Abstract
Synthetic phosphorothioate (PT) internucleotide linkages, in which a nonbridging oxygen is replaced by a sulphur atom, share similar physical and chemical properties with phosphodiesters but confer enhanced nuclease tolerance on DNA/RNA, making PTs a valuable biochemical and pharmacological tool. Interestingly, PT modification was recently found to occur naturally in bacteria in a sequence-selective and RP configuration-specific manner. This oxygen–sulphur swap is catalysed by the gene products of dndABCDE, which constitute a defence barrier with DndFGH in some bacterial strains that can distinguish and attack non-PT-modified foreign DNA, resembling DNA methylation-based restriction-modification (R-M) systems. Despite their similar defensive mechanisms, PT- and methylation-based R-M systems have evolved to target different consensus contexts in the host cell because when they share the same recognition sequences, the protective function of each can be impeded. The redox and nucleophilic properties of PT sulphur render PT modification a versatile player in the maintenance of cellular redox homeostasis, epigenetic regulation and environmental fitness. The widespread presence of dnd systems is considered a consequence of extensive horizontal gene transfer, whereas the lability of PT during oxidative stress and the susceptibility of PT to PT-dependent endonucleases provide possible explanations for the ubiquitous but sporadic distribution of PT modification in the bacterial world.
Keywords: DNA modification, DNA phosphorothioate modification, restriction modification, defence system, epigenetics, environmental fitness
DNA phosphorothioation is a sulphur-containing physiological modification on the DNA backbone with multiple functions.
INTRODUCTION
Naturally occurring epigenetic modifications have been found in the DNA of organisms and viruses from all domains of life. The variations in canonical A, T, C and G are attributable to the appending of a variety of chemical groups, ranging from methyl groups to polyamines, amino acids, saccharides, etc., to the nucleobase moiety of a nucleotide (Weigele and Raleigh 2016). These DNA structural changes do not interfere with the specificity of base-pairing but confer additional functions such as protection and genetic regulation. The diversity of DNA structural alteration surprises us once again with the discovery of DNA phosphorothioation, a sulphur-containing DNA modification to the sugar-phosphate backbone rather than to the nucleobase (Wang et al.2007).
In the 1980 s, Zhou et al. constantly encountered difficulties in obtaining good-quality DNA from the soil-dwelling microorganism Streptomyces lividans 1326 because its DNA degrades in agarose gels during electrophoresis (Zhou et al.1988). Although DNA degradation, generally caused by nuclease contamination or improper sample manipulation, is common in the laboratory, Zhou and others demonstrated that this DNA smear was attributable to the occurrence of an ‘unusual’ DNA modification in S. lividans 1326: this modification was distinct from known modifications such as methylation because it rendered DNA susceptible to a nucleolytic species accumulated in electrophoretic buffer (Zhou et al.1988). Ray et al. later identified the nucleolytic species as a peracid derivative of Tris that is generated at the anode during electrophoresis and mediates oxidative DNA strand scission specifically at the ‘unusual’ modification sites (Ray, Weaden and Dyson 1992; Ray, Mills and Dyson 1995). Therefore, if the Tris-acetate-ethylenediaminetetraacetic acid (EDTA) electrophoretic buffer is replaced with HEPES-acetate-EDTA or supplemented with a 5 μM or greater concentration of thiourea, a radical scavenger and reducing agent, non-degradative electrophoresis of S. lividans DNA can be achieved (Ray, Weaden and Dyson 1992; Ray, Mills and Dyson 1995). This susceptibility to a Tris derivative differentiates the ‘unusual’ DNA modification from the well-characterised types of DNA modification such as methylation and hydroxymethylation, pointing to an unprecedented DNA modification that is awaiting exploration.
However, the low abundance of this modified species and limited information made the chemical nature of this ‘unusual’ modification a mystery for nearly twenty years. In 2005, Zhou et al. determined that this ‘unusual’ DNA modification is conferred by gene products of the dndABCDE cluster in S. lividans 1326; deletion of the dnd cluster completely abolishes DNA degradation, even in the presence of Tris-containing electrophoretic buffer (Zhou et al.2005; Xu et al.2009). The association of the putative functions of Dnd proteins with sulphur metabolism, e.g. DndA and DndC proteins are homologous to cysteine desulphurase and 3΄-phosphoadenosine-5΄-phosphosulphate (PAPS) reductase, respectively, promoted Zhou et al. to perform 35S feeding experiments. Surprisingly, sulphur, one of the critical elements in proteins, is detected in DNA isolated from strains harbouring dnd clusters, including S. lividans, Streptomyces avermitilis and Pseudomonas fluorescens pf0-1. In sharp contrast, no trace of 35S was observed in the DNA of ▵dndA or ▵dndD mutants of S. lividans (Zhou et al.2005).
In 2007, Wang et al. coupled the high-performance liquid chromatography (HPLC) separation of 35S-containing DNA hydrolysates with mass spectrometric analysis to unravel the chemical nature of this sulphur-containing DNA modification (Wang et al.2007). Unlike methylation, this DNA modification occurs as phosphorothioation, in which a nonbridging oxygen in the DNA sugar-phosphate backbone is replaced by sulphur. This sulphur-for-oxygen substitution creates a chiral centre at phosphorus, leading to the formation of two chiral isomers, RP and SP (Eckstein 1985). All identified physiological phosphorothioate (PT) internucleotide linkages occur specifically in the RP configuration (Wang et al.2011). Owing to their nuclease tolerance, PT-linked dinucleotides are generated alongside canonical mononucleotides upon enzymatic hydrolysis. Therefore, PT modification is detected in the form of d(GPSG) dinucleotides (PS: P-S linkage) in S. lividans 1326 and P. fluorescens pf0-1 by liquid chromatography-mass spectrometry (LC-MS; Wang et al.2007, 2011).
Prior to the identification of physiological PT modifications, oligonucleotides bearing PT bonds had been chemically synthesised for decades (Eckstein 1985). As a phosphodiester analogue, the PT bond possesses similar physical and chemical properties but is resistant to nuclease digestion, prompting the consideration of this functional group for biochemical research and clinical applications (Gish and Eckstein 1988; Stein 1996; Dias and Stein 2002; Volk and Lokesh 2017). Take the antisense drug fomivirsen as an example: it is a 21-base synthetic oligonucleotide complementary to mRNA of the major immediate early region 2 of human cytomegalovirus and thus inhibits the replication of the virus. Replacing all the phosphodiesters in fomivirsen with PT analogues results in a prolonged half-life of approximately 55 h in humans (de Smet, Meenken and van den Horn 1999; Geary, Henry and Grillone 2002), whereas the half-life of an unmodified oligonucleotide is on the order of minutes (Monia et al.1996). When the chemical nature of the physiological PT modification was determined, two key questions immediately emerged: (i) what are the characteristics of PT modification in bacterial genomes, and (ii) what is its biological significance to bacterial hosts? These two questions have become the focus of this field and prompted a series of deeper investigations of PT physiology.
GENETIC AND ENZYMATIC STUDIES ON DND SYSTEMS
DndABCDE: DNA PT modification enzymes
In addition to their appearance in Streptomyces and Pseudomonas, PT-mediated DNA degradation phenomenon and dndABCDE homologues have been later observed in diverse bacterial genera such as Escherichia, Salmonella, Mycobacterium, Clostridium, Flavobacterium and Vibrio (Zhou et al.2005; Wang et al.2011; Barbier et al.2013; Ahlgren et al.2017). Typically, dndBCDE constitute a single operon, while dndA is either located adjacent to dndBCDE or is scattered elsewhere in different genomes (Fig. 1; Xu et al.2009; Tong et al.2018). DndA shares significant sequence identity and structural similarity with the well-characterised pyridoxal 5΄-phosphate (PLP)-dependent cysteine desulphurases IscS and NifS, which is consistent with the observation that IscS can functionally substitute for DndA to collaborate with DndBCDE in generating DNA PT modification in some bacterial strains (You et al.2007; An et al.2012). By using L-cysteine as a substrate, cysteine desulphurases catalyse the generation of L-alanine and sulphane sulphur via the formation of a protein-bound cysteine persulphide intermediate (R-S-SH; Fig. 1); the persulphide sulphur serves as the sulphur donor and is subsequently incorporated into many sulphur-containing biomolecules such as transfer RNA thionucleotides, biotin, thiamine and iron–sulphur (Fe-S) clusters (Mihara and Esaki 2002; Mueller 2006). For instance, IscS and ThiI proteins are sufficient for the biosynthesis of 4-thiouridine (s4U) at position 8 in tRNA (Lauhon and Kambampati 2000; Mueller, Palenchar and Buck 2001). IscS first forms a persulphide group on an active cysteine residue, and the terminal sulphur is transferred to the tRNA-binding protein ThiI (Kambampati and Lauhon 2000). ThiI probably plays two mixed functions in the following tRNA thiolation: (i) ThiI catalyses the adenylation of uridine-8 at the expense of ATP and (ii) ThiI persulphide performs a nucleophilic attack on the activated uridine-8 to generate s4U (Mueller, Palenchar and Buck 2001; You et al.2008). X-ray crystallographic characterisation of the DndA from Streptomyces lividans revealed structural similarities to the cysteine desulphurases IscS, NifS, CsdB and SufS (Fujii et al.2000; Kaiser et al.2000; Cupp-Vickery, Urbina and Vickery 2003; Tirupati et al.2004; Chen et al.2012). The structure of DndA shows a deep surface pocket recognizing its cofactor PLP, which forms a Schiff base covalent link with the amino group of a highly conserved lysine residue (Lys200). The catalytic cysteine residue (Cys327), located in the consensus motif ATGSACTS on a β strand, is far away from the substrate, suggesting that a conformational change occurs in DndA during catalysis (Chen et al.2012).
Figure 1.
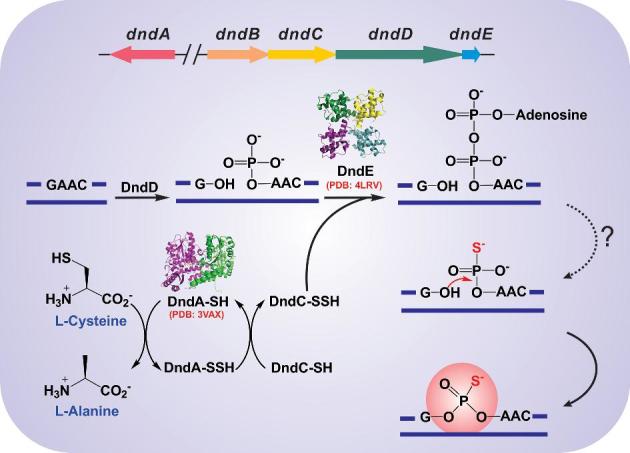
Proposed biochemical pathway of DNA PT modification. PT modification, conferred by DndACDE proteins, is predicted to start with the desulphurisation of cysteine by DndA. The sulphur atom is later transferred to the target DNA site, cleaved and activated, likely by DndD and DndC, to complete the sulphur incorporation.
DndC has sequence homology to PAPS reductase and exhibits ATP pyrophosphatase activity in vitro (Zhou et al.2005; You et al.2007). The [4Fe-4S] iron–sulphur centre of DndC, which is assembled by DndA in the presence of L-cysteine, is essential for its ATP pyrophosphatase activity, with a possible persulphide group formed between DndA and DndC (Fig. 1). DndC also shares an adenylation-specific P-loop motif, SGGKDS, with the PPi synthetase family, and this motif is critical for the adenylation of uridine-8 in the formation of tRNA s4U (Mueller and Palenchar 1999; You et al.2007). We propose that the DNA site to be PT modified might be activated by adenylation prior to the nucleophilic attack by DndC persulphide to generate site-specific DNA phosphorothioation, thus mimicking the functions of IscS and ThiI during tRNA modification of s4U.
DndD in Pseudomonas fluorescens pf0-1 shows ATPase activity with a conserved ATP-binding Walker A motif, A/GX4GKT/S (Yao et al.2009). DndD is predicted to provide the energy for stabilizing DNA secondary structure or for site-specific DNA nicking during sulphur incorporation (Fig. 1). DndE is the smallest Dnd protein, the length of which is generally no more than 141 residues. The DndE structure shows a tetramer conformer adopting a quadrate fold that resembles a four-leaf clover (Hu et al.2012). Two clefts (∼60° and ∼90°) in the surface are formed by the dimers, and the cleft-containing side is more hydrophobic than the opposite side. DndE displays preferential binding for nicked double-stranded DNA substrates in a sequence-independent manner (Fig. 1; Chen et al.2011; Hu et al.2012). Six positively charged lysine residues on the DndE surface are believed to mediate the binding affinity for negatively charged phosphorylated nicking sites (Hu et al.2012; Lai et al.2014). DndEi, an extended version of DndE, was recently observed in several bacterial strains, e.g. Riemerella anatipestifer strain ATCC 11845, Flavobacterium indicum CIP 109464T and Kordia algicida OT1, and harbours another domain in addition to the canonical DndE part (Barbier et al.2013; Zheng et al.2016). This additional domain was shown to possess DNA helicase and ATPase activity. Inactivating the DNA helicase activity abolished DNA PT modification in R. anatipestifer, indicating that DNA strands unwind during sulphur incorporation. The ATPase activity of DndEi is strongly stimulated by DNA substrates harbouring 5΄-GAAC-3΄/5΄-GTTC-3΄, which is the recognition site for PT modification in R. anatipestifer (Zheng et al.2016).
In sharp contrast to mutations in other dnd genes, the mutation of dndB aggravates DNA degradation and results in a two-fold increase in PT frequencies in Salmonella enterica serovar Cerro 87 without changing the specificity (He et al.2015). Based on a DNAase I footprinting assay, DndB was found to bind directly to the promoter region of the dndBCDE operon to regulate the transcription of dndCDE and itself, consequently affecting the PT frequency in the genome (Fig. 2; Cao et al.2014a; He et al.2015). Under physiological conditions, the dnd operon is still transcribed, even in the presence of DndB; it is thus likely that the control of dnd transcription and the resulting PT modification is a response of DndB to certain as yet unknown environmental or cellular cues.
Figure 2.
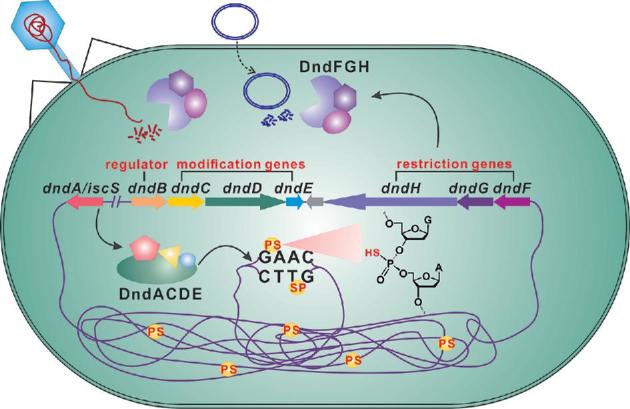
DNA PT modification-based R-M defence mechanism. PT-based R-M systems recognise the PT status of invading foreign DNA, e.g. phage genomes and plasmids. PT modification of the DNA backbone is performed by DndA, DndC, DndD and DndE, which form a large protein complex with equimolar stoichiometry in vitro. DndB functions as a negative regulator controlling the expression of dndCDE and itself via its affinity to their upstream promoter regions. DndFGH act as a cognate restriction enzyme that recognises foreign DNA lacking PT modification in consensus sequences such as the PT-modified 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ in E. coli B7A and cleaves double-stranded DNA.
In vitro co-purification experiments show that IscS, DndC, DndD and DndE form a large protein complex at equimolar ratios (Xiong et al.2015). Furthermore, DndA-C, C-D, D-E and C-D-E formed subcomplexes. Since DndE has been found to exist as a tetramer in vitro, it currently remains uncertain whether the equimolar stoichiometry changes during the dynamic process of PT modification. Functional and structural studies of key enzymes have provided insights into how enzymes cooperate with each other during the modification process (You et al.2007; Yao et al.2009; Chen et al.2012; Hu et al.2012; Lai et al.2014; Cheng et al.2015; He et al.2015). In light of the accumulated functional and structural data, a rough picture could be drawn to outline the DNA PT modification pathway. In one branch, L-cysteine is used as a substrate to generate a persulphide group on the active site cysteine residue of DndA. The terminal sulphur is next transferred to DndC to make a new persulphide group. In the other branch, once Dnd protein(s) binds the DNA consensus sequence, the target phosphodiester is likely nicked by DndD in the presence of ATP, and the nicked DNA strands are recognised and stabilised by DndE. The nicking sites might then be activated by adenylation by DndC, followed by nucleophilic attack by the persulphide group of DndC to generate a PT linkage. An intermediate of the modification process would be crucial to unveiling the biochemical pathway to this complicated modification. Multiple approaches should be exploited to unveil the role of Dnd proteins in PT modification; it is an attractive idea to characterise the Dnd protein complexes alone or with DNA substrates by crystallographic or cryo-electron microscopy analysis in the future. This structural analysis would provide insights into how the Dnd proteins cooperate with each other to incorporate sulphur into DNA in a sequence-selective and RP configuration-specific manner.
DndFGH: DNA PT modification-based restriction enzymes
Another three-gene cluster, dndFGH, was later identified in the neighbourhood of a PT modification gene cluster (Xu et al.2010; Fig. 2). In the presence of DndFGH, the efficiency of transforming a non-PT-containing plasmid into S. enterica serovar Cerro 87, possessing dndBCDE-dndFGH, is significantly lower than that of transforming a PT-containing plasmid (Xu et al.2010). Without PT protection, S. enterica serovar Cerro 87 suffers double-stranded DNA cleavage by DndFGH, which consequently triggers a cellular SOS response and prophage induction (Cao et al.2014a; He et al.2015). To date, there is no in vitro assay for the DndFGH DNA binding or cleavage. However, these features resemble the best-characterised ‘self-nonself discrimination’ mechanism of methylation-based restriction-modification (R-M) systems. R-M systems, which use two enzymatic activities, a methyltransferase (MTase) and a restriction endonuclease (REase), are considered primitive immune systems in prokaryotes and provide 102- to 108-fold protection for the hosting cell against bacteriophages or other invading DNA (Vasu, Nagamalleswari and Nagaraja 2012). The REases recognise and attack invading foreign DNA at a specific recognition sequence, whereas the MTases transfer a methyl group to the same DNA sequence within the host genome to protect self-DNA from cleavage by REases. This similar defensive mechanism makes DndABCDE-DndFGH a new member of the bacterial innate immune systems: DndABCDE function as the MTase to confer DNA PT modification in a sequence-selective manner, while DndFGH act as the cognate REase to recognise and destroy invading non-PT-modified foreign DNA for protection (Fig. 2).
Methylation-based R-M systems are classified into four major groups (type I–type IV) based on their architecture, sequence recognition, cofactor requirement and cleavage mechanism (Roberts et al.2003). In type I, II and III R-M systems, the host DNA is methylated within a specific sequence by the MTase for protection, while the cognate REase distinguishes and attacks unmethylated invading DNA. The type I R-M systems are the most complicated, consisting of three subunits (R (restriction), M (modification) and S (specificity)) that function as a holoenzyme requiring S-adenosyl-L-methionine (AdoMet), ATP and magnesium (Mg2+; Wilson and Murray 1991). Both type II and type III R-M systems consist of only two proteins, an REase and an MTase, though some type II systems also have regulatory proteins (Mruk and Kobayashi 2014). In type II R-M systems, the REase and MTase generally act independently; the MTase requires only AdoMet, and the restriction activity requires Mg2+ but not ATP (Roberts et al.2010). In type III R-M systems, the MTase can function independently to methylate target sequences, but the REase needs to pair with the MTase to accomplish restriction in the presence of Mg2+ and ATP (Humbelin et al.1988; Meisel et al.1995). In contrast, the type IV R-M systems cleave only DNA substrates with modifications such as methylation, hydroxymethylation or glucosyl-hydroxymethylation at specific sequences (Sutherland, Coe and Raleigh 1992; Bair and Black, 2007; Xu et al.2011). Considering that DndA, DndC, DndD and DndE function as a large complex and require ATP, DNA PT R-M systems are more similar to type I R-M systems. However, the Dnd system is far more complicated than all types of methylation-based R-M systems because it involves four proteins to confer DNA PT modification, one protein to regulate the PT level, and three proteins to recognise and restrict foreign non-PT DNA.
THE FEATURES OF PT MODIFICATION IN BACTERIAL GENOMES
The consensus sequences of DNA PT modification
Enzymatic DNA hydrolysis yields PT-linked dinucleotides in addition to canonical monodeoxynucleotides (Fig. 2), enabling the sequence identification and frequency quantitation of PT modifications by liquid chromatography-coupled tandem quadrupole mass spectrometry (LC-MS/MS; Wang et al.2011). Significant diversity in the sequence selectivity and quantity of PT modifications in different bacteria was immediately observed. For example, PT modifications occur between two guanines, denoted d(GPSG), in Streptomyces lividans and Pseudomonas fluorescens pf0-1; as d(GPSA) in Hahella chejuensis KCTC2396; and as d(GPSA) and d(GPST) at a 1:1 ratio in Escherichia coli B7A and Salmonella enterica serovar Cerro 87. Genomic PT modifications in these strains were quantified to occur at a frequency of 3–4 PT per 104 nt, suggesting a 5∼6-nt consensus motif (∼1 modification every 1024–4096 nt). Another, higher frequency of 22–31 PT per 104 nt was detected in a group of Vibrio isolates as d(CPSC), which is consistent with a 4 nt consensus sequence (∼1 modification every 256 nt; Wang et al.2011).
Mass spectrometry methods, however, require the decomposition of DNA to nucleotides, necessitating alternative means to explore long consensus sequences and genome-wide PT distribution. Three distinct approaches, primer extension assays, cloning and single-molecule real-time (SMRT) sequencing, were exploited by different groups to extend the understanding of PT features. The first two were based on the Tris-mediated DNA scission reaction because the chemical nature of PT was not known then and the ‘unusual’ modification was believed to be a base modification. Dyson et al. first inserted a 160-bp DNA fragment possessing a preferred modification site into the Streptomyces–E. coli shuttle vector pUCS75 (Dyson and Evans 1998). After the resultant plasmid was passaged through S. lividans to be modified, it was treated with the Tris peracid derivative and subsequent primer extension assays to locate the cleavage sites. The preferred modification site was believed to occur at guanine residues on both strands within a 6 bp 5΄-CGGCCG-3΄/5΄-CGGCCG-3΄ palindrome (the modified G is underlined; Dyson and Evans 1998). In comparison, single-stranded plasmid replication intermediates are unsusceptible to Tris-mediated strand scission, providing evidence for a post-replicative modification mechanism (Dyson and Evans 1998). The modification consensus contexts were further investigated by locating 14 modifying sites via the cloning and sequencing of DNA fragments resulting from the Tris-dependent cleavage of pHZ209, a pIJ101-derived plasmid isolated from S. lividans. In agreement with Dyson's observation, the PT recognition sites were positioned in a highly conserved core sequence, 5΄-GGCC-3΄/5΄-GGCC-3΄ (Liang et al.2007). After the realisation that the PT modification occurs in the d(GPSG) backbone instead of on a G base in S. lividans 1326, the modification consensus should have been updated to 5΄-GPSGCC-3΄/5΄-GPSGCC-3΄. However, this 4-bp 5΄-GPSGCC-3΄/5΄-GPSGCC-3΄ sequence is shorter than the 5∼6 nt-long consensus proposed based on the mass spectrometric calculation. Dyson and Liang et al. also observed that mutations in the sequence surrounding 5΄-GPSGCC-3΄/5΄-GPSGCC-3΄ sites influenced the occurrence of PT modifications to varying degrees, implying the requirement of a considerable flanking region or possible secondary structures.
The genomic distribution of DNA PT modifications
The emergence of SMRT sequencing enables the direct detection of PT modifications across bacterial genomes. SMRT sequencing is a sequencing-by-synthesis technology in which the progression of a single DNA polymerase molecule is monitored in real time, while individual base incorporation is catalysed using fluorescently tagged nucleotides (Flusberg et al.2010). The DNA polymerisation events, simultaneously taking place within thousands of arrayed zero-mode waveguides on a SMRT cell, can thus be recorded in the shape of a chain of fluorescent pulses (Flusberg et al.2010). The time that is taken to incorporate a single nucleotide (referred to as pulse width or PW) and the duration between the incorporation of two nucleotides (referred to as the inter-pulse duration or IPD) directly reflect DNA polymerase kinetics. The kinetic parameters are sensitive to many types of modified nucleotides in DNA templates, including N6-methyladenosine (6mA), 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) as well as damaged bases (8-oxoguanine, O6-methylguanine, 5-hydroxycytosine and thymine dimers, etc.), generating distinct kinetic signatures for each of them and enabling the identification of a wide range of epigenetic markers in nucleobases (Flusberg et al.2010; Clark et al.2011, 2012).
In consideration of the different characteristics between PT and methylation, Cao et al. first attempted to examine the influence of PT modification on DNA polymerase kinetics using synthetic oligonucleotides harbouring PT bonds. PT (RP)-induced IPD kinetic signals were captured, promoting the application of SMRT sequencing to mapping genomic PT sites in bacterial genomes (Cao et al.2014b). SMRT analysis showed that the d(GPSA) and d(GPST) formerly detected in E. coli B7A were located in 4-bp 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ motifs, which explains the 1:1 ratio of d(GPSA) and d(GPST) quantified by LC-MS/MS (Fig. 3A; Wang et al.2011; Cao et al.2014b). The d(CPSC) in Vibrio cyclitrophicus FF75, which occurs three times more frequently than total PT in E. coli B7A, was located in 3-bp 5΄-CPSCA-3΄ consensus contexts but not in 5΄-TGG-3΄ on the complementary strand, representing a single-stranded PT modification. Mapping the PT motifs on the two bacterial genomes did not reveal hotspot regions with enriched modifications. In total, 4855 PT-modified 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ sites were detected in the whole E. coli B7A genome, occurring with various spacings ranging from 0.1 to 10 kb; 4499 of 5384 open reading frames, 22 of 25 rRNA genes and 3 of 86 tRNA genes in E. coli B7A contained at least one PT site (Cao et al.2014b). The same situation holds for V. cyclitrophicus FF75 in that the 21778 detected 5΄-CPSCA-3΄ motifs were distributed sporadically throughout the genome. Surprisingly, sequence alignments revealed no strict sequence-context constraint beyond 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ or 5΄-CPSCA-3΄. As to the importance of the sequences surrounding a PT site observed by Dyson and Liang et al., a possible explanation is that these sites influence the effective interaction of Dnd proteins and recognition motifs rather than being part of the consensus sequences. This hypothesis is supported by the observation that the proximate bases of 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ or 5΄-CPSCA-3΄ still exhibit sequence biases (Cao et al.2014b). PT modifications exhibit strikingly low PT occurrence frequencies on 5΄-TGAAC(A/C)-3΄ and 5΄-CCAT-3΄ motifs in E. coli B7A and V. cyclitrophicus FF75, respectively.
Figure 3.
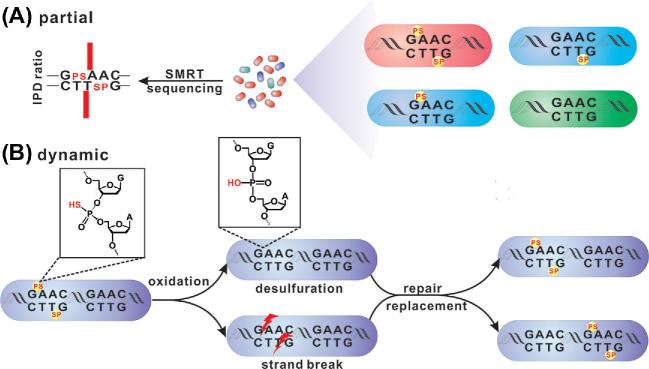
Partial and dynamic PT modification features. (A) In terms of a given 5΄-GAAC-3΄/5΄-GTTC-3΄ consensus in a population of cells, the kinetic signal of SMRT sequencing averaged over all molecules strongly represents 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄. However, single-molecule analysis reveals the presence of DNA molecules possessing 5΄-GAAC-3΄/5΄-GTTC-3΄, 5΄-GPSAAC-3΄/5΄-GTTC-3΄ and 5΄-GAAC-3΄/5΄-GPSTTC-3΄ sequences in addition to 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄, a result of partial modification. (B) Upon exposure to oxidants, e.g. HOCl, the PT backbone undergoes desulphuration, strand breakage and phosphonate formation. A repair/replacement mechanism is proposed to explain why the total PT level remains unchanged during oxidative stress even though PT-causing DNA damage is occurring; this mechanism provides an alternative explanation for the dynamic nature of PT modifications.
One possible mechanism for the exclusion of these sites from PT modification is that such sequences are poorly recognised by the DNA recognition Dnd protein, although its identity is not known, or by Dnd complexes. Another possibility is that the barely PT-modified motifs coincide with the DNA-binding consensus sequence of some regulatory proteins, i.e. TorR and RpoE. TorR is a response regulator of the E. coli torCAD operon that encodes the trimethylamine N-oxide reductase respiratory system under anaerobic conditions (Simon et al.1994). The binding consensus sequences of TorR, 5΄-CTGTTCATAT and 5΄-CCGTTCATCC-3΄, contain the PT recognition core sequence and especially the rarely modified 5΄-TGTTCA-3΄ motif. The rpoE gene encodes the σE heat shock sigma factor that recognises the –35 region that contains the conserved 5΄-GAACTT-3΄ binding consensus of the htrA promoter, the rpoH P3 promoter and its own P2 promoter to control gene expression (Raina, Missiakas and Georgopoulos 1995; Dai et al.2015). Both rpoH, encoding the major heat shock σ32 factor, and htrA, encoding a periplasmic endopeptidase, are essential for bacterial growth at high temperatures (Raina, Missiakas and Georgopoulos 1995). The ability of DNA PT modification to influence the interaction between RNA polymerase and promoters (discussed below) raises the possibility that Dnd modifying enzymes evolve to adopt recognition sites other than the binding consensus sites of the important regulatory proteins to avoid PT interference. It is also a plausible explanation that the occupation of the recognition consensus by the regulatory proteins blocks the binding of and modification by Dnd proteins.
PT is a partial and highly dynamic modification
Apart from the 4855 PT-modified 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ and 21778 5΄-CPSCA-3΄ sites, there are still 35846 and 138763 vacant 5΄-GAAC-3΄/5΄-GTTC-3΄ and 5΄-CCA-3΄ sites in the genomes of E. coli B7A and V. cyclitrophicus FF75, respectively. In addition, DNA PT modification does not occur consistently at a given site in a population of bacterial cells, i.e. there is partial or incomplete modification even in the presence of active DndFGH (Cao et al.2014b). For instance, regarding a selected genomic PT-modified 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ site in cells, single-molecule analysis of the SMRT sequencing data reveals the presence of some 5΄-GAAC-3΄/5΄-GTTC-3΄ molecules with no detectable PT kinetic signals. The reverse is also true; the kinetic signal of a given 5΄-GAAC-3΄/5΄-GTTC-3΄ site could be averaged over a mixture of molecules with 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄ and 5΄-GAAC-3΄/5΄-GTTC-3΄ (Fig. 3A). This partial modification suggests the heterogeneity of PT in the bacterial population, which is reminiscent of the ON/OFF phase variation of R-M systems. The Mod MTase associated with a type III R-M system in Haemophilus influenzae Rd is a well-studied example of phase variation; phase-variable MTase activity is mediated by a change in the number of tetranucleotide repeats within the MTase coding sequence resulting from slipped strand mispairing (Weiser, Williams and Moxon 1990; van Ham et al.1993). The mod gene can code for either a 72- or an 86-kDa MTase or can be switched to OFF with no MTase product due to mod repeat tract changes (Fox et al.2007; Atack et al.2018). The ON/OFF switching of methylation results in bacterial subpopulations with heterologous patterns of methylation; each methylation pattern has a different impact on global gene expression and generates bacteria in different physiological states (Fox et al.2007). Phase variation is thus regarded as a contingency mechanism for the adaptation of bacteria to fluctuating environments. However, no simple sequence repeats such as those in phase-variable R-M systems are observed within dnd genes or their promoter region, indicating a different phase variation mechanism if such variation occurs. Without repression by DndB, transcription of the dndBCDE operon is enhanced 15-fold, leading to a doubling of the PT level (He et al.2015). It is noteworthy that the newly generated PT modification still occurs only in 5΄-GPSAAC-3΄/5΄-GPSTTC-3΄, suggesting that the originally unmodified 5΄-GAAC-3΄/5΄-GTTC-3΄ sites in the genome are also substrates of Dnd modification enzymes, but are perhaps protected or have disfavoured flanking sequences. This result raises the possible scenario that the intracellular concentration of Dnd proteins allows only a certain amount of PT modification as long as physiological functions, such as the full protection against DndFGH, still occur independently of which 5΄-GAAC-3΄/5΄-GTTC-3΄ groups are selected.
This incomplete modification is one way in which standard R-M systems differ from the PT systems. In standard R-M systems, the R+-M− phenotype is generally lethal, with no need for additional stressors such as H2O2 (Kobayashi 1998). In contrast, in the absence of stressors, the R+-M− phenotype for at least one PT system is not lethal unless the R genes are overexpressed (Cao et al.2014a). In the absence of in vitro assays for DndFGH activity, it is hard to predict why R+-M− cells survive well, though one possible partial explanation is that the restriction activity requires the simultaneous binding of two unmodified sites, as with type IIE REases such as EcoRII (Takahashi et al.2008).
In later studies, Kellner et al. suggested a ‘PT turnover’ model, referring to PT loss and recovery based on coupling isotope labelling of PT with mass spectrometry (Fig. 3B; Kellner et al.2017). Escherichia coli B7A cells were first cultured in [15N]- and [34S]-containing nitrogen and sulphur sources. The resulting d(GPSA) and d(GPST) with [15N]-labelled base rings and a [34S]-labelled PT sulphur was regarded as the ‘parental’ PT. The cells were then transferred to [14N]/[32S]-containing medium, which could produce ‘new’ PT in the form of [14N]/[32S]-d(GPSA) and [14N]/[32S]-d(GPST). Indeed, after rounds of DNA replication, ‘new’ PT increased from being absent to replacing the heavy-isotope-labelled ‘parental’ d(GPSA) and d(GPST). The ‘turnover PT,’ possessing [15N]-labelled nucleobases like parental PT but an [32S]-labelled backbone like new PT, was detected at a frequency of 2% per hour (Kellner et al.2017). This observation demonstrated that PTs are highly dynamic modifications, which likely results from natural PT-linkage damage or desulphurisation mediated by endogenous oxidants (discussed below) and subsequent repair or replacement by bacterial DNA repair machineries or Dnd modification proteins, respectively (Fig. 3B).
THE PHYLOGENESIS OF DND SYSTEMS
Because the genetic organisation of dnd genes is highly conserved, Ou et al. developed a dndDB database (http://mml.sjtu.eu.cn/dndDB/) containing information about DNA degradation phenotypes, dnd gene clusters, Dnd proteins and conserved domains and other characteristics, facilitating the identification of dnd genes in a large number of taxonomically unrelated bacterial strains (Ou et al.2009). Metagenomic analysis of environmental samples, including those from the Sargasso Sea and Oregon coastal waters, revealed signature ecological PT distributions (Wang et al.2011). With regard to the occurrence of PT systems across large bacterial genomes, Tong et al. conducted a comparative genomic survey and found that only 734 out of 1349 dndBCDE clusters are accompanied by dndFGH as bona fide PT R-M pairs, while the rest (615, 45.6%) occur in the form of solitary dndBCDE clusters, denoted PT R−-M+, that lack the dndFGH restriction counterpart (Tong et al.2018). The overwhelming majority of PT R−-M+ systems raise two possibilities: either the solitary PT R−-M+ systems originate from an ancestor different from that of PT R-M systems or PT R−-M+ systems might result from gene loss (Albalat and Canestro 2016). However, no clear phylogenetic distinction between PT R-M and R−-M+ systems was observed based on DndBCD phylogenesis or significant DndFGH degradation intermediates (Tong et al.2018). These observations prompted Tong et al. to propose that even if gene loss from PT R-M to yield solitary PT R−-M+ occurs, it might not be a recent event. Nevertheless, the phylogenetic analysis shows apparent horizontal gene transfer events in both PT R-M and PT R−-M+ systems, which is consistent with the observation that dnd genes in Streptomyces lividans, Pseudomonas fluorescens pf0-1, Bacillus cereus E33L, Geobacter uraniumreducens Rf4, a group of Escherichia coli strains, etc., are harboured in diverse genomic islands (He et al.2007; Ho et al.2015). This result implies that horizontal gene transfer had a considerable impact on the distribution of dnd systems, explaining the widespread but sporadic occurrence of PT modification among bacteria.
PT R−-M+ is reminiscent of solitary DNA MTases, e.g. DNA adenine methylase (Dam) and DNA cytosine methylase (Dcm) of E. coli (Marinus and Lobner-Olesen 2014) and cell cycle-regulated methylase (CcrM) of Caulobacter crescentus (Gonzalez et al.2014), which are not associated with any restriction enzyme counterpart. In terms of the occurrence of solitary MTases, it is believed that when an entire R-M system or the MTase is disturbed, the DNA cleavage activity of REase becomes a lethal pressure that favours the selective mutation and ultimate loss of REase (Seshasayee, Singh and Krishna 2012). Given enough time, these solitary MTases have acquired new cellular functions such as the involvement of Dam in virulence and epigenetic control and the regulation of cell cycle progression by CcrM (Low, Weyand and Mahan 2001; Low and Casadesus, 2008; Kozdon et al.2013). These similar features may illuminate the occurrence of PT R-M and R−-M+ systems and indicate that PT modifications likely play a much larger role in supplementing defence mechanisms.
ADDITIONAL CELLULAR FUNCTIONS OF PT MODIFICATIONS
The responses of PT to oxidative stress
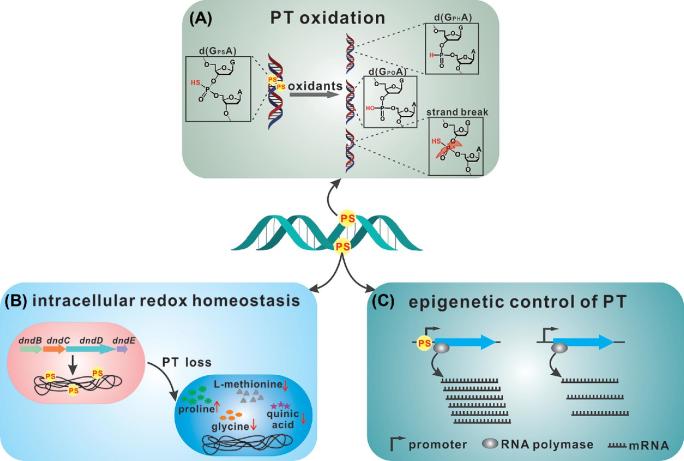
Considering the exceptional chemical nature of PT, several groups independently investigated the cellular roles of PT modification and found that PT is a versatile player involved in multiple cellular processes in addition to being a constituent of the PT R-M defence system. Xie et al. observed that the wild-type Salmonella enterica 87 showed a higher survival rate than PT-deficient mutants when exposed to hydrogen peroxide (H2O2; Xie et al.2012). Upon in vitro treatment with H2O2, PT consumed H2O2. During this reduction-oxidation reaction, d(GPSA) is oxidised, leading to a DNA strand break or the formation of a desulphurated d(GPOA) or a phosphonate-bridged d(GPHA; Fig. 4A; Xie et al.2012; Kellner et al.2017). Therefore, PT is regarded as a mild antioxidant in vivo. In addition, PT modification scavenges hydroxy radicals to protect genomic DNA and proteins against oxidative damage (Wu et al.2017). In addition to S. enterica Cerro 87 having a higher survival rate against H2O2 exposure than PT-deficient mutants, Yang et al. observed that PT modifications provide growth advantages to Escherichia coli and the extremophile Shewanella piezotolerans under multiple environmental stresses such as combinations of low temperature, low salinity level, high pressure, UV radiation, X-ray radiation and heavy-metal stresses (Yang et al.2017). As a plausible reaction mechanism of PT, Wu et al. proposed that PT is capable of donating an electron to hydroxy radicals, generating a PS• radical during the desulphurisation of PT to a normal phosphodiester. A PS• radical can instantly form a complex with OH− to release a highly reductive species, the hydrosulphide radical (HS•), which scavenges surrounding reactive oxygen species (ROS) such as H2O2 and O2−• to create a reductive atmosphere (Wu et al.2017).
Figure 4.
The functional diversity of DNA PT modification. DNA PT modification is a versatile player in environmental fitness (A), the maintenance of cellular redox homeostasis in addition to serving as a component of R-M defence barriers (B) and epigenetic regulation (C).
Similar to in vitro oxidation by H2O2, d(GPSA) also undergoes PT desulphurisation, d(GPHA) formation and DNA strand breaks after hypochlorous acid (HOCl) treatment (Kellner et al.2017). However, Kellner et al. observed that PT-modified S. enterica Cerro 87 was 4.8-fold more sensitive to HOCl than its ▵dndB-H mutant. In sharp contrast, under their experimental conditions, no growth difference was detected between S. enterica Cerro 87 and its ▵dndB-H mutant upon treatment with H2O2. This conflict resulted from the nonideal use of dnd deletion mutants in previous studies. Instead of using the ▵dndB-H mutant, Xie et al. employed individual dnd gene deletion mutants, i.e. ▵dndC, ▵dndD and ▵dndE, which have no PT modification but still possess the active restriction endonuclease DndFGH. The unconstrained DndFGH caused extensive DNA cleavage and thus led to heightened H2O2 susceptibility (Kellner et al.2017).
Because of the selective sensitivity to oxidation by HOCl, PT modifications compromise bacterial fitness as a result of lethal DNA breakage under commonly encountered chemical stresses. Environments offering HOCl exposure as well as exposure to other hypohalous acids (HOBr and HOI), which are potential PT oxidizing agents, are frequently encountered. For example, HOCl is produced and predominantly serves as a microbicide to defend against infectious agents during granulocyte activation (Prokopowicz et al.2010). Myeloperoxidase is capable of catalysing the production of HOCl, while eosinophil peroxidase utilises H2O2 to convert bromide to hypobromous acid (HOBr; Hampton, Kettle and Winterbourn 1998; Wang and Slungaard 2006). Such PT-mediated genomic instability under HOCl exposure could play a role in the virulence of PT-containing pathogens. Interestingly, no dnd systems have been found in Staphylococcus, which survives the microbicidal doses of HClO produced inside phagosomes (Chapman et al.2002). Currently, it is still uncertain whether there is a particular absence of dnd systems in pathogens such as Staphylococcus for better survival during phagocytosis; this possibility is worth addressing in future work. In addition, the types and levels of endogenous oxidants may also play a role in the ubiquitous but sporadic distribution of dnd systems in the bacterial kingdom by preventing the establishment of PT modification systems in new hosts.
Notably, under oxidative stresses, S. enterica Cerro 87 retains the same level of PT modifications as untreated cells, even though PT desulphurisation, phosphonate formation and strand breakage are ongoing (Kellner et al.2017). Taking into account that the PT lability after exposure to oxidants causes no PT loss in cells, one could envisage a scenario in which the damaged PT sites resulting from endogenous oxidants may be repaired by DNA repair machineries and reused by Dnd proteins to re-establish PT modification and thereby maintain a constant steady-state level of PT; meanwhile, Dnd proteins could also move to another vacant motif to generate new PT sites, causing the observed fractional PT modification and PT heterogeneity (Fig. 3B).
PT modification is involved in balancing intracellular redox homeostasis
The versatility of PT was proven again by the recent discovery that PT is involved in the maintenance of cellular redox homeostasis (Tong et al.2018). The deletion of PT in Pseudomonas fluorescens pf0-1, which harbours a solitary DndBCDE cluster, so no DndFGH-mediated damage occurred, the global metabolic profile significantly changed. This metabolic alteration is largely attributable to profound changes in a group of metabolites, including proline, quinic acid, methionine and glycine (Fig. 4B). Based on the correlation of these metabolites with ROS scavenging and protection against oxidative stress as well as significant changes in lipid metabolites (Zhang et al.2012; Liang et al.2013; Alhasawi et al.2015; Tong et al.2018), it is plausible that the PT loss in P. fluorescens disturbs the redox homeostasis and induces cellular oxidative stress. Organisms have developed a variety of defence mechanisms, including the use of antioxidant enzymes and free radical scavengers, to decompose, remove or neutralise oxygen radicals and their intermediates (Nimse and Pal 2015). For instance, superoxide dismutase is involved in the removal of superoxide (O2−), and catalase detoxifies H2O2, while ketoacids, glutathione, L-proline, ascorbic acid, etc. are well-documented low-molecular-weight free radical scavengers (Grune, Schröder and Biesalski 2005; Szabados and Savoure, 2010; Sheng et al.2014; Nimse and Pal 2015). Interestingly, PT-deficient P. fluorescens cells adapt to oxidative stress by metabolic reconfiguration rather than the upregulation of antioxidant genes. This behaviour explains the observation that the antioxidant genes, i.e. catalase and organic hydroperoxide resistance genes, in Streptomyces lividans are not induced even under exposure to H2O2 (Dai et al.2016). Therefore, the reducing ability of PT sulphur enables PT to contribute to balancing intracellular redox homeostasis.
Epigenetic control of DNA PT modification
Similar to its phosphodiester congener, the 5΄-CGPSGCCGCCGA-3΄/5΄-TCGGCGPSGCCG-3΄ duplex (with sulphur replacing oxygen) has a B-form conformation, but this modification reduces the melting temperature (Tm) by approximately 4.5°C, agreeing well with the theoretical prediction that PT modification in the RP configuration results in increased stiffness of the DNA backbone and consequent destabilisation of the B-type helical structure (Chen et al.2015; Lan et al.2016). More importantly for what it suggests about the biochemical impact of PT modification, the sulphur substitution in the DNA backbone is sufficient to change the kinetics of the DNA polymerase in SMRT sequencing, similar to the methylation of nucleobases (Cao et al.2014b). Methylation in bacteria can affect the affinity of regulatory elements for their DNA targets by steric hindrance or alterations in DNA structure, causing changes in gene expression. Is PT modification capable of affecting transcription by RNA polymerase, similar to epigenetic methylation? The dynamic changes in PT distribution suggest heterogeneity in the bacterial population, which prompted Tong et al. to use chemically synthesised oligonucleotides to ensure saturated PT modification at a particular site (Fig. 4C). Utilizing an in vitro transcriptional assay, they found that the presence of PT, either in the promoter region or within the coding region, can influence the transcriptional efficiency, reflecting the epigenetic regulation role of PT, which likely occurs by altering the interaction between RNA polymerase and DNA substrates (Tong et al.2018). This conclusion is conceivable because Stein and co-workers have observed nonspecific protein binding with oligonucleotides with a PT backbone one-to-two orders of magnitude greater than with natural phosphodiester oligonucleotides (Stein, Tonkinson and Yakubov 1991). PT modifications in some DNA sequences, however, showed no disturbance of transcription in vitro, indicating the association of PT epigenetic control with sequence composition. The PT heterogeneity in cell populations raises the possibility that each PT pattern might influence the expression of a different group of genes, generating phenotypically diverse pf0-1 cells (Fig. 4C). This diversity might be beneficial for bacteria coping with changing environments.
INTERACTION OF PT MODIFICATION WITH OTHER BACTERIAL DEFENCE SYSTEMS
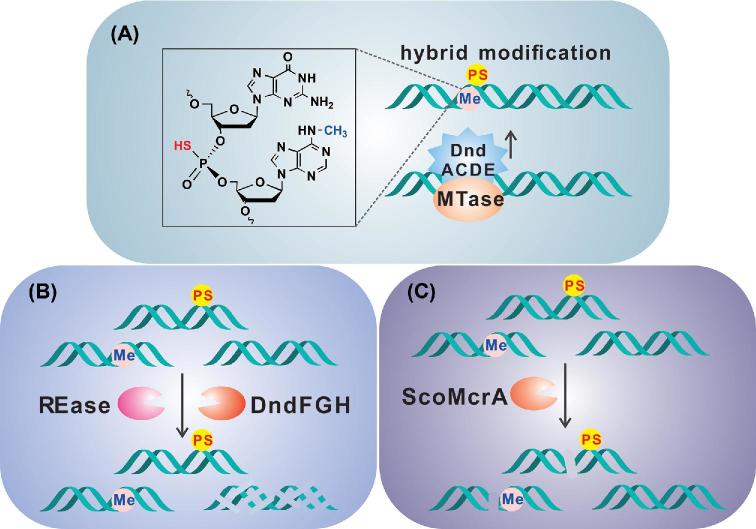
Despite having similar R-M defence functions, DNA methylation and PT modification have always been considered to function independently in prokaryotes because of their different enzymatic composition and distinctive modification features. However, Chen et al. recently demonstrated unexpected interaction and interference between these two modification systems. They first observed that DndBCDE in Hahella chejuensis KCTC2396, which confers double-stranded 5΄-GPSATC-3΄ PT modification, and Dam in Escherichia coli DH10B, which confers 5΄-G6mATC-3΄, share the 5΄-GATC-3΄ recognition target to produce the hybrid 5΄-GPS6mATC-3΄ (Fig. 5A; Chen et al.2017). Due to their nuclease resistance, PT modifications occurring in or adjacent to the cutting site can inhibit digestion by endonucleases to different extents. For instance, PT-modified 5΄-GGPSAATTCC-3΄ is recognised and cut by EcoRI but at a rate that is 1/15 that of the non-PT-modified DNA substrate (Connolly et al.1984). The work of Chen et al. found that methylated 5΄-G6mATC-3΄ is substituted for 5΄-GPSATC-3΄ to protect bacterial cells from the attack of the restriction counterpart DndFGH (Fig. 5B; Chen et al.2017). This interaction explains that in most cases, DNA PT modification and DNA methylation systems employ different consensus recognition sequences in a single host bacterial strain. Conversely, if the two systems share the same recognition motif, then they do not occur simultaneously in the same bacterial strain. This interaction between two representative defence systems suggests that such ‘conflict’ drives the evolution of different consensus sequences to avoid interference and to ensure the selective advantage of each R-M system.
Figure 5.
Interaction of DNA PT modifications with other defensive systems. (A) Dnd modification enzymes and Dam can share the same DNA motif, 5΄-GATC-3΄/5΄-GATC-3΄, to generate a hybrid 5΄-GPS6mATC-3΄/5΄-GPS6mATC-3΄ product modified with a PT in the DNA backbone and methylation of a nucleobase. (B) The methylated species 5΄-G6mATC-3΄/5΄-G6mATC-3΄ can be substituted with the PT-modified 5΄-GPSATC-3΄/5΄-GPSATC-3΄ to resist cleavage by DndFGH. (C) Bacteria have evolved the type IV ScoMcrA endonuclease to specifically recognise foreign DNA possessing 5mC and PT.
The extensive distribution of the dnd system in bacteria is believed to be the consequence of horizontal gene transfer. However, Liu et al. encountered difficulties in introducing the dnd cluster into Streptomyces coelicolor, a close relative of Streptomyces lividans (Liu et al.2010). These difficulties were later found to result from a PT-specific restriction barrier mediated by the type IV endonuclease ScoMcrA (Liu et al.2010). In the presence of Mn2+ or Co2+ at pH 9.0, ScoMcrA specifically recognises 5΄-C5mC(A/T)GG-3΄ (methylated by Dcm) and 5΄-GPSGCC-3΄ (PT modified by Dnd proteins) to perform double-stranded DNA cleavage 12–16 bp and 16–28 bp away from the methylation and PT sites, respectively (Fig. 5C; Liu et al.2010). Interestingly, a DNA sulphur recognition domain was characterised in SprMcrA from Streptomyces pristinaespiralis, a homolog of ScoMcrA. SprMcrA cuts both DNA strands 11 to 14 nucleotides away from the 5΄ side of the recognition PT site. Mutation of His residues in HRH motif of SprMcrA abolished restriction activity, whereas mutation of the R residue resulted in dramatically enhanced cleavage of phosphorothioate DNA (Yu et al.2018). It is not clear why bacteria have evolved such a defence barrier against the DNA PT system. However, this barrier provides an explanation for the sporadic distribution of DNA PT systems in the bacterial world. Is this phenomenon also a hint that a PT-containing bacteriophage exists and is a threat to bacteria? In the co-evolutionary arms race, bacteriophages have developed a variety of counter-resistance strategies to evade bacterial antiphage barriers (Labrie, Samson and Moineau 2010). The most striking example of an anti-restriction system is found in phage T4, which contains hydroxymethyl cytosine (HMC), which is further modified by glucosylation (Hattman 2009) rather than canonical cytosine. This modification allows the HMC-containing DNA to be impervious to R-M systems that recognise target sequences containing a cytosine (Labrie, Samson and Moineau 2010). On their side of co-evolutionary arms race, bacteria have evolved modification-dependent restriction enzymes to exclusively cleave methylated or hydroxymethylated DNA. To resist these new restriction enzymes, phage T4 adds glucose groups to HMC residues; the glucosylated DNA blocks the binding of the restriction enzymes (Hattman 2009; Labrie, Samson and Moineau 2010). It would be an attractive topic for future study to identify phages with PT modifications or to explore the strategies of phages for overcoming the defence barrier of PT R-M systems.
CONCLUSIONS
Sulphur is one of the main elements in methionine and cysteine, which are among the amino acid building blocks of proteins. The discovery of DNA PT modification makes sulphur the sixth elemental component of canonical DNA, joining carbon, nitrogen, phosphorus, oxygen and hydrogen. PT modification occurs in the DNA sugar-phosphate backbone rather than in the DNA nucleobases. Interestingly, prior to the identification of physiological PT modification, the PT bond was chemically synthesised and extensively applied in antisense therapy because it has properties similar to those of the phosphate analogue but with enhanced resistance to nucleases (Eckstein 2000, 2014; Lebedeva and Stein 2001). This nuclease tolerance enables DNA PT modification to serve as a constituent of the bacterial defence system in vivo, resembling methylation in methylation-based R-M systems. DNA PT modification is catalysed by DndABCDE proteins acting on a specific sequence consensus that is used as a recognition tag by the restriction counterpart DndFGH to recognise self versus nonself DNA and attack nonself invaders. Despite exhibiting similar defensive mechanisms against foreign DNA, PT- and methylation-based R-M systems might have co-evolved to use different recognition sequences in one host bacterial strain to ensure the protective function of each system (Tong et al.2018).
R-M systems are considered primitive immune systems that protect bacteria against bacteriophages. However, phages have evolved multiple tactics to ensure the evasion of restriction by their hosts (Labrie, Samson and Moineau 2010). Although T4 phages have evolved to use HMC instead of cytosine, some bacteria have acquired the ability to restrict HMC-modified phage DNA using modification-dependent R-M systems (Sutherland, Coe and Raleigh 1992). In turn, T4 phages acquire resistance again by the glycosylation of HMC residues, resulting in a co-evolutionary arms race (Labrie, Samson and Moineau 2010). Coliphage λ utilises Ral protein to enhance the methylase activity of type I R-M systems and thus alleviate the restriction activity of host cells (Zabeau et al.1980; Loenen and Murray 1986). Have phages evolved strategies to cope with PT-based R-M systems? The interference between PT- and methylation-based R-M systems suggests one possible counter-strategy: phages could utilise methylation sharing a consensus context identical to that of the PT R-M systems to mimic PT and evade the restriction activity of DndFGH. It is also possible that phages interfere with the DndB regulatory function to enhance the transcription of dndBCDE, resulting in the rapid PT modification of phage DNA, which is thereby protected from restriction by DndFGH. Moreover, the PT modification-dependent restriction of ScoMcrA (Liu et al.2010) suggests that at least some bacteria have evolved genetic barriers to inhibit the uptake of PT-containing DNA derived from other bacterial species or even phages possessing PT modification genes, if they exist. However, the existence of PT-modified phages could be expected with near certainty because phages that had propagated in Dnd+ cells would emerge with PT modifications.
Multi-layer defence systems might be important for bacteria to resist foreign DNA invasion. If a phage breaks through one defence system, another defence system might block it. It is not clear how defence tasks are divided between methylation and PT R-M systems or between R-M and CRISPR systems. It is also not clear whether they have overlapping targets or simply aim for their own particular targets. Diverse defence systems might attack the same pathogens; however, they could have differential target preferences against invaders. A type II methylation LlaDCHI R-M system from Lactococcus lactis was introduced into and found to be compatible with a type II CRISPR-Cas system (CR1) in Streptococcus thermophilus DGCC7710; moreover, these two systems worked together to achieve exponentially higher efficiency against phages (Dupuis et al.2013). Interestingly, this LlaDCHI R-M system targeted 35 5΄-GATC-3΄ sites on phage 2972, while CRISPR-Cas CR1 targeted only 3 bp upstream of the PAM site. The type II methylation R-M system and CRISPR-Cas system showed similar protection against phage 2972, whose efficiency of plaquing (EOP) was approximately 10−5–10−6; however, the combination of these two protection systems showed an EOP less than 10−9 (Dupuis et al.2013). As a new R-M system, the PT system could be involved in the multi-layer defence mechanism and provide a combination of protections in bacteria. Multi-layer defence systems might be essential against rapid phage genome mutation and evolution. The relationships between PT systems and CRISPR-Cas systems and other protection mechanisms remains to be explored.
It is still uncertain at this point whether the heterogeneity of PT modifications results from the unusual target selection mechanism used by Dnd proteins or from reactions with endogenous oxidants. In the former case, techniques could be developed to locate PT sites in individual cells, which enable the correlation between PT sites and epigenetic regulation to be identified in combination with the single-cell transcriptome analysis. If the PT turnover is attributed to modification by endogenous oxidants, alkylating agents or metals, it should be a random event and generate a diverse range of PT patterns in a cell population. The question is whether, if a given PT site is converted or repaired back to a regular phosphate diester, this site is reused for PT modification or the Dnd proteins choose another vacant genomic consensus sequence to generate new PT sites. Both scenarios might be true, considering that Dnd modification enzymes recognise only 4-bp consensus sequences for modification. Additionally, the dynamic nature of DNA PT modification is reminiscent of the methylation/demethylation pattern observed in eukaryotic chromosomes. The mammalian DNA methyltransferases establish and maintain 5mC modification, while ten-eleven translocation family enzymes can mediate DNA demethylation by converting 5mC to 5hmC, 5-formylcytosine and 5-carboxylcytosine (Ito et al.2011). Such dynamic methylation and demethylation processes are critical to embryonic development, chromatin structure, chromosome stability, genomic imprinting, etc. (Chen and Riggs 2011). We cannot currently rule out the possibility that PT heterogeneity results from enzyme-mediated PT removal or conversion until such an enzyme is identified. Nevertheless, epigenetic control and heterologous patterns of PT modification might be beneficial to bacteria coping with changing environments.
PT modifications have not been found in eukaryotes. A high-throughput screening method would enable further searching for potential PTs in eukaryotes. If PTs are present in eukaryotes, similar to methylation, the exploration of their function would be extremely interesting. It would also be attractive to determine whether PTs occur in RNAs. More than 140 different RNA modifications, including the sulphur modification of nucleobases (which forms such products as 4-thiourdine (s4U), 2-thiocytidine (s2C), 5-methylaminomethyl-2-thiouridine (mnm5s2U), 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) and N-6-isopentyl-2-methylthioadenosine (ms2i6A)), have been identified (Marbaniang and Vogel 2016). Almost all the modifications found in DNA are present in RNA, including m6A, which was recently found to be widespread in bacterial mRNA and potentially play roles in energy metabolism and small RNAs (Deng et al.2015). It is of great interest to determine whether RNA has PT modifications and their potential functions, including their possible impact on RNA stability, RNA localisation, translation rate, growth fitness and stress responses.
Although DNA PT systems are currently known to exist predominantly in the form of DndABCDE or DndABCDE-DndFGH, Dnd PT systems may actually be more diverse. The interaction of DNA PT and methylation shows the possible scenario that DNA methylation and DndFGH could constitute a hybrid R-M system (Tong et al.2018). Tong et al. observed that 63 of 651 PT R−-M+ systems are accompanied by genes encoding nucleases of the HNH family and the uma2 family as well as unclassified nucleases in the neighbourhood of solitary dndBCDE clusters (Tong et al.2018). Investigating whether solitary PTs have evolved to pair with nucleases other than DndFGH to yield new PT R-M modules is thus attractive. Surprisingly, 82 bacterial strains possess genes encoding DndFGH homologues but lack dndBCDE or genes for potential DNA modification, i.e. they have Dnd enzymes resembling REases but lack the corresponding MTases. Several reasons might explain the presence of solitary DndFGH: the solitary DndFGH are not expressed; they are expressed but nonfunctional due to mutations in their active sites; or DndFGH and DndBCDE or possibly MTase substituents are coded by distantly located genes in the genomes. Another possible explanation is that a bacterial genome contains no recognition sequences for the solitary DndFGH. However, this scenario is possible only if solitary DndFGH have wide recognition specificity. We should also expect the presence of other types of DNA PT modification systems, with enzymatic composition and modification features that are different from those of dnd systems. Clearly, multiple approaches are needed to fully appreciate this sulphur modification in the DNA backbone.
Acknowledgements
We thank Chao Chen, Xuan Zou, and Xingxiang Chen for their valuable comments on this manuscript.
FUNDING
This work was supported by grants from the National Science Foundation of China (31720103906, 31520103902, 31670072 and 31670086) and the Young One Thousand Talent Program of China.
Conflicts of interest. None declared.
REFERENCES
- Ahlgren NA, Chen Y, Needham DM et al.. Genome and epigenome of a novel marine Thaumarchaeota strain suggest viral infection, phosphorothioation DNA modification and multiple restriction systems. Environ Microbiol 2017;19:2434–52. [DOI] [PubMed] [Google Scholar]
- Albalat R, Canestro C. Evolution by gene loss. Nat Rev Genet 2016;17:379–91. [DOI] [PubMed] [Google Scholar]
- Alhasawi A, Castonguay Z, Appanna ND et al.. Glycine metabolism and anti-oxidative defence mechanisms in Pseudomonas fluorescens. Microbiol Res 2015;171:26–31. [DOI] [PubMed] [Google Scholar]
- An X, Xiong W, Yang Y et al.. A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. PLoS One 2012;7:e51265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atack JM, Tan A, Bakaletz LO et al.. Phasevarions of bacterial pathogens: methylomics sheds new light on old enemies. Trends Microbiol 2018;26:715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol 2007;366:768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier P, Lunazzi A, Fujiwara-Nagata E et al.. From the Flavobacterium genus to the phylum Bacteroidetes: genomic analysis of dnd gene clusters. FEMS Microbiol Lett 2013;348:26–35. [DOI] [PubMed] [Google Scholar]
- Cao B, Cheng QX, Gu C et al.. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate- based restriction system in Salmonella. Mol Microbiol 2014a;93:776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Chen C, DeMott MS et al.. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat Commun 2014b;5:3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AL, Hampton MB, Senthilmohan R et al.. Chlorination of bacterial and neutrophil proteins during phagocytosis and killing of Staphylococcus aureus. J Biol Chem 2002;277:9757–62. [DOI] [PubMed] [Google Scholar]
- Chen C, Wang L, Chen S et al.. Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes. P Natl Acad Sci U S A 2017;114:4501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Lin K, Zhang Z et al.. Purification, crystallization and preliminary X-ray analysis of the DndE protein from Salmonella enterica serovar Cerro 87, which is involved in DNA phosphorothioation. Acta Crystallogr F Struct Biol Cryst Commun 2011;67:1440–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Z, Lin K et al.. Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation. PLoS One 2012;7:e36635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang XL, Shi T et al.. Theoretical study on the relationship between Rp-phosphorothioation and base-step in S-DNA: Based on energetic and structural analysis. J Phys Chem B 2015;119:474–81. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem 2011;286:18347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Cao B, Yao F et al.. Regulation of DNA phosphorothioate modifications by the transcriptional regulator DptB in Salmonella. Mol Microbiol 2015;97:1186–94. [DOI] [PubMed] [Google Scholar]
- Clark TA, Spittle KE, Turner SW et al.. Direct detection and sequencing of damaged DNA bases. Genome Integr 2011;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Murray IA, Morgan RD et al.. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 2012;40:e29-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly BA, Potter BV, Eckstein F et al.. Synthesis and characterization of an octanucleotide containing the EcoRI recognition sequence with a phosphorothioate group at the cleavage site. Biochemistry 1984;23:3443–53. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery JR, Urbina H, Vickery LE. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J Mol Biol 2003;330:1049–59. [DOI] [PubMed] [Google Scholar]
- Dai D, Du A, Xiong K et al.. DNA Phosphorothioate modification plays a role in peroxides resistance in Streptomyces lividans. Front Microbiol 2016;7:1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wei H, Tian C et al.. An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol 2015;15:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smet MD, Meenken CJ, van den Horn GJ. Fomivirsen – a phosphorothioate oligonucleotide for the treatment of CMV retinitis. Ocul Immunol Inflamm 1999;7:189–98. [DOI] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ et al.. Widespread occurrence of N 6 -methyladenosine in bacterial mRNA. Nucleic Acids Res 2015;43:6557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias N, Stein CA. Antisense oligonucleotides: Basic concepts and mechanisms. Mol Cancer Ther 2002;1:347–55. [PubMed] [Google Scholar]
- Dupuis ME, Villion M, Magadan AH et al.. CRISPR-Cas and restriction–modification systems are compatible and increase phage resistance. Nat Commun 2013;4:2087. [DOI] [PubMed] [Google Scholar]
- Dyson P, Evans M. Novel post-replicative DNA modification in Streptomyces: Analysis of the preferred modification site of plasmid pIJ101. Nucleic Acids Res 1998;26:1248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem 1985;54:367–402. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev 2000;10:117–21. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Therapeutics 2014;24:374–87. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH et al.. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 2010;7:461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KL, Dowideit SJ, Erwin AL et al.. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res 2007;35:5242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Maeda M, Mihara H et al.. Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase. Biochemistry 2000;39:1263–73. [DOI] [PubMed] [Google Scholar]
- Geary RS, Henry SP, Grillone LR. Fomivirsen. Clin Pharmacokinet 2002;41:255–60. [DOI] [PubMed] [Google Scholar]
- Gish G, Eckstein F. DNA and RNA sequence determination based on phosphorothioate chemistry. Science 1988;240:1520–2. [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Kozdon JB, McAdams HH et al.. The functions of DNA methylation by CcrM in Caulobacter crescentus: A global approach. Nucleic Acids Res 2014;42:3720–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Schröder P, Biesalski HK. Low molecular weight antioxidants. In Grune T. (ed). Reactions, Processes: Oxidants and Antioxidant Defense Systems. Berlin, Heidelberg: Springer Berlin Heidelberg, 2005, 77–90. [Google Scholar]
- Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: Oxidants, myeloperoxidase, and bacterial killing. Blood 1998;92:3007–17. [PubMed] [Google Scholar]
- Hattman S. The first recognized epigenetic signal: DNA glucosylation of T-even bacteriophages. Epigenetics 2009;4:150–1. [DOI] [PubMed] [Google Scholar]
- He W, Huang T, Tang Y et al.. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci Rep 2015;5:12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ou HY, Yu Q et al.. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol 2007;65:1034–48. [DOI] [PubMed] [Google Scholar]
- Ho WS, Ou HY, Yeo CC et al.. The dnd operon for DNA phosphorothioation modification system in Escherichia coli is located in diverse genomic islands. BMC Genomics 2015;16:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Wang C, Liang J et al.. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Res 2012;22:1203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbelin M, Suri B, Rao DN et al.. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol 1988;200:23–29. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q et al.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011;333:1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser JT, Clausen T, Bourenkow GP et al.. Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly. J Mol Biol 2000;297:451–64. [DOI] [PubMed] [Google Scholar]
- Kambampati R, Lauhon CT. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J Biol Chem 2000;275:10727–30. [DOI] [PubMed] [Google Scholar]
- Kellner S, DeMott MS, Cheng CP et al.. Oxidation of phosphorothioate DNA modifications leads to lethal genomic instability. Nat Chem Biol 2017;13:888–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. Selfishness and death: raison d'être of restriction, recombination and mitochondria. Trends Genet 1998;14:368–74. [DOI] [PubMed] [Google Scholar]
- Kozdon JB, Melfi MD, Luong K et al.. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. P Natl Acad Sci U S A 2013;110:E4658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Micro 2010;8:317–27. [DOI] [PubMed] [Google Scholar]
- Lai C, Wu X, Chen C et al.. In vivo mutational characterization of DndE involved in DNA phosphorothioate modification. PLoS One 2014;9:e107981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Hu Z, Shen J et al.. Structural investigation into physiological DNA phosphorothioate modification. Sci Rep 2016;6:25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauhon CT, Kambampati R. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J Biol Chem 2000;275:20096–103. [DOI] [PubMed] [Google Scholar]
- Lebedeva I, Stein CA. Antisense oligonucleotides: promise and reality. Annu Rev Pharmacol Toxicol 2001;41:403–19. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang Z, He X et al.. DNA modification by sulfur: Analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res 2007;35:2944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhang L, Natarajan SK et al.. Proline mechanisms of stress survival. Antioxidants Redox Signaling 2013;19:998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Ou HY, Wang T et al.. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet 2010;6:e1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA, Murray NE. Modification enhancement by the restriction alleviation protein (Ral) of bacteriophage lambda. J Mol Biol 1986;190:11–22. [DOI] [PubMed] [Google Scholar]
- Low DA, Casadesus J. Clocks and switches: Bacterial gene regulation by DNA adenine methylation. Curr Opin Microbiol 2008;11:106–12. [DOI] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun 2001;69:7197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbaniang CN, Vogel J. Emerging roles of RNA modifications in bacteria. Curr Opin Microbiol 2016;30:50–57. [DOI] [PubMed] [Google Scholar]
- Marinus MG, Lobner-Olesen A. DNA Methylation. EcoSal Plus 2014, DOI: 10.1128/ecosalplus.ESP-0003-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel A, Mackeldanz P, Bickle TA et al.. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J 1995;14:2958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara H, Esaki N. Bacterial cysteine desulfurases: Their function and mechanisms. Appl Microbiol Biot 2002;60:12–23. [DOI] [PubMed] [Google Scholar]
- Monia BP, Johnston JF, Sasmor H et al.. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J Biol Chem 1996;271:14533–40. [DOI] [PubMed] [Google Scholar]
- Mruk I, Kobayashi I. To be or not to be: Regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res 2014;42:70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG. Trafficking in persulfides: Delivering sulfur in biosynthetic pathways. Nat Chem Biol 2006;2:185–94. [DOI] [PubMed] [Google Scholar]
- Mueller EG, Palenchar PM. Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci 1999;8:2424–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG, Palenchar PM, Buck CJ. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J Biol Chem 2001;276:33588–95. [DOI] [PubMed] [Google Scholar]
- Nimse SB, Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv 2015;5:27986–8006. [Google Scholar]
- Ou HY, He X, Shao Y et al.. dndDB: A database focused on phosphorothioation of the DNA backbone. PLoS One 2009;4:e5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopowicz ZM, Arce F, Biedron R et al.. Hypochlorous acid: A natural adjuvant that facilitates antigen processing, cross-priming, and the induction of adaptive immunity. J Immunol 2010;184:824–35. [DOI] [PubMed] [Google Scholar]
- Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J 1995;14:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray T, Weaden J, Dyson P. Tris-dependent site-specific cleavage of Streptomyces lividans DNA. FEMS Microbiol Lett 1992;96:247–52. [DOI] [PubMed] [Google Scholar]
- Ray T, Mills A, Dyson P. Tris-Dependent oxidative DNA strand scission during electrophoresis. Electrophoresis 1995;16:888–94. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J et al.. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 2010;38:D234–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Belfort M, Bestor T et al.. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 2003;31:1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshasayee AS, Singh P, Krishna S. Context-dependent conservation of DNA methyltransferases in bacteria. Nucleic Acids Res 2012;40:7066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Abreu IA, Cabelli DE et al.. Superoxide dismutases and superoxide reductases. Chem Rev 2014;114:3854–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, Mejean V, Jourlin C et al.. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J Bacteriol 1994;176:5601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA. Exploiting the potential of antisense: Beyond phosphorothioate oligodeoxynucleotides. Chem Biol 1996;3:319–23. [DOI] [PubMed] [Google Scholar]
- Stein CA, Tonkinson JL, Yakubov L. Phosphorothioate oligodeoxynucleotides—anti-sense inhibitors of gene expression? Pharmacol Therapeut 1991;52:365–84. [DOI] [PubMed] [Google Scholar]
- Sutherland E, Coe L, Raleigh EA. McrBC: A multisubunit GTP-dependent restriction endonuclease. J Mol Biol 1992;225:327–48. [DOI] [PubMed] [Google Scholar]
- Szabados L, Savoure A. Proline: A multifunctional amino acid. Trends Plant Sci 2010;15:89–97. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Matsuno H, Furusawa H et al.. Direct monitoring of allosteric recognition of type IIE restriction endonuclease EcoRII. J Biol Chem 2008;283:15023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirupati B, Vey JL, Drennan CL et al.. Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803. Biochemistry 2004;43:12210–9. [DOI] [PubMed] [Google Scholar]
- Tong T, Chen S, Wang L et al.. Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. P Natl Acad Sci U S A 2018;115:E2988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham SM, van Alphen L, Mooi FR et al.. Phase variation of H. influenzae fimbriae: Transcriptional control of two divergent genes through a variable combined promoter region. Cell 1993;73:1187–96. [DOI] [PubMed] [Google Scholar]
- Vasu K, Nagamalleswari E, Nagaraja V. Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. P Natl Acad Sci U S A 2012;109:E1287–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DE, Lokesh GLR. Development of phosphorothioate DNA and DNA thioaptamers. Biomedicines 2017;5:E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Slungaard A. Role of eosinophil peroxidase in host defense and disease pathology. Arch Biochem Biophys 2006;445:256–60. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Xu T et al.. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol 2007;3:709–10. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Vergin KL et al.. DNA phosphorothioation is widespread and quantized in bacterial genomes. P Natl Acad Sci U S A 2011;108:2963–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele P, Raleigh EA. Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 2016;116:12655–87. [DOI] [PubMed] [Google Scholar]
- Weiser JN, Williams A, Moxon ER. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect Immun 1990;58:3455–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet 1991;25:585–627. [DOI] [PubMed] [Google Scholar]
- Wu T, Huang Q, Wang XL et al.. Mechanistic investigation on ROS resistance of phosphorothioated DNA. Sci Rep 2017;7:42823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Liang J, Pu T et al.. Phosphorothioate DNA as an antioxidant in bacteria. Nucleic Acids Res 2012;40:9115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Zhao G, Yu H et al.. Interactions of Dnd proteins involved in bacterial DNA phosphorothioate modification. Front Microbiol 2015;6:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SY, Corvaglia AR, Chan SH et al.. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res 2011;39:5597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yao F, Zhou X et al.. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res 2010;38:7133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Liang J, Chen S et al.. DNA phosphorothioation in Streptomyces lividans: Mutational analysis of the dnd locus. BMC Microbiol 2009;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu GP, Liang JD et al.. DNA backbone sulfur-modification expands microbial growth range under multiple stresses by its antioxidation function. Sci Rep 2017;7:3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Xu T, Zhou X et al.. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett 2009;583:729–33. [DOI] [PubMed] [Google Scholar]
- You D, Wang L, Yao F et al.. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry 2007;46:6126–33. [DOI] [PubMed] [Google Scholar]
- You D, Xu T, Yao F et al.. Direct evidence that ThiI is an ATP pyrophosphatase for the adenylation of uridine in 4-thiouridine biosynthesis. ChemBioChem 2008;9:1879–82. [DOI] [PubMed] [Google Scholar]
- Yu H, Liu G, Zhao G et al.. Identification of a conserved DNA sulfur recognition domain by characterizing the phosphorothioate-specific endonuclease SprMcrA from Streptomyces pristinaespiralis. Mol Microbiol 2018, DOI: 10.1111/mmi.14118. [DOI] [PubMed] [Google Scholar]
- Zabeau M, Friedman S, Van Montagu M et al.. The ral gene of phage lambda. I. Identification of a non-essential gene that modulates restriction and modification in E. coli. Molec Gen Genet 1980;179:63–73. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang J, Zhao B et al.. Quinic acid could be a potential rejuvenating natural compound by improving survival of Caenorhabditis elegans under deleterious conditions. Rejuvenation Res 2012;15:573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Jiang P, Cao B et al.. DndEi exhibits helicase activity essential for DNA phosphorothioate modification and ATPase activity strongly stimulated by DNA substrate with a GAAC/GTTC Motif. J Biol Chem 2016;291:1492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Deng Z, Firmin JL et al.. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucl Acids Res 1988;16:4341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, He X, Liang J et al.. A novel DNA modification by sulphur. Mol Microbiol 2005;57:1428–38. [DOI] [PubMed] [Google Scholar]