Abstract
One of the treatment failures for colorectal cancer (CRC) is resistance to chemotherapy drugs. miRNAs have been demonstrated to be a new regulator of pathobiological processes in various tumors. While few studies have explored the specific role of miR-141 in mediating 5-fluorouracil (5-FU) sensitivity of CRC cells, the present study aimed to detect the contribution of miR-141 in 5-FU sensitivity. The CRC cells viability was measured by MTS assay and cell colony forming. The expression of miR-141 and its downstream targets were assessed by reverse transcription quantitative PCR, Western blotting, and immunohistochemistry. The functional assays were conducted using CRC cells and nude mice. At the present study, we found overexpression of miR-141 could inhibit proliferation, migration, tumor-forming and invasive potential of CRC cells in vitro and mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) was verified as a directed target of miR-141. The combination treatment of miR-141 with 5-FU, directly targetting MAP4K4, could better inhibit invasion and metastasis of CRC cells colony than either one alone. Furthermore, overexpression of miR-141, targetting MAP4K4, enhanced the effected of 5-FU and suppressed the malignant biological behaviors, in vivo. Our findings showed that 5-FU inhibited malignant behavior of human CRC cells in vitro and in vivo by enhancing the efficiency of miR-141. Our data suggested that targetting the miR-141/MAP4K4 signaling pathway could be a potential molecular target that may enhance chemotherapeutic efficacy in the treatment of CRC.
Keywords: Colorectal cancer, miR-141, MAP4K4, 5-FU
Introduction
In China, colorectal cancer (CRC) is the third and fourth most common high mortality cancer of males and females respectively [1]. Surgery including chemotherapy and/or radiotherapy is the most basic treatment method in the stage patients. Approximately 25–50% of patients with metastasis CRC miss the optimal treatment opportunity [2]. Therefore, the effective diagnosis and treatment for CRC are urgent needed. Recently, identifying the molecular target in early diagnosis, therapy and prognosis of cancer have become the focus of anticancer research field. However, the formation mechanism, prevention, and treatment modality of CRC has not been fully illuminated. Therefore, understanding the pathological mechanism of CRC and searching for novel targets for preventing CRC remains to be needed.
miRNAs are a class of noncoding RNA, which play an important role in regulating gene-expression programs, by binding to target genes [3]. Some miRNAs act as tumor suppressors, whereas others, when deregulated overexpressed, can result in the tumor initiation [4,5]. Our previous research has concluded the expression of miR-141 in the CRC tissues is lower than that of the normal colonic mucosa [6]. Moreover, the up-regulation of mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) may be associated with the down-regulation of miR-141 in the CRC [6]. MAP4K4 belongs to the sterile-20 protein kinase family and is involved in many cellular processes, including cell transformation, adhesion, and motility [7–10]. To explore the functional mechanism of miR-141 in the CRC, we have performed further research. Meanwhile, we hypothesized miR-141 affected the role of 5-fluorouracil (5-FU) in inhibiting the progression of CRC. Moreover, we supposed that the chemotherapeutic drugs could play their suppressive impact on CRC cell via disturbing the miR-141/ MAP4K4 signal pathway.
Materials and methods
Ethics statement
The present experimental methods were performed in accordance with the approved guidelines, which was approved by the Research Center, the Fourth Hospital of Hebei Medical University, Shijiazhuang, China.
Chemicals
RPMI-1640 medium, FBS and PBS were obtained from Gibco-BRL (Life Technologies, Paisley, Scotland). 5-FU (Sigma Chemical Co., Poole, U.K.) was diluted to 0, 12.5, and 25.0 µg/ml with RPMI-1640 medium. The nucleotide sequences (mimics, mimics control, inhibitor and inhibitor control) of miRNA-141 were designed and synthesized by RIBOBIO (Shanghai, China). 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) were obtained from Promega Corporation(Madison, WI, U.S.A.).
MTS and transfections
Human CRC cells of HCT-116 and HCT-8 are kind gift from Dr. Shi Juan (Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China). Cell viability was detected by MTS. Cells were seeded in a 96-well plate at a density of 10,000 cells/well for MTS assays cultured RPMI-1640 medium with 10% FBS at 37°C in a 5% CO2 cell culture incubator. Twenty microliter of MTS solution was added into each well and incubated for 0, 0.5, 1, 2, and 3 days. Then the optical density absorbance at 492 nm was measured using the microplate reader. HCT-116 and HCT-8 cells were cultured in a six-well plate at 40–60% confluence the day before transfection. miR-141 was transiently transfected to CRC cells by Lipofectmine 2000 reagent (Invitrogen) based on the manufacturer’s instructions.
Reverse transcription quantitative PCR and Western blot analysis
To detect the expression levels of miRNA-141 and MAP4K4 in CRC cells, total RNA was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, U.S.A.). For the detection of MAP4K4, the primer sequences were listed as follows: MAP4K4 forward, 5′-AAG GAG AGA GCG GGA AGC TA-3′, and reverse, 5′-TTG TTG CAA CTG CCT CTG GA-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for the internal reference and the primer sequences were listed as follows: GAPDH forward, 5′-GTT GGA GGT CGG AGT CAA CGGA-3′, and reverse, 5′-GAG GGA TCT CGC TCC TGG AGGA-3′. The PCR condition consisted of denaturation at 94°C for 5 min, followed by 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s, for a total of 35 cycles. For the detection of miRNA-141, the primer sequences were shown as follows: miR-141, 5′-CCG GTA ACA CTG TCT GGT AA-3′. U6 was utilized for the internal reference and the primer sequences were shown as follows: U6, 5′-GCT TCG GCA GCA CAT ATA CTA AAAT-3′. The PCR condition was as follows: denaturation at 95°C for 5 min, followed by 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s, for 40 cycles. The relative quantitation was calculated using the 2−ΔΔCT method.
Colon cancer cells were lysed on ice with lysis RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China), containing protease inhibitor. Protein concentration was determined using the BCA kit (Beyotime Institute of Biotechnology). Protein sample (50 ug) was subjected to 12% SDS/PAGE and transferred to a PVDF (Amersham Biosciences, Chicago, IL, U.S.A.). The blot was incubated with 5% skim milk at room temperature for 1 h and then incubated with primary antibodies: MAP4K4(1:1,000; cat. no. ab155583; Abcam, Cambridge, MA, U.S.A.), GAPDH (1:5,000; cat. no. ab9485; Abcam) at 4°C overnight. After washing with Tris Buffered Saline containing Tween (TBST) for three times, the membranes were incubated with goat antirabbit immunoglobulin G (1:3,000; cat. no. ab6721; Abcam) at room temperature for 1 h. Membranes were imaged using the Odyssey imaging system (U.S.A., LI-COR).
Colony-forming assay
Two hundred cells/well were seeded in a six-well plate and treated with compounds and cultured for 2 weeks in medium containing 10% FBS. After removing the medium, cells were washed with PBS for three times and then fixed with pure methanol for 15 min, and last stained in crystal violet for 25 min. Colony-forming unit of more than 50 cells was counted using the inverted microscope.
Cell migration and invasion assay
Cell invasion assay was evaluated using Transwell inserts (Corning Costar) with matrigel-coated membrane matrix (8 µm pore sizes). The HCT-116 and HCT-8 cells were treated with miR-141 mimics, 5-FU, or combination of miR-141 with 5-FU and then seeded in the upper chamber in the 24-well plate. The medium with FBS was added to the lower chamber. Serum-free medium was added in the lower chamber. The cells were cultured in the incubator at 37°C with 5% CO2 for 24 h and then removed with a cotton swab. After being fixed, stained, washed, and air dried, invading cells were counted under a microscope.
Wound-healing assay was conducted to evaluate the migration ability of colon cells. After treated by compounds, colon cells were cultured in a six-well plate at a density of 105 cells each well. At last, the cell monolayer was scratched with a 10-µl pipette tip. Images of the colon cancer cells with different treatment were taken using a microscope every 24 h. The cell-healing rate was calculated.
Target prediction of miR-141 and dual-luciferase reporter assay
The target of miR-141 was analyzed by bioinformatics as previous description. The mRNA 3′-UTR of MAP4K4 containing the predicted binding region or mutated binding region was subcloned into a basicluciferase reporter vector (Promega Corporation, WI, U.S.A.). Vector containing the wild type (WT) miR-141-MAP4K4 response element (MAP4K4-WT) and the corresponding mutant (MAP4K4-MUT) were purchase from RiboBio Co Ltd.(Guangzhou, China). The vectors containing WT or mimics controls were cotransfected into HCT-116 cells using Lipofectamine 2000. After 48 h, luciferase activities were detected by the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer’s protocol.
Animal experiment
All above animal experimental procedures were conducted in accordance with National Institutes protocols Health Guide for Care and Use of Laboratory Animals and Ethics Committee of the Fourth Hospital of Hebei Medical University. To conduct xenograft tumor experiment, HCT-116 cells were transfected with lenti-miR-41 and lenti-NC. After 24 h, cells were harvested and diluted in PBS, and then injected subcutaneously (2 × 106 cells/100 ml/mouse) [11] into the right limb of nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd). On day 7, the tumor volume was measured using a vernier caliper every 3 days. Once palpable tumors had developed (the 14 days), experimental group mice were treated with 5-FU, 5 mg/kg, every 3 days. The tumor volume was calculated using the following formulate: volume (mm3) = 1/2 × length × (width)2 [12] The nude mice were sacrificed and the tumors were dissected after 29 days.
Immunohistochemistry
Immunohistochemistry was performed to detect the expression levels of Ki-67, MAP4K4, and MMP9 in tumor tissue, whose processes are accordance with previous study [13]. The slides of tumor tissue were deparaffinized with xylene and rehydrated through a series of ethanol concentrations. Antigens were retrieved by boiling under pressure in EDTA buffer (pH = 9.0) for 3 min. Sections were incubated with 0.3% H2O2 for 20 min and blocked with goat serum for 45 min followed by washing with PBS. Sections were then incubated with primary antibodies of Ki-67 (1:100; cat. no. 27309-1-AP; Proteintech, U.S.A.), MAP4K4 (1:100), and MMP9 (1:100; cat no. ab38898; Abcam) at 4°C overnight. The next day, sections were incubated with the secondary antibody at 37°C for 30 min which followed by incubating with the HRP labeled streptavidin solution for 30 min. PBS was used for washing after each step. After being visualized by incubating with 3, 3-diaminobenzidine-tetrachloride (DAB) for 5 min, sections were counterstained with hematoxylin and evaluated by light microscopy.
Statistical analysis
Data were analyzed by the two-tailed Student’s t test using SPSS software (version 18.0; SPSS, Inc., Chicago, IL, U.S.A.) and represented as the mean ± S.D. Statistical significance was ascribed to P<0.05.
Results
The overexpression of miR-141 decreased cell proliferation and colony formation in CRC cells
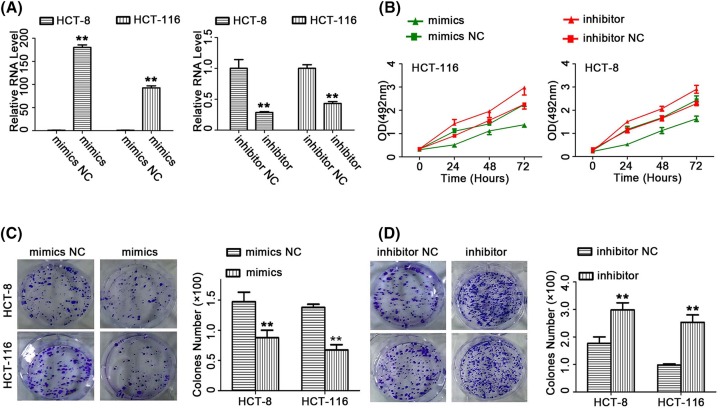
To elucidate the potential role of miR-141 in CRC cells, we respectively used control, miR-141 mimics, and miR-141 inhibitor to transfect CRC cells (HCT-116; HCT-8). Transfection efficiency and expression level of miR-141 were first evaluated by reverse transcription quantitative PCR (RT-qPCR). The results indicated that the expression of miR-141 is higher in cells transfected with miR-141 mimics than that of control, while the expression of miR-141 is lower in cells transfected with miR-141 inhibitor than that of control (Figure 1A). We evaluated the effect of miR-141 over expression and low expression on the proliferation of CRC cells. The low expression of miR-141 promoted the proliferation of CRC cells (Figure 1B). Up-regulation of miR-141 inhibited the proliferation of CRC cells. It is reported that miR-141 is involved in malignant biological behaviors of cancer cells. In the present study, we performed the colony formation assay. Up-regulation of miR-141 reduced the number of colonies in CRC cells. Depletions of miR-141 promoted the capability of colony formation in CRC cells (Figure 1C,D).
Figure 1. The overexpression of miR-141 decreased cell proliferation and colony formation in CRC cells.
HCT-116 and HCT-8 cells were transfected with miR-141 mimics, miR-141 inhibitor and control (NC). (A and B) 24 h after the transfection, cells were harvested for qRT-PCR analysis. The effect of miR-141 on the viability of CRC cells. Cell viability was determined by MTS assays. (C and D) representative images from colony formation assay of HCT-116 and HCT-8 cells after transfection of miRNA interference; n=3, **P<0.01.
miR-141 inhibited the cell migration and invasion of CRC cells
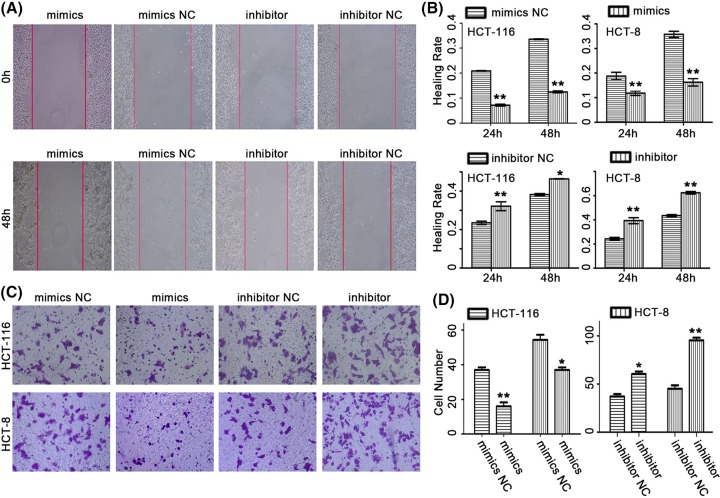
The previous studies have reported miR-141 is associated with migration. In view of this, we performed the experiments of miR-141 overexpression and depletion in CRC cells. Next, we conducted the wound-healing assay and transwell experiment. Wound images were captured at 0, 24, and 48 h after scratching. The results demonstrated that overexpression of miR-141 inhibited the migration of CRC cells, while lowexpression of miR-141 accelerated the migration of CRC cells (Figure 2A,B). The transwell assay results indicated that overexpression of miR-141 inhibited the CRC cells invasion. Inversely, lowexpression of miR-141 accelerated the CRC cells invasion (Figure 2C,D).
Figure 2. Effects of miR-141 on CRC cells migration and invasion.
For the wound-healing and invasion assay, CRC cells were seeded and transfected with miR-141 mimics, miR-141 inhibitor and the NC for 24 h. (A) Wounds healing from a representative experiment of the HCT-116 cell. (B) The wound-healing rate in CRC cells transfected with miR-141 mimics was significantly decreased, while accelerated in CRC cells transfected with miR-141 inhibitor compared with the NC. (C and D) Representative images of the transwell invasion assay. The normal ratio of invasive cells is shown. Up-regulation of miR-141 in CRC cells reduced cell invasion, while inhibition of miR-141 expression increased cell invasion; n=3, *P<0.05, **P<0.01.
MAP4K4 was a direct target of miR-141
To find potential target genes of miR-141, we performed the bioinformatics analysis as described before [6]. The result suggested that MAP4K4 could be the important target of miR-141. Based on the above result, we further explored the mechanism of miR-141 in tumor suppression.
To further determine whether miR-141 suppressed CRC cells by targetting MAP4K4, we conducted MAP4K4-3′-UTR luciferase reporter assays. First, we constructed MAP4K4-3′-UTR-luciferase reporter plasmids [11] for CRC cells transfection. CRC cells were cotransfected with miR-141 mimics and luciferase reporter plasmid MAP4K4-3′-UTR-luc. In line with the bioinformatics prediction, the transfection of miR-141 mimics significantly reduced the luciferase activity in CRC cells transfected with MAP4K4-3′-UTR-luc (Figure 3B).
Figure 3. miR-141 regulated MAP4K4 expression.
(A) predicted miR-141 target sequence in the 3′-UTR of MAP4K4. (B) Dual-luciferasereporter assay of the HCT-116 cells transfected with the MAP4K4-3′-UTR reporter, the MAP4K4-3′-UTR-mutation reporter, miR-141 mimics, inhibitor, and mimic NC. Being linked to the segment containing the target sequence within the 3′-UTR in MAP4K4 mRNA, miR-141 expression reduced the luciferase activity. (C) 24 h h after the transfection, cells were harvested for RT-qPCR analysis using miR-141 primers. Overexpression of miR-141 significantly reduced the MAP4K4 mRNA expression levels in CRC cells. (D) MAP4K4 proteins levels in CRC cells treated with miR-141 mimics, inhibitors and NC; n=3, *P<0.05.
To determine whether miR-141 could regulate the expression of MAP4K4, we conducted the transient transfection study in CRC cells. Our RT-qPCR assay revealed that the expression of MAP4K4 was significantly decreased (P<0.05) in cells transfected with miR-141 mimics compared with the control. On the contrary, the expression of MAP4K4 was observably induced (P<0.05) in cells transfected with miR-141 inhibitor (Figure 3C).
To further explore the function of miR-141 in regulating CRC progression, we performed transfection study in CRC cells. Our Western blot assay indicated that overexpression of miR-141 significantly decreased the protein level of MAP4K4 (Figure 3D). At the same time, down-regulation of miR-141 significantly increased the protein level of MAP4K4 (Figure 3D).
5-FU suppressed CRC cells movement via regulating miR-141-mediated MAP4K4 axis
Invasion, metastasis, proliferation, and colony forming are the markers of cancer cells. We found that 5-FU inhibited cell colony formation, migration and invasion, and its inhibitory effects overexpressed cells were stronger than the control cells (Figure 4A–F). We also found that the 5-FU could inhibit the proliferation of CRC cells and decrease the expression of MAP4K4, in dose dependent manners (Figure 5).
Figure 4. 5-FU treatment suppressed CRC cells movement via regulating miR-141-mediated MAP4K4 axis.
(A and B) representative results of colony formation of 5-FU, miR-141 mimics, combination of the two and NC. (C and D) Wound-healing assay was conducted and phase-contrast images were obtained immediately after wounding and at 48 h. (E and F) Invasion assay was performed. Representative images were recorded. (G and H) Expression level of MAP4K4 mRNA and proteins were assessed in CRC cells treated by 5-FU, miR-141 mimics, combination of the two and NC; n=3, *P<0.05, **P<0.01.
Figure 5. After treatment of CRC cells with 5-FU, levels of MAP4K4 genes following miR-141 also subsequently changed.
(A) CRC cell viability was detected with MTS assays. (B) The expression level of miR-141 was detected by RT-qPCR assays in the CC cells treated with different concentration of 5-FU. (C) The expression level of MAP4K4 genes was detected by PCR in CRC cells treated with different concentration of 5-FU; n = 3, *P<0.05.
The present study indicated that miR-141 could regulate the expression of MAP4K4 in CRC cells. To explore the effective and potential mechanism of miR-141 in CRC chemoresistance, we performed the Western blot assay and PCR. We found that MAP4K4 was also regulated by miR-141 in the process of chemosensitivity. The PCR and Western blot assays results showed that combined miR-141 overexpression and 5-FU significantly decreased the expression of MAP4K4, compared with the control (Figure 4G,H).
miR-141 sensitized CRC cells to 5-FU treatment in vivo
To further confirm the above findings, we performed tumor formation in nude mice to validate the effect of miR-141 on 5-FU-sensitivity. Lenti-miR-141 and Lenti-NC cells were respectively injected into the right limb of nude mice. After treatment with 5-FU, tumors derived from miR-141-overexpressing cells grew more slowly and had lower tumor weight than the control (Figure 6D,C).
Figure 6. miR-141 and 5-FU could reverse the colon cancer tumorigenesis.
Each representative images treatment group are presented the effect of 5-FU, miR-141, and combined the two on colon tumors in the nude mice. (A) Phase-contrast and flurescence images of HCT-116 transfected with lenti-miR-141. (B) RT-qPCR analysis of miR-141 in HCT-116 transfected with Lenti-miR-141 or Lenti-NC. (C, D, and E) Tumor weight and volume of lenti-NC+5-FU, lenti-NC, lenti-miR-141 + 5FU, and lenti-miR-141 cells were recorded. (F) The expression levels of MAP4K4, Ki-67, and MMP9 in the tumors tissues of mice in different groups were determined by immunohistochemistry (×200); n=3, **P<0.01.
To explore the molecular change in each of the treatment group, we conducted the immunohistochemistry staining. The expression analysis of Ki-67, MMP9 and MAP4K4 indicated that miR-141 can sensitize CRC cells to 5-FU (Figure 6F).
Discussion
Because of the introduction of the currently used FOLFOX and FOLFIRI regiments in metastatic CRC, the response rates have been improved to about 30–40% [14]; however, the resistance to chemotherapy is still a major barrier in the CRC treatment. Most deaths in CRC are attributed to the resistance to chemotherapeutic drugs. Varied mechanisms are explored to find the novel target. miR-21 functions as the chemical resistance for cyclo-oxygenase-2 inhibitor, which prevents gastric carcinoma [15]. miR-141 is highly overexpressed and PTEN is inhibited in 5-FU and oxaliplatin chemo-resistance to esophageal cancer cell [16]. Shota’s study showed that the expression levels of miR-200c and miR-141 were significantly reduced in oxaliplatin-resistant CRC cells [17]. miR-20b reduces 5-FU resistance by regulating the expression of ADAM9/EGFR in CRC [18]. In the present study, the major finding was that miR-141 enhanced the CRC chemosensitivity of 5-FU in vitro and tumor xenografts in vivo by inhibiting MAP4K4 expression.
miRNAs, 18–25 nts long, are abundant and evolutionary conserved single-stranded RNAs. It is reported that miRNAs account for 1–2% of the human genome and regulate more than 50% protein-coding genes. Compared with the normal counterpart, miRNAs are up- or down-regulated in malignant tissues, which can be considered as oncogenes or tumor-suppressors, respectively [19]. Some studies showed that miRNAs dysregulation in the human is relevant to clinical course cancer. The present study showed that miR-141 suppressed CRC cell migration, invasion, and proliferation, which indicated that miR-141 was associated with CRC progression. Depending on the subcellular localization, the function of miR-141 (tumor suppressor or oncogene) is different. Liu’s research showed that miR-141 suppressed the proliferation, invasion, and metastasis of the prostate cancer cell by multiple mechanisms [20]. Expression of miR-141 and MEG3 can abolish the expression of E2F3 and inhibit the process of gastric cancer [21]. Our previous study showed that the expression of miR-141 was dramatically decreased in tumor tissues and positive lymph nodes [6]. This indicated that miR-141 was an important tumor suppressor. Deregulation of miR-141 has been showed in human tumors. miR-141 could suppress CRC cells proliferation and migration [22]. A previous research indicated that ELF3 promotes epithelial-mesenchymal transition (EMT) by regulating miR-141-3p leading to the enhancement of ZEB1, in hepatocellular carcinoma [23]. Here, our results showed that comparison with down-regulated CRC cells, up-regulated miR-141 expression inhibited tumor progression, in vivo and in vitro.
We conducted luciferase reporter assays and the result indicated a dramatic reduction in MAP4K4 protein levels in the CRC cells treated with miR-141 mimics, but down-regulation of miR-141 had a contrary result. The present study revealed that MAP4K4 was a novel target gene of miR-141, and the deregulation of miR-141/MAP4K4 contributed to CRC progression. MAP4K4 participate in the maintenance of malignant phenotype of many human cancers. A study showed knockdown of MAP4K4 inhibited progression of lung adenocarcinoma in vivo and in vitro [24]. EMT and invasion were facilitated by MAP4K4-activating JNK and NF-κB signaling, in hepatocellular carcinoma cells [25]. Our previous data also demonstrated that the MAP4K4 were significantly up-regulated in the tumor and lymph nodes of CRC patients [6]. Recent researches indicate that the Ki-67 protein exists in the cell nucleus during mitosis, regulates the cell cycle. MMP9 promotes tumor metastasis by facilitating tumor cell migration and invasion. Furthermore, our results indicated that the combination of up-regulated miR-141 with 5-FU could inhibit the expression of Ki-67 and MMP9 proteins. In the present study, cell proliferation, migration, and invasion were inhibited when miR-141/MAP4K4 was elevated. In summary, the present data certified a correlation between the miR-141 and both sensitive to 5-FU and MAP4K4.
In the present study, the miR-141-mediated MAP4K4 axis was the novel target of 5-FU in CRC cells. The present study showed that MAP4K4 was the target gene of miR-141. Downexpression of MAP4K4 prevented the growth of tumor. MAP4K4 regulates the expression of kinase, transcription factor, transmembrane receptor that is important for cell–cell communication and matrix metalloproteinases in tumor [26,27]. However, the function of MMP9 and Ki-67 in the process of 5-FU anticancer has not been illuminated. And large effectors or signaling mediators are not explored. Our study showed that MAP4K4 play an important role in CRC progression.
The present study showed that the synergistic role of overexpressed miR-141 and 5-FU lead to the decreased expression of MAP4K4. In conclusion, our results demonstrated loss of miR-141/MAP4K4 contributed to the progression of CRC. Up-regulating miR-141 expression may represent a novel therapeutic method approach and prevention for CRC.
Abbreviations
- 5-FU
5-fluorouracil
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- E2F3
E2F transcription factor 3
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Ki-67
proliferating antigen Ki-67
- MAP4K4
mitogen-activated protein kinase kinase kinase kinase 4
- MMP9
matrix metalloproteinase-9
- PTEN
phosphatase and tensin
- RT-qPCR
reverse transcription quantitative PCR
- WT
wild type
- U6
snRNA internal control gene
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The present study was funded by The Health and Family Planning Commission of Hebei Province [grant number 20150362] and National Natural Science Foundation of China [grant numbers 81472661, 81490753, 81230047, 81672743]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contribution
F.W., J.Z., and Z.M. were responsible for date collection and drafted the paper. C.Z., G.W., Y.L., M.L., and J.X. were responsible for the analysis of data. L.Z., W.N., and G.W. were responsible for revising the article critically for important intellectual content.
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F.. et al. (2015) Cancer statistics in China. CA Cancer J. Clin. 66, 115–132 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J. and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4.Croce C.M. (2009) Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams B.D., Kasinski A.L. and Slack F.J. (2014) Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 24, R762–R776 10.1016/j.cub.2014.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng L., Ma H., Chang L., Zhou X., Wang N., Zhao L.. et al. (2016) Role of microRNA-141 in colorectal cancer with lymph node metastasis. Exp. Ther. Med. 12, 3405–3410 10.3892/etm.2016.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright J.H., Wang X., Manning G., LaMere B.J., Le P., Zhu S.. et al. (2003) The STE20 kinase HGK is broadly expressed in human tumour cells and can modulate cellular transformation, invasion, and adhesion. Mol. Cell. Biol. 23, 2068–2082 10.1128/MCB.23.6.2068-2082.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang J.J., Wang H., Rashid A., Tan T.H., Hwang R.F., Hamilton S.R.. et al. (2008) Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin. Cancer Res. 14, 7043–7049 10.1158/1078-0432.CCR-08-0381 [DOI] [PubMed] [Google Scholar]
- 9.Vitorino P., Yeung S., Crow A., Bakke J., Smyczek T., West K.. et al. (2015) MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature 519, 425–430 10.1038/nature14323 [DOI] [PubMed] [Google Scholar]
- 10.Collins C.S., Hong J., Sapinoso L., Zhou Y., Liu Z., Micklash K.. et al. (2006) A small interfering RNA screen for modulators of tumour cell motility identifies MAP4K4 as a promigratory kinase. Proc. Natl. Acad. Sci. U.S.A. 103, 3775–3780 10.1073/pnas.0600040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T., Hou J., Li Z., Zheng Z., Wei J., Song D.. et al. (2017) miR-15a-3p and miR-16-1-3p negatively regulate twist1 to repress gastric cancer cell invasion and metastasis. Int. J. Biol. Sci. 13, 122–134 10.7150/ijbs.14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J., Zheng D., Liu Y., Sham M.H., Tam P., Farzaneh F.. et al. (2005) Overexpression of soluble TRAIL induces apoptosis in human lung adenocarcinoma and inhibits growth of tumor xenografts in nude mice. Cancer Res. 65, 1687–1692 10.1158/0008-5472.CAN-04-2749 [DOI] [PubMed] [Google Scholar]
- 13.Li L., Zhao L.M., Dai S.L., Cui W.X., Lv H.L., Chen L.. et al. (2016) Periplocin extracted from cortex periplocae induced apoptosis of gastric cancer cells via the ERK1/2-EGR1 pathway. Cell. Physiol. Biochem. 38, 1939–1951 10.1159/000445555 [DOI] [PubMed] [Google Scholar]
- 14.Prenen H. and Cutsem E.V. (2013) Role of targeted agents in metastatic colorectal cancer. Targ. Oncol. 8, 83–96 10.1007/s11523-013-0281-x [DOI] [PubMed] [Google Scholar]
- 15.Li H., Cheng J., Mao Y., Jiang M. and Fan X. (2015) miR-21 inhibits the effects of cyclooxygenase-2 inhibitor NS398 on apoptosis and invasion in gastric cancer cells. Onco. Targets Ther. 8, 3245–3253 10.2147/OTT.S90012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y.Y., Chen Q.J., Xu K., Ren H.T., Bao X., Ma Y.N.. et al. (2016) Involvement of microRNA-141-3p in 5-fluorouracil and oxaliplatin chemo-resistance in esophageal cancer cells via regulation of PTEN. Mol. Cell. Biochem. 422, 161–170 10.1007/s11010-016-2816-9 [DOI] [PubMed] [Google Scholar]
- 17.Tanaka S., Hosokawa M., Yonezawa T., Hayashi W., Ueda K. and Iwakawa S. (2015) Induction of epithelial-mesenchymal transition and down-regulation of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol. Pharm. Bull. 38, 435–440 10.1248/bpb.b14-00695 [DOI] [PubMed] [Google Scholar]
- 18.Fu Q., Cheng J., Zhang J., Zhang Y., Chen X., Luo S.. et al. (2017) miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol. Rep. 37, 123–130 10.3892/or.2016.5259 [DOI] [PubMed] [Google Scholar]
- 19.Di Leva G., Garofalo M. and Croce C.M. (2014) microRNAs in cancer. Annu. Rev. Pathol. 9, 287–314 10.1146/annurev-pathol-012513-104715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Liu R., Zhang D., Deng Q., Liu B., Chao H.P.. et al. (2017) microRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 8, 14270 10.1038/ncomms14270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Ji G., Ke X. and Zhang G. (2015) miR-141 inhibits gastric cancer proliferation by interacting with long noncoding RNA MEG3 and down-regulating E2F3 expression. Dig. Dis. Sci. 60, 3271–3282 10.1007/s10620-015-3782-x [DOI] [PubMed] [Google Scholar]
- 22.Long Z.H., Bai Z.G., Song J.N., Zheng Z., Li J., Zhang J.. et al. (2017) miR-141 inhibits proliferation and migration of colorectal cancer SW480 cells. Anticancer Res. 37, 4345–4352 [DOI] [PubMed] [Google Scholar]
- 23.Zheng L., Xu M., Xu J., Wu K., Fang Q., Liang Y.. et al. (2018) ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 9, 387 10.1038/s41419-018-0399-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X., Chen G., Gao C., Zhang D.H., Kuan S.F., Stabile L.P.. et al. (2017) MAP4K4 is a novel MAPK/ERK pathway regulator required for lung adenocarcinoma maintenance. Mol. Oncol. 11, 628–639 10.1002/1878-0261.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng X.J., Pan Q., Wang S.M., Pan Y.C., Wang Q., Zhang H.H.. et al. (2016) MAP4K4 promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. Tumour Biol. 37, 11457–11467 10.1007/s13277-016-5022-1 [DOI] [PubMed] [Google Scholar]
- 26.Yang N., Wang Y., Hui L., Li X. and Jiang X. (2015) Silencing SOX2 expression by rna interference inhibits proliferation, invasion and metastasis, and induces apoptosis through MAP4K4/JNK signaling pathway in human laryngeal cancer TU212 cells. J. Histochem. Cytochem. 63, 721–733 10.1369/0022155415590829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y.F., Qu G.Q., Lu Y.M., Kong W.M., Liu Y., Chen W.X.. et al. (2015) Silencing of MAP4K4 by short hairpin RNA suppresses proliferation, induces G1 cell cycle arrest and induces apoptosis in gastric cancer cells. Mol. Med. Rep. 13, 41–48 10.3892/mmr.2015.4510 [DOI] [PMC free article] [PubMed] [Google Scholar]