Abstract
Background: We performed the present study to better elucidate the correlations of methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) gene polymorphisms with the risk of congenital heart diseases (CHD).
Methods: Eligible articles were searched in PubMed, Medline, Embase and CNKI. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to detect any potential associations of MTHFR and MTRR gene polymorphisms with CHD.
Results: A total of 47 eligible studies were finally included in our meta-analysis. Our overall analyses suggested that MTRR rs1801394, MTRR rs1532268, MTHFR rs1801131 and MTHFR rs1801133 polymorphisms were all significantly associated with the risk of CHD in certain genetic models. Further subgroup analyses according to ethnicity of study participants demonstrated that the MTRR rs1801394 polymorphism was significantly correlated with the risk of CHD only in Asians, whereas MTRR rs1532268, MTHFR rs1801133 and MTHFR rs1801131 polymorphisms were significantly correlated with the risk of CHD in both Asians and Caucasians.
Conclusions: Our findings indicated that MTRR rs1532268, MTHFR rs1801131 and MTHFR rs1801133 polymorphisms may affect the risk of CHD in Asians and Caucasians, while the MTRR rs1801394 polymorphism may only affect in risk of CHD in Asians.
Keywords: Congenital heart diseases (CHD), Gene polymorphisms, Methylenetetrahydrofolate reductase (MTHFR), Methionine synthase reductase (MTRR), Meta-analysis
Introduction
Congenital heart diseases (CHD) refer to a group of structural heart defects that are resulted from abnormal cardiac development. The incidence of CHD is estimated to be approximately 1% in newborns, and despite rapid advances in surgical treatments and interventional therapies over the past few decades, CHD is still the primary non-infectious cause of infant mortality worldwide [1]. Moreover, its associated complications such as heart failure, arrhythmia and sudden cardiac death may occur even after effective correction of cardiac abnormalities [2,3]. Until now, the exact cause of CHD is still largely unclear despite extensive investigations. Nevertheless, mounting evidence supports that genetic factors play a crucial part in its development. First, family clustering of CHD with variable phenotypes is not uncommon, and descendants of CHD patients suffer a higher risk of developing cardiac malformations compared with the general population [4,5]. Second, multiple genetic variants have been found to be associated with an increased risk of CHD [6–9]. Overall, these findings jointly indicate that genetic predisposition to CHD is vital for its occurrence and development.
Methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) play central roles in the regulation of folate metabolism and homocysteine synthesis [10]. Previous studies have shown that taking folate supplements during pregnancy could significantly reduce the risk of cardiovascular congenital malformations in newborns [11,12]. Consequently, functional MTHFR and MTRR polymorphisms, which were known to affect plasma folate levels, were considered to be ideal candidate genetic biomarkers of CHD.
So far, numerous studies have been conducted to assess the roles of MTHFR and MTRR gene polymorphisms in CHD, but the results of these studies were controversial [13–16]. Therefore, we conducted the present meta-analysis to better evaluate potential associations of MTHFR and MTRR gene polymorphisms with the risk of CHD.
Materials and methods
Literature search and inclusion criteria
The current meta-analysis was adhered to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement [17]. A systematic literature search of PubMed, Medline, Embase and China National Knowledge Infrastructure (CNKI) was performed to retrieve all relevant articles. The key words used in this literature search included: ‘5-methyltetrahydrofolate-homocysteine methyltransferase reductase’, ‘methionine synthase reductase’, ‘MTRR’, ‘MSR’, ‘methylenetetrahydrofolate reductase’, ‘MTHFR’, ‘polymorphism’, ‘variant’, ‘mutation’, ‘genotype’, ‘allele’, ‘congenital heart disease‘, ‘congenital heart defect’ and ‘congenital cardiovascular malformation’ (see Supplementary File S1). To identify other potentially relevant publications, we also reviewed the reference lists of all retrieved articles.
Eligible studies of the current meta-analysis must met all the following criteria: (1) evaluate potential associations of MTRR and/or MTHFR gene polymorphisms with the risk of CHD; (2) provide sufficient data to calculate odds ratios (ORs) and 95% confidence intervals (CIs); (3) full text in Chinese or English available. For duplicate reports, only the study with the largest sample size was included. Reviews, comments, letters and family-based association studies were excluded.
Data extraction and quality assessment
The following information was extracted from each included study: name of the first author, year of publication, country and ethnicity of study subjects, type of CHD, genotypic frequencies of MTRR and/or MTHFR gene polymorphisms in cases and controls, and whether the distributions of investigated gene polymorphisms in the control group violated Hardy–Weinberg equilibrium (HWE).
The Newcastle–Ottawa scale (NOS), a classical assessment tool of observational studies that evaluates the quality of articles from three dimensions: selection, comparability and exposure, was adopted to assess the quality of included studies [18]. The NOS has a score range of 0 to 9, and studies with a score of more than 7 were considered to be of high quality.
Two reviewers (Aiping Xu and Weiping Wang) conducted data extraction and quality assessment independently. When necessary, the reviewers wrote to the corresponding authors for extra information or raw data. Disagreements between two reviewers were solved by discussion with the third reviewer (Xiaolei Jiang) until a consensus was reached.
Statistical analysis
All data analyses in the present study were carried out using Review Manager Version 5.3.3 (The Cochrane Collaboration, Software Update, Oxford, United Kingdom). The probability value (P value) of HWE in the control group was calculated with the chi-square test. ORs and 95% CIs were used to estimate potential associations of MTRR and/or MTHFR gene polymorphisms with the risk of CHD in the dominant, recessive, additive and allele models, and a P value of 0.05 or less was considered as statistically significant. The Q test and I2 statistic were adopted to assess between-study heterogeneity. If P value of Q test was less than 0.1 or I2 was greater than 50%, random-effect models would be applied for analyses due to the existence of obvious heterogeneity. Otherwise, fixed-effect models would be employed for analyses. Subgroup analyses were subsequently performed according to ethnicity of study participants and type of disease. Sensitivity analyses were conducted to test the stability of the results. Publication bias was evaluated with funnel plots.
Results
Characteristics of included studies
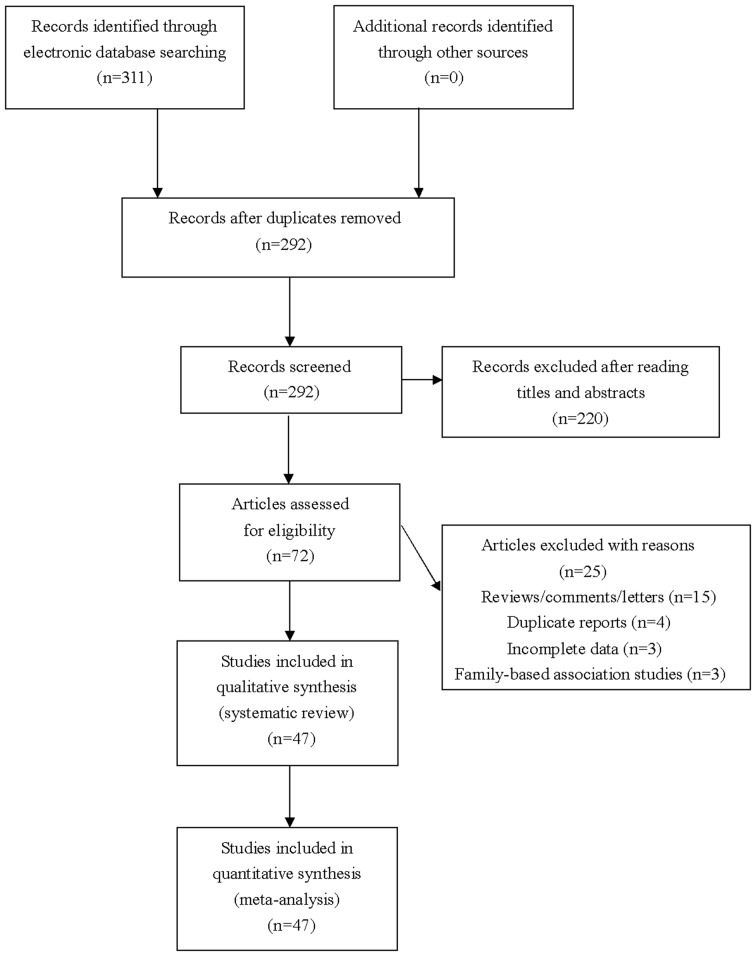
The literature search identified 311 citations. After exclusion of irrelevant and duplicate articles by reading titles and abstracts, 72 articles were selected for further evaluation. Another 25 articles were subsequently excluded after reading full texts, and a total of 47 studies that met the inclusion criteria were finally included in our meta-analysis (see Figure 1). Characteristics of included studies were summarized in Table 1.
Figure 1. Flowchart of study selection for the present study.
Table 1. The characteristics of included studies.
| First author, year | Country | Ethnicity | Sex, male (%) Case/ Control | Age (years) Case/ Control | Type of disease | Sample size | Genotype distribution | P-value for HWE | NOS score | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||
| rs1801394 | ||||||||||
| Benke, 2015 | Hungry | Caucasian | 59.2/61.5 | 2.42/3.08 | CHD | 72/117 | 64/7/1 | 110/6/1 | 0.016 | 7 |
| Christensen, 2013 | Canada | Mixed | NA | NA | CHD | 245/65 | 68/123/54 | 22/32/11 | 0.912 | 7 |
| Gong, 2010 | China | Asian | NA | NA | CHD | 60/60 | 38/21/1 | 52/6/2 | 0.007 | 7 |
| Guo, 2017 | China | Asian | NA | 2.31/2.48 | CHD | 99/114 | 44/46/9 | 67/44/3 | 0.174 | 8 |
| Guo, 2017 | China | Asian | NA | 2.33/2.47 | VSD | 21/114 | 7/11/3 | 67/44/3 | 0.174 | 8 |
| Hassan, 2017 | Egypt | Caucasian | 36.0/32.0 | 1.30/1.28 | CHD | 100/100 | 26/32/42 | 48/36/16 | 0.048 | 8 |
| Liu, 2007 | China | Asian | 48.5/NA | 6.50/NA | CHD | 132/107 | 33/84/15 | 52/45/10 | 0.953 | 7 |
| Locke, 2010 | U.S.A. | Mixed | NA | NA | CHD | 92/94 | 27/50/15 | 31/46/17 | 0.993 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | CHD | 153/147 | 46/74/33 | 61/63/23 | 0.323 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | VSD | 74/147 | 24/32/18 | 61/63/23 | 0.323 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | TOF | 79/147 | 22/42/15 | 61/63/23 | 0.323 | 7 |
| Pishva, 2013 | Iran | Caucasian | 46.3/44.8 | 4.51/5.43 | VSD | 123/125 | 41/54/28 | 62/53/10 | 0.776 | 7 |
| Su, 2017 | China | Asian | NA | NA | VSD | 183/201 | 68/97/18 | 107/85/9 | 0.120 | 8 |
| van Beynum, 2006 | Netherlands | Caucasian | NA | NA | CHD | 159/245 | 51/83/25 | 74/124/47 | 0.699 | 7 |
| Verkleij-Hagoort, 2008 | Netherlands | Caucasian | NA | 1.40/1.39 | CHD | 229/251 | 79/112/38 | 77/122/52 | 0.774 | 7 |
| Wang, 2013 | China | Asian | NA | NA | CHD | 160/188 | 90/59/11 | 105/71/12 | 0.999 | 7 |
| Wang, 2018 | China | Asian | NA | 2.18/1.81 | CHD | 102/100 | 51/39/12 | 75/21/4 | 0.126 | 7 |
| Weine, 2012 | Russia | Caucasian | NA | 2.15/2.11 | CHD | 51/390 | 19/23/9 | 128/191/71 | 0.986 | 7 |
| Zeng, 2011 | China | Asian | 43.4/47.2 | NA | CHD | 599/672 | 309/234/56 | 375/253/44 | 0.880 | 8 |
| Zhao, 2012 | China | Asian | 52.4/54.4 | 6.59/6.59 | CHD | 2340/2270 | 1308/860/172 | 1294/818/158 | 0.067 | 8 |
| rs1532268 | ||||||||||
| Hassan, 2017 | Egypt | Caucasian | 36.0/32.0 | 1.30/1.28 | CHD | 100/100 | 14/40/46 | 38/36/26 | 0.007 | 8 |
| Pishva, 2013 | Iran | Caucasian | 46.3/44.8 | 4.51/5.43 | VSD | 123/125 | 53/50/20 | 66/54/5 | 0.134 | 7 |
| Su, 2017 | China | Asian | NA | NA | VSD | 183/201 | 66/96/21 | 105/80/16 | 0.889 | 8 |
| Zeng, 2011 | China | Asian | 43.4/47.2 | NA | CHD | 599/672 | 383/201/15 | 476/176/20 | 0.450 | 8 |
| rs1801131 | ||||||||||
| Božović, 2011 | Croatia | Caucasian | 49.1/49.3 | 1.03/2.78 | CHD | 54/221 | 30/22/2 | 101/98/22 | 0.803 | 7 |
| Brandalize, 2009 | Brazil | African | NA | NA | CHD | 239/197 | 143/84/12 | 113/76/8 | 0.275 | 8 |
| Chao, 2014 | Taiwan | Asian | 11.8/38.2 | 46.7/50.9 | PDA | 17/34 | 13/2/2 | 15/19/0 | 0.024 | 8 |
| Christensen, 2013 | Canada | Mixed | NA | NA | CHD | 246/65 | 133/93/20 | 36/22/7 | 0.212 | 7 |
| Feng, 2016 | China | Asian | 46.3/63.2 | 1.3/1.9 | CHD | 257/49 | 194/51/12 | 35/14/0 | 0.243 | 7 |
| Galdieri, 2007 | Brazil | African | NA | NA | CHD | 57/38 | 35/21/1 | 19/16/3 | 0.884 | 7 |
| Guo, 2017 | China | Asian | NA | 2.31/2.48 | CHD | 99/114 | 71/28/0 | 89/24/1 | 0.655 | 8 |
| Guo, 2017 | China | Asian | NA | 2.33/2.48 | VSD | 21/114 | 14/7/0 | 89/24/1 | 0.655 | 8 |
| Huang, 2014 | China | Asian | 56.1/57.5 | 2.54/2.70 | TOF | 170/206 | 111/56/3 | 146/54/6 | 0.712 | 8 |
| Koshy, 2015 | India | Caucasian | NA | NA | CHD | 96/100 | 27/32/37 | 58/20/22 | <0.001 | 7 |
| Locke, 2010 | U.S.A. | Mixed | NA | NA | CHD | 87/88 | 42/39/6 | 30/49/9 | 0.090 | 7 |
| Obermann-Borst, 2011 | Netherlands | Caucasian | 60.8/55.9 | 17.0/17.3 | CHD | 139/183 | 69/57/13 | 75/90/18 | 0.227 | 8 |
| Sahiner, 2014 | Turkey | Caucasian | 57.1/NA | 7.63/NA | CHD | 137/93 | 45/68/24 | 31/54/8 | 0.022 | 8 |
| Sayin Kocakap, 2015 | Turkey | Caucasian | NA | NA | CHD | 69/99 | 20/36/13 | 51/37/11 | 0.288 | 8 |
| Shi, 2015 | China | Asian | 38.85/57.41 | 2.18/2.12 | CHD | 153/216 | 95/39/19 | 157/53/6 | 0.555 | 7 |
| Storti, 2003 | Italy | Caucasian | NA | 2.50/2.58 | CHD | 100/100 | 43/46/11 | 50/43/7 | 0.582 | 7 |
| van Driel, 2008 | Netherlands | Caucasian | 58.0/57.0 | 1.40/1.39 | CHD | 230/251 | 104/102/24 | 116/104/31 | 0.311 | 8 |
| Wang, 2018 | China | Asian | NA | 2.18/1.81 | CHD | 102/100 | 57/40/5 | 60/36/4 | 0.624 | 7 |
| Xu, 2010 | China | Asian | 53.7/53.0 | 6.50/6.69 | CHD | 502/527 | 316/168/18 | 326/185/16 | 0.091 | 8 |
| Xu, 2010 | China | Asian | NA | NA | VSD | 257/527 | 169/86/2 | 326/185/16 | 0.091 | 8 |
| Xu, 2010 | China | Asian | NA | NA | ASD | 41/527 | 21/16/4 | 326/185/16 | 0.091 | 8 |
| Zidan, 2013 | Egypt | Caucasian | NA | NA | CHD | 80/80 | 16/27/37 | 30/26/24 | 0.002 | 7 |
| rs1801133 | ||||||||||
| Božović, 2011 | Croatia | Caucasian | 49.1/49.3 | 1.03/2.78 | CHD | 54/221 | 20/28/6 | 101/97/23 | 0.968 | 7 |
| Brandalize, 2009 | Brazil | African | NA | NA | CHD | 239/197 | 94/113/32 | 86/93/18 | 0.313 | 8 |
| Chao, 2014 | Taiwan | Asian | 11.8/38.2 | 46.7/50.9 | PDA | 17/34 | 10/5/2 | 19/12/3 | 0.586 | 8 |
| Christensen, 2013 | Canada | Mixed | NA | NA | CHD | 246/65 | 94/117/35 | 27/29/9 | 0.787 | 7 |
| Feng, 2016 | China | Asian | 46.3/63.2 | 1.3/1.9 | CHD | 257/49 | 122/114/21 | 21/22/6 | 0.949 | 7 |
| Galdieri, 2007 | Brazil | African | NA | NA | CHD | 58/38 | 30/21/7 | 18/14/6 | 0.263 | 7 |
| Gong, 2012 | China | Asian | 65.5/61.8 | 1.91/1.58 | CHD | 244/136 | 45/123/76 | 43/72/21 | 0.309 | 9 |
| Gong, 2012 | China | Asian | 61.6/61.8 | 1.55/1.58 | TOF | 120/136 | 21/59/40 | 43/72/21 | 0.309 | 9 |
| Gong, 2012 | China | Asian | 69.4/61.8 | 2.27/1.58 | TGA | 124/136 | 24/64/36 | 43/72/21 | 0.309 | 9 |
| Guo, 2017 | China | Asian | NA | 2.31/2.48 | CHD | 99/114 | 20/41/38 | 36/48/30 | 0.097 | 8 |
| Guo, 2017 | China | Asian | NA | 2.33/2.48 | VSD | 21/114 | 8/8/5 | 36/48/30 | 0.097 | 8 |
| Huang, 2014 | China | Asian | 56.1/57.5 | 2.54/2.70 | TOF | 168/204 | 63/45/60 | 84/72/48 | <0.001 | 8 |
| Jiang, 2015 | China | Asian | NA | 2.34/2.35 | CHD | 100/100 | 38/46/16 | 41/48/11 | 0.523 | 7 |
| Jing, 2013 | China | Asian | NA | NA | CHD | 104/208 | 16/42/46 | 55/114/39 | 0.139 | 7 |
| Junker, 2001 | Germany | Caucasian | 53.0/NA | 16.0/NA | CHD | 114/228 | 51/42/21 | 129/78/21 | 0.075 | 7 |
| Koshy, 2015 | India | Caucasian | 63.5/49.0 | 6.51/7.61 | CHD | 96/90 | 95/1/0 | 83/7/0 | 0.701 | 7 |
| Kuehl, 2010 | U.S.A. | Mixed | 50.4/56.0 | NA | CHD | 55/300 | 12/33/10 | 134/134/32 | 0.861 | 8 |
| Lee, 2005 | Taiwan | Asian | NA | NA | CHD | 213/195 | 110/89/14 | 114/68/13 | 0.513 | 7 |
| Li, 2005 | China | Asian | 48.4/57.2 | NA | CHD | 183/103 | 30/95/58 | 22/57/24 | 0.277 | 7 |
| Li, 2013 | China | Asian | 54.2/57.1 | 2.68/2.79 | CHD | 144/168 | 26/52/66 | 49/84/35 | 0.928 | 7 |
| Liu, 2007 | China | Asian | 48.5/NA | 6.5/NA | CHD | 132/107 | 30/68/34 | 46/48/13 | 0.930 | 7 |
| Locke, 2010 | U.S.A. | Mixed | NA | NA | CHD | 91/94 | 38/39/14 | 49/37/8 | 0.787 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | CHD | 153/147 | 95/51/7 | 100/46/1 | 0.078 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | VSD | 74/147 | 24/32/18 | 100/46/1 | 0.078 | 7 |
| Noori, 2017 | Iran | Caucasian | NA | NA | TOF | 79/147 | 22/42/15 | 100/46/1 | 0.078 | 7 |
| Obermann-Borst, 2011 | Netherlands | Caucasian | 60.8/55.9 | 1.41/1.44 | CHD | 139/183 | 64/66/9 | 92/76/15 | 0.900 | 8 |
| Sahiner, 2014 | Turkey | Caucasian | 57.1/NA | 7.63/NA | CHD | 136/93 | 69/53/14 | 47/39/7 | 0.779 | 8 |
| Sayin Kocakap, 2015 | Turkey | Caucasian | NA | NA | CHD | 75/95 | 40/33/2 | 43/44/8 | 0.484 | 8 |
| Shaw, 2005 | China | Asian | NA | NA | CHD | 153/434 | 69/68/16 | 202/180/52 | 0.227 | 7 |
| Shi, 2015 | China | Asian | 38.85/57.41 | 2.18/2.12 | CHD | 153/216 | 55/68/30 | 70/101/45 | 0.444 | 7 |
| Storti, 2003 | Italy | Caucasian | NA | 2.50/2.58 | CHD | 100/100 | 27/53/20 | 26/54/20 | 0.401 | 7 |
| van Beynum, 2006 | Netherlands | Caucasian | 55.0/49.0 | 3.4/9.4 | CHD | 158/261 | 72/68/18 | 131/107/23 | 0.863 | 7 |
| van Driel, 2008 | Netherlands | Caucasian | 58.0/57.0 | 1.40/1.39 | CHD | 229/251 | 99/103/27 | 119/107/25 | 0.895 | 8 |
| Wang, 2013 | China | Asian | NA | NA | CHD | 160/188 | 59/76/25 | 53/100/35 | 0.312 | 7 |
| Wang, 2016 | China | Asian | NA | NA | CHD | 147/168 | 14/73/60 | 49/84/35 | 0.928 | 8 |
| Wang, 2018 | China | Asian | NA | 2.18/1.83 | CHD | 102/100 | 31/58/13 | 55/42/3 | 0.130 | 7 |
| Xu, 2010 | China | Asian | 53.7/53.0 | 6.50/6.69 | CHD | 502/527 | 162/244/96 | 151/261/115 | 0.911 | 8 |
| Xu, 2010 | China | Asian | NA | NA | VSD | 257/527 | 83/130/44 | 151/261/115 | 0.911 | 8 |
| Xu, 2010 | China | Asian | NA | NA | ASD | 41/527 | 12/17/12 | 151/261/115 | 0.911 | 8 |
| Xu, 2013 | China | Asian | 64.8/52.4 | NA | CHD | 228/230 | 73/106/49 | 124/74/32 | <0.001 | 8 |
| Yan, 2003 | China | Asian | NA | NA | CHD | 187/103 | 32/97/58 | 22/57/24 | 0.277 | 7 |
| Zhou, 2012 | China | Asian | 48.5/57.8 | NA | TOF | 136/277 | 23/60/53 | 88/126/63 | 0.168 | 8 |
| Zhu, 2006 | China | Asian | 35.1/57.7 | 6.2/8.4 | CHD | 56/103 | 7/22/27 | 22/57/24 | 0.277 | 7 |
| Zhu, 2006 | China | Asian | NA | NA | ASD | 22/103 | 3/7/12 | 22/57/24 | 0.277 | 7 |
| Zhu, 2006 | China | Asian | NA | NA | PDA | 34/103 | 4/15/15 | 22/57/24 | 0.277 | 7 |
| Zidan, 2013 | Egypt | Caucasian | NA | NA | CHD | 80/80 | 18/21/41 | 32/21/27 | <0.001 | 7 |
Abbreviations: ASD, atrial septal defect; CHD, congenital heart disease; HWE, Hardy–Weinberg equilibrium; NA, not available; NOS, Newcastle–Ottawa scale; PDA, patent ductus arteriosus; TGA, transposition of the great arteries; TOF, tetralogy of fallot; VSD, ventricular septal defect.
Overall and subgroup analyses for MTRR polymorphisms
To investigate potential associations between MTRR gene polymorphisms and the risk of CHD, 17 studies about rs1801394 polymorphism and 4 studies about rs1532268 polymorphism were enrolled for overall analyses. Significant associations with the risk of CHD were detected for rs1801394 (dominant model: P=0.0001, OR = 0.68, 95%CI 0.56–0.83; recessive model: P=0.009, OR = 1.40, 95%CI 1.09–1.79; additive model: P=0.008, OR = 1.12, 95%CI 1.03-1.21; allele model: P=0.0001, OR = 0.73, 95%CI 0.63–0.86) and rs1532268 (dominant model: P=0.001, OR = 0.56, 95%CI 0.39–0.80; additive model: P=0.0009, OR = 1.36, 95%CI 1.13–1.63; allele model: P=0.0006, OR = 0.61, 95%CI 0.47–0.81) polymorphisms in overall analyses. Further subgroup analyses according to ethnicity of study participants demonstrated that the rs1801394 polymorphism was significantly correlated with the risk of CHD only in Asians, whereas the rs1532268 polymorphism was significantly correlated with the risk of CHD in both Asians and Caucasians. When we stratified data based on type of disease, we found that both rs1801394 and rs1532268 polymorphisms were significantly associated with the risk of VSD (see Table 2 and Supplementary Figure S1).
Table 2. Results of overall and subgroup analyses.
| Population | Sample size | Dominant comparison | Recessive comparison | Additive comparison | Allele comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95%CI) | I2 statistic | P value | OR (95%CI) | I2 statistic | P value | OR (95%CI) | I2 statistic | P value | OR (95%CI) | I2 statistic | ||
| rs1801394 | |||||||||||||
| Overall | 4899/5246 | 0.0001 | 0.68 (0.56–0.83) | 72% | 0.009 | 1.40 (1.09–1.79) | 56% | 0.008 | 1.12 (1.03–1.21) | 48% | 0.0001 | 0.73 (0.63–0.86) | 77% |
| Caucasian | 887/1375 | 0.11 | 0.75 (0.52–1.07) | 68% | 0.18 | 1.45 (0.85–2.49) | 76% | 0.62 | 1.05 (0.87–1.26) | 0% | 0.10 | 0.75 (0.53–1.06) | 84% |
| Asian | 3675/3712 | 0.0008 | 0.60 (0.45–0.81) | 82% | 0.02 | 1.24 (1.04–1.48) | 34% | 0.006 | 1.43 (1.11–1.85) | 74% | 0.0007 | 0.69 (0.55–0.85) | 79% |
| VSD | 558/587 | <0.0001 | 0.55 (0.43–0.69) | 0% | <0.0001 | 2.22 (1.51–3.26) | 15% | 0.02 | 1.32 (1.04–1.66) | 0% | <0.0001 | 0.60 (0.51–0.72) | 0% |
| rs1532268 | |||||||||||||
| Overall | 1005/1098 | 0.001 | 0.56 (0.39–0.80) | 64% | 0.06 | 1.83 (0.96–3.48) | 68% | 0.0009 | 1.36 (1.13–1.63) | 23% | 0.0006 | 0.61 (0.47–0.81) | 68% |
| Caucasian | 223/225 | 0.08 | 0.44 (0.18–1.09) | 78% | <0.0001 | 2.93 (1.77–4.88) | 16% | 0.94 | 1.02 (0.70–1.48) | 0% | <0.0001 | 0.50 (0.38–0.66) | 46% |
| Asian | 782/873 | 0.008 | 0.64 (0.46–0.89) | 52% | 0.65 | 1.12 (0.69–1.80) | 29% | 0.0002 | 1.48 (1.21–1.83) | 0% | 0.0007 | 0.75 (0.63–0.88) | 33% |
| VSD | 306/326 | <0.0001 | 0.58 (0.42–0.79) | 0% | 0.11 | 2.48 (0.82–7.52) | 70% | 0.47 | 1.25 (0.68–2.28) | 71% | <0.0001 | 0.62 (0.49–0.79) | 0% |
| rs1801131 | |||||||||||||
| Overall | 2834/2761 | 0.44 | 0.93 (0.76–1.13) | 63% | 0.003 | 1.36 (1.11–1.67) | 42% | 0.88 | 0.99 (0.88–1.11) | 38% | 0.23 | 0.90 (0.75–1.07) | 72% |
| Caucasian | 905/1127 | 0.14 | 0.74 (0.49–1.10) | 78% | 0.01 | 1.40 (1.08–1.81) | 45% | 0.62 | 1.05 (0.87–1.26) | 43% | 0.10 | 0.77 (0.57–1.05) | 81% |
| Asian | 1300/1246 | 0.75 | 0.97 (0.82–1.15) | 23% | 0.009 | 1.78 (1.15–2.75) | 47% | 0.97 | 0.99 (0.74–1.34) | 55% | 0.42 | 0.89 (0.68–1.18) | 64% |
| VSD | 601/641 | 0.95 | 0.99 (0.79–1.25) | 21% | 0.74 | 1.12 (0.58–2.18) | 0% | 0.96 | 0.99 (0.78–1.26) | 44% | 0.88 | 0.98 (0.81–1.20) | 0% |
| rs1801133 | |||||||||||||
| Overall | 5508/6207 | <0.0001 | 0.73 (0.63–0.84) | 62% | <0.0001 | 1.54 (1.30–1.83) | 59% | 0.86 | 1.01 (0.93–1.09) | 86% | <0.0001 | 0.75 (0.67–0.84) | 73% |
| Caucasian | 1334/1749 | 0.02 | 0.83 (0.72–0.97) | 26% | 0.01 | 1.35 (1.06–1.72) | 28% | 0.42 | 1.06 (0.91–1.24) | 0% | 0.004 | 0.84 (0.75–0.95) | 50% |
| Asian | 3485/3764 | <0.0001 | 0.67 (0.55–0.83) | 73% | <0.0001 | 1.66 (1.32–2.10) | 71% | 0.43 | 0.96 (0.87–1.06) | 48% | <0.0001 | 0.70 (0.60–0.83) | 81% |
| TOF | 701/764 | 0.0001 | 0.63 (0.50–0.80) | 48% | <0.0001 | 2.17 (1.66–2.84) | 0% | 0.28 | 0.89 (0.71–1.10) | 0% | <0.0001 | 0.62 (0.53–0.72) | 4% |
| VSD | 754/788 | 0.48 | 0.85 (0.54–1.33) | 68% | 0.37 | 1.43 (0.65–3.14) | 75% | 0.90 | 0.99 (0.81–1.21) | 0% | 0.36 | 0.82 (0.54–1.25) | 80% |
Abbreviations: ASD, atrial septal defect; CHD, congenital heart disease; CI, confidence interval; NA, not available; OR, odds ratio; PDA, patent ductus arteriosus; TOF, tetralogy of fallot; VSD, ventricular septal defect.
The values in bold represent there is statistically significant differences between cases and controls.
Overall and subgroup analyses for MTHFR polymorphisms
To investigate potential associations between MTHFR gene polymorphisms and the risk of CHD, 19 studies about rs1801131 polymorphism and 37 studies about rs1801133 polymorphism were enrolled for overall analyses. Significant associations with the risk of CHD were detected for rs1801131 (recessive model: P=0.003, OR = 1.36, 95%CI 1.11–1.67) and rs1801133 (dominant model: P<0.0001, OR = 0.73, 95%CI 0.63–0.84; additive model: P<0.0001, OR = 1.54, 95%CI 1.30–1.83; allele model: P<0.0001, OR = 0.75, 95%CI 0.67–0.84) polymorphisms in overall analyses. Further subgroup analyses according to ethnicity of study participants demonstrated that rs1801133 and rs1801131 polymorphisms were significantly correlated with the risk of CHD in both Asians and Caucasians. When we stratified data based on type of disease, we found that the rs1801133 polymorphism was significantly associated with the risk of TOF (see Table 2 and Supplementary Figure S1).
Sensitivity analyses
To examine stabilities of synthetic results, sensitivity analyses were further performed by removing studies that departed from HWE. No changes of results were detected for investigated gene polymorphisms in any comparisons, which indicated that our findings were quite statistically stable.
Publication biases
Funnel plots were used to assess potential publication biases in the present study. No apparent asymmetry of funnel plots was observed in any comparisons, which suggested that our findings were unlikely to be influenced by obvious publication biases (see Supplementary Figure S2).
Discussion
CHD contain various structural cardiovascular malformations that are actually or potentially of functional significances [19]. Historically, few CHD patients reached adulthood, but thanks to enormous advances in interventional therapies and surgical treatments over the past few years, the average life expectancy of CHD patients has been significantly improved [20]. However, despite substantially improved prognosis, CHD remains to be the leading cause of infant deaths all over the world.
MTHFR and MTRR are fundamental regulatory enzymes of folate and homocysteine metabolism. Considering the consistently observed association between folic acid consumption and a reduced risk of cardiac deformity, functional polymorphisms of MTHFR and MTRR, which were known to be associated with altered enzymatic activities, were thought to be correlated with the risk of CHD [11,12]. Recently, several studies have tried to explore the potential associations of functional MTHFR and MTRR gene polymorphisms with the risk of CHD, but the results of these studies were inconsistent. Therefore, we conducted the present meta-analysis to obtain a more conclusive result. Our overall analyses suggested that MTRR rs1801394, MTRR rs1532268, MTHFR rs1801131 and MTHFR rs1801133 polymorphisms were all significantly associated with the risk of CHD in certain genetic models. Further subgroup analyses according to ethnicity of study participants demonstrated that the MTRR rs1801394 polymorphism was significantly correlated with the risk of CHD only in Asians, whereas MTRR rs1532268, MTHFR rs1801133 and MTHFR rs1801131 polymorphisms were significantly correlated with the risk of CHD in both Asians and Caucasians. When we stratified data based on type of disease, we found that both MTRR rs1801394 and MTRR rs1532268 polymorphisms were significantly associated with the risk of VSD, whereas the MTHFR rs1801133 polymorphism was significantly associated with the risk of TOF. The stabilities of synthetic results were subsequently evaluated in sensitivity analyses, and no changes of results were observed in any comparisons, which indicated that our findings were quite stable and reliable. It is noteworthy that obvious between-study heterogeneities were detected in several comparisons. However, a great reduction in heterogeneities was found in further stratified analyses, which suggested that differences in ethnic background and type of disease could partially explain the observed heterogeneities.
Our meta-analysis is certainly not without limitations. First, our results were based on unadjusted estimations, and lack of analyses adjusted for potential confounding factors such as age, sex and co-morbidity conditions may impact the reliability of our findings. Second, heterogeneity remained significant in certain subgroups, which suggested that the conflicting results of eligible studies could not be fully explained by differences in ethnicity of study population or type of CHD, and other unmeasured characteristics of study participants may also attribute to the observed between-study heterogeneities. Third, associations between investigated polymorphisms and the risk of CHD may also be influenced by gene–gene and gene–environmental interactions. However, we failed to analyze the effect of these interactions in our study because only very little relevant data were provided by enrolled literatures. Taken these limitations into consideration, the results obtained by the present study should be interpreted with caution.
In conclusion, the current meta-analysis indicated that MTRR rs1801394, MTRR rs1532268, MTHFR rs1801131 and MTHFR rs1801133 polymorphisms may affect the risk of CHD in Asians and Caucasians, while the MTRR rs1801394 polymorphism may only affect in risk of CHD in Asians. However, it is notable that relevant studies were still at the early stage and further well-designed studies are still warranted to confirm our findings.
Supporting information
Funnel plots of investigated polymorphisms.
Funnel plots of investigated polymorphisms.
Acknowledgments
None.
Abbreviations
- CHD
congenital heart diseases
- CI
confidence interval
- HWE
Hardy–Weinberg equilibrium
- MTHFR
methylenetetrahydrofolate reductase
- MTRR
methionine synthase reductase
- NOS
Newcastle–Ottawa scale
- OR
odds ratio
Author Contribution
Aiping Xu and Weiping Wang conceived the study and participated in its design. Aiping Xu and Weiping Wang conducted the systematic literature review. Aiping Xu, Weiping Wang and Xiaolei Jiang performed data analyses. Aiping Xu and Weiping Wang drafted the manuscript. All authors have read and approved the final manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Hoffman J.I. and Kaplan S. (2002) The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 10.1016/S0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- 2.Van der Bom T., Zomer A.C., Zwinderman A.H., Meijboom F.J., Bouma B.J. and Mulder BJ. (2011) The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 8, 50–60 10.1038/nrcardio.2010.166 [DOI] [PubMed] [Google Scholar]
- 3.Verheugt C.L., Uiterwaal C.S., van der Velde E.T., Meijboom F.J., Pieper P.G., van Dijk A.P.. et al. (2010) Mortality in adult congenital heart disease. Eur. Heart J. 31, 1220–1229 10.1093/eurheartj/ehq032 [DOI] [PubMed] [Google Scholar]
- 4.Samanek M. (2000) Congenital heart malformations: prevalence, severity, survival, and quality of life. Cardiol. Young 10, 179–185 10.1017/S1047951100009082 [DOI] [PubMed] [Google Scholar]
- 5.Pierpont M.E., Basson C.T., Benson D.W. Jr, Gelb B.D., Giglia T.M., Goldmuntz E.. et al. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115, 3015–3038 10.1161/CIRCULATIONAHA.106.183056 [DOI] [PubMed] [Google Scholar]
- 6.Garg V., Muth A.N., Ransom J.F., Schluterman M.K., Barnes R., King I.N.. et al. (2005) Mutations in NOTCH1 cause aortic valve disease. Nature 437, 270–274 10.1038/nature03940 [DOI] [PubMed] [Google Scholar]
- 7.Muncke N., Jung C., Rudiger H., Ulmer H., Roeth R., Hubert A.. et al. (2003) Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation 108, 2843–2850 10.1161/01.CIR.0000103684.77636.CD [DOI] [PubMed] [Google Scholar]
- 8.Robinson S.W., Morris C.D., Goldmuntz E., Ulmer H., Roeth R., Hubert A.. et al. (2003) Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am. J. Hum. Genet. 72, 1047–1052 10.1086/374319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A.. et al. (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 10.1038/nature01827 [DOI] [PubMed] [Google Scholar]
- 10.Moll S. and Varga EA. (2015) Homocysteine and MTHFR mutations. Circulation 132, e6–e9 10.1161/CIRCULATIONAHA.114.013311 [DOI] [PubMed] [Google Scholar]
- 11.Botto L.D., Khoury M.J., Mulinare J. and Erickson J.D. (2010) Periconceptional nutrient intakes and risks of conotruncal heart defects. Birth Defects Res. A Clin. Mol. Teratol. 88, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botto L.D., Khoury M.J., Mulinare J. and Erickson J.D. (1996) Periconceptional multivitamin use and the occurrence of conotruncal heart defects: results from a population-based, case-control study. Pediatrics 98, 911–917 [PubMed] [Google Scholar]
- 13.Koshy T., Venkatesan V., Perumal V., Hegde S. and Paul S.F. (2015) The A1298C methylenetetrahydrofolate reductase gene variant as a susceptibility gene for non-syndromic conotruncal heart defects in an Indian population. Pediatr. Cardiol. 36, 1470–1475 10.1007/s00246-015-1188-3 [DOI] [PubMed] [Google Scholar]
- 14.Obermann-Borst S.A., van Driel L.M., Helbing W.A., de Jonge R., Wildhagen M.F., Steegers E.A.. et al. (2011) Congenital heart defects and biomarkers of methylation in children: a case-control study. Eur. J. Clin. Invest. 41, 143–150 10.1111/j.1365-2362.2010.02388.x [DOI] [PubMed] [Google Scholar]
- 15.Guo Q.N., Wang H.D., Tie L.Z., Li T., Xiao H., Long J.G.. et al. (2017) Parental genetic variants, MTHFR 677C>T and MTRR 66A>G, associated differently with fetal congenital heart defect. Biomed. Res. Int. 2017, 3043476 10.1155/2017/3043476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahiner U.M., Alanay Y., Alehan D., Tuncbilek E. and Alikasifoglu M. (2014) Methylene tetrahydrofolate reductase polymorphisms and homocysteine level in heart defects. Pediatr. Int. 56, 167–172 10.1111/ped.12222 [DOI] [PubMed] [Google Scholar]
- 17.Moher D., Liberati A., Tetzlaff J. and Altman D.G.. PRISMA group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151, 264–269 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- 18.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 19.Sun R., Liu M., Lu L., Zheng Y. and Zhang P. (2015) Congenital heart disease: causes, diagnosis, symptoms, and treatments. Cell Biochem. Biophys. 72, 857–860 10.1007/s12013-015-0551-6 [DOI] [PubMed] [Google Scholar]
- 20.Warnes C.A. (2005) The adult with congenital heart disease: born to be bad? J. Am. Coll. Cardiol. 46, 1–8 10.1016/j.jacc.2005.02.083 [DOI] [PubMed] [Google Scholar]