Abstract
Defensins are important components of innate host defence system against bacteria, fungi, parasites and viruses. Here, we predicted six potential defensin genes from the genome of the scorpion Mesobuthus martensii and then validated four genes from them via the combination of PCR and genomic sequence analysis. These four scorpion defensin genes share the same gene organization and structure of two exons and one phase-I intron with the GT-AG rule. Conserved motif and phylogenetic analysis showed that they belonged to the members of the invertebrate cysteine-stabilized α-helix/β-sheet motif defensin (CSαβ) defensin family. All these four CSαβ defensin genes have the expression feature of constitutive transcription (CON) by the whole scorpion infection model, promoter sequence analysis and dual luciferase assays. Further evolution and comparison analysis found that the invertebrate CSαβ defensin genes from most of arachnids and mollusks appear to share the expression pattern of CON, but those from insects and lower invertebrates (nematodes, annelids, cnidarians and sponges) seem to have identical inducible transcription (IND) after being challenged by microorganisms. Together, we identified four scorpion CSαβ defensin genes with the expression feature of CON, and characterized the diversified expression patterns of the invertebrate CSαβ defensin genes, which will shed insights into the evolution of the invertebrate CSαβ defensin genes and their expression patterns.
Keywords: CSαβ defensin genes, constitutive transcription, expression pattern, inducible transcription, scorpion
Introduction
Defensins are small cationic antimicrobial peptides containing three or four intramolecular disulphide bonds formed by six or eight cysteine residues in a complex folded arrangement of two or three antiparallel β-sheets with or without an α-helix structure [1–4]. Defensins are important components of the host immune system and produced by a wide range of organisms including vertebrates, invertebrates, plants and fungi, which have defensive functions against a broad array of infectious pathogens like bacteria, fungi, parasites and viruses [3,5,6]. Generally, the spatial structures of defensins differ according to the arrangement of conserved cysteine residues [1–3,7].
Vertebrates’ defensins have six cysteine residues and are divided into three distinct families: α, β and θ defensins, which have lengths of 29–35, 35–45 and 18 amino acid residues respectively [8]. Defensins have ubiquitously been identified among invertebrates, predominantly in mollusks [9,10], nematodes [11–13] and arthropods [14]. The classification of invertebrate defensins differs based on the complex criteria [3]. Invertebrate defensins can primarily be classified into two classes. One class is the invertebrate cysteine-stabilized α-helix/β-sheet motif defensin (CSαβ defensin), containing an α-helix linked to an antiparallel two-stranded β-sheet by disulphide bridges and is found in mollusks, nematodes and arthropods [15,16]. The other class is the invertebrate big defensin, which has a disulphide array stabilized β-sheet structure different from the invertebrate CSαβ motif [17,18] and exists mainly in mollusks [19] but also in arthropods [20].
Scorpions are one of the most ancient terrestrial venomous animal lineages [21]. Scorpions have more than 400 million years of evolutionary time, and so are considered to be living fossils [22]. Scorpions have a strong ability to adapt to different environments and are widely distributed on all continents except Antarctica [23]. More than 2329 scorpion species classified into 17 families have been recorded to date (http://www.ntnu.no/ub/scorpion-files/specieslist.php). The scorpion M. martensii belonged to the family Buthidae and was used to be an important raw material in Chinese Traditional Medicine for the treatment of some nervous system diseases [24].
As one kind of arachnid animals, scorpions mainly rely on innate immune system against infectious microorganisms like insects [25]. Especially, defensins are a class of ubiquitously expressed cationic antimicrobial peptides (CAPs) that play an important role in innate defence [26]. Scorpion defensin research can be traced back as early as 1993, when Cociancich et al. [27] isolated and structurally characterized a defensin from the scorpion Leiurus quinquestriatus. In 1996, Ehret-Sabatier et al. [28] identified Androctonus defensin with three disulphide bridges from the haemolymph of the scorpion Androctonus australis. Insect defensins are considered to be a group of inducible small-sized antibacterial peptides [29], but chelicerate defensins are implicated to have complicated alternative defensive systems: up-regulation dependent on inducible (IND) or constitutive transcription (CON) and constitutive production [30]. Scorpion is one of chelicerate animals, but only one scorpion defensin Cll-dlp was demonstrated to accumulate in the haemolymph in response to septic injury, independent of transcriptional regulation [30]. Expressive characteristics of defensin genes from scorpions and invertebrates still need to be systematically studied and analysed, which provides insights into the evolution of defensins and innate immune system.
Our group previously determined the draft-genome sequence of the scorpion M. martensii (Scorpions: Buthidae) [31]. In the present work, we predicted six potential defensin genes from the scorpion M. martensii genome, and then experimentally validated four defensin genes. Gene organization and evolutionary analyses confirmed that they are the members of the invertebrate CSαβ defensin gene family. Their expression features were identified by an intact scorpion infection challenge, promoter sequence analysis and dual luciferase assay. Finally, we summarized and compared the expression patterns of almost all CSαβ defensins from the kingdom of invertebrates.
Materials and methods
Annotation of defensin genes
The NCBI database (http://www.ncbi.nlm.nih.gov/protein/) was used as a search engine for nucleotide sequences of scorpion defensins. Then, the downloaded nucleotide sequences of scorpion defensins were used to search in the scorpion M. martensii genome (GenBank number BioProject PRJNA171479) by BLAST as previously reported [31] and obtained several candidate defensin genes. These predicated defensin genes were further confirmed in the transcriptome of the scorpion M. martensii followed by ORF forecasting using a NCBI software (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi) to find the corresponding proteins. The signal peptides of the corresponding proteins were predicted by SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/). Multiple alignments were achieved by GeneDoc.
Preparation of genomic DNA and PCR amplification
Genomic DNA was isolated as previously described [31]. Simply intact individuals of the scorpion M. martensii were washed three times with 75% ethanol and grounded to fine powder in liquid nitrogen. Then genomic DNA extraction was performed using TIANamp Genomic DNA Kit (Tiangen, China) according to the manufacturer’s instructions.
Primers (Supplementary Table S1) from 5′-UTR and 3′-UTR regions of the predicted defensin genes from the scorpion M. martensii genome were picked up to amplify the corresponding genomic DNA by PCR. Amplification was performed with one cycle of 5 min at 95°C, 30 cycles of 40 s at 95°C, 40 s at 58°C, 150 s at 72°C, and a final cycle of 10 min at 72°C using Ex Taq (TaKaRa, China). PCR products were purified using the DNA Clean-up Kit (CWBio, China) and ligated to pGEM-T Easy Vector (TaKaRa, China). Sequencing was performed by Tsingke Biological Technology.
Phylogenetic analysis
Mature peptide sequences of defensins from the scorpion M. martensii and the other species including vertebrates and invertebrates were aligned by ClustalX 1.83. Phylogenetic tree of defensin superfamily was constructed using Neighbor-Joining method in MEGA 5.1 (http://www.megasoftware.net). Bootstrap sampling was reiterated 10000 times. The detailed sequence information for the analysis of defensins is in Supplementary Table S2.
Preparation of bacterial materials
The bacterial strains Escherichia coli (AB94012) and Staphylococcus aureus (AB94004) were purchased from China Center for Type Culture Collection (CCTCC). The bacterial materials were acquired as following. Bacteria cultured in LB medium to OD600 =0.6 at 37°C was collected by centrifuging for 3 min at 6000 rpm. Then the collections were suspended in PBS for intact scorpion challenge. For cell stimulation, the supernatant was further smashed by ultrasonic and cleaned through a 0.22-μm filter. The filtrate was quantified by Pierce BCA Protein Assay Kit (Thermo Scientific, U.S.A.) according to the manufacturer’s instructions and stored at −20°C at a final concentration of 50 mg/ml.
Intact scorpion infection model and real-time PCR analysis
The scorpion M. martensii individuals were challenged according to the modified protocol as previously described [30,32,33]. Simply, adult male M. martensii were maintained in the laboratory with sufficient water and food for 2 weeks before experimentation. Individuals of the test group were pricked with a fine needle with 5 μl of 200 ng/ml lipopolysaccharides (LPS, Sigma), 500 ng/ml lipoteichoic acid (LTA, Sigma), 50 μg/ml E. coli or 50 μg/ml S. aureus by a puncture between the second and third segments of the scorpion metasoma. Individuals of the control group were injected with 5 μl PBS; 36 h post injection, scorpions were washed for RNA extraction followed by cDNA synthesis and real-time PCR assay as previously described [31]. cDNA was synthesized using the First Strand cDNA Synthesis Kit (Thermo Scientific, U.S.A.). Real-time PCR was performed using the SYBR Green PCR assay and an ABI 7500 system according to the manufacturer’s instructions. The relative mRNA expression of four validated defensin genes from the scorpion M. martensii was normalized to β-actin. Primers used in the real-time PCR were summarized in Supplementary Table S3.
Cloning, sequencing and analysis of promoter regions
According to the whole genome sequence of the scorpion M. martensii, gene-specific primers (Supplementary Table S4), located at ~1–2 thousand nucleotides upstream the translation start site of the validated four scorpion defensin genes, was used to clone their corresponding potential promoter regions. Amplification was performed with one cycle of 5 min at 95°C, 30 cycles of 40 s at 95°C, 40 s at 55°C, 90 s at 72°C, and a final cycle of 10 min at 72°C using ExTaq. PCR products were then purified, cloned and sequenced as above. Potential transcription factor binding sites upstream the initiation codon were searched using the P-Match program (http://www.generegulation.com/pub/programs.html#pmatch) and the AliBaba2 program (http://www.cs.uni-magdeburg.de/grabe/alibaba2) against the TRANSFAC database.
Cell culture and dual luciferase assay of promoter activity
Human embryonic kidney (HEK293T) cells and Drosophila S2 cells were obtained from CCTCC. HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco/Life Technologies, U.S.A.) supplemented with 10% FBS (Gibco/Life Technologies, U.S.A.) and 1% penicillin/streptomycin in a humidified 5% CO2 incubator at 37°C. Drosophila S2 cells were cultured in Schneider’s Insect medium (Sigma, U.S.A.), supplemented with 10% FBS and 1% penicillin/streptomycin at 28°C.
The firefly luciferase reporter vectors were constructed by using the recognition sequence of MluI and XhoI to insert the cloned promoter region into pGL3 basic promoter vector. Primers used for dual luciferase analysis are described in Supplementary Table S5. HERK293T or Drosophila S2 cells were seeded in 24-well plates 24 h before transfection. And then 1 μg constructed firefly luciferase reporter vector and 50 ng Renilla luciferase plasmid pRL-TK (Promega, U.S.A.) as an internal control were transfected using Lipofectamine 2000 Transfection Reagent (Invitrogen, U.S.A.) following the manufacturer’s instructions. Twenty-four hours post transfection, cells were stimulated with bacterial materials E. coli (50 μg/ml), S. aureus (50 μg/ml), LPS (200 ng/ml) and LTA (500 ng/ml) respectively. Cells were assayed 48 h later with Dual Luciferase Assay (Promega, U.S.A.) according to the manufacturer’s instructions.
Statistical analysis
All assays were performed in triplicate. Data from repeated experiments are expressed as the means ± S.D.
Results
Annotation of defensin genes in the scorpion M. martensii genome
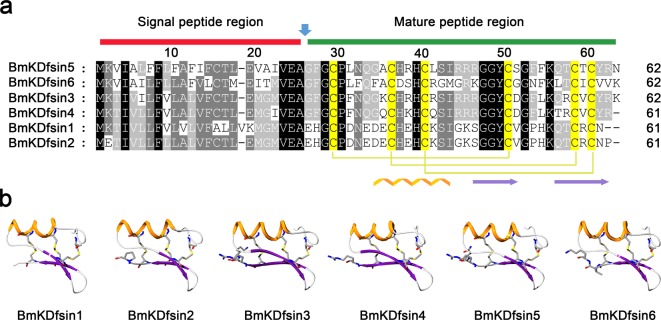
Defensins from scorpion species were first identified in L. quinquestriatus, followed by A. australis [27,28]. A total of 14 scorpion defensins were retrieved from online databases. A scorpion defensin query set was constructed and used in a tblastn search against the M. martensii genome. E-values less than 10−4 were used as a cut-off for significant hits of candidate defensin genes. Each hit was further verified manually based on similarities in sequence, cysteine patterns and gene structural organization. Six defensin genes were annotated in the scorpion M. martensii genome and named as BmKDfsin1, BmKDfsin2, BmKDfsin3, BmKDfsin4, BmKDfsin5 and BmKDfsin6 (Supplementary Table S6) respectively. These six defensin precursors consist of 61 or 62 amino acid residues, including 24 or 25 signal peptide residues and 37 or 38 mature peptide residues (Figure 1a), which is consistent with previously reported lengths of defensin precursors and mature peptides [34,35]. Each of these six defensins has six conserved cysteine residues which probably form three pairs of intramolecular disulphide bridges to induce the CSαβ fold. Their 3D structures were modelled using the arthropod defensin as a template (PDB code: 2ru0) on the SWISS-MODEL server (http://swissmodel.expasy.org/). The 3D structures display that six putative defensins from the scorpion M. martensii are composed of an α-helix linked to an antiparallel two-stranded β-sheet by three disulphide bridges (Figure 1b), suggesting that they all belong to the members of the invertebrate CSαβ defensin family.
Figure 1. Six potential defensins annotated from the genome of the scorpion M. martensii.
(a) Multiple sequence alignment of six potential defensins from the scorpion M. martensii genome. The red line indicates the deduced signal peptide regions of the defensins, and the green line shows the deduced mature peptide regions of the defensins. The six conserved cysteine residues are highlighted with yellow background and form three pairs of intramolecular disulphide bridges that induce the CSαβ fold. (b) The predicted 3D structures of the six potential defensins from the scorpion M. martensii. Their 3D structures were modelled using the arthropod defensin (PDB code: 2ru0) as a template by the SWISS-MODEL server (http://swissmodel.expasy.org/). α-helical and β-parallel regions are highlighted with orange and purple colours respectively.
Validation of defensin genes from the genome of the scorpion M. martensii
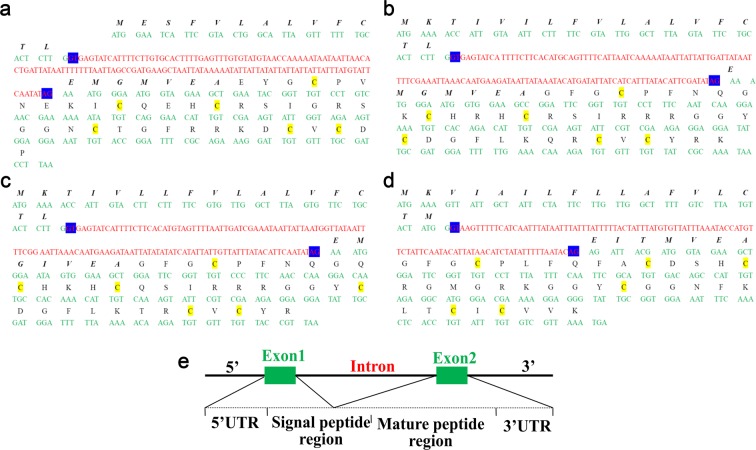
BioProject PRJNA171479 was a draft genome sequence of the scorpion M. martensii. To validate whether the six newly discovered putative defensin genes truly existed in the genome of the scorpion M. martensii, PCR using 5′-UTR and 3′-UTR primers was performed and five amplified products were cloned. Four of these five PCR products were close to their predicted lengths (1449 bp for BmKDfsin2, 1492 bp for BmKDfsin3, 1265 bp for BmKDfsin4 and 991 bp for BmKDfsin6), while the 600-bp fragment for BmKDfsin1 was much shorter than the predicted size (945 bp) and no fragment for BmKDfsin5 was acquired (Supplementary Figure S1). Unfortunately, although we tried our best to design several pairs of primers and amplify, we still failed to get the predicated genomic fragments of BmKDfsin1 and BmKDfsin5. We thought that the existence of some wrong sequence information in the current draft genome version of M. martensii could account for our failure to achieve genomic sequences of BmKDfsin1 and BmKDfsin5. Finally, sequencing analysis of four DNA fragments with right lengths showed identical amino acid coding sequences to their counterparts annotated from the M. martensii genome (Figures 1a and 2a–d). Thus, we confirmed the existence of four CSαβ defensin genes (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) in the scorpion M. martensii genome.
Figure 2. Sequence analysis of the four defensin genes validated from the scorpion M. martensii.
(a) BmKDfsin2. (b) BmKDfsin3. (c) BmKDfsin4. (d) BmKDfsin6. (e) Gene organization and structure analysis of four defensins from the scorpion M. martensii. Green letters represent exon sequences, and red letters are intron sequences. GT-AG splicing sites are shaded with blue background. The letters above the nucleotides are the corresponding amino acid sequences, the signal peptide regions are indicated by italics and bold letters, and the mature peptide regions are indicated with black letters. Cysteine residues are highlighted with yellow background.
Genomic organization and classification of defensin genes from the scorpion M. martensii
Based on PCR validated sequence information, the gene organizations and structures of BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6 were further analysed. These four scorpion CSαβ defensins share identical gene organization of two exons and one intron, where the first exon encodes the 5′-UTR region and part of the signal peptide, whereas the other exon encodes the remainder of the signal peptide, the mature peptide and the 3′-UTR region (Figure 2a–d). The intron has a consensus splice junction following the GT-AG rule [36]. Moreover, the gene organization of the four putative defensins closely resembles other mollusk and arthropod defensin genes: (i) the exon encoding the mature defensin is not split by any intron and (ii) the intron flanking the exon encoding the mature defensin shares a strict level of phase conservation (phase I) (Figure 2a-e). The mature defensin encoded by the second exon likely displays folding autonomy as one structural entity [35,37]. These four putative defensins also contain six cysteine residues in a CX4–15CX2HCX6–9GX1CX4–9CX1C distribution, the typical cysteine pattern of the invertebrate CSαβ defensins that induces the CSαβ fold [35,36]. These features further suggest that the four PCR-validated defensin genes from the scorpion M. martensii belong to the invertebrate CSαβ defensin family.
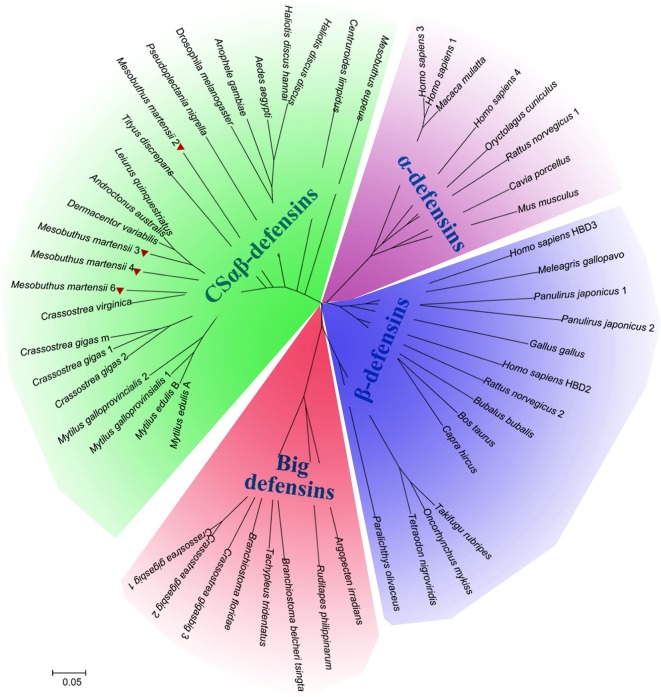
After confirming that their gene organizations and distribution of cysteine residues were consistent with those of the other invertebrate CSαβ defensins, we further estimated the evolutionary status of these four putative scorpion defensins by building a phylogenetic tree including sequences from other defensin families belonging to a variety of species and phyla. Phylogenetic analysis of vertebrate α- and β-defensin, invertebrate CSαβ defensin, and big defensin sequences revealed that the four PCR-validated putative defensins (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) from the scorpion M. martensii were clustered into the invertebrate CSαβ defensin family (Figure 3).
Figure 3. Phylogenetic analysis and classification of defensin genes from the scorpion M. martensii.
The tree was constructed using the Neighbor-Joining method in MEGA 5.1. Bootstrap sampling was reiterated 10000 times. We obtained a similar topology tree with the Maximum-Parsimony method in MEGA 5.1. The sequence information used in the defensin analysis is included in the Supplementary Table S2. The phylogenetic tree showed that the animal defensin superfamily clusters into four families: CSαβ-defensins, big defensins, α-defensins and β-defensins. The four defensins from the scorpion M. martensii are highlighted with red triangles and clustered into the invertebrate CSαβ defensin family clade only.
CON of CSαβ defensin genes from the scorpion M. martensii
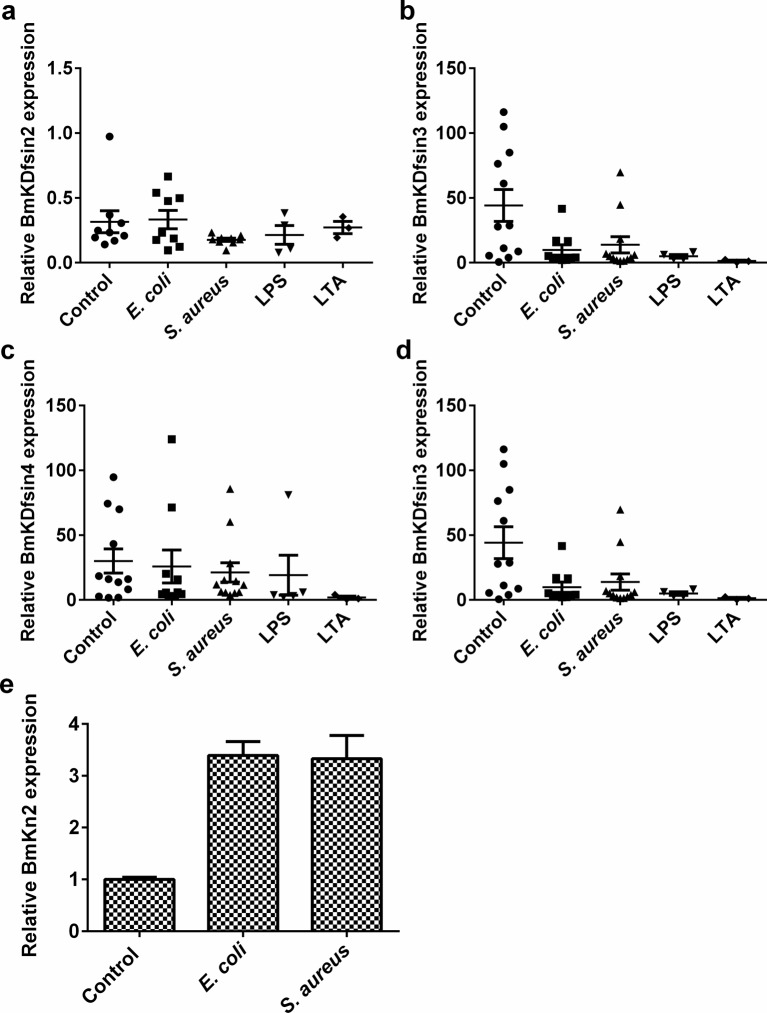
Defensins are important components of host innate immune systems with defensive function against pathogens. The transcription of defensin gene is either inducible or constitutive when challenged by a pathogenic microorganism. We developed an intact scorpion (M. martensii) challenge model using E. coli, S. aureus, LPS or LTA to determine whether the four scorpion defensin genes (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) undergo inducible or constitutive mRNA expression in vivo. Scorpions were injected with bacteria or bacterial components. After 36 h, the scorpions were ground to analyse the mRNA expression of the four scorpion defensin genes. Real-time PCR indicated no obvious difference of their mRNA expression between the treated and untreated groups (Figure 4a–d). However, we found that some genes encoding antibacterial peptides like BmKn2 from the scorpion M. martensii can be induced using the same sample (Figure 4e). This solidly proved the validity of our intact scorpion challenge model. Thus, BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6 from the scorpion M. martensii were revealed to constitutively express at the transcriptional level.
Figure 4. In vivo transcriptional activity analysis of four defensins from the scorpion M. martensii.
The mRNA expression of four defensins (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) from the scorpion M. martensii was analysed. The scorpion M. martensii was challenged with 5 μl of S. aureus (OD600 =0.6), E. coli (OD600 =0.6), LPS (200 ng/ml) or LTA (200 ng/ml). PBS was used as an unchallenged control. After 36 h, the whole scorpions were ground and defensin mRNA expression was analysed. The antibacterial peptide gene BmKn2 was detected as the positive control. (a) BmKDfsin2. (b) BmKDfsin3. (c) BmKDfsin4. (d) BmKDfsin6. (e) BmKn2.
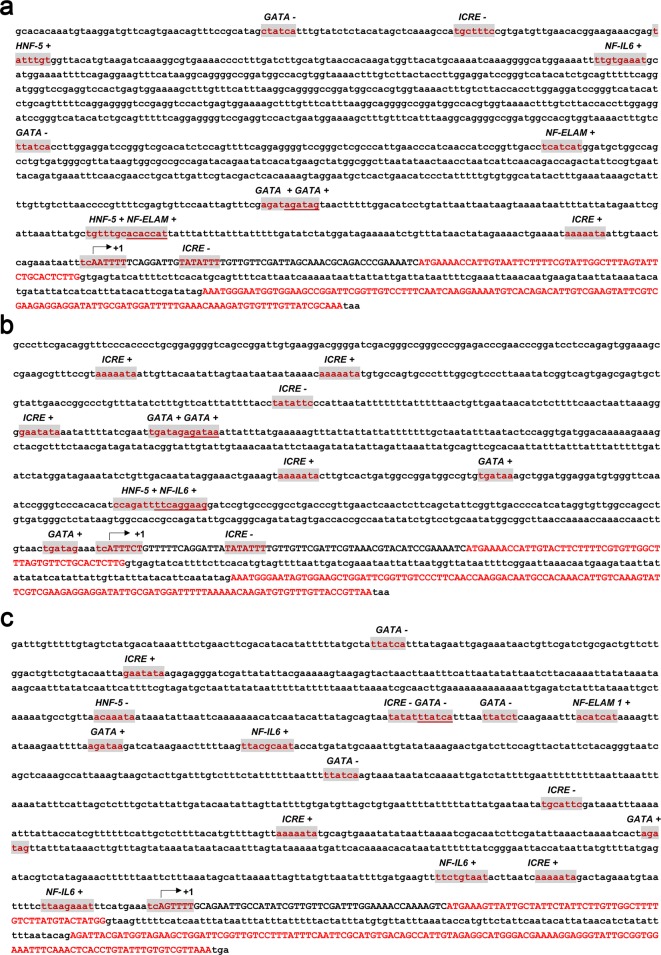
Constitutive mRNA expression of the four scorpion defensins observed in the challenge experiment suggests a lack of immune response elements in the upstream regulatory regions of their promoters. Consequently, we attempted to clone and characterize their promoter regions based on the sequence of the scorpion M. martensii genome. Among the PCR-validated four scorpion defensins, the putative promoter regions of only three defensin genes (BmKDfsin3, BmKDfsin4 and BmKDfsin6) were successfully amplified. They all covered ~1500 nts upstream of the initiation codon (Supplementary Figure S2). We tried hard and designed several pairs of primers and attempted a number of PCR methods but we failed to successfully amplify the promoter sequence of BmKDfsin2. We attributed it to that the the current assembled genome of the scorpion M. martensii that it is a draft genome and the upstream promoter sequence of BmKDfsin2 is probably wrongly assembled. Those three promoter regions were sequenced and first analysed to detect potential initiator elements according to the eukaryotic initiator element consensus [(TC)CA+1N(TA)(TC)(TC)(TC)] (N could be any of the four bases and A+1 is the first base of the transcription start site) [33,36]. BmKDfsin3, BmKDfsin4 and BmKDfsin6 have initiator elements with conserved sequences of TCA+1ATTTT, TCA+1TTTCT and TCA+1GTTTT respectively (Figure 5). The initiator element is an alternative promoter element capable of replacing the function of the TATA box, and its presence potentially explains the absence of a TATA box ~30 nts upstream of the transcription start site [38]. Next, the P-Match program and AliBaba2 program were used in conjunction with the TRANSFAC database to identify other putative transcription factor binding sites in the promoter regions of BmKDfsin3, BmKDfsin4 and BmKDfsin6 [39,33]. The result showed that their promoter regions have a variety of transcription factor binding sites, including nuclear factor interleukin 6 (NF-IL6) consensus TKNNGNAAK (K is G or C), GATA factor consensus WGATAR (W is A or T, and R is A or G), interferon consensus response element (ICRE) motif RAAWRYA (Y is C or T), hepatic nuclear factor 5 (HNF-5) consensus TRTTTGY, and nuclear factor endothelial leucocyte adhesion molecule 1 (NF-ELAM1) element WCAKCAK (Figure 5). Although the number of transcription factor binding sites varies between BmKDfsin3, BmKDfsin4 and BmKDfsin6, the types of response elements are similar. Briefly, BmKDfsin3 promoter region contains one NF-IL6, four GATA factor, three ICRE, one HNF-5 and two NF-ELAM1 sites (Figure 5a). BmKDfsin4 promoter region has four copies of GATA factor, one copy of NF-IL6, six copies of ICRE, and one copy of HNF-5, but no copy of NF-ELAM1 (Figure 5b). The promoter region of BmKDfsin6 covers six copies of GATA factor motif, two copies of NF-IL6 element, five copies of ICRE, one copy of HNF-5 consensus and one copy of NF-ELAM1 site (Figure 5c). All these elements are homologous to transcription factor binding sequences previously characterized in Drosophila [40], mosquitoes [39] and scorpions [33,36]. Thus, these conserved elements likely play similar roles in the regulation of BmKDfsin3, BmKDfsin4 and BmKDfsin6 genes during immune response. Their lack of the nuclear factor κB (NF-κB) binding site, which is a critical member of inducible immune factors and has been found in promoter regions of almost inducible SCAMP genes in animals [41], suggests that BmKDfsin3, BmKDfsin4 and BmKDfsin6 have the expression feature of CON.
Figure 5. Promoter sequence analysis of three defensin genes from the scorpion M. martensii.
The cloned promoter regions of BmKDfsin3, BmKDfsin4 and BmKDfsin6 were sequenced and analysed to identify potential transcriptional regulating elements. The potential transcriptional regulating elements are highlighted with red letters and grey background. The sequences of the ORFs are presented in red capital letters. (a) BmKDfsin3. (b) BmKDfsin4. (c) BmKDfsin6.
Complementary to the promoter sequence analysis, their promoter activities were measured by a luciferase assay. Upstream promoter regions of BmKDfsin3, BmKDfsin4 and BmKDfsin6 were subcloned into luciferase reporter vectors called pBmKDfsin-3, 4 and 6 respectively. Because there is no cultured scorpion cell lines to date, the constructs were transfected into the human HEK293T cells which have high transfection efficiency or the S2 cells from Drosophila which have closer relationship with scorpions than humans. Their promoter activities, as indicated by the ratio of firefly luciferase and Renilla luciferase activities, did not change upon stimulation with bacterial components (Figure 6). Taken together, the result was consistent with their in vivo mRNA expression and promoter sequence characters, which firmly concluded that BmKDfsin3, BmKDfsin4 and BmKDfsin6 are constitutive at the transcriptional level.
Figure 6. Dual luciferase assay of the promoter activity of three defensin genes in HEK293T and Drosophila S2 cells.
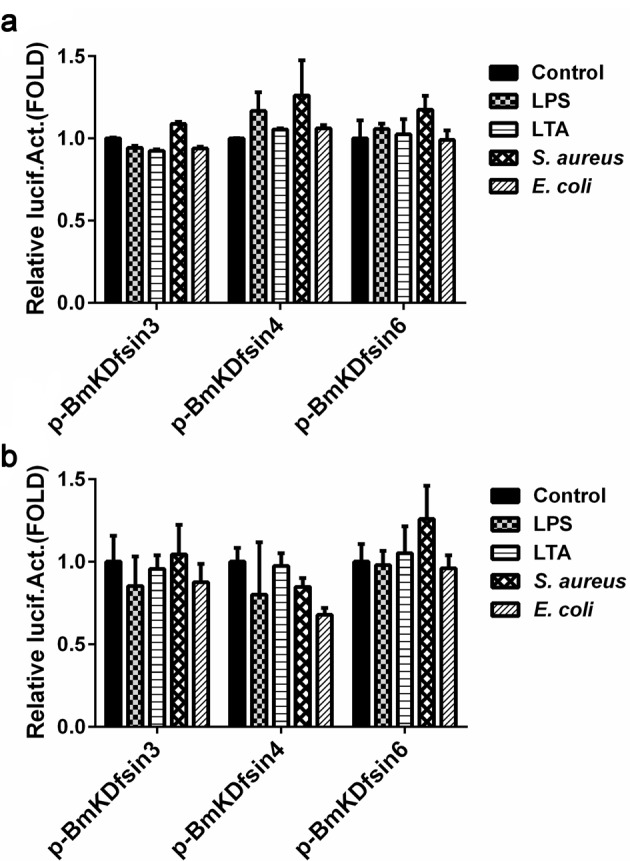
HEK293T or Drosophila S2 cells were transfected with pBmKDfsin-3, 4 or 6 vector respectively. At 6 h post transfection, the cells were treated with 200 ng/ml LPS, 500 ng/ml LTA, 50 μg/ml S. aureus or E. coli bacterial materials. PBS treatment was used as the negative control. The dual luciferase assay was performed at 48 h post transfection. (a) HEK293T cell line. (b) Drosophila S2 cell line.
Discussion
CON of defensins or defensin-like genes from scorpions
We validated four defensin genes (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) from the scorpion M. martensii at both the genome and cDNA levels. BmKDfsin4 was recently identified at the protein level by our research group [42]. The four defensins (BmKDfsin2, BmKDfsin3, BmKDfsin4 and BmKDfsin6) were classified into the invertebrate CSαβ defensin family based on their genomic organization, predicted amino acid sequences and phylogenetic analysis. Using whole-scorpion bacterial infection model, sequencing of the potential promoter region and a dual luciferase promoter activity assay, we concluded that BmKDfsin3, BmKDfsin4 and BmKDfsin6 are constitutively expressed at the mRNA level, which was consistent with the expression of previously investigated scorpion defensins. The defensins LqDef [27] and AaDef [28] from the scorpions L. quinquestriatus and A. australis respectively, are both constitutively expressed independent of immune stimulus. Another scorpion defensin, Cll-dlp [30], exhibits an inducible liberation of stored peptides following CON. Additionally, some scorpion defensin-like peptides, such as scorpine [43], opiscorpines 1–4 [36] and Heteroscorpine-1 [44], exhibit no response to immune stimuli and have identical patterns of constitutive expression. All these data suggest that most defensins or defensin-like genes from scorpions are constitutively expressed.
Diversification and evolution of defensin expression patterns in invertebrates
Defensins are a large family of antimicrobial peptides that belong to a variety of categories and that exhibit highly complex expression patterns in response to immune challenge [45]. Vertebrate α-defensins are frequently confirmed to be constitutively expressed in paneth cells and neutrophils of humans, monkeys and rodents [18]. By contrast, almost all vertebrate β-defensins exhibit inducible expression, except HBD1, which has high levels of basal expression but can also be induced [18,46,47]. The expression patterns of invertebrate CSαβ defensins are most complex and include the following three types: CON independent of exogenous immune stimulus, IND in response to an immune challenge or inducible protein release from storage in haemocyte granules stimulated by immune challenge but without transcriptional regulation [30]. The two main expression patterns of vertebrate defensins and three types of invertebrate defensins belong to two types of expression at the transcriptional level: one is CON independent of exogenous immune stimulus and the other is IND in response to an immune challenge.
In our study, we characterized the diversity and evolution of the expression patterns of defensin genes by analysing the most widespread group of defensins, the invertebrate CSαβ defensin family. As was shown in Table 1 and Figure 7, CSαβ defensin genes from insects, lower invertebrates (including nematodes, annelids, cnidarians and sponges), and some of arachnids and mollusks were found to be IND, whereas those from most of arachnids and mollusks are CON (Table 1 and Figure 7). The ancient species of invertebrates clearly had the IND type of CSαβ defensins.
Table 1. Typical invertebrate CSαβ defensin genes and their expression patterns.
| Abbreviated name and category | Species | Expression pattern* | Accession number |
|---|---|---|---|
| Insects | |||
| AaeA | Aedes aegypti | IND | P91793 |
| AaeB | Aedes aegypti | IND | AAD40114 |
| AaeC | Aedes aegypti | IND | AAD40116 |
| Adi | Allomyrina dichotoma | IND | Q10745 |
| Aga | Anopheles gambiae | IND | Q17027 |
| Ame1 | Apis mellifera | IND | C55392 |
| Bpa | Bombus pascuorum | IND | P81462 |
| Dme | Drosophila melanogaster | IND | P36192 |
| Ete | Eristalis tenax | IND | CAM92111 |
| Orh | Oryctes rhinoceros | IND | O96049 |
| Pap | Pyrrhocoris apterus | IND | P37364 |
| PteA | Phormia terraenovae | IND | 1ICA |
| PteB | Phormia terraenovae | IND | P10891 |
| Sbu | Sarcophaga bullata | IND | 1L4V |
| Sca1 | Stomoxys calcitrans | IND | O16136 |
| Sca2 | Stomoxys calcitrans | IND | O16137 |
| Tmo | Tenebrio molitor | IND | BAA04552 |
| ZatA | Zophobas atratus | IND | AAB20745 |
| ZatB | Zophobas atratus | IND | AAB20746 |
| Ace | Apis cerana | NC | ACH96385 |
| Ame2 | Apis mellifera | NC | P17722 |
| Big | Bombus ignites | NC | AAQ94318 |
| Cvi | Calliphora vicina | NC | C0HJX7 |
| Faq | Formica aquilonia | NC | AY875720 |
| Fru | Formica rufa | NC | 9672756 |
| Hdi | Holotrichia diomphalia | NC | JC2554 |
| HrhA | Rhodnius prolixus | NC | AAO74624 |
| HrhB | Rhodnius prolixus | NC | AAO74625 |
| HrhC | Rhodnius prolixus | NC | AAO74626 |
| Mde1 | Mayetiola destructor | NC | AAY82237 |
| Ppr | Palomena prasina | NC | P80407 |
| SpeA | Sarcophaga peregrina | NC | P18313 |
| SpeC | Sarcophaga peregrina | NC | P31530 |
| Arachnids | |||
| Ahe1 | Amblyomma hebraeum | IND | AY437137 |
| Hlo2 | Haemaphysalis longicornis | IND | ABO28925 |
| Hlo3 | Haemaphysalis longicornis | IND | ABO28926 |
| OmoA | Ornithodoros moubata | IND | BAB41028 |
| OmoB | Ornithodoros moubata | IND | BAB41027 |
| OmoC | Ornithodoros moubata | IND | BAC10303 |
| OmoD | Ornithodoros moubata | IND | BAC10304 |
| Aau | Androctonus australis | CON | P56686 |
| Bmi | Boophilus microplus | CON | AAO48943 |
| Csa | Cupiennius salei | CON | |
| Cli | Centruroides limpidus | CON | P83738 |
| Hlo1 | Haemaphysalis longicornis | CON | AB105544 |
| Ipe | Ixodes persulcatus | CON | BAH09304 |
| Isc1 | Ixodes scapularis | CON | AAV74387 |
| Lqu | Leiurus quinquestriatus | CON | P41965 |
| Mma2 | Mesobuthus martensii | CON | |
| Mma3 | Mesobuthus martensii | CON | |
| Mma4 | Mesobuthus martensii | CON | |
| Mma6 | Mesobuthus martensii | CON | |
| Ahe2 | Amblyomma hebraeum | NC | AY437138 |
| Asp | Argiope spp | NC | AAW01790 |
| Dva | Dermacentor variabilis | NC | AAO24323 |
| Iri1 | Ixodes ricinus | NC | DQ361064 |
| Iri2 | Ixodes ricinus | NC | AY335442 |
| Isc2 | Ixodes scapularis | NC | EEC17844 |
| Isc3 | Ixodes scapularis | NC | EEC08554 |
| Meu | Mesobuthus eupeus | NC | ABR21037 |
| Mgi | Mesobuthus gibbosus | NC | CAL48845 |
| Tdi | Tityus discrepans | NC | P0CF77 |
| Mollusks | |||
| Rph | Ruditapes philippinarum | IND | ALO24364 |
| MedA | Mytilus edulis | CON | P81610 |
| MedB | Mytilus edulis | CON | P81611 |
| Cgi | Crassostrea gigas | NC | ACQ76287 |
| Cvi | Crassostrea virginica | NC | P85008 |
| Mga1 | Mytilus galloprovincialis | NC | AAD45117 |
| Mga2 | Mytilus galloprovincialis | NC | AAD45118 |
| Mga2b | Mytilus galloprovincialis | NC | AAD52660 |
| Nematodes | |||
| Asu-α | Ascaris suum | IND | BAA89497 |
| Asu-β | Ascaris suum | IND | BAC00497 |
| Asu-γ | Ascaris suum | IND | BAC00498 |
| Asu-δ | Ascaris suum | IND | BAC00499 |
| Asu-ε | Ascaris suum | NC | BAC41495 |
| Asu-ζ | Ascaris suum | NC | BAC57992 |
| Asu-6Cys-α | Ascaris suum | NC | AB086059 |
| Cel1 | Caenorhabditis elegans | NC | BAA89489 |
| Cel2 | Caenorhabditis elegans | NC | BAA89490 |
| Annelids | |||
| Hme | Hirudo medicinalis | IND | EU156754 |
| Tte | Theromyzon tessulatum | IND | AY434032 |
| Cnidarians | |||
| Hvu | Hydra vulgaris | IND | B3RFR8 |
| Sponges | |||
| Sdo | Suberites domuncula | IND | CCC55928 |
| Lba | Lubomirskia baicalensis | NC | CCC55929 |
NC indicates not clear.
*CON indicates constitutive transcription
*IND indicates inducible transcription
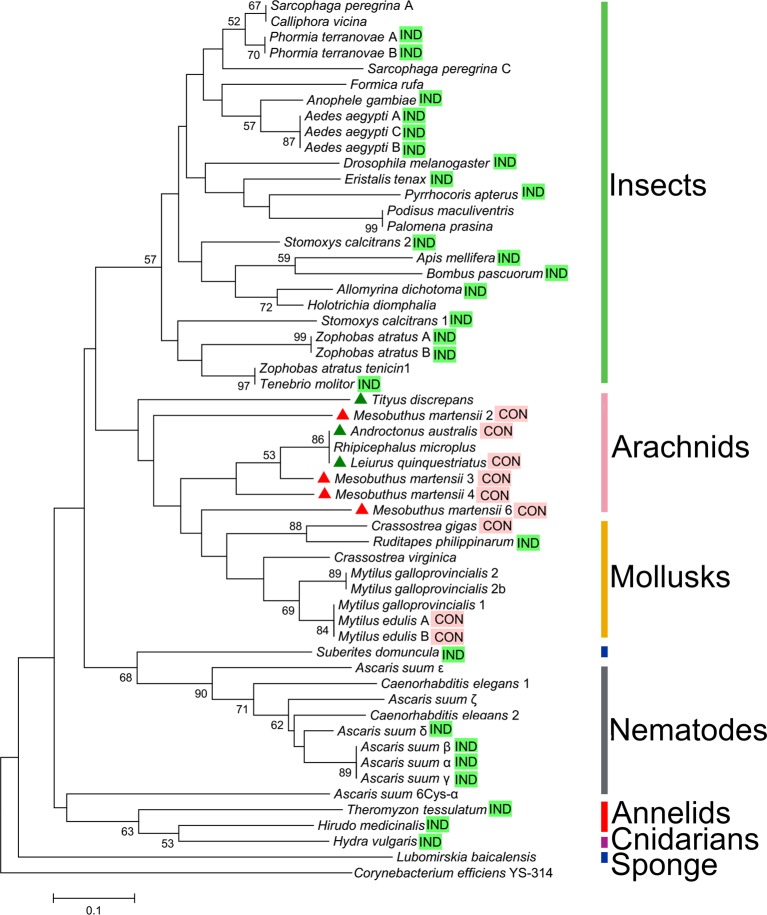
Figure 7. Diversification of CSαβ defensin expression patterns in invertebrates.
The phylogenetic tree was constructed using the Neighbor-Joining method in MEGA 5.1. Bootstrap sampling was reiterated 10000 times. Numbers above nodes represent bootstrap values greater than 50% (values below 50% not shown). The defensin of the bacterium Corynebacterium efficiens YS-314 was used as the outgroup. Evolutionary analysis using the Maximum-Parsimony method yielded a similar topology tree. The sequence information included in the analysis of the defensins is presented in Table 1 and Supplementary Table S2. CSαβ defensins with the expression pattern of IND are indicated with bright green background ‘IND’, while CSαβ defensins with the expression mode of CON are indicated with pink background ‘CON’. The four defensins from the scorpion M. martensii are highlighted with red triangles, while defensins from other scorpion species are indicated with green triangles.
Evolutionary driving force and possible biological role of invertebrate CSαβ defensin expression types
Since CSαβ defensins from invertebrates have undergone unique routes of diversification and evolution along with the evolution of species from low to high, the driving force and potential biological role behind the evolution and diversification of the CSαβ defensin family are of interest. As shown in Figure 7, the IND type of CSαβ defensins is found to exist in all insects and lower invertebrates, and some mollusks and arachnids, whereas the CON expression type of CSαβ defensins occurs only in both mollusks and arachnids. In addition, CSαβ defensins of insects and arachnids exhibit significantly different features of molecular evolution. CSαβ defensins from the same species of insects (such as Zophobas atratus, Phormia terraenovae and Aedes aegypti) cluster together and share the same expression type with each other, suggesting that the CSαβ defensin genes in these species are the product of a recent gene duplication event and that the expression type was established before this event. Most of the CSαβ defensins arising from gene duplication exhibit IND expression. In species other than insects, CSαβ defensins from the same species (such as the scorpion M. martensii and the tick Haemaphysalis longicornis) do not cluster together, but most have the same expression type as arachnids, which implies that CSαβ defensin genes from these species underwent gene duplication before the split of scorpions, spiders and ticks and that the CON expression type of CSαβ defensins was formed after the gene duplication. This result was also consistent with the occurrence of CSαβ defensin CON expression type in mollusks. The use of the same CSαβ defensin expression mode within species and between closely genetically related species implies that the driving force behind the evolution of CSαβ defensin expression modes in invertebrates may be the similar immune challenges faced by these species. Most mollusk and arachnid species, such as snails and scorpions, usually live in an environment rich of a wide variety of microbial populations. Evolutionary CON expression type of CSαβ defensin genes in mollusks and arachnids possibly help them to defend microbial infection.
Supporting information
Fig S1.
PCR validation of defensin genes from the scorpion M. martensii
The genomic DNA of the scorpion M. martensii was used as the PCR template. M, 1 Kb DNA Ladder. 1, BmKDfsin1. 2, BmKDfsin2. 3, BmKDfsin3. 4, BmKDfsin4. 5, BmKDfsin5. 6, BmKDfsin6.
Fig S2.
PCR amplification of promoter regions of three defensin genes the scorpion M. martensii
Three defensins (BmKDfsin3, BmKDfsin4 and BmKDfsin6) from the scorpion M. martensii were selected to clone their promoter regions. M, 1Kb DNA Ladder. 1, BmKDfsin3. 2, BmKDfsin4. 3, BmKDfsin6.
Table S1. PCR primers for the validation of potential defensin genes.
Table S2. Amino acid sequence information of defensins used in Figure 3 and Figure 7.
Table S3. Real-time PCR primers for M. martensii defensin and actin genes.
Table S4. Primers for cloning the promoter regions of defensin genes.
Table S5. Primers for inserting the promoter regions of into pGL3-Basic vector.
Table S6. Six potential defensin genes characterized from the scorpion M. martensii genome.
Abbreviations
- CCTCC
China Center for Type Culture Collection
- CON
constitutive transcription
- CSαβ
cysteine-stabilized α-helix/β-sheet motif defensin
- HEK293T
human embryonic kidney
- HNF-5
hepatic nuclear factor 5
- ICRE
interferon consensus response element
- IND
inducible transcription
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- NF-ELAM1
nuclear factor endothelial leucocyte adhesion molecule 1
- NF-IL6
nuclear factor interleukin 6
Funding
This work was supported by the National Science Fund of China [grant numbers 31422049, 31572289, 81630091]; the International S&T Cooperation Program of China [grant number S2016G3110]; the Hubei Science Fund [grant numbers 2015CFA042, 2016CFA018]; the China-Kazakhstan Cooperation Program [grant number CK-07-09]; and the Fundamental Research Funds for the Central Universities in China [grant number 2042017kf0242].
Author contribution
Y.L. and Z.C. designed the experiments and analysed the data. Y.L. and X.P. performed most of the experiments. X.P., Z.D. and Q.Z. cloned the defensin genes and collected the defensin sequence. H.W., B.S., F.L. and G.L. helped to perform experiments. Y.Y. and X.L. performed the genomic sequence. Y.W. and W.L. helped to design the study. Y.L. and Z.C. wrote the manuscript.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bulet P., Stocklin R. and Menin L. (2004) Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 198, 169–184 [DOI] [PubMed] [Google Scholar]
- 2.Froy O. (2005) Convergent evolution of invertebrate defensins and nematode antibacterial factors. Trends Microbiol. 13, 314–319 [DOI] [PubMed] [Google Scholar]
- 3.Tassanakajon A., Somboonwiwat K. and Amparyup P. (2015) Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 48, 324–341 [DOI] [PubMed] [Google Scholar]
- 4.Shafee T.M., Lay F.T., Phan T.K., Anderson M.A. and Hulett M.D. (2016) Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 74, 663–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehrer R.I. (2007) Multispecific myeloid defensins. Curr. Opin. Hematol. 14, 16–21 [DOI] [PubMed] [Google Scholar]
- 6.Wong J.H., Xia L. and Ng T.B. (2007) A review of defensins of diverse origins. Curr. Protein Pept. Sci. 8, 446–459 [DOI] [PubMed] [Google Scholar]
- 7.Hazlett L. and Wu M. (2011) Defensins in innate immunity. Cell Tissue Res. 343, 175–188 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen T.X., Cole A.M. and Lehrer R.I. (2003) Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides 24, 1647–1654 [DOI] [PubMed] [Google Scholar]
- 9.Charlet M., Chernysh S., Philippe H., Hetru C., Hoffmann J.A. and Bulet P. (1996) Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc, Mytilus edulis. J. Biol. Chem. 271, 21808–21813 [DOI] [PubMed] [Google Scholar]
- 10.Hubert F., Noel T. and Roch P. (1996) A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur. J. Biochem. 240, 302–306 [DOI] [PubMed] [Google Scholar]
- 11.Andersson M., Boman A. and Boman H.G. (2003) Ascaris nematodes from pig and human make three antibacterial peptides: isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 60, 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato Y., Aizawa T., Hoshino H., Kawano K., Nitta K. and Zhang H. (2002) abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato Y. and Komatsu S. (1996) ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum. Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem. 271, 30493–30498 [DOI] [PubMed] [Google Scholar]
- 14.Dimarcq J.L., Bulet P., Hetru C. and Hoffmann J. (1998) Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers 47, 465–477 [DOI] [PubMed] [Google Scholar]
- 15.Carvalho Ade O. and Gomes V.M. (2009) Plant defensins-prospects for the biological functions and biotechnological properties. Peptides 30, 1007–1020 [DOI] [PubMed] [Google Scholar]
- 16.Zhu S. (2008) Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol. Immunol. 45, 828–838 [DOI] [PubMed] [Google Scholar]
- 17.Kouno T., Fujitani N., Mizuguchi M., Osaki T., Nishimura S., Kawabata S. et al. (2008) A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria. Biochemistry 47, 10611–10619 [DOI] [PubMed] [Google Scholar]
- 18.Zhu S. and Gao B. (2013) Evolutionary origin of beta-defensins. Dev. Comp. Immunol. 39, 79–84 [DOI] [PubMed] [Google Scholar]
- 19.Saito T., Kawabata S., Shigenaga T., Takayenoki Y., Cho J., Nakajima H. et al. (1995) A novel big defensin identified in horseshoe crab hemocytes: isolation, amino acid sequence, and antibacterial activity. J. Biochem. 117, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 20.Teng L., Gao B. and Zhang S. (2012) The first chordate big defensin: identification, expression and bioactivity. Fish Shellfish Immunol. 32, 572–577 [DOI] [PubMed] [Google Scholar]
- 21.Sunagar K., Undheim E.A., Chan A.H., Koludarov I., Muñoz-Gómez S.A., Antunes A. et al. (2013) Evolution stings: the origin and diversification of scorpion toxin peptide scaffolds. Toxins (Basel) 5, 2456–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunlop J.A. and Selden P.A. (2009) Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Exp. Appl. Acarol. 48, 183–197 [DOI] [PubMed] [Google Scholar]
- 23.Lourenço W.R. (2014) Scorpion diversity and distribution: Past and present patterns. In Scorpion Venoms, Toxinology, vol. 1, pp. 1–20, Springer Science+Business Media, Dordrecht [Google Scholar]
- 24.Wang Z., Wang W., Shao Z., Gao B., Li J., Ma J. et al. (2009) Eukaryotic expression and purification of anti-epilepsy peptide of Buthus martensii Karsch and its protein interactions. Mol. Cell. Biochem. 330, 97–104 [DOI] [PubMed] [Google Scholar]
- 25.Charles H.M. and Killian K.A. (2015) Response of the insect immune system to three different immune challenges. J. Insect Physiol. 81, 97–108 [DOI] [PubMed] [Google Scholar]
- 26.Baxter A.A., Richter V., Lay F.T., Poon I.K., Adda C.G., Veneer P.K. et al. (2015) The tomato defensin TPP3 binds phosphatidylinositol (4,5)-bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol. Cell. Biol. 35, 1964–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cociancich S., Goyffon M., Bontems F., Bulet P., Bouet F., Menez A. et al. (1993) Purification and characterization of a scorpion defensin, a 4kDa antibacterial peptide presenting structural similarities with insect defensins and scorpion toxins. Biochem. Biophys. Res. Commun. 194, 17–22 [DOI] [PubMed] [Google Scholar]
- 28.Ehret-Sabatier L., Loew D., Goyffon M., Fehlbaum P., Hoffmann J.A., van Dorsselaer A. et al. (1996) Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. J. Biol. Chem. 271, 29537–29544 [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann J.A. and Hetru C. (1992) Insect defensins: inducible antibacterial peptides. Immunol. Today 13, 411–415 [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez de la Vega R.C., Garcia B.I., D’Ambrosio C., Diego-Garcia E., Scaloni A. and Possani L.D. (2004) Antimicrobial peptide induction in the haemolymph of the Mexican scorpion Centruroides limpidus limpidus in response to septic injury. Cell. Mol. Life Sci. 61, 1507–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Z., Yu Y., Wu Y., Hao P., Di Z., He Y. et al. (2013) The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 4, 2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima Y., Saido-Sakanaka H., Taylor D. and Yamakawa M. (2003) Up-regulated humoral immune response in the soft tick, Ornithodoros moubata (Acari: Argasidae). Parasitol. Res. 91, 476–481 [DOI] [PubMed] [Google Scholar]
- 33.Gao B., Tian C. and Zhu S. (2007) Inducible antibacterial response of scorpion venom gland. Peptides 28, 2299–2305 [DOI] [PubMed] [Google Scholar]
- 34.Baumann T., Kuhn-Nentwig L., Largiader C.R. and Nentwig W. (2010) Expression of defensins in non-infected araneomorph spiders. Cell. Mol. Life Sci. 67, 2643–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froy O. and Gurevitz M. (2003) Arthropod and mollusk defensins–evolution by exon-shuffling. Trends Genet. 19, 684–687 [DOI] [PubMed] [Google Scholar]
- 36.Zhu S. and Tytgat J. (2004) The scorpine family of defensins: gene structure, alternative polyadenylation and fold recognition. Cell. Mol. Life Sci. 61, 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gueguen Y., Herpin A., Aumelas A., Garnier J., Fievet J., Escoubas J.M. et al. (2006) Characterization of a defensin from the oyster Crassostrea gigas. Recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J. Biol. Chem. 281, 313–323 [DOI] [PubMed] [Google Scholar]
- 38.Cherbas L. and Cherbas P. (1993) The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem. Mol. Biol. 23, 81–90 [DOI] [PubMed] [Google Scholar]
- 39.Eggleston P., Lu W. and Zhao Y. (2000) Genomic organization and immune regulation of the defensin gene from the mosquito, Anopheles gambiae. Insect Mol. Biol. 9, 481–490 [DOI] [PubMed] [Google Scholar]
- 40.Dimarcq J.L., Hoffmann D., Meister M., Bulet P., Lanot R., Reichhart J.M. et al. (1994) Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur. J. Biochem. 221, 201–209 [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann J.A. and Reichhart J.M. (2002) Drosophila innate immunity: an evolutionary perspective. Nat. Immun. 3, 121–126 [DOI] [PubMed] [Google Scholar]
- 42.Meng L., Xie Z., Zhang Q., Li Y., Yang F., Chen Z. et al. (2016) Scorpion potassium channel-blocking defensin highlights a functional link with neurotoxin. J. Biol. Chem. 291, 7097–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conde R., Zamudio F.Z., Rodriguez M.H. and Possani L.D. (2000) Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett. 471, 165–168 [DOI] [PubMed] [Google Scholar]
- 44.Uawonggul N., Thammasirirak S., Chaveerach A., Arkaravichien T., Bunyatratchata W., Ruangjirachuporn W. et al. (2007) Purification and characterization of Heteroscorpine-1 (HS-1) toxin from Heterometrus laoticus scorpion venom. Toxicon 49, 19–29 [DOI] [PubMed] [Google Scholar]
- 45.Dimarcq J.L. and Hunneyball I. (2003) Pharma-entomology: when bugs become drugs. Drug Discov. Today 8, 107–110 [DOI] [PubMed] [Google Scholar]
- 46.Pazgier M., Hoover D.M., Yang D., Lu W. and Lubkowski J. (2006) Human beta-defensins. Cell. Mol. Life Sci. 63, 1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu B.D., Feng Y., Huang N., Wu Q. and Wang B.Y. (2003) Mycobacterium bovis bacille Calmette-Guerin (BCG) enhances human beta-defensin-1 gene transcription in human pulmonary gland epithelial cells. Acta Pharmacol. Sin. 24, 907–912 [PubMed] [Google Scholar]