniae is currently unknown.
A common limitation of these models is that the optimal temperature for maintaining them is 28°C, whereas the optimum temperature for K. pneumoniae is 37°C. Since virulence gene expression is frequently regulated by temperature, it is likely that temperature requirements may affect the interaction of K. pneumonaie strains causing human infections with these hosts. Nonetheless, it is important to note that the impact of environmental cues on the regulation of Klebsiella virulence factors is poorly understood. The larvae of the wax moth Galleria mellonella is emerging as a suitable model to study the virulence of many human pathogens because, among other advantages, it grows at 37°C (Table 1) (Glavis-Bloom, Muhammed and Mylonakis 2012; Tsai, Loh and Proft 2016). G. mellonella defence against bacterial infections consists of cellular and humoral immunity (Glavis-Bloom, Muhammed and Mylonakis 2012; Browne, Heelan and Kavanagh 2013; Wojda 2017). The cellular response is mediated by phagocytic cells, termed hemocytes, found within the haemolymph. Hemocytes govern the clotting reaction to trap pathogens, and the melanisation response consisting of the deposition of melanin to encapsulate pathogens at the site of infection followed by the coagulation of hemolymph. Melanisation can be considered analogous to abscess formation in mammalian infections. The humoral response is orchestrated by soluble effector molecules that immobilise or kill the pathogen and includes complement-like proteins, and antimicrobial peptides. The suitability of G. mellonella as a model to investigate K. pneumoniae pathogenesis has only been recently demonstrated (Insua et al.2013). This infection model discriminates the pathogenic potential of K. pneumoniae strains (Insua et al.2013; Wand et al.2013), and there is a strong correlation with the virulence previously determined in the mouse pneumonia model (Insua et al.2013). Furthermore, K. pneumoniae infection of G. mellonella results in responses similar to those reported in the mouse pneumonia model including cell death-associated with bacterial replication, inhibition of phagocytosis and attenuation of host defence responses, chiefly the production of antimicrobial peptides (Insua et al.2013). Interestingly, virulence factors necessary in the mouse pneumonia model, CPS, LPS and OMPs, are also required for K. pneumoniae survival in G. mellonella (Insua et al.2013). The fact that all attenuated Klebsiella mutants activate G. mellonella defensive responses (Insua et al.2013) supports the notion that prevention of host responses is an important feature of K. pneumoniae pathogenesis. Despite the clear utility of G. mellonella as a surrogate host to assess infections with K. pneumoniae, it is important to note that the model only reflects early features of the interaction between the pathogen and ancient innate immune mechanisms of defence. These mechanisms are indeed conserved in evolution; however, the evolutionary distance between insects, mice and humans makes that many host-specific phenomena are likely to exist.
The larvae of Danio rerio (zebrafish) is another non-mammalian infection model receiving increasing attention because it is genetically tractable, optically accessible and present a fully functional innate immune system with macrophages and neutrophils that mimic their mammalian counterparts (Torraca and Mostowy 2018). Although a wide variety of pathogenic bacteria have been already investigated using zebrafish, only recently it has been assessed whether zebrafish larvae are a suitable model to investigate K. pneumoniae virulence (Cheepurupalli et al.2017; Marcoleta et al.2018). These studies report the optimisation of the model to investigate K. pneumoniae pathogenesis. Injection of larvae seems to be the most reliable inoculation method to ensure consistent colonisation of the gastrointestinal tract (Cheepurupalli et al.2017). Further studies are warranted to validate whether the model is useful to identify Klebsiella virulence factors and to uncover features of the interaction between K. pneumoniae and the immune system.
CONTRIBUTION OF HOST SIGNALLING IN DEFENCE AGAINST K. PNEUMONIAE INFECTIONS
The published evidence during more than 20 years clearly establishes that pro-inflammatory signalling is crucial to K. pneumoniae clearance (Fig. 1). One of the first conclusive piece of evidence showed that mice deficient in the TNFα receptors (TNFR1) are susceptible to K. pneumoniae pneumonia (Laichalk et al.1996, 1998; Moore et al.2005). Subsequent studies reported that mice lacking the chemokines CXCL15 (Chen et al.2001) and CCL3 (Lindell et al.2001), and unable to synthesise leukotrienes (Bailie et al.1996; Mancuso et al.1998) and nitric oxide (Tsai et al.1997) are also exquisitely susceptible to Klebsiella pneumonia. All these are markers associated with human pneumonia and are part of the common host response to infections (Jenner and Young 2005; Lachmandas et al.2016; Li et al.2016; Martinez et al.2017). In turn, strategies to boost pro-inflammatory signalling in the airways have proven to be successful to limit K. pneumoniae infections. Intrapulmonary expression of CCL3 (Zeng et al.2003), and KC (Tsai et al.1998; Cai et al.2010) leads to improved clearance of K. pneumoniae. Intratracheal instillation of CpG increases the production of TNFα, and Th1 cytokines, including IL-12, IFNγ and the IFNγ-dependent ELR-CXC chemokines (Deng et al.2004a) in a TLR9-dependent manner (Bhan et al.2007), enhancing bacterial clearance. Cyclic di-GMP, a molecule sensed by STING (Burdette et al.2011), also triggers a vigorous expression of chemokines and Th1 cytokines (Karaolis et al.2007). These treatments also result in the increased recruitment of neutrophils, αβ and γδ T cells, and activated NK cells to the site of infection, suggesting that these cells are crucial in host defence against K. pneumoniae. Indeed, γδ T cells and NK cells play a pivotal role in the resolution of Klebsiella infections by controlling early production of pro-inflammatory cytokines (Moore et al.2000; Xu et al.2014).
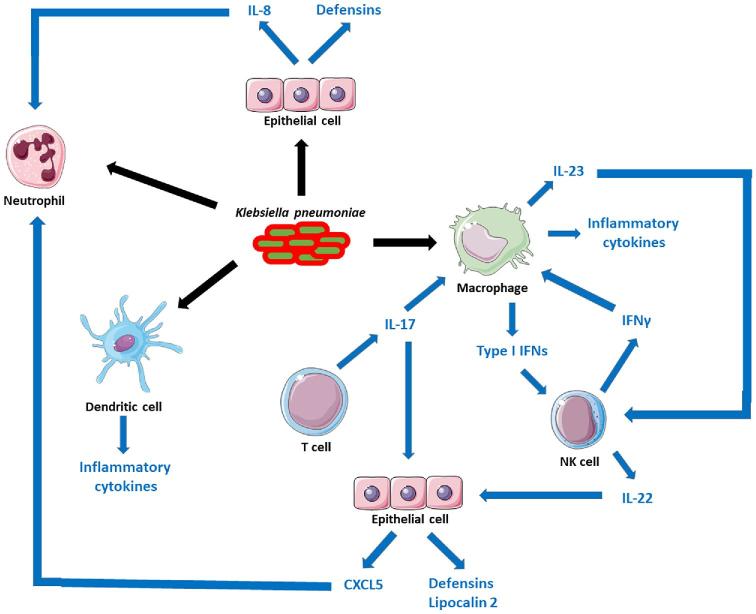
Figure 1.
Mechanisms of innate immunity to K. pneumoniae infections. The figure depicts the cells implicated in containing K. pneumoniae infection. There is conclusive evidence demonstrating the interaction of K. pneumoniae with neutrophils, macrophages (and monocytes [not shown]), dendritic cells and epithelial cells. These interactions are marked with black arrows. The interaction with different subset of T cells, NK cells and other lymphocytes has not been investigated yet, although these cells participate in bacterial clearance. The network of connections between cells, and the role played by different cytokines activating host responses are depicted with blue arrows.
The role of different cytokines in host defence against K. pneumoniae has been also investigated (Fig. 1). Early studies demonstrated the importance of IFNγ and IFNγ-dependent cytokines to control the progression of Klebsiella-induced pneumonia (Yoshida et al. 2001; Moore et al.2002; Zeng et al.2005a,b). IL23 drives the production of IFNγ and IL17 (Happel et al.2003, 2005); however, the fact that IL17 administration restores bacterial control in mice deficient on IL23 production indicates an independent role for the IL17-governed axis on host defence against K. pneumoniae (Happel et al.2005). Adding further evidence to this notion, IL17 signalling is critical for the induction of Th1 responses, neutrophil recruitment and local control of pulmonary infection (Ye et al.2001a,b). IL17 signalling is also augmented via IL12 production through IFNγ (Happel et al.2005). The contribution of IL12 signalling to control Klebsiella pneumonia is exemplified by the fact that STAT4−/− knock-out mice displayed impaired production of IFNγ and Th1 cytokines and greater bacterial burden compared to wild-type infected mice (Deng et al.2004b). STAT4 is a critical transcriptional factor in the IL12 signalling pathway (Bacon et al.1995). IL22, produced in an IL23-dependent manner, is also important in host defence against K. pneumoniae (Aujla et al.2008; Zheng et al.2016). Administration of an anti-IL22 blocking antibody results in higher bacterial loads in the lungs and dissemination of bacteria to spleen (Aujla et al.2008), whereas therapeutic administration of IL22 attenuates Klebsiella-triggered peritonitis (Zheng et al.2016). IL22 regulates the levels of IL6 and CCL3 upon Klebsiella infection, and its role is predominant over IL17 in regulating these cytokines (Aujla et al.2008). The synergistic effect of both cytokines governing host defences against Klebsiella is marked by the fact that neutralisation of IL22 in Il17a−/− mice results in greater bacterial growth in the lung and significantly more bacterial dissemination to the spleen than in those observed in infected Il17a−/− (Aujla et al.2008).
Collectively, the summarised evidence strongly suggests that the IL23/IL17 and IL12/IFNγ axes are essential for the generation of an effective innate immune response in the lungs against K. pneumoniae. However, two questions require additional investigations: Which are the cell(s) responsible for the production of these cytokines? and Which are the main innate defence mechanisms (humoral and cellular) activated by these cytokine networks responsible for the clearance of K. pneumoniae? To set the framework for future studies, we next discuss the available evidence. Initial data suggest that γδ T and NK cells could be the major source of IL17 and IL22, respectively, in Klebsiella-infected mice (Xu et al.2014; Murakami et al.2016), whereas alveolar macrophages could be the initial source of IL23 (Happel et al.2005). Only recently, it has been also suggested that type 3 innate lymphocytes could be another source of IL17 in vivo (Xiong et al.2016). Altogether, there is need to dissect the source of IL17 during K. pneumoniae infections. Dendritic cells have been shown to orchestrate the production of IL12, IL23 and IL17 in vivo (Bhan et al.2007), although the specific singular role of dendritic cells in Klebsiella infections and the connection with other immune cells have not yet been fully defined. There is evidence showing that NK cells are the source of IFNγ in Klebsiella-infected mice (Van Elssen et al.2010; Ivin et al.2017), although it cannot be ruled out that other cell types such as CD4 and CD8 T cells might contribute as well. Alveolar macrophages and recruited inflammatory monocytes are considered the main cellular target for IFNγ and IL17, respectively (Xiong et al.2015, 2016; Ivin et al.2017). These cytokines enhance the microbicidal activity of alveolar macrophages and inflammatory monocytes by increasing phagocytosis and facilitating bacterial killing (Xiong et al.2015, 2016; Ivin et al.2017). Whether IFNγ and IL17 trigger other antimicrobial activity on these cells remains to be investigated. IL17 and IL22 also aid in the clearance of Klebsiella by regulating the antimicrobial activity of the lung epithelium (Aujla et al.2008; Chen et al.2016). Both cytokines activate antimicrobial programmes in epithelial cells having in common the production of CXCL5 and lipocalin 2 (Aujla et al.2008; Chen et al.2016). Ablation of this programme results in higher bacterial loads in the lungs of infected animals. CXCL5 participates in the recruitment of neutrophils to the site of infection (Chen et al.2016), whereas lipocalin 2 inhibits the growth of some strains of Klebsiella in vitro and in vivo by preventing bacterial iron acquisition (Bachman, Miller and Weiser 2009; Chan et al.2009; Bachman et al.2012). Additionally, lipocalin 2 may also promote the induction of pro-inflammatory responses, which facilitates the recruitment of neutrophils like CXCL5 does. However, it is important to be aware that there are conflicting reports on the role of neutrophils in vivo to clear Klebsiella infections (Greenberger et al.1996; Broug-Holub et al.1997; Xiong et al.2015). Therefore, the recruitment of neutrophils to the lungs of Klebsiella-infected mice cannot be rigorously taken as conclusive evidence of these cells being a major player in host defence against the pathogen. In vivo mechanistic studies involving selective depletion of neutrophils together with ex vivo experiments testing isolated mouse neutrophils should be the norm in these type of studies.
Only recently, the role of type I IFN and type I IFN-governed signalling in host defence against Klebsiella infections has been investigated (Fig. 1). Alveolar macrophages are one of the sources of type I IFN in vivo, and in vitro experiments revealed that K. pneumoniae activates a TLR4-TRAM-TRIF-IRF3 signalling pathway to induce type I IFN- and type I IFN-dependent genes (Ivin et al.2017). To assess the importance of type I IFN signalling in host defence, researchers infected mice lacking type I IFN 1 receptor-deficient (Ifnar1−/−) mice (Ivin et al.2017). Ifnar1 is one of the subunits of the type I IFN receptor which mediates type I IFN responses in innate and acquired immunity to infection. Ifnar1−/− are exquisitely sensitive to Klebsiella infection exhibiting a markedly decreased survival, higher bacterial lung burden, increased dissemination to spleen and liver and severe bronchopneumonia (Ivin et al.2017). The lack of type I IFN signalling results in defect in the production of IFNγ, IL-12 and CXCL10 in K. pneumoniae-infected lungs, but it has no impact on the number of alveolar macrophages, inflammatory monocytes and neutrophils recruited to the site of infection (Ivin et al.2017). In contrast, the number of NK cells was lower in the lungs of Ifnar1−/−-infected mice than in the wild-type ones (Ivin et al.2017). This is consistent with the reduced levels of the NK chemoattractant chemokine CXCL10 in the lungs of Ifnar1−/−-infected mice (Ivin et al.2017). Type I IFN signalling is crucial for NK cells to produce IFNγ, which is required for enhancing the bactericidal action and the production of the NK cell response-amplifying IL-12 and CXCL10 by alveolar macrophages (Ivin et al.2017). Remarkably, type I IFN signalling is dispensable in myeloid cells including alveolar macrophages, monocytes and neutrophils for host defence and IFNγ activation (Ivin et al.2017), uncovering a hitherto unknown crosstalk between alveolar macrophages and NK cells based on type I IFN and IFNγ in Klebsiella infection.
Few studies have addressed the contribution of PRR-governed signalling to control Klebsiella infections. As with other bacterial infections, TLR4 signalling plays a prominent role in antibacterial defence against Klebsiella infection (Branger et al.2004; Wieland et al.2011; Standiford et al.2012). TLR4−/− mice show reduced survival upon infection with increased bacterial loads in lungs and bronchopneumonia (Branger et al.2004; Wieland et al.2011; Standiford et al.2012). The lack of TLR4 signalling is associated with a decrease in the levels of IL17 and IL23 in the lungs of infected TLR4−/− mice (Happel et al.2003), which may explain the susceptibility of these mice to Klebsiella infection. It remains an open question which cells are more affected by the lack of TLR4 signalling. Initial observations suggest that TLR4 signalling is indispensable in cells of myeloid origin for the clearance of Klebsiella (Wieland et al.2011); however, it cannot be ruled out that other cell types may require TLR4 signalling to aid in the elimination of Klebsiella infections. Supporting this notion, TLR4 signalling is required to protect the lung epithelium from Klebsiella-induced pathophysiology (Standiford et al.2012). TLR9-controlled signalling is also required for protective immunity against Klebsiella-induced pneumonia (Bhan et al.2007, 2010). Mice deficient in TLR9 fail to generate an effective Th1 cytokine response, resulting in increased bacterial loads in the lungs and dissemination to other organs (Bhan et al.2007). TLR9−/−-infected mice present no major defects on the accumulation of immune cells except on the influx and maturation of conventional dendritic cells (Bhan et al.2007). This reduced accumulation and activation of dendritic cells explains the impaired bacterial clearance because adaptive transfer of dendritic cells from wild-type mice reconstitutes the protective immunity in TLR9−/− mice (Bhan et al.2007). Since TLR9 is located in endosomes and it recognises DNA oligonucleotides with unmethylated CpG base pairs (Hemmi et al.2000), it is intriguing to consider how Klebsiella infection may lead to the activation of this intracellular receptor. Data obtained in other infection models indicate that TLR9 can be activated by bacterial and host DNA released into the airways during pneumonia (van der Meer et al.2016), as well as by intracellular bacteria and DNA of mitochondrial origin released to the cytosol upon infection (Zhang et al.2010; Arpaia et al.2011). Future studies should address this knowledge gap. TLR2 signalling has a dual role in host defence against Klebsiella (Wieland et al.2011). In the early phase of infection, TLR2 signalling has an anti-inflammatory role (Wieland et al.2011), perhaps to prevent a detrimental overwhelming inflammation as a result of the activation of other PRRs. Similar observation has been done in Acinetobacter baumannii-triggered pneumonia (Knapp et al.2006), suggesting that the dampening function of TLR2 during pneumonia is not bacterial species-specific. In the later stage of infection, TLR2 contributes to antibacterial defence (Wieland et al.2011). Interestingly, cooperative roles of TLR4 and TLR2 signalling are involved in controlling Klebsiella infection because TLR4−/−xTLR2−/− mice are more susceptible to the infection than each of the single knock-out mice (Wieland et al.2011).
The role of TLR signalling during K. pneumoniae infection has been further probed by demonstrating the contribution of TLR adaptors in host defence. MyD88 is the universal adaptor for all TLRs except TLR3 (O’Neill and Bowie et al. 2007), and it has been shown to be important for pulmonary host defence against several respiratory pathogens (Baral et al.2014). TRIF is the sole adaptor for TLR3 and also contributes to TLR4 signalling (O’Neill and Bowie et al.2007). Infections of MyD88−/− and TRIF−/− mice demonstrated that both adaptors are required to restrict K. pneumoniae growth in the lungs (Cai et al.2009; van Lieshout et al.2012). MyD88-dependent protection during Klebsiella pneumonia is mediated by both hematopoietic and resident cells excluding endothelial cells, whereas TRIF-mediated protection is driven by hematopoietic cells (van Lieshout et al.2012, 2014; Anas et al.2017). MyD88 and TRIF deficiencies limit the production of Th1 cytokines and the activation of signalling pathways controlling host defence mechanisms (Cai et al.2009). Interestingly, the characteristic bronchopneumonia of Klebsiella infections is virtually absent in infected MyD88−/− mice and significantly reduced in TRIF−/− mice despite high bacterial loads in both mice (Cai et al.2009). This evidence indicates that the histopathological changes associated with Klebsiella infection are dependent on the host inflammatory response to the infection. TIRAP/MAL is another adaptor linking MyD88 to the activated TLR2 and TLR4 receptors (O’Neill and Bowie et al.2007). In this context, it is not surprising that MAL−/− mice have substantial mortality, higher bacterial burden in the lungs, enhanced bacterial dissemination, attenuated production of Th1 cytokines and no lung histopathology following K. pneumoniae infection (Jeyaseelan et al.2006). At present, the mechanisms underlying MyD88-MAL-mediated defence against Klebsiella are ill-defined. However, it is important to note here that MyD88-MAL are also required for the activation of other signalling pathways such as those governed by IL1β and IFNγ (Cohen 2014; Ni Cheallaigh et al.2016). Therefore, MyD88-MAL-dependent protective immunity is most likely also mediated by IL1β- and IFNγ-governed host antibacterial responses. Likewise IFNγ-deficient mice, IL1R−/− mice are exquisitely susceptible to Klebsiella infection demonstrating the importance of IL1β-controlled responses for host survival and bacterial clearance (Cai et al.2012). On the other hand, the impaired antibacterial defence of TRIF−/-mice is associated with the lack of IFNγ in the lungs of infected mice (van Lieshout et al.2015). The fact that TRIF is required for type I IFN production following Klebsiella infection (Ivin et al.2017) suggests that the impairment of IFNγ production in TRIF−/–mice is secondary to the deficient type I IFN production in these mice.
A small number of studies have investigated the contribution of NLR signalling to defence against Klebsiella. NLRP3−/− mice demonstrate an increase in mortality following Klebsiella infection albeit the protective role of NLRP3 is not as important as those of TLR4 and TLR2, and any of the TLR adaptors (Willingham et al.2009). In vitro experiments confirmed the contribution of NLRP3 to caspase-1 activation and IL1β release following Klebsiella infection (Willingham et al.2009). In good agreement, Klebsiella-induced IL1β is reduced in NLRP3−/− mice (Willingham et al.2009). Recent evidence supports that the CPS and the LPS are the Klebsiella components responsible for priming NLRP3, whereas Klebsiella-triggered ROS may be responsible for the activation (Hua et al.2015). The NLRC4 inflammasome also contributes to Klebsiella-triggered IL1β production in vitro and in vivo (Cai et al.2012). However, there are contradictory results on the importance of NLRC4 to confer protection against Klebsiella infection (Willingham et al.2009; Cai et al.2012). The main apparent difference between studies is the different bacterial inoculum, with the study using an inoculum closer to the LD50 showing a contribution of NLRC4 on host immunity against Klebsiella (Cai et al.2012). Nonetheless, an open question is the identification of the Klebsiella component(s) inducing the activation of NLRC4. This receptor senses bacterial flagellin and the type III secretion system apparatus (Zhao et al.2011; Duncan and Canna 2018). Notably, Klebsiella is not flagellated and in silico analysis of more than 700 genomes confirms that this pathogen does not encode any type III secretion system. It is then tempting to speculate that any of the secretion systems encoded by Klebsiella, including the type II and type VI secretion systems, might be sensed by NLRC4. Intriguingly, Klebsiella-induced pyroptosis requires NLRP3 but not NLRC4 (Willingham et al.2009; Cai et al.2012). Whether pyroptosis is one of the bona fide host defence mechanisms against Klebsiella infection is yet unknown. In fact, the specific importance of pyroptosis in host immunity remains a challenging question. Initial evidence shows that NLRC4-dependent pyroptosis mediates the clearance of the intracellular pathogen Salmonella typhimurium by generating a structure that entraps the previously intracellular bacteria and drives their elimination by containing the bacteria and elaborating signals that promote efferocytosis (Miao et al.2010; Jorgensen et al.2016). However, it is unlikely that this mechanism operates in the context of Klebsiella infections because, as discussed before, the lack of NLRC4 does not affect Klebsiella-triggered pyroptosis.
MICROBIOME PROTECTION AGAINST KLEBSIELLA INFECTIONS
There is a wealth of evidence demonstrating the role of the intestinal microbiota to prevent infection by pathogenic bacteria. This is achieved by interactions within the microbial community and by shaping the tissue immune responses to limit infection (McKenney and Pamer 2015). Surprisingly, there is virtually no data on the impact of the gut microbiome on Klebsiella gut colonisation and/or orogastric infection. This knowledge gap is particularly relevant considering the recent clinical evidence demonstrating that gastrointestinal carriage is a major reservoir of K. pneumoniae infections in the healthcare environment (Martin et al.2016; Gorrie et al.2017). On the other hand, the gut microbiota has been shown to protect against Klebsiella pneumonia. Experiments infecting germfree mice revealed that these animals are susceptible to Klebsiella in an IL-10–dependent manner (Fagundes et al.2012). In germfree mice, IL-10 in the lungs restrains pro-inflammatory mediator production and favours Klebsiella growth and dissemination (Fagundes et al.2012). Neutralisation of IL10, or transient TLR4 activation with LPS, restores germfree mice resistance to K. pneumoniae infection (Fagundes et al.2012). Subsequent studies provided compelling evidence on the members of the gut microbiota that drive protection against Klebsiella infection and the signalling pathways responsible for this microbiota-controlled immune protection. A consortium of bacterial species common to the rodent and human intestinal microbiota formed by Lactobacillus reuteri, Enterococcus faecalis, Lactobacillus crispatus and Clostridium orbiscindens induces potent NOD2 activation to trigger IL17-GM-CSF in the lung which, in turn, stimulates pathogen killing and clearance by alveolar macrophages through MAPK extracellular signal-regulated kinase signalling (Clarke 2014; Brown, Sequeira and Clarke 2017). The source of IL17 in the lung and how the microbiota governs the production of this cytokine remain open questions. Nevertheless, these findings further highlight the critical role of IL17 in host defence against Klebsiella, and the crucial role played by alveolar macrophages promoting antibacterial defence in the lung. Although the contribution of the upper airway microbiota, either permanent resident or aspirated from the oropharynx, to limit respiratory infections has not been demonstrated yet, it is notable that intranasal inoculation of bacteria colonising the upper airway of humans and mice (Lactobacillus crispatus, Staphylococcus aureus, S. epidermidis) enhances lung immunity against Klebsiella by the same IL17-GM-CSF-alveolar macrophage axis (Brown, Sequeira and Clarke 2017). Collectively, this evidence demonstrates the facility of the lung immune system to integrate microbial signals from different mucosal sites to launch antibacterial defence mechanisms.
KLEBSIELLA EVASION STRATEGIES OF HOST DEFENCES
The widely held belief is that K. pneumoniae is a stealth pathogen, which fails to stimulate innate immune responses (Paczosa and Mecsas 2016). Essentially, Klebsiella shields its pathogen-associated molecular patterns from detection by PRRs and soluble effectors of the immune system, and avoids the interaction with hematopoietic and non-hematopoietic cells to prevent the activation of host antimicrobial responses (Table 2). However, there is now enough evidence demonstrating that Klebsiella also actively subverts host defences (Table 2). We will review both immune evasion strategies in the context of the interaction of Klebsiella with different effectors of the immune system.
Table 2.
Immune evasion strategies of K. pneumoniae.
| Immune evasion strategies | Mechanism | Bacterial factor | References |
|---|---|---|---|
| (i) Stealth pathogen | |||
| Preventing the antimicrobial action of soluble innate immune effectors | |||
| Preventing complement bactericidal effect, and opsonisation | Limiting C3b deposition | CPS, LPS O-polysaccharide | Merino et al.1992; Alvarez et al.2000; de Astorza et al.2004 |
| Limiting antimicrobial activity of collectins | Blunting interaction with SP-A and SP-D | CPS | Kabha et al.1997; Ofek et al.2001; Kostina et al.2005 |
| Counteracting bactericidal action CAMPs and polymyxins | Limiting the interaction with the bacterial surface. Efllux of CAMPs. | CPS, LPS lipid A decorations, AcrAB | Campos et al.2004; Llobet et al.2011; Kidd et al.2017; Mills et al.2017; Padilla et al.2010 |
| Attenuating the interaction with immune cells | |||
| Attenuating engulfment by epithelial cells | CPS | Cortes et al.2002; Regueiro et al.2006 | |
| Avoiding phagocytosis by neutrophils | CPS, OmpK36 | Regueiro et al.2006; Pan et al.2011 | |
| Avoiding phagocytosis by macrophages | CPS, LPS lipid A decorations, OmpA, OmpK36 | March et al.2013 | |
| Limiting the activation of PPRs | Limiting the recognition of LPS by TLR4 | LPS lipid A 2-hydroxylation | Llobet et al.2015 |
| (ii) Subversion host defences | |||
| Attenuating cell-intrinsic immunity | |||
| Controlling maturation dendritic cells | CPS, LPS O-polysaccharide | Evrard et al.2010 | |
| Manipulation phagosome maturation | Activation PI3K-AKT-Rab14 axis | Unknown | Cano et al.2015 |
| Controlling cell death | Cytotoxicity in epithelial cells. Triggering apoptosis in macrophages. | CPS Unknown | Cano et al.2009; Leone et al.2016 |
| Abrogating TLR-controlled inflammatory responses: | |||
| Abolishing TLR signalling | CPS, LPS O-polysaccharide, OmpA, T2SS | March et al.2011; Frank et al.2013; Tomas et al.2015 | |
| Blunting NF-κB signalling | Upregulation deubiquitinase CYLD by targeting NOD1 and EGFR. | CPS, and other unknown factor(s) | Regueiro et al.2011; Frank et al.2013 |
| Blunting MAPKs | Upregulation MAPKs phosphatase MKP-1 via NOD1 activation. | Unknown | Regueiro et al.2011 |
| Manipulating mucosal immunity | Induction of IL10. | Unknown | Greenberger et al.1995; Yoshida et al.2001 |
| Counteracting nutritional immunity | Secretion of several siderophores | Yersiniabactin, salmochelin, aerobactin | Lawlor, O’connor and Miller 2007; Bachman et al.2011; Russo et al.2011 |
Counteracting soluble effectors of the immune system
Early research focused on investigating the interplay between complement and K. pneumoniae. The OMPs and LPS of K. pneumoniae are known to activate the classical pathway (Alberti et al.1996a,b). OmpK36 and OmpK35, homologues to OmpF and OmpC, respectively, and two of the most abundant porins in the outer membrane of K. pneumoniae, bind Cq1 in an antibody-independent manner triggering complement activation (Alberti et al.1993, 1996a). K. pneumoniae LPS without O-polysaccharide also activates the classical pathway, although less efficiently than the OMPs (Alberti et al.1996b). C3b deposition on the bacterial surface upon complement activation results in increased internalisation of Klebsiella by human lung epithelial cells promoting bacterial clearance (de Astorza et al.2004), as well as opsonophagocytosis by neutrophils and macrophages (Domenico et al.1994; Salo et al.1995; Regueiro et al.2006). Not surprisingly, the main complement evasion strategy of Klebsiella is based on preventing C3b deposition by exploiting Klebsiella surface polysaccharides. Whether K. pneumoniae may exploit other complement evasion strategies, such as targeting factor H, has not been described yet. The CPS is the main factor protecting Klebsiella from complement; cps mutants are susceptible to the bactericidal action of complement and show increased deposition of C3b on the surface (Merino et al.1992; Alvarez et al.2000). The protection conferred by CPS is more dependent on the thickness of the polysaccharide than the chemical composition of the polysaccharide (de Astorza et al.2004), although CPS containing manno(rhamno)biose may activate the lectin complement pathway (Sahly, Keisari and Ofek 2009). The LPS O-polysaccharide also protects Klebsiella from complement by limiting the deposition of C3b on the bacterial surface (Merino et al.1992). In those strains lacking the LPS O-polysaccharide, the CPS is then the main factor protecting Klebsiella from the bactericidal action of complement (Alvarez et al.2000).
The CPS also confers protection against lung collectins SP-A and SP-D, components of the lung surfactant, by preventing the binding of the collectins to the LPS (Kabha et al.1997; Ofek et al.2001; Kostina et al.2005). The binding of both collectins to the bacterial surface triggers bacterial agglutination and facilitates phagocytosis by macrophages (Kabha et al.1997; Ofek et al.2001; McCormack and Whitsett 2002). Interestingly, pulmonary surfactant challenge shapes the transcriptome of K. pneumoniae, inducing a programme strongly associated with virulence in the pneumonia mouse model (Willsey et al.2018). The CPS is one of the systems induced by pulmonary surfactant, further emphasising the importance of this polysaccharide to protect Klebsiella against collectins.
Like many other bacterial pathogens, K. pneumoniae has developed strategies to counteract host cationic antimicrobial peptides (CAMPs), chiefly defensins. Importantly, CAMPs and antibiotics such as quinolones and polymyxins share the same initial target in the outer membrane of Gram-negative bacteria (Nikaido 2003). Therefore, there is a relationship between resistance to CAMPs and polymyxins (Campos et al.2004; Campos, Morey and Bengoechea 2006; Nizet 2006; Llobet et al.2011; Kidd et al.2017). To counteract the bactericidal action of CAMPs and polymyxins, K. pneumoniae exploits the versatility of the CPS and the LPS. CPS limits the interaction of CAMPs and polymyxins with Klebsiella surface, and, in fact, there is a correlation between the amount of CPS expressed by a given strain and the resistance to polymyxin B (Campos et al.2004). Furthermore, free CPS, which may be released from the bacterial surface by CAMPs and polymyxins, binds CAMPs, neutralising their bactericidal effect (Llobet, Tomas and Bengoechea 2008). Therefore, the CPS acts as a bacterial decoy for CAMPs. Notably, this trait is shared by anionic CPS expressed by Pseudomonas aeruginosa and Streptococcus pneumoniae (Llobet, Tomas and Bengoechea 2008), strongly suggesting that trapping CAMPs is a general feature of anionic CPS.
K. pneumoniae also remodels its LPS lipid A domain to counteract CAMPs and polymyxins (Llobet et al.2011, 2015; Kidd et al.2017; Mills et al.2017). Klebsiella pneumoniae lipid A can be decorated with palmitate, 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and 2-hydroxymyristate (Llobet et al.2011, 2015; Kidd et al.2017). These decorations provide resistance to CAMPs, and K. pneumoniae mutants lacking these lipid A decorations are attenuated for virulence in the mouse pneumonia model (Llobet et al.2011, 2015; Kidd et al.2017). There are reports showing that the lipid A acylation also mediates resistance to CAMPs (Clements et al.2007; Mills et al.2017). However, this role could be indirect since mutants deficient in the late acyltransferases lpxM and lpxL display changes in the CPS levels and the 2-hydroxylation of the lipid A, respectively (Clements et al.2007; Mills et al.2017).
The lipid A of K. pneumoniae shows a remarkable plasticity. Just a brief incubation with CAMPs upregulates the expression of the loci required to modify the lipid A with a concomitant increase in the lipid A species containing such modifications (Llobet et al.2011). The regulatory network controlling these transcriptional changes is complex and involves, at least, the PhoPQ, PmrAB and the Rcs systems (Llobet et al.2011). Of note, and in contrast to Salmonella, PhoPQ and PmrAB control independently the regulatory architecture governing these CAMPs-induced changes (Llobet et al.2011). Intriguingly, OmpA is also part of the regulatory network controlling systems required to ameliorate CAMPs bactericidal action (Llobet et al.2009). Whether there is any connection between the OmpA and the PhoPQ and PmrAB-regulated networks is currently unknown.
Counteracting immune cells
Although it is well appreciated that the airway epithelium plays a central role orchestrating pulmonary inflammatory and immune responses against infections (Whitsett and Alenghat 2015), few studies detail the interaction of Klebsiella with these cells. Initial research argued that Klebsiella entry into epithelial cells may protect the pathogen from the actions of antibiotics and the immune system (Sahly et al.2000). These studies clearly demonstrated the role of the CPS limiting the attachment and internalisation to epithelial cells. However, subsequent studies have shown that epithelial cells engulfment of Klebsiella is most likely a host defence mechanism (Cortes et al.2002; Cano et al.2009). This interpretation is consistent with the observations that bacterial internalisation triggers an inflammatory response due to the activation of NF-κB signalling (Regueiro et al.2006), and that K. pneumoniae triggers a cytotoxic effect on epithelial cells (Cano et al.2009; Leone et al.2016). Klebsiella-induced cytotoxicity requires the presence of live bacteria and of CPS since it is observed with isolates expressing different amounts of CPS and/or different serotypes but not with non-capsulated bacteria (Cano et al.2009). K. pneumoniae infection also increases the levels of TLR4 and TLR2 in human airway epithelial cells which results in enhanced inflammatory response upon stimulation with TLR agonists (Regueiro et al.2009). TLR upregulation upon infection is dependent on the activation of NF-κB-governed signalling pathway by the CPS (Regueiro et al.2009). Evidence demonstrates that Klebsiella CPS is sensed by TLR4 (Regueiro et al.2009; Yang et al.2011; Frank et al.2013).
There is scarce data on the interaction between Klebsiella and neutrophils, although the recruitment of neutrophils to the lung is one of the hallmarks of Klebsiella-triggered pneumonia. In vitro experiments indicate that neutrophil-dependent clearance of Klebsiella occurs after phagocytosis. In turn, CPS abrogates killing by neutrophils due to its anti-phagocytic activity (Regueiro et al.2006; Pan et al.2011). Not surprisingly, antibodies against the CPS empower neutrophil-mediated killing (Diago-Navarro et al.2018), showing promise as new therapeutics to treat Klebsiella infections. Interestingly, K. pneumoniae does not induce NETosis (Branzk et al.2014), although anti-CPS antibodies enhance the release of neutrophil extracellular traps (NETs), which may contribute to extracellular killing of Klebsiella (Diago-Navarro et al.2017, 2018). NETs are large, extracellular, web-like structures composed of decondensed chromatin and neutrophil antimicrobial factors. NETs trap and kill a variety of microbes (Amulic et al.2012). NETs are released primarily via a cell death program that requires ROS, and the granule proteins myeloperoxidase and neutrophil elastase (Amulic et al.2012). Recently, it has been shown that K. pneumoniae interferes with clearance of neutrophils by efferocytosis (Jondle et al.2018). Efferocytosis is a regulated process which facilitates the elimination of apoptotic neutrophils by phagocytic cells, mainly macrophages, which prevents heightened inflammation triggered by dead cells (Ariel and Serhan 2012; Poon et al.2014; Angsana et al.2016). Mechanistically, Klebsiella infection of neutrophils results in a drastic decrease in the exposure of phosphatidylserine, which is recognised as ‘eat-me’ signal by macrophages to initiate the engulfment of neutrophils (Jondle et al.2018). The Klebsiella factor(s) responsible for this phenotype is currently unknown. Interestingly, evidence suggests that Klebsiella not only limits the efferocytosis of neutrophils, but it may also programme their cell death towards necroptosis (Jondle et al.2018). In contrast to apoptosis, necroptosis is a cell death triggering inflammation (Pasparakis and Vandenabeele 2015). Therefore, Klebsiella-triggered necroptosis may also contribute to the exuberant inflammation observed in this infection. It is then tempting to speculate that inhibition of necroptosis may improve Klebsiella-induced immunopathology. Preliminary observations indicate that this might be the case (Jondle et al.2018).
The specific role of dendritic cells in the clearance of K. pneumoniae has been poorly characterised, although there are data indicating that Klebsiella may activate different subsets of dendritic cells (Hackstein et al.2013). An outer membrane fraction of K. pneumoniae, containing CPS, LPS and porins, induces dendritic cell maturation (Van Elssen et al.2010). However, K. pneumoniae CPS and LPS attenuate dendritic maturation by hampering bacterial binding and internalisation (Evrard et al.2010). It should be mentioned that the latter results were obtained testing UV-killed bacteria. This technical approach is useful to assess the impact of an intact bacterial surface on cell activation/maturation but hampers any investigation on anti-immune systems deployed by live bacteria such as the injection of effector proteins by secretion systems. Nonetheless, future investigations are warranted to deconstruct the interaction between dendritic cells and K. pneumoniae to address whether the pathogen is able to survive intracellularly, the impact of Klebsiella infection on signalling governing cell maturation and whether Klebsiella may interfere with the processing of antigens and presentation of antigen-derived peptides to T cells, among other questions.
Historically, K. pneumoniae is considered an extracellular pathogen, yet there are reports suggesting that K. pneumoniae can survive in macrophages (Willingham et al.2009; Greco et al.2012; Fevre et al.2013; Fodah et al.2014). Providing compelling evidence for this hypothesis, Cano and co-workers (Cano et al.2015) have demonstrated that K. pneumoniae survives within human and mouse macrophages in a unique vacuolar intracellular compartment which deviates from the canonical endocytic pathway and it does not fuse with lysosomes. Furthermore, data suggest that K. pneumoniae triggers a programmed cell death in macrophages displaying features of apoptosis 10 h post infection (Cano et al.2015). One of the hallmarks of the Klebsiella containing vacuole (KCV) is its acidic pH, and, likewise for other pathogens (Yu et al.2010; Martinez, Siadous and Bonazzi 2018), phagosome acidification is essential for the intracellular survival of K. pneumoniae (Cano et al.2015). Klebsiella pneumoniae targets the PI3K–AKT–Rab14 axis to control phagosome maturation to survive inside macrophages (Cano et al.2015), strategy shared with S. typhimurium and Mycobacterium tuberculosis (Kuijl et al.2007), two classical intracellular pathogens. Interestingly, this axis is suitable for therapeutic manipulation to develop new anti-infective drugs. AKT inhibitors have already proven useful to eliminate intracellular Salmonella and M. tuberculosis (Kuijl et al.2007; Lo et al.2014), suggesting these inhibitors might provide selective alternatives to manage K. pneumoniae infections. Providing initial support to this hypothesis, in vitro experiments showed that AKT inhibition abrogates Klebsiella intracellular survival (Cano et al.2015). At present, the Klebsiella factors interfering with the phagosomal maturation pathway are unknown. Unexpectedly, the CPS does not play a large role, if any, in intracellular survival of Klebsiella as a cps mutant does not display any loss of viability upon phagocytosis (Cano et al.2015). In fact, Klebsiella downregulates the expression of the cps once inside the KCV (Cano et al.2015). It is tempting to speculate that Klebsiella may downregulate capsule expression to better survive in the intracellular environment which is poor in nutrients or because CPS may interfere with other factors required to manipulate phagosome maturation, among other possibilities.
Despite the crucial role of macrophages to clear Klebsiella infections in vivo, the macrophage responses following Klebsiella infection are poorly characterised. This is particularly relevant considering the plasticity of these cells, which can adjust their phenotype and physiology in response to environmental cues (Lavin et al.2014; Davies and Taylor 2015; Liddiard and Taylor 2015). These functional phenotypes led to classify macrophages as either classically activated M1 macrophages or alternatively activated M2 macrophages (Mills et al.2000; Murray et al.2014). M1 phenotype is characterised by the expression of high levels of pro-inflammatory cytokines, high production of reactive oxygen intermediates and iNOS-dependent reactive nitrogen intermediates, and promotion of Th1 response by IL12 production (Mills et al.2000; Murray et al.2014). The released cytokines and chemokines play a crucial role dictating cell-to-cell communications, hence regulating global tissue responses to infection. M2 macrophages are characterised by little to no secretion of pro-inflammatory cytokines, increased secretion of anti-inflammatory cytokines, enhanced scavenging of cellular debris, and promotion of tissue remodelling and repair (Mills et al.2000; Murray et al.2014). M1 macrophages are generally considered responsible for resistance against intracellular pathogens. However, uncontrolled M1 responses associated with acute infections may lead to immunopathology (Benoit, Desnues and Mege 2008). The M1-M2 transition may provide protection against overwhelming inflammation; however, a phenotypic switch may also favour pathogen survival. Indeed, a growing number of studies show that some pathogens have evolved different strategies to interfere with M1 polarisation, whereas chronic evolution of infectious diseases is thought to be associated with macrophage reprogramming toward a M2 profile (Benoit, Desnues and Mege 2008). Of note, reports exist showing the presence of M2 macrophages in Klebsiella-infected mouse models recapitulating human diseases, and the improvement in bacterial clearance when this macrophage population is eliminated in vivo (Dolgachev et al.2014; Ohama et al.2015; Tsuchimoto et al.2015). These observations suggest that K. pneumoniae might induce the polarisation of macrophages towards an M2-like phenotype. This hypothesis is initially supported by the fact that macrophages expressing high levels of IL10, a classical marker of M2 macrophages, are required to establish a macrophage/monocyte polarising tissue microenvironment (Fevre et al.2013). Conversely, treating mice with GM-CSF to stimulate M1 polarisation enhances K. pneumoniae clearance in vivo (Standiford et al.2012).
Ablating host defence signalling
Early studies showed that Klebsiella-triggered pneumonia is characterised by high levels of the anti-inflammatory cytokine IL10 (Yoshida et al.2000, 2001). This cytokine is expressed by many cells of the immune system, and impacts on many cell types controlling the activation of innate immune responses (Gabrysova et al.2014). The induction of the anti-inflammatory response is mediated through the IL-10 receptor (IL-10R) and activation of signal transducer and activator of transcription 3 (STAT3) (Gabrysova et al.2014). The facts that neutralisation of IL10 in vivo results in prolonged survival of Klebsiella-infected mice with an enhancement of bacterial clearance in the lungs and blood, and an upregulation of inflammation (Greenberger et al.1995) suggests that Klebsiella exploits IL10 to attenuate immune responses. In agreement with this idea, IL10 overexpression causes a more pronounced bacteraemia and accelerated mortality in intratracheally infected mice (Dolgachev et al.2014). In turn, IFNγ plays an important role counteracting Klebsiella-induced IL10-dependent immune evasion because production of IL10 is significantly upregulated in infected IFNγ−/− with a concomitant increase in bacterial burden and decreased in inflammatory mediators (Moore et al.2002). Collectively, this evidence strongly supports the notion that induction of IL10 is part of the arsenal of K. pneumoniae immune evasion strategies. Adding additional weight to this notion, IL10 production is associated with K. pneumoniae pathogenicity because high levels of IL10 are only detected in mice infected with the wild-type strain but not in those mice infected with a cps mutant (Yoshida et al. 2001; Lawlor, Handley and Miller 2006). These results may suggest that the CPS is necessary for induction of IL10, although this has not been rigorously addressed yet. The identification of the cellular source of IL10 in vivo, the signalling pathway controlling Klebsiella induction of IL10 and the bacterial factor(s) needed for induction of IL10 are questions warranting future investigations.
In vitro and in vivo evidence demonstrates that a significant number of anti-Klebsiella responses are controlled by the transcriptional factor NF-κB upon activation of a TLR4/2-MyD88 signalling pathway (Regueiro et al.2006; Moranta et al.2010; Wieland et al.2011; Frank et al.2013; Tomas et al.2015). By limiting Klebsiella internalisation by epithelial cells, the CPS limits the activation of NF-κB and, hence, the production of IL8, ICAM1 and human defensins (Regueiro et al.2006; Moranta et al.2010). The reduced production of defensins by epithelial cells following Klebsiella infection can be considered another mechanism of resistance against these antimicrobial agents. However, Klebsiella also actively supresses NF-κB signalling by hijacking the host deubiquitinase cylindromatosis (CYLD) (Regueiro et al.2011; Frank et al.2013). CYLD deconjugates K63-linked ubiquitin chains to factors of the NF-κB signalling pathway, thereby abrogating the activation of the NF-κB signalling pathway (Sun 2008). In cyld knock-down cells, Klebsiella is no longer able to blunt the activation of TLR4/2-MyD88, leading to the production of IL8 following Klebsiella infection (Frank et al.2013). To upregulate the levels of CYLD, K. pneumoniae activates a NOD1 and an EGF receptor (EGFR)-phosphatidylinositol 3-OH kinase (PI3K)-AKT-PAK4-ERK-GSK3β signalling pathways (Regueiro et al.2011; Frank et al.2013). To the best of our knowledge, K. pneumoniae is the first pathogen to date activating a NLR receptor to blunt inflammation. The activation of NOD1 is mediated by the inhibition of the Rho GTPase Rac1, although the Klebsiella factor(s) responsible is unknown (Regueiro et al.2011). Whilst other bacterial pathogens are known to activate EGFR (Zhang et al.2004; Keates et al.2007; Choi et al.2011; Xu et al.2011), only Klebsiella manipulates this receptor to ablate the production of inflammatory cytokines. Klebsiella-dependent activation of the EGFR pathway is mediated by the CPS and, therefore, a cps mutant does not activate EGFR (Frank et al.2013). Interestingly, CPS-mediated EGFR activation is indirect and requires the cSRC kinase, which is activated upon recognition of the CPS by TLR4 (Frank et al.2013). NOD1 and EGFR do not play redundant roles in Klebsiella-triggered block of NF-κB activation because in cells in which EGFR and NOD1 expressions are downregulated by siRNA the anti-inflammatory effect is completely abolished in contrast to what happens in the single knockdown cells (Regueiro et al.2011; Frank et al.2013).
The production of inflammatory mediators and defensins by epithelial cells following Klebsiella infection is also governed by the MAPKs p38, ERK and JNK (Wu et al.2006; Moranta et al.2010; Regueiro et al.2011). Klebsiella inhibits MAPKs activation via the MAPK phosphatase-1 (MKP-1) (Regueiro et al.2011). Klebsiella upregulates the levels of MKP-1 by activating NOD1, and both MKP-1 and CYLD play synergistic roles to abrogate the production of IL8 by infected epithelial cells (Regueiro et al.2011). It is remarkable that Klebsiella hijacks two host proteins, CYLD and MKP-1, involved in immune homeostasis after inflammation to protect the host from an overwhelming inflammatory response (Liu, Shepherd and Nelin 2007; Sun 2008). This immune evasion strategy is radically different to that deployed by other pathogens based on exploiting bacterial effectors to ablate NF-κB and MAPKs activation.
As a result of a high-throughput screen interrogating a library of 5320 K. pneumoniae transposon mutants using as a readout a gain of NF-κB activation (Tomas et al.2015), 114 mutants no longer blunting NF-κB signalling were identified. Metabolism and envelope-related genes are the gene ontology categories accounting for half of the loci identified in the screening (Tomas et al.2015). Follow-up characterisation conclusively established that the LPS O-polysaccharide and the pullulanase (PulA) type 2 secretion system (T2SS) are required for full effectiveness of the immune evasion (Tomas et al.2015). Importantly, the CPS, the LPS O-polysaccharide and the PulA T2SS do not play a redundant role attenuating inflammation (Tomas et al.2015). In good agreement with the in vitro results, the LPS O-polysaccharide and pulA mutants induce higher inflammation in vivo than the wild-type strain (Tomas et al.2015). Furthermore, they are attenuated in the pneumonia mouse model (Tomas et al.2015). The fact that LPS O-polysaccharide and T2SS mutant-induced responses are dependent on TLR2-TLR4-MyD88 activation suggests that LPS O-polysaccharide and PulA perturb TLR-dependent recognition of K. pneumoniae.
OmpA has also been shown to be implicated in preventing TLR activation to limit inflammatory responses in vitro and in vivo (March et al.2011). The contribution of OmpA to Klebsiella immune evasion is independent of CPS because a double mutant lacking cps and ompA induces higher inflammatory response than any of the single mutants (March et al.2011). An ompA mutant is also attenuated in the pneumonia mouse model (March et al.2011), further highlighting the importance of immune evasion for Klebsiella virulence. However, studies testing OmpA purified from K. pneumoniae have yielded opposite results. Purified OmpA activates TLR2 signalling leading to enhanced cytokine production (Jeannin et al.2000; Soulas et al.2000; Pichavant et al.2003), and instillation of the purified protein in vivo also results in upregulation of inflammation (Jeannin et al.2000; Soulas et al.2000; Pichavant et al.2003). Of note, an E. coli ompA mutant strain induces higher expression of pro-inflammatory mediators (Selvaraj and Prasadarao 2005), strongly suggesting that indeed OmpA plays a role in immune evasion. The contradiction between the results observed assessing the role of OmpA in the biological context of the bacterial membrane and those testing the protein as ligand reflects the importance of investigating the interplay between pathogens and the immune system interrogating whole live bacteria instead of purified ligands.
Subversion of nutritional immunity
To limit infections, humans and other mammals restrict access to essential metals in a process termed ‘nutritional immunity’. Originally referring to restriction of iron availability, the term now also applies to mechanisms for withholding other metals in addition to Fe such as Zn, Mn and Cu, or directing the toxicity of these metals against pathogens (Palmer and Skaar 2016). In the case of Klebsiella infections, most of the research has focused on understanding how Klebsiella subverts iron nutritional immunity. Like many other Enterobacteriaceae, Klebsiella secretes the siderophore enterobactin to compete iron off of iron-loaded host proteins (Wooldridge and Williams 1993). Notably, and as a perfect example of a host–pathogen arms race, the host employs lipocalin 2 to compete out bacteria from binding the siderophore (Bachman, Miller and Weiser 2009; Bachman et al.2011, 2012). It is then not surprising that Klebsiella strains secrete additional siderophores, namely aerobactin, salmochelin and yersiniabactin (Bachman et al.2011; Russo et al.2011, 2014). Yersiniabactin promotes Klebsiella pneumonia by evading lipocalin 2 (Lawlor, O’connor and Miller 2007; Bachman et al.2011). Furthermore, epidemiological studies demonstrate that the acquisition of yersiniabactin is one of the traits of Klebsiella strains causing invasive infections (Bachman et al.2011; Holt et al.2015), highlighting the importance of overcoming nutritional immunity in Klebsiella infection biology. It is interesting to consider the regulation of the expression of these siderophores during infection but also in the context of survival in the environment. Initial findings suggest that enterobactin plays a major role under more iron-restricted conditions than any of the other siderophores (Lawlor, O’connor and Miller 2007).
Intriguingly, recent evidence argues that in Klebsiella-triggered pneumonia siderophores contribute to bacteria dissemination by stabilising the transcriptional factor HIF-1α (Holden et al.2016). This transcriptional factor governs mucosal immunity and cellular intrinsic immunity (Palazon et al.2014). However, in the case of Klebsiella infections the data are consistent with the hypothesis that the pathogen exploits HIF-1α to promote infection (Holden et al.2016). Further studies are warranted to validate this hypothesis, and to identify the HIF-1α-governed responses facilitating Klebsiella dissemination.
ANTIBIOTIC RESISTANCE AND KLEBSIELLA IMMUNE EVASION
Whilst the impact of antibiotic resistance in bacterial physiology is well appreciated, it is still poorly understood the relationship between virulence and antibiotic resistance in the absence of antibiotic challenge, and whether antibiotic resistance mechanisms affect the interaction of pathogens with innate host defence mechanisms. In the case of Klebsiella infections, recent studies have investigated these questions by comparing infection outcomes between strains with different patterns of antibiotic resistance. For example, the inflammatory responses induced by one strain of each of the two clades of the globally disseminated K. pneumoniae ST258 clonal group have been compared. Preliminary results might support that one of the clades triggers more inflammation than the other (Castronovo et al.2017; Clemente et al.2017). The ability of few clinical isolates of the same clonal group to avoid phagocytosis by neutrophils and to survive in serum has been also investigated (Kobayashi et al.2016; DeLeo et al.2017). However, the number of strains tested is small and it is not evident how representative these strains are. Furthermore, there is limited information on the Klebsiella factor(s) responsible for the phenotypes described. One possible explanation is the challenge to construct mutants in multidrug-resistant clinical isolates. This is exemplified in the recent work from the Prince laboratory in which authors investigated the evolution of a local outbreak of a clone of the ST258 group (Ahn et al.2016). The locally predominant clone, represented by strain KP35, outcompeted related ST258 strains by becoming more competent supressing inflammatory responses (Ahn et al.2016). Mechanistically, KP35 triggered the recruitment to the lung of Ly6Chi monocytic myeloid-derived suppressor cells that lacked phagocytic capabilities and contributed to create an anti-inflammatory microenvironment (Ahn et al.2016). Authors identified the acquisition of four new orthologues by KP35 in comparison to other ST258 strains (Ahn et al.2016), suggesting that the acquisition of these novel genes contributed to its fitness and persistence. Unfortunately, the lack of genetic tools to manipulate KP35 made impossible to pinpoint any of the genetic changes as responsible for the enhanced fitness in vivo. Nonetheless, this study demonstrates how the challenge imposed by the immune system drives the adaptation of strains to the host microenvironment enhancing fitness independently of the antimicrobial selective pressure.
Few studies have addressed the contribution of antibiotic resistance mechanisms to virulence in the absence of antibiotic pressure. However, the emerging scenario supports a strong relationship between immune evasion, virulence and antibiotic resistance mechanisms in K. pneumoniae. OmpK35 and OmpK36 are two porins whose expression is downregulated in many clinical isolates including ESBL-producing and carbapenem-resistant strains (Ardanuy et al.1998; Hernandez-Alles et al.1999, 2000; Mena et al.2006; Shin et al.2012). Downregulation of OmpK35 and OmpK36 limits the influx of antibiotics to the bacterium (Ardanuy et al.1998; Hernandez-Alles et al.1999, 2000; Mena et al.2006; Shin et al.2012), whereas restoration of expression decreases antibiotic resistance (Martinez-Martinez et al.1999; Doumith et al.2009; Tsai et al.2011). OmpK36 contributes to Klebsiella virulence in the peritonitis and pneumonia mouse model (Chen et al.2010; Tsai et al.2011; March et al.2013). Mechanistically, OmpK36 is not required for serum resistance (Tsai et al.2011), and does not play any role counteracting the bactericidal action of CAMPs (Llobet et al.2009). However, OmpK36 prevents phagocytosis by neutrophils and alveolar macrophages (Tsai et al.2011; March et al.2013) which may explain the attenuation of the ompK36 mutant. Interestingly, the antiphagocytic effect of OmpK36 is also observed in the absence of CPS because a double mutant lacking cps and ompK36 is phagocytosed in higher numbers than the single mutants (March et al.2013). Whether OmpK36 or any other porin shapes the inflammatory response following Klebsiella infection warrants investigation.
AcrAB is an efflux pump implicated in antibiotic resistance in K. pneumoniae and other Enterobacteriaceae (Li, Plesiat and Nikaido 2015). Increased expression of AcrAB contributes to resistance against several antibiotics in clinical isolates (Bialek-Davenet et al.2015; Wang et al.2015) which is consistent with the wide range of substrates that this pump transports (Li, Plesiat and Nikaido 2015). However, AcrAB is also necessary for Klebsiella virulence because an acrAB mutant is attenuated in the pneumonia mouse model (Padilla et al.2010). Moreover, the acrAB mutant is more susceptible to CAMPs present in human bronchoalveolar lavage fluid and to human antimicrobial peptides than the wild-type strain (Padilla et al.2010), demonstrating a role for this efflux pump counteracting innate defences in addition to contributing to a multidrug resistance phenotype.
Several regulators have been demonstrated to influence acrAB expression in Klebsiella and other Enterobacteriaceae (Weston et al.2017). For example, the AraC family transcriptional regulator RamA is involved in tigecycline resistance via upregulation of AcrAB (Rosenblum et al.2011). This antibiotic has been introduced into clinical practice for the treatment of community-acquired Gram-negative infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae. Interestingly, RamA overexpression, a feature observed in clinical isolates even before the introduction of tigecycline (Rosenblum et al.2011), results in a multidrug-resistant strain with enhanced virulence (De Majumdar et al.2015). Increased levels of RamA decrease susceptibility of Klebsiella to polymyxins and the human CAMP LL-37, and reduce bacterial adhesion and uptake into macrophages (De Majumdar et al.2015). Infection with a strain overexpressing RamA results in higher bacterial burden in the lungs and increase systemic dissemination (De Majumdar et al.2015), demonstrating the relevance of these phenotypes to heightened fitness in vivo. RamA overexpression triggers significant changes in Klebsiella transcriptome making then difficult the identification of the RamA-controlled loci responsible for the increase virulence (De Majumdar et al.2015). Initial experimental evidence demonstrates that RamA directly activates acrAB and loci implicated in lipid A remodelling (De Majumdar et al.2015). Based on the discussed evidence, it is tempting to postulate that these loci may underline RamA-linked phenotypes.
lpxO is one of the lipid A loci regulated by RamA (De Majumdar et al.2015). LpxO mediates the 2-hydroxylation of the lipid A in Klebsiella and other Gram-negative pathogens including Salmonella and Pseudomonas (Gibbons et al.2000, 2008; Moskowitz and Ernst 2010). Remarkably, K. pneumoniae switches on this lipid A modification in the lungs of infected mice in a PhoPQ-dependent manner (Llobet et al.2015). This modified lipid A does not activate inflammatory responses in vivo and in vitro to the same extent as the lipid A produced by Klebsiella in normal laboratory medium (Llobet et al.2015), demonstrating that the 2-hydroxylation of the lipid A contributes to immune evasion. Not surprisingly, an lpxO mutant is attenuated in the pneumonia mouse model (Insua et al.2013; Llobet et al.2015). Furthermore, this lipid A modification also mediates resistance to human antimicrobial peptides and colistin (Llobet et al.2015; Mills et al.2017), one of the last options to treat multidrug-resistant Klebsiella (Li et al.2006). Changes in the lipid A acylation pattern have also been linked to resistance to colistin and virulence independently of the lipid A decorations (Clements et al.2007; Halaby et al.2016; Mills et al.2017).
Resistance to colistin is of significant concern in those settings with high number of ESBL and carbapenem-producing Klebsiella strains. Unfortunately, colistin resistance arises frequently upon treatment and even following decontamination protocols to limit infections in the healthcare setting. Several recent studies highlight the emergence of colistin resistance in multidrug-resistant K. pneumoniae arising from loss of function mutations of the mgrB gene, a negative regulator of the PhoPQ signalling system (Lippa and Goulian 2009; Cannatelli et al.2013, 2014). Alarmingly, mgrB-dependent colistin resistance is not associated with any fitness cost in vitro and in vivo and it is stably maintained in the absence of selective pressure (Cannatelli et al.2015; Kidd et al.2017). This may explain the rapid dissemination of strains carrying this resistance mechanism in the clinical setting. Further complicating the health problem posed by these infections, inactivation of mgrB enhances K. pneumoniae virulence (Kidd et al.2017). In fact, the heightened virulence of these strains might be one of the explanations underlying the increased mortality associated with these infections (Capone et al.2013; Falcone et al.2016). Mechanistically, mgrB mutation induces PhoPQ-governed lipid A remodelling which confers not only resistance to polymyxins, but also enhances K. pneumoniae virulence by decreasing CAMPs susceptibility and attenuating the activation of early host defence responses in macrophages (Kidd et al.2017). These findings also illustrate that the development of antibiotic resistance is not inexorably linked with subdued bacterial fitness and virulence. Overall, this research stresses the importance of considering antimicrobial resistance and virulence together, and the urgent need to include the identification of virulent clones in clinical microbiology laboratories.
CONCLUSIONS AND PERSPECTIVES
The World Health Organization has recently included Klebsiella in the critical list of microorganisms for which new therapeutics are urgently needed. The increasing isolation of strains resistant to ‘last resort’ antimicrobials has significantly narrowed, or in some settings completely removed, the therapeutic options for the treatment of Klebsiella infections. Not surprisingly, several international organisations including the United Nations have regarded multidrug-resistant Klebsiella as an ‘urgent threat to human health’. Whilst there are several new therapeutic approaches under investigation including the use of bacteriophages, enzybiotics (phage-derived lytic enzymes) and antibodies against Klebsiella surface molecules; unfortunately, at present, we cannot identify candidate compounds in late-stage development for treatment of multidrug Klebsiella infections. This pathogen is exemplary of the mismatch between unmet medical needs and the current antimicrobial research and development pipeline. Furthermore, our understanding of Klebsiella pathogenesis still contains considerable gaps. Therefore, understanding the various Achilles heels of host defence against Klebsiella is a high priority and timely for the development of preventative and novel therapeutic measures to combat these infections.
K. pneumoniae has been traditionally considered a formidable pathogen evading defence mechanisms. The roles of CPS limiting the activation of inflammatory responses, preventing the bactericidal action of complement and CAMPs, and abrogating phagocytosis by neutrophils and macrophages are perfect examples of Klebsiella stealth strategies. The lack of expression of porins to avoid complement activation, and the role of the LPS O-polysaccharide to limit complement deposition on the bacterial surface are other examples of this Klebsiella stealth behaviour.
However, a body of evidence mostly recently obtained strongly suggests that Klebsiella has also evolved mechanisms to actively supress innate immune responses, illustrating the diversity and sophistication of Klebsiella immune evasion strategies. The manipulation of phagosome maturation to establish the KCV, the upregulation of lipid A decorations upon sensing the host microenvironment to counteract the action of CAMPs and to avoid the activation of NF-κB-controlled inflammation are examples of these Klebsiella subversion strategies. At the cellular level, the evidence clearly demonstrates that an essential aspect of K. pneumoniae infection biology is to thwart the TLR-dependent activation of host defence responses controlled by NF-κB and MAPKs. To do so, Klebsiella hijacks the deubiquitinase CYLD and the MAPKs phosphatase MKP-1. Both proteins play a pivotal role in host homeostasis by limiting overwhelming inflammatory responses to avoid immunopathology. This is an exquisite example of how a bacterial pathogen exploits the host machinery to avoid immune activation. This immune evasion strategy is different to those deployed by other bacterial pathogens such as Listeria, Legionella, E. coli, Salmonella or M. tuberculosis based on deploying bacterial proteins and effectors to blunt host defence signalling pathways. Considering the increasing number of host proteins contributing to balance host responses, it is tempting to speculate that Klebsiella may hijack other proteins as well. Interestingly, Klebsiella activates TLR4 and NOD1 signalling to increase the levels of CYLD and MKP-1. Thus, Klebsiella infection biology is largely based on the balance between avoiding PRR detection/activating PRR for immune evasion, hence being a true pathogen-host arms race. At the tissue level, Klebsiella exploits the anti-inflammatory properties of IL10 to shape the local microenvironment. Collectively, the data strongly suggest that Klebsiella hijacks host effectors devoted to prevent overwhelming inflammatory responses as virulence strategy.
The CPS is necessary but not sufficient for Klebsiella immune evasion, and Klebsiella exploits few other factors acting synergistically to abrogate immune responses. The importance of avoiding immune responses in Klebsiella pathogenesis is marked by the fact that mutants lacking these factors are attenuated in vivo. However, we believe there is much left to be uncovered about which K. pneumoniae factors are required during infection. In this regard, recent epidemiological studies have demonstrated the wide spectrum of genetic diversity within the genus (Holt et al.2015; MC Lam et al.2018). It is important to note that the epidemiological studies are biased towards strains of clinical origin. There is limited information on the attributes of strains isolated from the environment, and the potential transmission route between the environment, and the healthcare setting. The immune evasion strategies described so far rely on factors belonging to the Klebsiella core genome, indicating that Klebsiella anti-immunology is at the fulcrum of the species biology. However, the Klebsiella accessory genome is close to 30 000 genes, more than the human genome (Holt et al.2015). Moreover, around 50% of the genes of an individual strain belong to the accessory genome (Holt et al.2015), illustrating the considerable genomic plasticity that is contained within this species. Notably, there is virtually no information on the contribution of the accessory genome to Klebsiella immune evasion, although recent findings suggest that the accessory genome may also facilitate the adaptation of Klebsiella to host microenvironments (Ahn et al.2016). Future studies should address the relative contribution of the core and accessory genome to Klebsiella infection biology, and uncover the regulatory networks coordinating the spatial-temporal expression of these factors. Additionally, there is a significant knowledge gap on the virulence of other Klebsiella species including K. oxytoca and K. variicola. To obtain comprehensive information, it will be necessary to assess infection phenotypes interrogating large collection of strains from different environments and genomic backgrounds to capture Klebsiella diversity. Ideally, these studies will yield a catalogue of virulence factors and host pathways subverted across different infections and common to many strains, leading to the definition of the Klebsiella signature of infection. This knowledge may help to better stratify patients and to tailor treatments not only based on the antibiotic resistance profile but also taking into consideration virulence features of the infecting strain. Nonetheless, it remains a challenge to develop efficient genetic tools to construct mutants and to complement multidrug-resistant strains to provide mechanistic information on the specific factors responsible for the investigated virulence phenotypes.
It is also important to understand how Klebsiella evades the immunological challenges it faces when colonising/infecting the GI tract and the oropharyngeal sites. This may require developing new infection models to follow bacterial colonisation for at least several days. It is exciting to consider that the immune evasion strategies of Klebsiella may differ in different mucosae. There is much to be learnt about how Klebsiella disseminates from the primary infection site, either the lung or the gut, to other sites. For example, a yet poorly investigated question is the interaction between Klebsiella and endothelial cells lining vascular and lymphatic vessels. This interaction is clinically relevant considering the number of sepsis cases associated with Klebsiella infections (Girometti et al.2014). We do foresee that the interplay between Klebsiella-endothelial cells could be another host–pathogen battleground playing an important role in Klebsiella infection biology.
As mentioned above, a crucial virulence strategy of Klebsiella is to subdue TLR-mediated inflammatory responses. Substantial data also confirm the relative importance of IL23/IL17 and IL12/IFNγ-activated signalling pathways to generate an effective innate immune response against Klebsiella. It can be then argued that Klebsiella lack of immunostimulatory activities may limit the activation of these signalling axes. However, and considering the sophisticated strategies deployed by the pathogen to perturb TLR recognition and activation, we postulate that Klebsiella has devised specific strategies to attenuate IL23/IL17 and IL12/IFNγ signalling. Among other possibilities, it is tempting to speculate that Klebsiella may target the cells responsible for the production of these cytokines, ablate the signalling pathways needed for the production of these cytokines, and even subvert the receptors and signalling pathways activated by these cytokines. Future studies combining in vivo and in vitro approaches are warranted to investigate these questions.
The concept of targeted specific antibiotic therapy for Klebsiella infections is plausible with the availability of real-time PCR-based methods for rapid detection. This will vastly reduce the time required to direct appropriate therapy and limit the use of empirical broad range treatments. However, the global emergence of multidrug-resistant Klebsiella strains significantly reduces the available options to treat these infections, making it imperative to consider other options. It is evident that therapeutic strategies to improve innate immune mechanisms may enhance clearance of Klebsiella. This could be achieved through pro-inflammatory agents or by boosting TLR-governed defences, or by abrogating Klebsiella immune evasion strategies. In this context, it would appear that targeting major Klebsiella immune evasins such as the CPS could be a successful avenue to counteract Klebsiella anti-immune strategies. However, given the genetic diversity of this pathogen we consider likely the selection of clones able to evade this therapy focused on a single or even a limited number of gene products. Instead, we favour the approach of targeting the host factors manipulated by Klebsiella to subvert host defence responses (Zumla et al.2016). This host-directed therapeutics approach is thought to apply less selective pressure for the development of resistance than traditional strategies, which are aimed at killing pathogens or preventing their growth. Interestingly, there might be drugs already available targeting the host factors hijacked by Klebsiella, which might be even in clinical use to treat other diseases. From the drug discovery point of view, this significantly circumvents the drug development process, hence allowing a fast-track transition from pre-clinical research to clinical development.
Despite our significant advances on our understanding of Klebsiella pathogenesis during the last years, we still have a fragmented picture of the interaction of K. pneumoniae with the immune system, and there is a significant knowledge gap on the repertoire of virulence factors enabling Klebsiella to overcome defences to multiply in the tissues. Further studies on the fascinating infection biology of Klebsiella will help to shore-up more precisely the vulnerable hot spots of our immune system while uncovering new means exploited by a human pathogen to counteract the challenge of an activated immune system.
Acknowledgement
We apologise to all colleagues whose work was not discussed in this review. We thank present and former members of the J.A.B. laboratory for their thoughtful discussions on the Klebsiella saga.
FUNDING
This work was supported by the Marie Curie Career Integration Grant U-KARE (PCIG13-GA-2013-618162), Biotechnology and Biological Sciences Research Council (BBSRC, BB/L007223/1, BB/N00700X/1, BB/P006078/1, and BB/P020194/1), Medical Research Council (MRC, MR/R005893/1), and Queen's University Belfast start-up funds.
Conflict of interest. None declared.
REFERENCES
- Ahn D, Penaloza H, Wang Z et al.. Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection. JCI Insight 2016;1:e89704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Alvarez D, Merino S et al.. Analysis of complement C3 deposition and degradation on Klebsiella pneumoniae. Infect Immun 1996a;64:4726–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Marques G, Camprubi S et al.. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun 1993;61:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Marques G, Hernandez-Alles S et al.. Interaction between complement subcomponent C1q and the Klebsiella pneumoniae porin OmpK36. Infect Immun 1996b;64:4719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D, Merino S, Tomas JM et al.. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun 2000;68:953–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amulic B, Cazalet C, Hayes GL et al.. Neutrophil function: from mechanisms to disease. Annu Rev Immunol 2012;30:459–89. [DOI] [PubMed] [Google Scholar]
- Anas AA, Claushuis TAM, Mohan RA et al.. Epithelial myeloid-differentiation factor 88 is dispensable during Klebsiella Pneumonia. Am J Respir Cell Mol Biol 2017;56:648–56. [DOI] [PubMed] [Google Scholar]
- Angsana J, Chen J, Liu L et al.. Efferocytosis as a regulator of macrophage chemokine receptor expression and polarization. Eur J Immunol 2016;46:1592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardanuy C, Linares J, Dominguez MA et al.. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob Agents Chemother 1998;42:1636–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel A, Serhan CN. New lives given by cell death: macrophage differentiation following their encounter with apoptotic leukocytes during the resolution of inflammation. Front Immun 2012;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Godec J, Lau L et al.. TLR signaling is required for Salmonella typhimurium virulence. Cell 2011;144:675–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla SJ, Chan YR, Zheng M et al.. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med 2008;14:275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Lenio S, Schmidt L et al.. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 2012;3:e00224–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Miller VL, Weiser JN. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 2009;5:e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Oyler JE, Burns SH et al.. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 2011;79:3309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon CM, Petricoin EF 3rd, Ortaldo JR et al.. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci 1995;92:7307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailie MB, Standiford TJ, Laichalk LL et al.. Leukotriene-deficient mice manifest enhanced lethality from Klebsiella pneumonia in association with decreased alveolar macrophage phagocytic and bactericidal activities. J Immunol 1996;157:5221–4. [PubMed] [Google Scholar]
- Ballinger MN, Standiford TJ. Postinfluenza bacterial pneumonia: host defenses gone awry. J Interferon Cytokine Res 2010;30:643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P, Batra S, Zemans RL et al.. Divergent functions of toll-like receptors during bacterial lung infections. Am J Respir Crit Care Med 2014;190:722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Fauvarque MO, Tournebize R et al.. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol 2006;8:139–48. [DOI] [PubMed] [Google Scholar]
- Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol 2008;181:3733–9. [DOI] [PubMed] [Google Scholar]
- Bhan U, Ballinger MN, Zeng X et al.. Cooperative interactions between TLR4 and TLR9 regulate interleukin 23 and 17 production in a murine model of gram negative bacterial pneumonia. PLoS One 2010;5:e9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan U, Lukacs NW, Osterholzer JJ et al.. TLR9 is required for protective innate immunity in Gram-negative bacterial pneumonia: role of dendritic cells. J Immunol 2007;179:3937–46. [DOI] [PubMed] [Google Scholar]
- Bialek-Davenet S, Lavigne JP, Guyot K et al.. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 2015;70:81–8. [DOI] [PubMed] [Google Scholar]
- Bowdish DM, Davidson DJ, Hancock RE. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol 2006;306:27–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxx GM, Cheng G. The roles of type I interferon in bacterial infection. Cell Host Microbe 2016;19:760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branger J, Knapp S, Weijer S et al.. Role of Toll-like receptor 4 in gram-positive and gram-negative pneumonia in mice. Infect Immun 2004;72:788–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzk N, Lubojemska A, Hardison SE et al.. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 2014;15:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broug-Holub E, Toews GB, van Iwaarden JF et al.. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infect Immun 1997;65:1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun 2017;8:1512–017–01803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne N, Heelan M, Kavanagh K. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 2013;4:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K et al.. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011;478:515–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Batra S, Lira SA et al.. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-kappaB, and MAPKs. J Immunol 2010;185:6214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Batra S, Shen L et al.. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J Immunol 2009;183:6629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Batra S, Wakamatsu N et al.. NLRC4 inflammasome-mediated production of IL-1beta modulates mucosal immunity in the lung against gram-negative bacterial infection. J Immunol 2012;188:5623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MA, Morey P, Bengoechea JA. Quinolones sensitize gram-negative bacteria to antimicrobial peptides. Antimicrob Agents Chemother 2006;50:2361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MA, Vargas MA, Regueiro V et al.. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 2004;72:7107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A, D’Andrea MM, Giani T et al.. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 2013;57:5521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A, Giani T, D’Andrea MM et al.. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 2014;58:5696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A, Santos-Lopez A, Giani T et al.. Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob Agents Chemother 2015;59:2898–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano V, March C, Insua JL et al.. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol 2015;17:1537–60. [DOI] [PubMed] [Google Scholar]
- Cano V, Moranta D, Llobet-Brossa E et al.. Klebsiella pneumoniae triggers a cytotoxic effect on airway epithelial cells. BMC Microbiol 2009;9:156–2180–9–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone A, Giannella M, Fortini D et al.. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect 2013;19:E23–30. [DOI] [PubMed] [Google Scholar]
- Castronovo G, Clemente AM, Antonelli A et al.. Differences in inflammatory response induced by two representatives of clades of the pandemic ST258 Klebsiella pneumoniae clonal lineage producing KPC-type carbapenemases. PLoS One 2017;12:e0170125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YR, Liu JS, Pociask DA et al.. Lipocalin 2 is required for pulmonary host defense against Klebsiella infection. J Immunol 2009;182:4947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheepurupalli L, Raman T, Rathore SS et al.. Bioactive molecule from streptomyces sp. mitigates MDR Klebsiella pneumoniae in zebrafish infection model. Front Microbiol 2017;8:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Siu LK, Fung CP et al.. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 2010;65:986–90. [DOI] [PubMed] [Google Scholar]
- Chen K, Eddens T, Trevejo-Nunez G et al.. IL-17 receptor signaling in the lung epithelium is required for mucosal chemokine gradients and pulmonary host defense against K. pneumoniae. Cell Host Microbe 2016;20:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Mehrad B, Deng JC et al.. Impaired pulmonary host defense in mice lacking expression of the CXC chemokine lungkine. J Immunol 2001;166:3362–8. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Seo CH, Park SH et al.. Involvement of epidermal growth factor receptor-linked signaling responses in Pseudomonas fluorescens-infected alveolar epithelial cells. Infect Immun 2011;79:1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun 2014;82:4596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente AM, Castronovo G, Antonelli A et al.. Differential Th17 response induced by the two clades of the pandemic ST258 Klebsiella pneumoniae clonal lineages producing KPC-type carbapenemase. PLoS One 2017;12:e0178847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A, Tull D, Jenney AW et al.. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem 2007;282:15569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The TLR and IL-1 signalling network at a glance. J Cell Sci 2014;127:2383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes G, Alvarez D, Saus C et al.. Role of lung epithelial cells in defense against Klebsiella pneumoniae pneumonia. Infect Immun 2002;70:1075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies LC, Taylor PR. Tissue-resident macrophages: then and now. Immunology 2015;144:541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Astorza B, Cortes G, Crespi C et al.. C3 promotes clearance of Klebsiella pneumoniae by A549 epithelial cells. Infect Immun 2004;72:1767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Majumdar S, Yu J, Fookes M et al.. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 2015;11:e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Kobayashi SD, Porter AR et al.. Survival of carbapenem-resistant Klebsiella pneumoniae sequence type 258 in human blood. Antimicrob Agents Chemother 2017;61: e02533–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JC, Moore TA, Newstead MW et al.. CpG oligodeoxynucleotides stimulate protective innate immunity against pulmonary Klebsiella infection. J Immunol 2004;173:5148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JC, Zeng X, Newstead M et al.. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol 2004;173:4075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diago-Navarro E, Calatayud-Baselga I, Sun D et al.. Antibody-based immunotherapy to treat and prevent infection with hypervirulent Klebsiella pneumoniae. Clin Vaccine Immunol 2017;24: e00456–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diago-Navarro E, Motley MP, Ruiz-Perez G et al.. novel, broadly reactive anticapsular antibodies against carbapenem-resistant Klebsiella pneumoniae protect from infection. mBio 2018;9:e00091–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgachev VA, Yu B, Sun L et al.. Interleukin 10 overexpression alters survival in the setting of gram-negative pneumonia following lung contusion. Shock 2014;41:301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenico P, Salo RJ, Cross AS et al.. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect Immun 1994;62:4495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M, Ellington MJ, Livermore DM et al.. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 2009;63:659–67. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Canna SW. The NLRC4 inflammasome. Immunol Rev 2018;281:115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JD, Bosmani C, Barisch C et al.. Eat prey, live: Dictyostelium discoideum as a model for cell-autonomous defenses. Front Immunol 2018;8:1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer PB, Lehrer RI. Mouse neutrophils lack defensins. Infect Immun 1992;60:3446–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control. Antimicrobial resistance (EARS-Net). ECDC. Annual Epidemiological Report for 2014. Stockholm: ECDC; 2018. [Google Scholar]
- Evrard B, Balestrino D, Dosgilbert A et al.. Roles of capsule and lipopolysaccharide O antigen in interactions of human monocyte-derived dendritic cells and Klebsiella pneumoniae. Infect Immun 2010;78:210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes CT, Amaral FA, Vieira AT et al.. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol 2012;188:1411–20. [DOI] [PubMed] [Google Scholar]
- Falcone M, Russo A, Iacovelli A et al.. Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Clin Microbiol Infect 2016;22:444–50. [DOI] [PubMed] [Google Scholar]
- Fevre C, Almeida AS, Taront S et al.. A novel murine model of rhinoscleroma identifies Mikulicz cells, the disease signature, as IL-10 dependent derivatives of inflammatory monocytes. EMBO Mol Med 2013;5:516–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodah RA, Scott JB, Tam HH et al.. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS One 2014;9:e107394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CG, Reguerio V, Rother M et al.. Klebsiella pneumoniae targets an EGF receptor-dependent pathway to subvert inflammation. Cell Microbiol 2013;15:1212–33. [DOI] [PubMed] [Google Scholar]
- Friedlander C. Ueber die Schizomyceten bei der acuten fibrösen Pneumonie. Archiv F Pathol Anat 1882;87:319–24. [Google Scholar]
- Gabrysova L, Howes A, Saraiva M et al.. The regulation of IL-10 expression. Curr Top Microbiol Immunol 2014;380:157–90. [DOI] [PubMed] [Google Scholar]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 2003;3:710–20. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Lin S, Cotter RJ et al.. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem 2000;275:32940–9. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Reynolds CM, Guan Z et al.. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella; lipid A. Biochemistry 2008;47:2814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girometti N, Lewis RE, Giannella M et al.. Klebsiella pneumoniae bloodstream infection: epidemiology and impact of inappropriate empirical therapy. Medicine (Baltimore) 2014;93:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavis-Bloom J, Muhammed M, Mylonakis E. Of model hosts and man: using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv Exp Med Biol 2012;710:11–17. [DOI] [PubMed] [Google Scholar]
- Gorrie CL, Mirceta M, Wick RR et al.. Gastrointestinal carriage is a major reservoir of klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 2017;65:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco E, Quintiliani G, Santucci MB et al.. Janus-faced liposomes enhance antimicrobial innate immune response in Mycobacterium tuberculosis infection. Proc Natl Acad Sci 2012;109:E1360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberger MJ, Strieter RM, Kunkel SL et al.. Neutralization of IL-10 increases survival in a murine model of Klebsiella pneumonia. J Immunol 1995;155:722–9. [PubMed] [Google Scholar]
- Greenberger MJ, Strieter RM, Kunkel SL et al.. Neutralization of macrophage inflammatory protein-2 attenuates neutrophil recruitment and bacterial clearance in murine Klebsiella pneumonia. J Infect Dis 1996;173:159–65. [DOI] [PubMed] [Google Scholar]
- Gu D, Dong N, Zheng Z et al.. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2018;18:37–46. [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease and therapeutics. Nat Med 2015;21:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein H, Kranz S, Lippitsch A et al.. Modulation of respiratory dendritic cells during Klebsiella pneumonia infection. Respir Res 2013;14:91–9921–14–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y et al.. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 2013;341:1250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar AM, Ernst RK, Tsai JH et al.. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol 2002;3:354–9. [DOI] [PubMed] [Google Scholar]
- Halaby T, Kucukkose E, Janssen AB et al.. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob Agents Chemother 2016;60:6837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley PJ. Species differences in the structure and function of the immune system. Toxicology 2003;188:49–71. [DOI] [PubMed] [Google Scholar]
- Happel KI, Dubin PJ, Zheng M et al.. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med 2005;202:761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel KI, Odden AR, Zhang P et al.. Acute alcohol intoxication suppresses the interleukin 23 response to Klebsiella pneumoniae infection. Alcoholism Clin Exp Res 2006;30:1200–7. [DOI] [PubMed] [Google Scholar]
- Happel KI, Zheng M, Young E et al.. Cutting edge: roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J Immunol 2003;170:4432–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T et al.. A Toll-like receptor recognizes bacterial DNA. Nature 2000;408:740–5. [DOI] [PubMed] [Google Scholar]
- Hernandez-Alles S, Alberti S, Alvarez D et al.. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 1999;145:673–9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Alles S, Conejo M, Pascual A et al.. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J Antimicrob Chemother 2000;46:273–7. [DOI] [PubMed] [Google Scholar]
- Holden VI, Breen P, Houle S et al.. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1 alpha stabilization during pneumonia. mBio 2016;7: e01397–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holers VM. Complement and its receptors: new insights into human disease. Annu Rev Immunol 2014;32:433–59. [DOI] [PubMed] [Google Scholar]
- Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol 2003;21:547–78. [DOI] [PubMed] [Google Scholar]
- Holt KE, Wertheim H, Zadoks RN et al.. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 2015;112:E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua KF, Yang FL, Chiu HW et al.. Capsular Polysaccharide is involved in NLRP3 inflammasome activation by Klebsiella pneumoniae serotype K1. Infect Immun 2015;83:3396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insua JL, Llobet E, Moranta D et al.. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun 2013;81:3552–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivin M, Dumigan A, de Vasconcelos FN et al.. Natural killer cell-intrinsic type I IFN signaling controls Klebsiella pneumoniae growth during lung infection. PLoS Pathog 2017;13:e1006696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannin P, Renno T, Goetsch L et al.. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat Immunol 2000;1:502–9. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microb 2005;3:281–94. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan S, Young SK, Yamamoto M et al.. Toll/IL-1R domain-containing adaptor protein (TIRAP) is a critical mediator of antibacterial defense in the lung against Klebsiella pneumoniae but not Pseudomonas aeruginosa. J Immunol 2006;177:538–47. [DOI] [PubMed] [Google Scholar]
- Jondle CN, Gupta K, Mishra BB et al.. Klebsiella pneumoniae infection of murine neutrophils impairs their efferocytic clearance by modulating cell death machinery. PLoS Pathog 2018;14:e1007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Zhang Y, Krantz BA et al.. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med 2016;213:2113–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabha K, Schmegner J, Keisari Y et al.. SP-A enhances phagocytosis of Klebsiella by interaction with capsular polysaccharides and alveolar macrophages. Am J Physiol 1997;272:L344–52. [DOI] [PubMed] [Google Scholar]
- Kamaladevi A, Balamurugan K. Role of PMK-1/p38 MAPK defense in Caenorhabditis elegans against Klebsiella pneumoniae infection during host-pathogen interaction. Pathog Dis 2015;73: ftv021. [DOI] [PubMed] [Google Scholar]
- Kamaladevi A, Balamurugan K. Global proteomics revealed Klebsiella pneumoniae induced autophagy and oxidative stress in caenorhabditis elegans by inhibiting PI3K/AKT/mTOR pathway during infection. Front Cell Infect Microbiol 2017;7:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Newstead MW, Zeng X et al.. Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infect Immun 2007;75:4942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keates S, Keates AC, Katchar K et al.. Helicobacter pylori induces up-regulation of the epidermal growth factor receptor in AGS gastric epithelial cells. J Infect Dis 2007;196:95–103. [DOI] [PubMed] [Google Scholar]
- Kidd TJ, Mills G, Sa-Pessoa J et al.. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 2017;9:430–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Wieland CW, Florquin S et al.. Differential roles of CD14 and toll-like receptors 4 and 2 in murine Acinetobacter pneumonia. Am J Respir Crit Care Med 2006;173:122–9. [DOI] [PubMed] [Google Scholar]
- Ko WC, Paterson DL, Sagnimeni AJ et al.. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 2002;8:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Porter AR, Dorward DW et al.. Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J Infect Dis 2016;213:1615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostina E, Ofek I, Crouch E et al.. Noncapsulated Klebsiella pneumoniae bearing mannose-containing O antigens is rapidly eradicated from mouse lung and triggers cytokine production by macrophages following opsonization with surfactant protein D. Infect Immun 2005;73:8282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik P, Castiglia V, Ivin M et al.. Type I interferons in bacterial infections: A balancing act. Front Immunol 2016;7:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp F, Morris AR, Ozer EA et al.. Virulence characteristics of carbapenem-resistant Klebsiella pneumoniae strains from patients with necrotizing skin and soft tissue infections. Sci Rep 2017;7:13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijl C, Savage ND, Marsman M et al.. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 2007;450:725–30. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Takahashi M, Nishitani C. Pulmonary collectins in innate immunity of the lung. Cell Microbiol 2007;9:1871–9. [DOI] [PubMed] [Google Scholar]
- Lachmandas E, Boutens L, Ratter JM et al.. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat Microbiol 2017;2:16246. [DOI] [PubMed] [Google Scholar]
- Laichalk LL, Bucknell KA, Huffnagle GB et al.. Intrapulmonary delivery of tumor necrosis factor agonist peptide augments host defense in murine gram-negative bacterial pneumonia. Infect Immun 1998;66:2822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichalk LL, Kunkel SL, Strieter RM et al.. Tumor necrosis factor mediates lung antibacterial host defense in murine Klebsiella pneumonia. Infect Immun 1996;64:5211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MMC, Wick RR, Wyres KL et al.. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 2018;4:e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol 2013;13:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R et al.. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014;159:1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 2006;74:5402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MS, O’connor C, Miller VL. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun 2007;75:1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone L, Mazzetta F, Martinelli D et al.. Klebsiella pneumoniae is able to trigger epithelial-mesenchymal transition process in cultured airway epithelial cells. PLoS One 2016;11:e0146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nation RL, Turnidge JD et al.. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 2006;6:589–601. [DOI] [PubMed] [Google Scholar]
- Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 2015;28:337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Oosting M, Deelen P et al.. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med 2016;22:952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddiard K, Taylor PR. Understanding local macrophage phenotypes in disease: shape-shifting macrophages. Nat Med 2015;21:119–20. [DOI] [PubMed] [Google Scholar]
- Lima WC, Balestrino D, Forestier C et al.. Two distinct sensing pathways allow recognition of Klebsiella pneumoniae by Dictyostelium amoebae. Cell Microbiol 2014;16:311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell DM, Standiford TJ, Mancuso P et al.. Macrophage inflammatory protein 1alpha/CCL3 is required for clearance of an acute Klebsiella pneumoniae pulmonary infection. Infect Immun 2001;69:6364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet 2009;5:e1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases – regulating the immune response. Nat Rev Immunol 2007;7:202–12. [DOI] [PubMed] [Google Scholar]
- Llobet E, Campos MA, Gimenez P et al.. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect Immun 2011;79:3718–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E, March C, Gimenez P et al.. Klebsiella pneumoniae OmpA confers resistance to antimicrobial peptides. Antimicrob Agents Chemother 2009;53:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E, Martinez-Moliner V, Moranta D et al.. Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proc Natl Acad Sci USA 2015;112:E6369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology 2008;154:3877–86. [DOI] [PubMed] [Google Scholar]
- Lo JH, Kulp SK, Chen CS et al.. Sensitization of intracellular Salmonella enterica serovar Typhimurium to aminoglycosides in vitro and in vivo by a host-targeted antimicrobial agent. Antimicrob Agents Chemother 2014;58:7375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill SS, Edwards JR, Bamberg W et al.. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014;370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso P, Gottschalk A, Phare SM et al.. Leptin-deficient mice exhibit impaired host defense in Gram-negative pneumonia. J Immunol 2002;168:4018–24. [DOI] [PubMed] [Google Scholar]
- Mancuso P, Standiford TJ, Marshall T et al.. 5-Lipoxygenase reaction products modulate alveolar macrophage phagocytosis of Klebsiella pneumoniae. Infect Immun 1998;66:5140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March C, Cano V, Moranta D et al.. Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One 2013;8:e56847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March C, Moranta D, Regueiro V et al.. Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J Biol Chem 2011;286:9956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoleta AE, Varas MA, Ortiz-Severin J et al.. Evaluating different virulence traits of Klebsiella pneumoniae using Dictyostelium discoideum and zebrafish larvae as host models. Front Cell Infect Microbiol 2018;8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho FV, Benmerzoug S, Oliveira SC et al.. The emerging roles of STING in bacterial infections. Trends Microbiol 2017;25:906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Cao J, Brisse S et al.. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 2016;1:e00261–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Siadous FA, Bonazzi M. Tiny architects: biogenesis of intracellular replicative niches by bacterial pathogens. FEMS Microbiol Rev 2018;42:425–47. [DOI] [PubMed] [Google Scholar]
- Martinez I, Oliveros JC, Cuesta I et al.. Apoptosis, Toll-like, RIG-I-like and NOD-like receptors are pathways jointly induced by diverse respiratory bacterial and viral pathogens. Front Microbiol 2017;8:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martinez L, Pascual A, Hernandez-Alles S et al.. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother 1999;43:1669–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MC Lam M, Wyres KL, Duchene S et al.. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018;9:2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest 2002;109:707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell 2015;163:1326–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrad B, Standiford TJ. Role of cytokines in pulmonary antimicrobial host defense. Immunol Res 1999;20:15–27. [DOI] [PubMed] [Google Scholar]
- Mena A, Plasencia V, Garcia L et al.. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol 2006;44:2831–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino S, Camprubi S, Alberti S et al.. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect Immun 1992;60:2529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM et al.. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 2010;11:1136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CD, Kincaid K, Alt JM et al.. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000;164:6166–73.10843666 [Google Scholar]
- Mills G, Dumigan A, Kidd T et al.. Identification and characterization of two Klebsiella pneumoniae lpxL lipid a late acyltransferases and their role in virulence. Infect Immun 2017;85: e00068–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgerd JP, Skerrett SJ. Animal models of human pneumonia. Am J Physiol Lung Cell Mol Physiol 2008;294:L387–98. [DOI] [PubMed] [Google Scholar]
- Montminy SW, Khan N, McGrath S et al.. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol 2006;7:1066–73. [DOI] [PubMed] [Google Scholar]
- Moore TA, Lau HY, Cogen AL et al.. Defective innate antibacterial host responses during murine Klebsiella pneumoniae bacteremia: tumor necrosis factor (TNF) receptor 1 deficiency versus therapy with anti-TNF-alpha. Clin Infect Dis 2005;41:S213–7. [DOI] [PubMed] [Google Scholar]
- Moore TA, Moore BB, Newstead MW et al.. Gamma delta-T cells are critical for survival and early proinflammatory cytokine gene expression during murine Klebsiella pneumonia. J Immunol 2000;165:2643–50. [DOI] [PubMed] [Google Scholar]
- Moore TA, Perry ML, Getsoian AG et al.. Divergent role of gamma interferon in a murine model of pulmonary versus systemic Klebsiella pneumoniae infection. Infect Immun 2002;70:6310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moranta D, Regueiro V, March C et al.. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect Immun 2010;78:1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell Biochem 2010;53:241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Hatano S, Yamada H et al.. Two types of interleukin 17A-producing gammadelta T cells in protection against pulmonary infection with Klebsiella pneumoniae. J Infect Dis 2016;214:1752–61. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK et al.. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014;41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017;41:252–75. [DOI] [PubMed] [Google Scholar]
- Ni Cheallaigh C, Sheedy FJ, Harris J et al.. A common variant in the adaptor mal regulates interferon gamma signaling. Immunity 2016;44:368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003;67:593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 2006;8:11–26. [PubMed] [Google Scholar]
- Ofek I, Mesika A, Kalina M et al.. Surfactant protein D enhances phagocytosis and killing of unencapsulated phase variants of Klebsiella pneumoniae. Infect Immun 2001;69:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama H, Asai A, Ito I et al.. M2b macrophage elimination and improved resistance of mice with chronic alcohol consumption to opportunistic infections. Am J Pathol 2015;185:420–31. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007;7:353–64. [DOI] [PubMed] [Google Scholar]
- Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 2016;80:629–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla E, Llobet E, Domenech-Sanchez A et al.. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 2010;54:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V et al.. HIF transcription factors, inflammation, and immunity. Immunity 2014;41:518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LD, Skaar EP. Transition metals and virulence in bacteria. Annu Rev Genet 2016;50:67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity 2013;38:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YJ, Lin TL, Hsu CR et al.. Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae. Infect Immun 2011;79:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015;517:311–20. [DOI] [PubMed] [Google Scholar]
- Pichavant M, Delneste Y, Jeannin P et al.. Outer membrane protein A from Klebsiella pneumoniae activates bronchial epithelial cells: implication in neutrophil recruitment. J Immunol 2003;171:6697–705. [DOI] [PubMed] [Google Scholar]
- Podschun R, Pietsch S, Holler C et al.. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbiol 2001;67:3325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998;11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG et al.. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014;14:166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro V, Campos MA, Pons J et al.. The uptake of a Klebsiella pneumoniae capsule polysaccharide mutant triggers an inflammatory response by human airway epithelial cells. Microbiology 2006;152:555–66. [DOI] [PubMed] [Google Scholar]
- Regueiro V, Moranta D, Campos MA et al.. Klebsiella pneumoniae increases the levels of Toll-like receptors 2 and 4 in human airway epithelial cells. Infect Immun 2009;77:714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro V, Moranta D, Frank CG et al.. Klebsiella pneumoniae subverts the activation of inflammatory responses in a NOD1-dependent manner. Cell Microbiol 2011;13:135–53. [DOI] [PubMed] [Google Scholar]
- Rehli M. Of mice and men: species variations of Toll-like receptor expression. Trends Immunol 2002;23:375–8. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K et al.. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010;11:785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Reis ES, Mastellos DC et al.. Complement component C3 - The “Swiss Army Knife” of innate immunity and host defense. Immunol Rev 2016;274:33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum R, Khan E, Gonzalez G et al.. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents 2011;38:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo TA, Olson R, Macdonald U et al.. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 2014;82:2356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo TA, Shon AS, Beanan JM et al.. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 2011;6:e26734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly H, Keisari Y, Ofek I. Manno(rhamno)biose-containing capsular polysaccharides of Klebsiella pneumoniae enhance opsono-stimulation of human polymorphonuclear leukocytes. J Innate Immune 2009;1:136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly H, Podschun R, Oelschlaeger TA et al.. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immune 2000;68:6744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale L, Smith MR, Wood WB. Studies on the Mechanism of Recovery in Pneumonia due to Friedlander's Bacillus: Ii. the Effect of Sulfonamide Chemotherapy upon the Pulmonary Lesion of Experimental Friedlander's Bacillus Pneumonia. J Exp Med 1947;86: 249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale L, Wood WB. Studies on the mechanism of recovery in pneumonia due to Friedlander's Bacillus: I. the pathogenesis of experimental Friedlander's Bacillus pneumonia. J Exp Med 1947;86: 239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo RJ, Domenico P, Tomas JM et al.. Salicylate-enhanced exposure ofKlebsiella pneumoniae subcapsular components. Infection 1995;23:371–7. [DOI] [PubMed] [Google Scholar]
- Sano H, Kuroki Y. The lung collectins, SP-A and SP-D, modulate pulmonary innate immunity. Mol Immunol 2005;42:279–87. [DOI] [PubMed] [Google Scholar]
- Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 2016;16:566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj SK, Prasadarao NV. Escherichia coli K1 inhibits proinflammatory cytokine induction in monocytes by preventing NF-kappaB activation. J Leukoc Biol 2005;78:544–54. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y et al.. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014;514:187–92. [DOI] [PubMed] [Google Scholar]
- Shin SY, Bae IK, Kim J et al.. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol 2012;61:239–45. [DOI] [PubMed] [Google Scholar]
- Siu LK, Yeh KM, Lin JC et al.. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 2012;12:881–7. [DOI] [PubMed] [Google Scholar]
- Soulas C, Baussant T, Aubry JP et al.. Outer membrane protein A (OmpA) binds to and activates human macrophages. J Immunol 2000;165:2335–40. [DOI] [PubMed] [Google Scholar]
- Standiford LR, Standiford TJ, Newstead MJ et al.. TLR4-dependent GM-CSF protects against lung injury in Gram-negative bacterial pneumonia. Am J Physiol Lung Cell Mol Physiol 2012;302:L447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieter RM, Standiford TJ, Huffnagle GB et al.. “The good, the bad, and the ugly.” The role of chemokines in models of human disease. J Immunol 1996;156:3583–6. [PubMed] [Google Scholar]
- Sun J, Li N, Oh KS et al.. Comprehensive RNAi-based screening of human and mouse TLR pathways identifies species-specific preferences in signaling protein use. Sci Signal 2016;9:ra3-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol 2008;8:501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805–20. [DOI] [PubMed] [Google Scholar]
- Tomas A, Lery L, Regueiro V et al.. Functional genomic screen identifies Klebsiella pneumoniae factors implicated in blocking nuclear factor kappaB (NF-kappaB) signaling. J Biol Chem 2015;290:16678–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca V, Mostowy S. Zebrafish infection: From pathogenesis to cell biology. Trends Cell Biol 2018;28:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016;7:214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Strieter RM, Wilkowski JM et al.. Lung-specific transgenic expression of KC enhances resistance to Klebsiella pneumoniae in mice. J Immunol 1998;161:2435–40. [PubMed] [Google Scholar]
- Tsai WC, Strieter RM, Zisman DA et al.. Nitric oxide is required for effective innate immunity against Klebsiella pneumoniae. Infect Immun 1997;65:1870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YK, Fung CP, Lin JC et al.. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 2011;55:1485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto Y, Asai A, Tsuda Y et al.. M2b monocytes provoke bacterial pneumonia and gut bacteria-associated sepsis in alcoholics. J Immunol 2015;195:5169–77. [DOI] [PubMed] [Google Scholar]
- Tu YC, Lu MC, Chiang MK et al.. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 2009;77:2657–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer AJ, Achouiti A, van der Ende A et al.. Toll-like receptor 9 enhances bacterial clearance and limits lung consolidation in murine pneumonia caused by methicillin-resistant staphylococcus aureus. Mol Med 2016;22:292–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elssen CH, Vanderlocht J, Frings PW et al.. Klebsiella pneumoniae-triggered DC recruit human NK cells in a CCR5-dependent manner leading to increased CCL19-responsiveness and activation of NK cells. Eur J Immunol 2010;40:3138–49. [DOI] [PubMed] [Google Scholar]
- van Lieshout MH, Anas AA, Florquin S et al.. Hematopoietic but not endothelial cell MyD88 contributes to host defense during gram-negative pneumonia derived sepsis. PLoS Pathog 2014;10:e1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lieshout MH, Blok DC, Wieland CW et al.. Differential roles of MyD88 and TRIF in hematopoietic and resident cells during murine gram-negative pneumonia. J Infect Dis 2012;206:1415–23. [DOI] [PubMed] [Google Scholar]
- van Lieshout MH, Florquin S, Van’t Veer C et al.. TIR-Domain-Containing Adaptor-Inducing Interferon-beta (TRIF) Mediates Antibacterial Defense during Gram-Negative Pneumonia by Inducing Interferon-gamma. J Innate Immun 2015;7:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand ME, McCowen JW, Nugent PG et al.. Complex interactions of Klebsiella pneumoniae with the host immune system in a Galleria mellonella infection model. J Med Microbiol 2013;62:1790–8. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen H, Zhang Y et al.. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: Role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents 2015;45:635–40. [DOI] [PubMed] [Google Scholar]
- Weston N, Sharma P, Ricci V et al.. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol 2017;169:425–31. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 2015;16:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland CW, van Lieshout MH, Hoogendijk AJ et al.. Host defence during Klebsiella pneumonia relies on haematopoietic-expressed Toll-like receptors 4 and 2. Eur Respir J 2011;37:848–57. [DOI] [PubMed] [Google Scholar]
- Willingham SB, Allen IC, Bergstralh DT et al.. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol 2009;183:2008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey GG, Ventrone S, Schutz KC et al.. Pulmonary surfactant promotes virulence gene expression and biofilm formation in Klebsiella pneumoniae. Infect Immun 2018;86:e00135–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojda I. Immunity of the greater wax moth Galleria mellonella. Insect Sci 2017;24:342–57. [DOI] [PubMed] [Google Scholar]
- Wooldridge KG, Williams PH. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev 1993;12:325–48. [DOI] [PubMed] [Google Scholar]
- Wu JH, Hong LC, Tsai YY et al.. Mitogen-activated protein kinase (MAPK) signalling pathways in HepG2 cells infected with a virulent strain of Klebsiella pneumoniae. Cell Microbiol 2006;8:1467–74. [DOI] [PubMed] [Google Scholar]
- Xiong H, Carter RA, Leiner IM et al.. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 2015;83:3418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Keith JW, Samilo DW et al.. Innate lymphocyte/Ly6C(hi) monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell 2016;165:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Steere RR, Fedorchuk CA et al.. Activation of epidermal growth factor receptor is required for NTHi-induced NF-kappaB-dependent inflammation. PLoS One 2011;6:e28216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weiss ID, Zhang HH et al.. Conventional NK cells can produce IL-22 and promote host defense in Klebsiella pneumoniae pneumonia. J Immunol 2014;192:1778–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FL, Yang YL, Liao PC et al.. Structure and immunological characterization of the capsular polysaccharide of a pyrogenic liver abscess caused by Klebsiella pneumoniae: activation of macrophages through Toll-like receptor 4. J Biol Chem 2011;286:21041–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Qin S, Chen S et al.. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 2018;18:25–3099(17)30628-X. [DOI] [PubMed] [Google Scholar]
- Ye P, Garvey PB, Zhang P et al.. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am J Respir Cell Mol Biol 2001a;25:335–40. [DOI] [PubMed] [Google Scholar]
- Ye P, Rodriguez FH, Kanaly S et al.. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med 2001b;194:519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Matsumoto T, Tateda K et al.. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J Med Microbiol 2000;49:1003–10. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Matsumoto T, Tateda K et al.. Induction of interleukin-10 and down-regulation of cytokine production by Klebsiella pneumoniae capsule in mice with pulmonary infection. J Med Microbiol 2001a;50:456–61. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Matsumoto T, Tateda K et al.. Protection against pulmonary infection with Klebsiella pneumoniae in mice by interferon-gamma through activation of phagocytic cells and stimulation of production of other cytokines. J Med Microbiol 2001b;50:959–64. [DOI] [PubMed] [Google Scholar]
- Yu XJ, McGourty K, Liu M et al.. pH sensing by intracellular Salmonella induces effector translocation. Science 2010;328:1040–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Moore TA, Newstead MW et al.. Interferon-inducible protein 10, but not monokine induced by gamma interferon, promotes protective type 1 immunity in murine Klebsiella pneumoniae pneumonia. Infect Immun 2005a;73:8226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Moore TA, Newstead MW et al.. IP-10 mediates selective mononuclear cell accumulation and activation in response to intrapulmonary transgenic expression and during adenovirus-induced pulmonary inflammation. J Interferon Cytokine Res 2005b;25:103–12. [DOI] [PubMed] [Google Scholar]
- Zeng X, Moore TA, Newstead MW et al.. Intrapulmonary expression of macrophage inflammatory protein 1alpha (CCL3) induces neutrophil and NK cell accumulation and stimulates innate immunity in murine bacterial pneumonia. Infect Immun 2003;71:1306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li H, Wang J et al.. Role of EGFR transactivation in preventing apoptosis in Pseudomonas aeruginosa-infected human corneal epithelial cells. Invest Ophthalmol Vis Sci 2004;45:2569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y et al.. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010;464:104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zeng J, Liu W et al.. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015;71:553–60. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao C, Wang Q et al.. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: Geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother 2016;60:6115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J et al.. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011;477:596–600. [DOI] [PubMed] [Google Scholar]
- Zheng M, Horne W, McAleer JP et al.. Therapeutic role of interleukin 22 in experimental Intra-abdominal Klebsiella pneumoniae infection in mice. Infect Immun 2016;84:782–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Rao M, Wallis RS et al.. Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect Dis 2016;16:e47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]