Abstract
Gastric cancer (GC) is the second most frequent cause of cancer-related mortality in the world, with Eastern Asia having the highest incidence rates. E2F is a family of transcription factor proteins that has a variety of functions, which include control of cell cycle, cell differentiation, DNA damage response and cell death. E2F transcription factors are divided into two subfamilies: transcription activators (E2F transcription factors 1 (E2F1), 2 (E2F2) and 3a (E2F3a)) and repressors (E2F3b, E2F transcription factors 4 (E2F4), 5 (E2F5), 6 (E2F6), 7 (E2F7) and 8 (E2F8)). Studies have demonstrated that E2F had prognostic significance in a number of cancers. However, the entirety of the prognostic roles of E2F mRNA expression in GC has not yet been apparently determined. In the present study, the prognostic value of individual family members of E2F mRNA expression for overall survival (OS) was evaluated by using online Kaplan–Meier Plotter (KM Plotter) database. Our result demonstrated that high expressions of three family members of E2F (E2F1, E2F3, E2F4) mRNA were significantly associated with unfavourable OS in all GC patients. However, increased expressions of E2F2, E2F5, E2F6 and E2F7 were significantly associated with favourable OS, especially for higher clinical stages in GC patients. These results provided a better insight into the prognostic functions of E2F mRNA genes in GC. Although the results should be further verified in clinical trials, our findings may be a favourable prognostic predictor for the development of newer therapeutic drugs in the treatment of GC.
Keywords: E2F, gastric cancer, KM plotter, mRNA(mrna s), Prognostic values
Introduction
Gastric cancer (GC) is the second most frequent cause of cancer-related mortality in the world, with Eastern Asia having the highest incidence rates [1]. Gastric adenocarcinoma (GAC) is the most common type of GC and according to the Lauren Classification, it is classified into two histological types: intestinal and diffuse [2]. Investigations and attempts at identifying molecular target therapy to improve patients’ outcomes still continue, thus GC remains a challenge to cure [3]. There have been various chemotherapeutic agents, which have been found to improve survival, however the median survival still remains less than a year [4]. Therefore, the identification of prognostic markers in GC is fundamental in improving clinical outcomes for GC patients.
E2F is a family of transcription factor proteins that has a variety of functions and has earned its title as a master of regulators of cell proliferation. Its variety of functions include, control of cell cycle, cell differentiation and DNA damage response and cell death [5,6]. This transcription factor family also consists of DNA-binding domains (DBD), which binds to specific target promoters and regulates their expressions [7,8]. E2F transcription factors are divided into two subfamilies: transcription activators (E2F transcription factors 1 (E2F1), 2 (E2F2) and 3a (E2F3a)) and repressors (E2F transcription factors 3b (E2F3b), 4 (E2F4), 5 (E2F5), 6 (E2F6), 7 (E2F7) and 8 (E2F8)) [6]. The first six E2F transcription factors also bind to DNA as heterodimers with the related dimerisation proteins (DP), such as DP1 and DP2 [5,6]. Transcription factors E2F7 and E2F8 are unique, because they consist of two unique DNA-binding subdomains [9]. E2F repressors that function independently from retinoblastoma (Rb), may form a new class of E2Fs (E2F6 and E2F7), and this is due to the evolution of these repressors [10]. Quiescent cells can be driven into S-phase via activator transcription factors, E2F1, E2F2 and E2F3a, by activating target genes required for G1/S transition [11]. During G0 phase, repressors E2F4 and E2F5 that are bound to Rb-related pocket proteins and associated co-repressors, supress target gene transcription. E2F6 contains MAX gene associated (Mga) and MYC-associated factor X (Max) which are part of a multimeric protein complex that represses G1/S gene transcription, independent from Rb Family members [12,13].
In most human cancers E2F transcription factors are altered and deregulated, via different molecular mechanisms that inactivate the Rb family [14]. E2F family member activators could possess oncogenic behaviour in human carcinogenesis. Repressors from the E2F family could be associated with tumour suppressing functions in human carcinogenesis [6].
Research has revealed that E2F1’s overexpression in GC prompted an outspread of cell death through various mechanisms, therefore proving the role of E2F1 in tumour suppression in GC [15]. However, the entirety of the prognostic roles of E2F mRNA expression in GC has not been determined apparently. In the current study, we will investigate whether E2F genes are of prognostic significance in human GC patients. We will evaluate clinical data, which include clinical stages, Lauren classification, differentiation degree, human epidermal growth factor receptor-2 (HER2) status and gender and treatment strategies. In the present study, we comprehensively investigated the prognostic values of seven E2F family members using the Kaplan–Meier Plotter (KM Plotter).
Methods
The prognostic values for individual E2Fs members’ mRNA expressions for overall survival (OS) were evaluated by using online KM Plotter (http://kmplot.com/analysis) database. This database was established using gene expression data and survival information from Gene Expression Omnibus (GEO) [16], including Genet Sel Evol (GSE)14210 [17], GSE22377 [18], GSE51105 [19], GSE15459 [20] and GSE29272 [21]. Currently, the database has been established with 54675 genes that have been identified in GC [16], breast cancer [22], ovarian cancer [23], lung cancer [24] and liver cancer [25]. The database consists of a collection of clinical data including Lauren classification, clinical stages, differentiation degree, gender, HER2 status and treatment of GC patients. In the present study, using the available data source, we gathered the clinical data. We entered seven individual members of the E2F family (E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, E2F7), to obtain Kaplan–Meier survival plots. Hazard ratio (HR), 95% confidence interval (CI) and log rank P were determined and displayed on the webpage. A value of P<0.05 was considered statistically significant.
Results
Prognostic values of E2F mRNA expression in all GC patients
In the current study, seven out of the eight E2F members’ data were obtained using the Kaplan–Meier survival plots (http://www.kmplot.com).
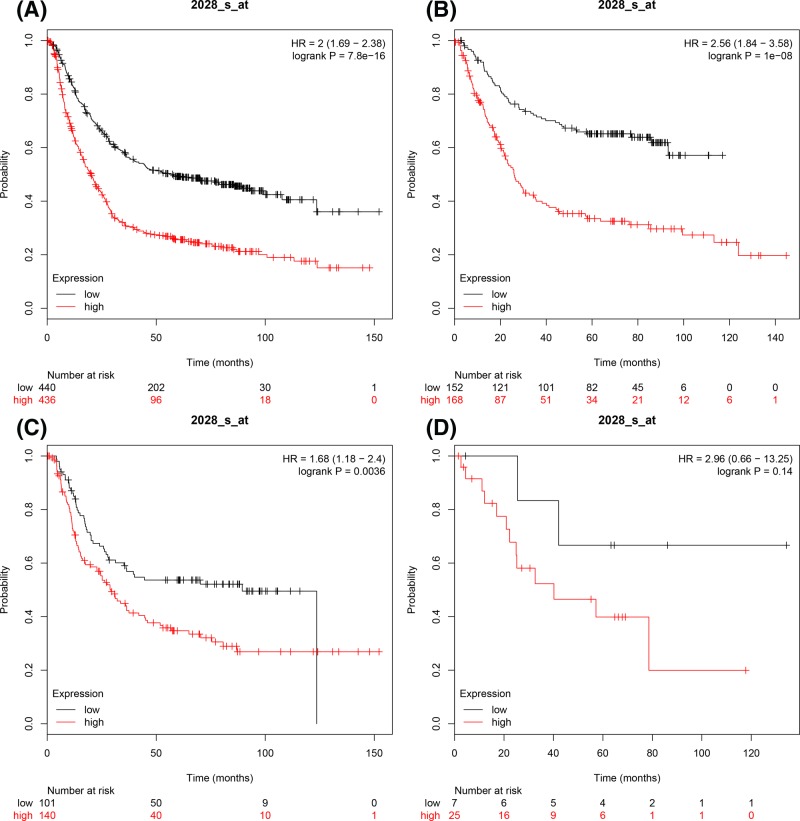
The prognostic values of E2F1 mRNA expression were evaluated in the database. ID 2028_s_at. OS curves were plotted for GC patients. Increase in E2F1 mRNA expression level revealed a significant association with poor OS, for all GC patients, (n=876), HR = 2 (1.69–-2.38), P=7.8 × 10−16 (Figure 1A). The Lauren classification subtype results revealed that increased E2F1 mRNA expression was correlated with unfavourable OS for patients with intestinal GC, HR = 2.56 (1.84–3.58), P=1 × 10−8 (Figure 1B) and patients with diffuse GC, HR = 1.68 (1.18–2.4), P=0.0036 (Figure 1C). However, the expression level of E2F1 mRNA in patients with mixed-type GC, did not show a significant correlation, HR = 2.96 (0.66–13.25), P=0.14 (Figure 1D).
Figure 1. The prognostic values of E2F1 expression in GC.
The prognostic of E2F1 expression in www.kmplot.com ID. 2028_s_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
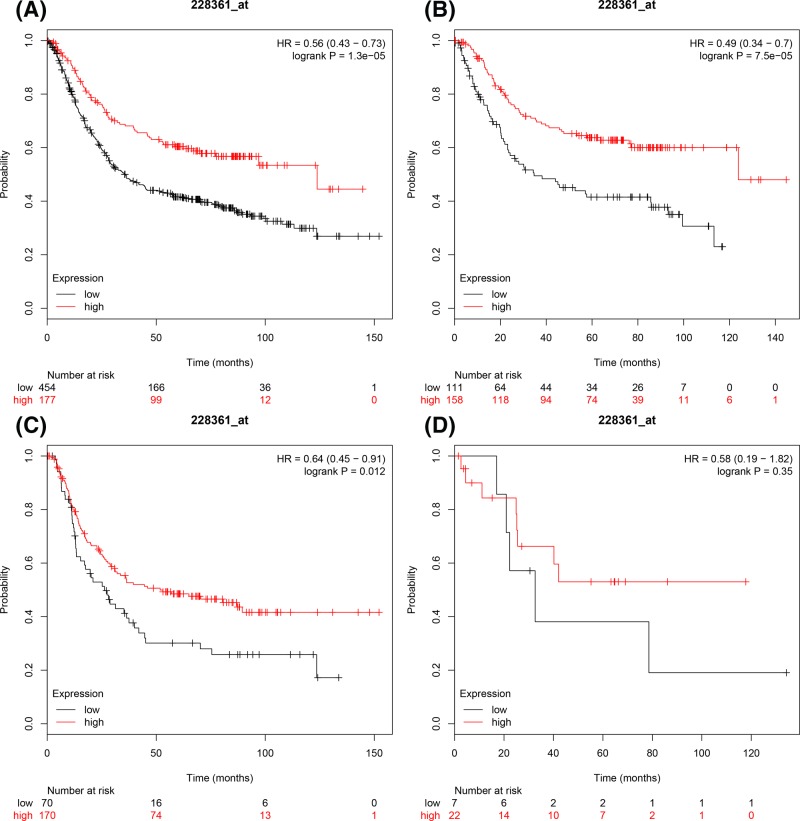
The next set of prognostic values for E2F2 mRNA expression were evaluated in the database. ID228361_s_at. E2F2 mRNA expression levels were significantly associated with favourable OS for all GC patients, HR = 0.56 (0.43–0.73), P=1.3 × 10−5 (Figure 2A), intestinal GC patients, HR = 0.49 (0.34–0.7), P=7.5 × 10−5 (Figure 2B), and diffuse GC patients, HR = 0.64 (0.45–0.91), P=0.012 (Figure 2C). Whereas the expression level of E2F2 mRNA in mixed-type GC patients did not show any association with OS, HR = 0.58 (0.19–1.82), P=0.35 (Figure 2D).
Figure 2. The prognostic values of E2F2 expression in GC.
The prognostic of E2F2 expression in www.kmplot.com ID.228361_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
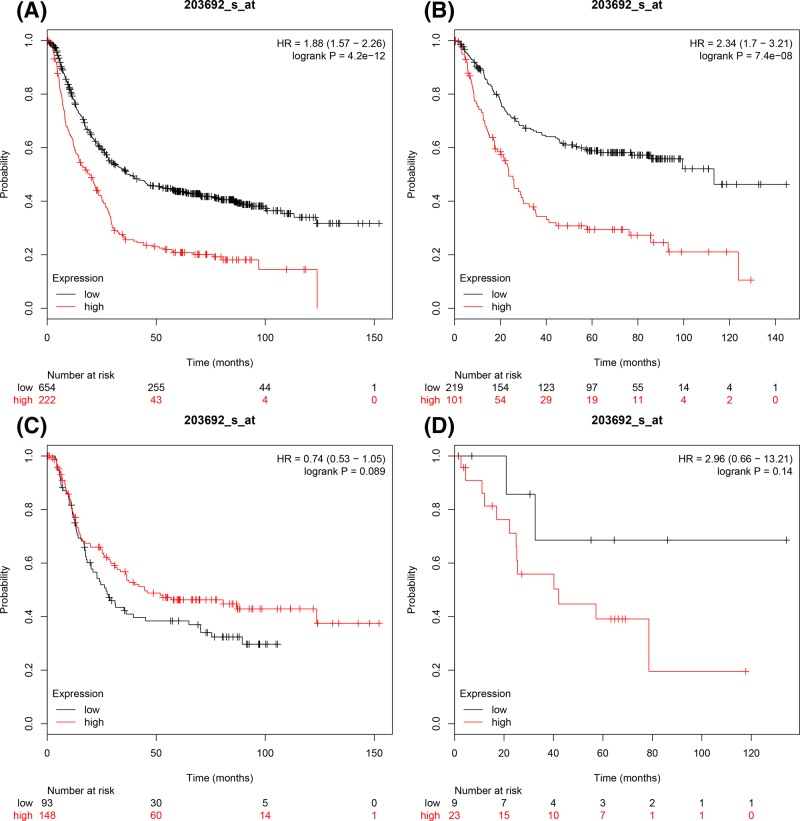
Figure 3 showed the prognostic values of E2F3 in the database. ID 203692_s_at. E2F3 mRNA expression was significantly associated with poor OS for all GC patients and intestinal cancer patients, HR = 1.88 (1.57–2.26), P=4.2 × 10−12 (Figure 3A), HR = 2.34 (1.7–3.21), P=7.4 × 10−8 (Figure 3B) respectively. However, there was no significant association in OS of diffuse GC patients and mixed type GC patients, HR = 0.74 (0.53–1.05), P=0.089 (Figure 3C), HR = 2.96 (0.66–13.21), P=0.14 (Figure 3D) respectively.
Figure 3. The prognostic values of E2F3 expression in GC.
The prognostic of E2F3 expression in www.kmplot.com ID. 203692_s_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
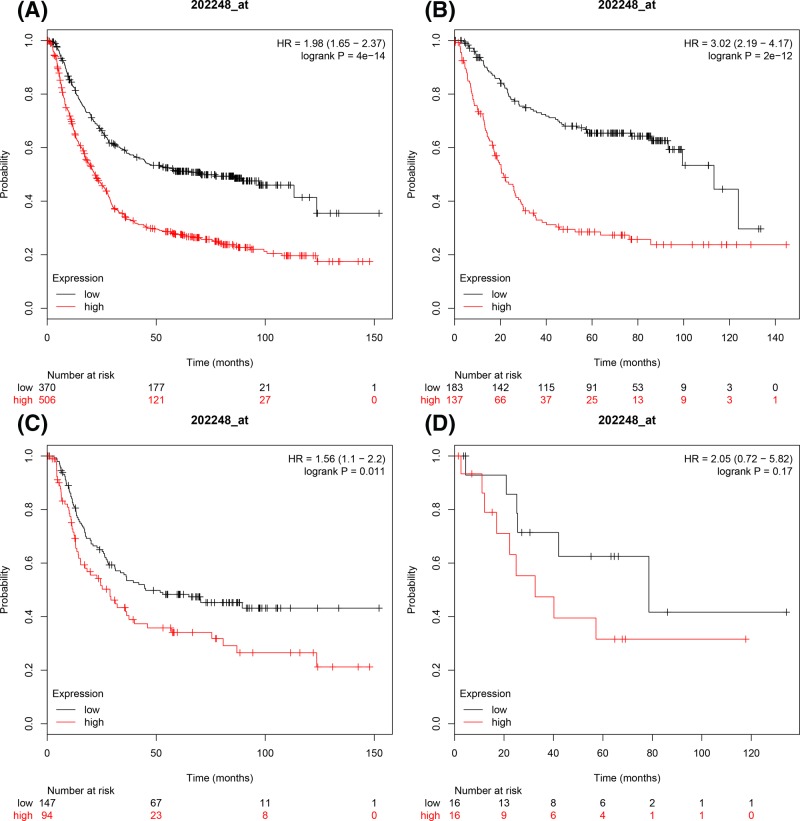
Figure 4 presented prognostic significance of E2F4 mRNA expression in the database. ID 202248_s_at. High E2F4 mRNA levels were significantly correlated with unfavourable OS in all GC patients, intestinal GC patients and diffuse GC patients, HR = 1.98 (1.65–2.37), P=4 × 10−14 (Figure 4A), HR = 3.02 (2.19–4.17), P=2 × 10−12 (Figure 4B), HR = 1.56 (1.1–2.2), P=0.011 (Figure 4C) respectively. But there was not any association with OS of mixed type GC, HR = 2.05 (0.72–5.82), P=0.17 (Figure 4 D).
Figure 4. The prognostic values of E2F4 expression in GC.
The prognostic of E2F4 expression in www.kmplot.com ID. 202248_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
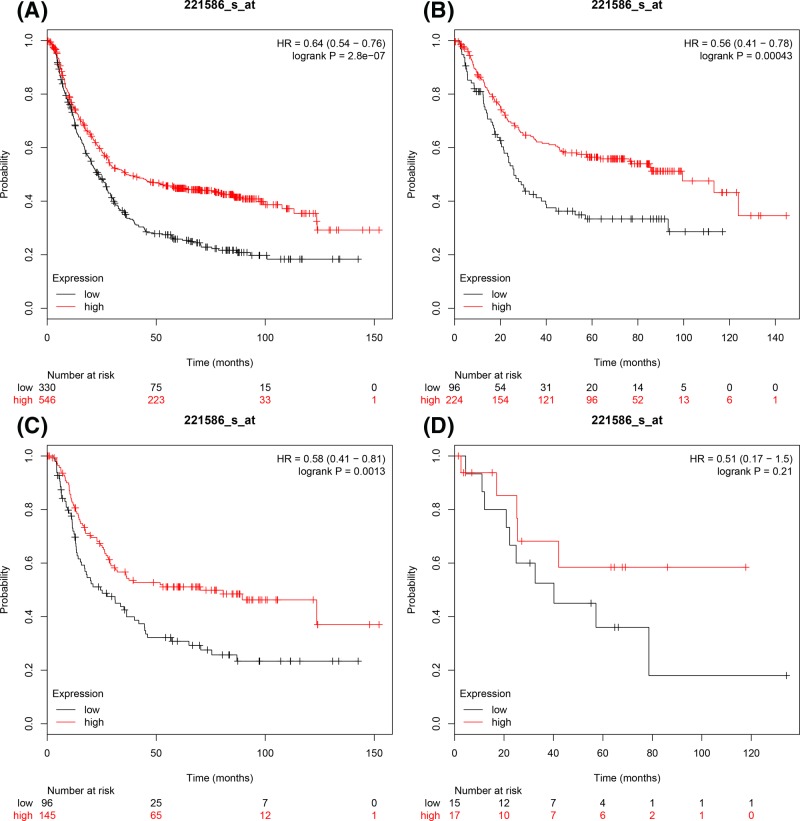
Figure 5 illustrated prognostic association of E2F5 mRNA expression in the database. ID 221586_s_at. Increased expression of E2F5 was correlated with favourable OS in all GC patients, HR = 0.64 (0.54–0.076), P=2.8 × 10−7 (Figure 5A), intestinal cancer patients, HR = 0.56 (0.41–0.76), P=0.00043 (Figure 5B) and diffuse GC patients, HR = 0.58 (0.41–0.81), P=0.0013 (Figure 5C). However, increased E2F5 mRNA in mixed-type mRNA was not correlated with OS, HR = 0.51 (0.17–1.5), P=0.21 (Figure 5D).
Figure 5. The prognostic values of E2F5 expression in GC.
The prognostic of E2F5 expression in www.kmplot.com ID. 221586_s_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
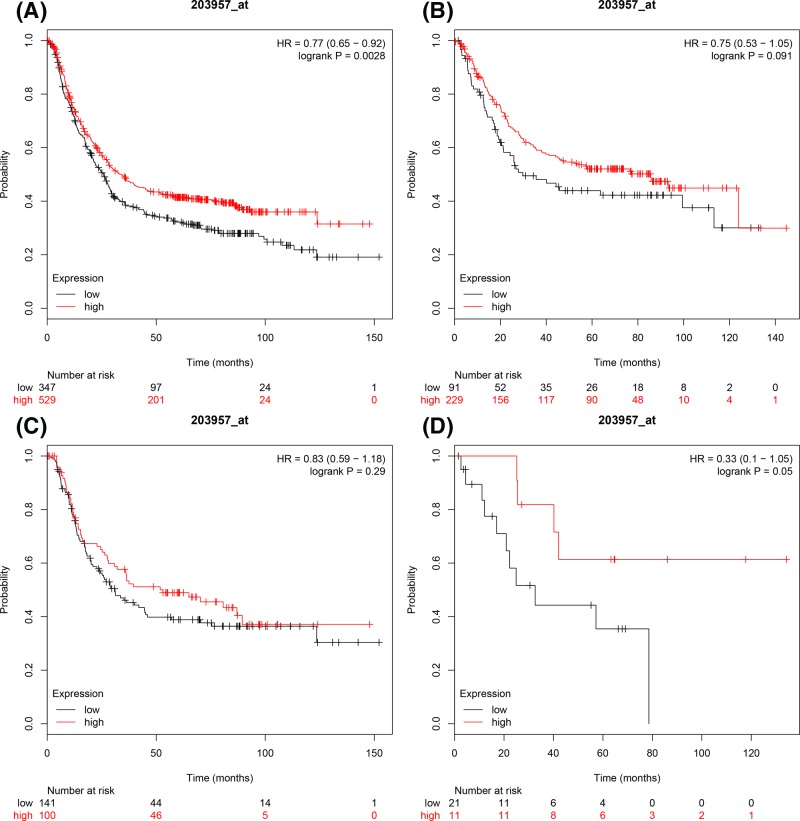
Prognostic values for E2F6 mRNA expression were evaluated in the database. ID 203957_s_at. Overexpression of E2F6 mRNA was found to be associated with favourable OS of all GC patients, HR = 0.77 (0.65–0.92), P=0.0028 (Figure 6A) and mixed-type GC, HR = 0.33 (0.1–1.05), P=0.05 (Figure 6D). Whereas, increased E2F6 mRNA expression showed no correlation with OS in neither intestinal GC, HR = 0.75 (0.53–1.05), P=0.091 (Figure 6B) nor diffuse GC, HR = 0.83 (0.59–1.18), P=0.29 (Figure 6C).
Figure 6. The prognostic values of E2F6 expression in GC.
The prognostic of E2F6 expression in www.kmplot.com ID. 203957_at. OS curves were plotted for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients, (D) mixed cancer patients.
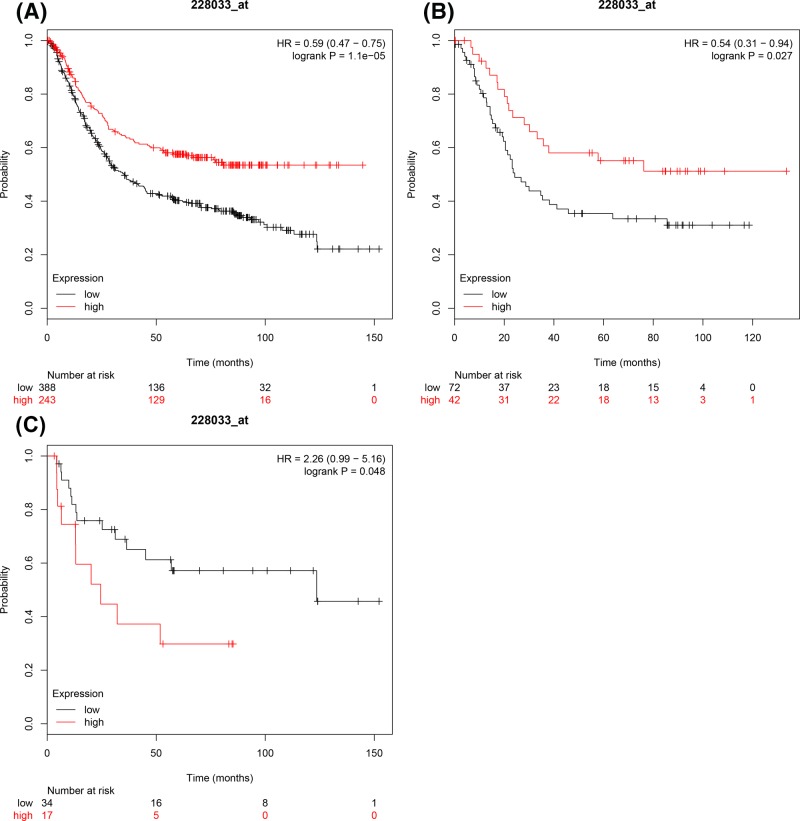
Figure 7 demonstrated prognostic significance of E2F7 mRNA expression in the database. ID 228033_s_at. High expression of E2F7 mRNA levels showed a significant correlation with favourable OS in all GC patients, HR = 0.59 (0.47–0.75), P=1.1 × 10−5 (Figure 7A) and intestinal GC patients, HR = 0.54 (0.31–0.94), P=0.027 (Figure 7B). However, overexpression of E2F7 mRNA showed a signification association with unfavourable OS in diffuse GC patients, HR = 2.26 (0.99–5.16), P=0.048 (Figure 7C) and mixed-type GC patients’ sample size was too small to demonstrate any significant results.
Figure 7. The prognostic values of E2F7 expression in GC.
The prognostic of E2F7 expression in www.kmplot.com ID.228033_s_at. OS curves were for (A) all the patients (n=876), (B) intestinal cancer patients, (C) diffuse cancer patients.
In addition to our investigations of the prognostic values of E2F mRNA expression, we evaluated the association with other clinicopathological characteristics, including correlation of E2Fs with clinical stages, HER2 status, treatment strategies and gender status and differentiation degree of GC patients.
As illustrated in Table 1, we found that overexpressions of E2F2, E2F5 and E2F6 were correlated with favourable OS in stage III GC patients. Consecutive mRNA expressions were associated with better OS in E2F5 stages (II, III and IV) and E2F7 stage IV GC patients. However, increased expressions in E2F1 all stages (I, II, III, IV), E2F3 stage III and E2F4 stage (III, IV) were correlated with poor OS in GC patients.
Table 1. Correlation of E2Fs’ genes expression with OS in GC patients at different clinical stages.
| E2Fs | Clinical stage | Cases | HR (95% CI) | P-value |
|---|---|---|---|---|
| E2F1 | I | 67 | 5.18 (1.45−18.45) | 0.0049* |
| II | 140 | 2.19 (1.14−4.21) | 0.016* | |
| III | 305 | 2.29 (1.57−3.33) | 9.9e−06* | |
| IV | 148 | 1.55 (1.05−2.3) | 0.027* | |
| E2F2 | I | 62 | 0.43 (0.14−1.34) | 0.13 |
| II | 135 | 0.67 (0.36−1.24) | 0.2 | |
| III | 297 | 0.53 (0.36−0.8) | 0.0018* | |
| IV | 135 | 0.67 (0.36−1.24) | 0.2 | |
| E2F3 | I | 67 | 2.41 (0.89−6.49) | 0.074 |
| II | 140 | 1.77 (0.95−3.32) | 0.07 | |
| III | 148 | 1.81 (1.36−2.41) | 4e−05* | |
| IV | 148 | 1.24 (0.83−1.84) | 0.29 | |
| E2F4 | I | 67 | 2.48 (0.92−6.66) | 0.064 |
| II | 140 | 1.73 (0.93−3.21) | 0.078 | |
| III | 305 | 2.23 (1.6−3.11) | 1.1e−06* | |
| IV | 148 | 1.64 (1.11−2.43) | 0.012* | |
| E2F5 | I | 67 | 0.57 (0.2−1.64) | 0.29 |
| II | 140 | 0.52 (0.28−0.97) | 0.036* | |
| III | 305 | 0.63 (0.47−0.84) | 0.0016* | |
| IV | 148 | 0.55 (0.35−0.84) | 0.0051* | |
| E2F6 | I | 67 | 0.44 (0.16−1.18) | 0.095 |
| II | 140 | 1.77 (0.96−3.24) | 0.063 | |
| III | 305 | 0.74 (0.56−0.98) | 0.037* | |
| IV | 148 | 0.73 (0.49−1.07) | 0.1 | |
| E2F7 | I | 62 | 0.3 (0.08−1.11) | 0.056 |
| II | 135 | 0.55 (0.29−1.04) | 0.063 | |
| III | 197 | 0.63 (0.41−0.96) | 0.032* | |
| IV | 140 | 0.57 (0.37−0.86) | 0.007* |
P<0.05.
In Table 2 we further investigated the association between E2Fs expression and HER2 status in GC patients. E2F2 and E2F5 showed correlations with better OS when HER2 expression was both positive and negative in GC patients. A negative expression of HER2 in E2F6 and E2F7 was correlated with favourable OS in GC patients. However, both positive and negative HER2 expressions were associated with unfavourable OS in E2F1, E2F3 and E2F4 mRNA expressions in GC patients. On the other hand, E2F6 and E2F7 had no correlation with positive HER2 status in GC patients.
Table 2. Correlation of E2Fs’ genes expression with OS in GC patients with HER2 expression status.
| E2Fs | HER2 status | Cases | Low | High | HR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| E2F1 | Negative | 532 | 292 | 240 | 1.98 (1.58−2.48) | 1.7e−09* |
| Positive | 344 | 131 | 213 | 1.93 (1.46−2.55) | 2.7e−06* | |
| E2F2 | Negative | 429 | 222 | 207 | 0.6 (0.46−0.79) | 0.00018* |
| Positive | 202 | 115 | 87 | 0.6 (0.4−0.88) | 0.0083* | |
| E2F3 | Negative | 532 | 361 | 171 | 1.64 (1.3−2.07) | 2.1e−05* |
| Positive | 344 | 185 | 159 | 1.76 (1.36−2.29) | 1.6e−05* | |
| E2F4 | Negative | 532 | 292 | 240 | 1.84 (1.47−2.3) | 8.1e−08* |
| Positive | 344 | 120 | 224 | 2.01 (1.49−2.7) | 2.5e−06* | |
| E2F5 | Negative | 532 | 192 | 340 | 0.58 (0.46−0.72) | 1.5e−06* |
| Positive | 344 | 126 | 218 | 0.71 (0.55−0.93) | 0.011* | |
| E2F6 | Negative | 532 | 201 | 331 | 0.69 (0.55−0.86) | 0.0012* |
| Positive | 344 | 156 | 188 | 0.89 (0.68−1.15) | 0.36 | |
| E2F7 | Negative | 429 | 154 | 275 | 0.54 (0.41−0.7) | 3.7e−06* |
| Positive | 202 | 142 | 60 | 0.65 (0.43−1.01) | 0.051 |
P<0.05.
The results in Table 3 showed the correlation of E2F mRNA expression with OS using different treatment strategies in GC patients. The results revealed that expressions of E2F2 and E2F7 showed better OS for GC patients treated with surgery alone. However, increased mRNA expressions of E2F1, E2F3 and E2F4 were significantly correlated with poor OS for GC patients treated with surgery alone. Consecutive expressions of all E2Fs except E2F2 showed unfavourable OS in GC patients treated with 5-Fluorouracil (5FU) adjuvant treatment. Subsequent expressions of E2F2, E2F4 and E2F5 were significantly associated with favourable OS, when GC patients were treated with other adjuvant treatments.
Table 3. Correlation of E2Fs’ genes expression with OS in GC patients with different treatment strategies.
| E2Fs | Treatment | Cases | HR (95% CI) | P-value |
|---|---|---|---|---|
| E2F1 | Surgery alone | 380 | 1.45 (1.06−1.97) | 0.019* |
| 5FU-based adjuvant | 153 | 2.06 (1.36−3.13) | 0.00054* | |
| Other adjuvant | 76 | 0.56 (0.23−1.37) | 0.2 | |
| E2F2 | Surgery alone | 380 | 0.59 (0.42−0.83) | 0.0022* |
| 5FU-based adjuvant | 34 | 2.47 (0.94−6.44) | 0.058 | |
| Other adjuvant | 76 | 0.4 (0.16−0.99) | 0.041* | |
| E2F3 | Surgery alone | 380 | 1.43 (1.05−1.95) | 0.023* |
| 5FU-based adjuvant | 153 | 1.8 (1.24−2.62) | 0.0018* | |
| Other adjuvant | 76 | 0.44 (0.18−1.08) | 0.064 | |
| E2F4 | Surgery alone | 380 | 1.41 (1.02−1.93) | 0.034* |
| 5FU-based adjuvant | 153 | 1.81 (1.22−2.68) | 0.0026* | |
| Other adjuvant | 76 | 0.26 (0.1−0.73) | 0.0056* | |
| E2F5 | Surgery alone | 380 | 0.76 (0.56−1.04) | 0.085 |
| 5FU-based adjuvant | 153 | 1.85 (1.27−2.7) | 0.0013* | |
| Other adjuvant | 76 | 0.26 (0.1−0.67) | 0.0028* | |
| E2F6 | Surgery alone | 380 | 0.85 (0.62−1.17) | 0.31 |
| 5FU-based adjuvant | 153 | 1.78 (1.24−2.55) | 0.0015* | |
| Other adjuvant | 76 | 3.33 (0.77−14.37) | 0.087 | |
| E2F7 | Surgery alone | 380 | 0.64 (0.47−0.86) | 0.0033* |
| 5FU-based adjuvant | 34 | 3.24 (1.26−8.28) | 0.01* | |
| Other adjuvant | 76 | 0.51 (0.2−1.27) | 0.14 |
P<0.05.
Table 4 showed prognostic significance between the E2F mRNA expression and gender in GC patients. Increased expressions of E2F2, E2F5 and E2F7 were correlated with favourable OS in both male and female GC patients. However, overexpressions of E2F1 and E2F4 were associated with poor OS for both male and female GC patients. Subsequent expression of E2F6 was significantly correlated with favourable OS for male patients, whereas E2F3 expression was correlated with unfavourable OS for female GC patients.
Table 4. Correlation of E2Fs’ genes expression with OS in GC patients with gender expression status.
| E2Fs | Gender | Cases | HR (95% CI) | P-value |
|---|---|---|---|---|
| E2F1 | Male | 545 | 2.33 (1.84−2.95) | 5.1e−13* |
| Female | 876 | 2 (1.69−2.38) | 7.8e−16* | |
| E2F2 | Male | 349 | 0.53 (0.39−0.72) | 2.6e−05* |
| Female | 187 | 0.53 (0.31−0.91) | 0.02* | |
| E2F3 | Male | 545 | 2.08 (1.68−2.58) | 9e−12* |
| Female | 236 | 1.54 (1.05−2.25) | 0.026 | |
| E2F4 | Male | 545 | 2.24 (1.79−2.81) | 7.2e−13* |
| Female | 236 | 2.17 (1.53−3.09) | 8.6e−06* | |
| E2F5 | Male | 545 | 0.65 (0.52−0.81) | 9e−05* |
| Female | 236 | 0.59 (0.42−0.84) | 0.003* | |
| E2F6 | Male | 545 | 0.89 (0.71−1.11) | 0.28 |
| Female | 236 | 0.56 (0.39−0.79) | 0.00097* | |
| E2F7 | Male | 349 | 0.6 (0.45−0.82) | 0.0011* |
| Female | 187 | 0.48 (0.3−0.75) | 0.0011* |
P<0.05.
Lastly, Table 5 demonstrated correlations of E2Fs expression with OS according to their differentiation degree in GC patients. We found that GC patients with high expressions of E2F2 and E2F3 with moderate differentiation had better OS. Whereas, E2F3 and E2F7 expressions correlated with worse OS for poor differentiation and E2F4 expression was also correlated with worse OS for moderate differentiation in GC patients. All other gene expressions showed no significance in OS with differentiation degree in GC patients.
Table 5. Correlation of E2Fs’ genes expression with OS in GC patients with differentiation degree.
| E2Fs | Differentiation | Cases | HR (95% CI) | P-value |
|---|---|---|---|---|
| E2F1 | Poor | 165 | 1.43 (0.91−2.26) | 0.12 |
| Moderate | 67 | 0.63 (0.29−1.39) | 0.25 | |
| Good | 32 | 1.87 (0.77−4.54) | 0.16 | |
| E2F2 | Poor | 121 | 0.65 (0.34−1.24) | 0.19 |
| Moderate | 67 | 0.37 (0.19−0.74) | 0.0037* | |
| Good | _ | _ | _ | |
| E2F3 | Poor | 165 | 1.53 (1.01−2.3) | 0.042* |
| Moderate | 67 | 0.49 (0.26−0.95) | 0.031* | |
| Good | 32 | 2.47 (0.72−8.47) | 0.14 | |
| E2F4 | Poor | 165 | 1.3 (0.86−1.95) | 0.21 |
| Moderate | 67 | 3.98 (1.51−10.48) | 0.0028* | |
| Good | 32 | 3.06 (0.9−10.42) | 0.06 | |
| E2F5 | Poor | 165 | 1.45 (0.97−2.15) | 0.066 |
| Moderate | 67 | 0.75 (0.37−1.55) | 0.44 | |
| Good | 32 | 0.59 (0.25−1.39) | 0.22 | |
| E2F6 | Poor | 165 | 1.43 (0.95−2.15) | 0.084 |
| Moderate | 67 | 2.05 (1.05−4.01) | 0.032 | |
| Good | 32 | 0.71 (0.3−1.7) | 0.44 | |
| E2F7 | Poor | 121 | 2.12 (1.27−3.52) | 0.0032* |
| Moderate | 67 | 0.68 (0.35−1.35) | 0.27 | |
| Good | _ | _ | _ |
P<0.05.
Discussion
In the current study, we investigated the prognostic significance of E2F mRNA expression in human GC and we gathered all our data by using the KM Plotter. Our results revealed that, E2F2, E2F5, E2F6 and E2F7 family members were significantly associated with favourable survival in all GC patients. However, overexpressions of E2F1, E2F3 and E2F4 revealed unfavourable OS in all GC patients.
Among all the E2F family members E2F1 has been the most researched and investigated member in human cancers [26]. Numerous studies have reported that E2F1 expression was significant for poor prognosis in malignancies such as oesophageal carcinoma [27], hepatic cellular carcinoma (HCC) [28], pancreatic cancer [29], non-small cell lung cancer [30] and breast cancer [31]. However, previous studies indicated that E2F1 played a tumour suppressing role in GC, and this was due to apoptosis being induced by adenovirus-mediated overexpression of E2F1. The cell death is probably due to a combination of immunologic signalling pathways and cyclin-dependent kinase (CDK) inhibitors [15,32–33]. In another study done by Xu et al. [34], it was discovered that overexpression of E2F1 played a role in cell growth and tumorigenicity. These patients’ higher expression of E2F1 was shown to have a poor survival, due to increased tumour sizes and higher tumour stages. This was similar to discoveries made in our study, in which overexpression of E2F1 mRNA was associated with unfavourable OS in all clinical stages and both male and female GC patients.
E2F2 was critical to many cell processes, including the cell cycle, proliferation, differentiation and cancer development [35–37]. It has recently been reported that in colorectal adenocarcinoma [38], E2F2 expression at the tissue level was low. Li et al. [46] indicated that E2F2 promoter polymorphisms, which affected the expression of E2F2, were significantly associated with increased risk squamous cell carcinoma of the oropharynx and many other cancers. E2F2 has not been investigated in GC. In our current analysis, we have found that high E2F2 expression was significantly correlated with favourable survival, especially with higher clinical stages (stages III) in GC patients.
According to previous studies, high expression of E2F3 has been known to show poor prognosis in many human cancers such as human bladder and prostate cancer. E2F3 has been known to be significant with tumour stages, grades and cell proliferation in bladder cancer and significant with tumour aggressive in prostate cancer [39]. Zeng et al. [40] found that E2F3 was also correlated with an unfavourable biomarker in HCC patients and high expression indicated a poor prognosis. In the present study, we have discovered that E2F3 had also been correlated with the evidence of unfavourable prognosis, especially with higher clinical stages (stages III) in GC patients.
In earlier investigations it has been demonstrated that E2F4 mutations had been associated with GACs, ulcerative colitis-associated neoplasms, colorectal carcinomas, endometrial cancers and prostatic carcinomas and that E2F4 expression did not assist in apoptosis [41]. Patients with breast cancer exhibiting increased expression of E2F4 target genes displayed a more severe cancer and shorter survival [44]. The changes in the levels of E2F4 in prostate cancer were thought to promote increased sensitivity, via inactivation of E2F4 by siRNA, and prevent ionising radiation-induced apoptosis [42,43]. Farman et al. [45] displayed that E2F4 methylation status had a noteworthy influence on its expression, and that there might be some prognostic values in breast carcinogenesis. In our study’s results, we have discovered that high E2F4 expression was significantly correlated with poor OS in all the patients with GC.
Recent studies have shown that E2F5 expression was associated with several tumours, such as glioblastoma [47], prostate cancer [48] and Rb [49]. Fang et al. [47] reported that due to the down-regulation of E2F5 expression, miR-129-3p was inhibited in glioblastoma. Zhang et al. [49] indicated that miR-613 functioned as a tumour suppressor in Rb through down-regulation of E2F5. In human lung carcinoma E2F5 expression was reportedly increased and was significantly associated with worse OS [50]. Nevertheless, it has not been reported in GC. In the present study, we demonstrated that higher expression of E2F5 in GC patients had better prognosis, and this expression was associated with clinical stages (stages II, III, IV) in patients.
Numerous studies have reported that E2F6 expression was significant for prognosis in malignancies such as pancreatic cancer [51], breast cancer [52] and nasopharyngeal carcinoma [53]. What is more, E2F6 functions as a tumour suppressor in nasopharyngeal carcinoma. Restoring E2F6 expression in nasopharyngeal carcinoma impairs proliferation [53]. However, the prognostic role of E2F6 in GC has yet to be investigated. We have also discovered similar findings in that high E2F6 expression was significantly correlated with favourable OS in all the patients with GC, and this expression was apparently correlated with clinical stages (stage III) in patients.
Various previous studies have shown that E2F7 was associated with several tumours, such as gallbladder cancer [54], gliomas [55] and squamous cell carcinoma [56]. E2F7 was also associated with breast cancer, and increased expression of E2F7 was significantly correlated with worse prognosis in patients being treated with tamoxifen [57]. In GC, the prognostic role of E2F7 is yet to be investigated. Therefore, in the present study, we demonstrated that high E2F7 expression was significantly correlated with better OS in all the patients with GC. This expression was markedly correlated with higher clinical stages (stages III, IV) in GC patients.
HER2 belongs to the epidermal growth factor receptor (EGFR) family. HER2 expression is associated with poor prognosis in GC, which is also a predictive factor of poor response to hormonal therapy and chemotherapy [58]. HER2 positive tumours resulted from E2Fs activity and involvement, therefore it could be used as a tool to predict relapse-free survival [59]. According to our tests, negative HER2 expression of E2F6 and E2F7 was associated with favourable prognosis in GC patients. Besides, the theory needs to be further tested. 5FU has been used for a number of clinical applications including cancer therapy and antiproliferative treatment, and it is most commonly used to treat gastrointestinal cancers such as colorectal cancer, GAC and pancreatic cancer [60]. According to a study by Yu et al. [61], GC cells have some resistance to 5FU in the presence of certain molecular mechanisms and high expressions of certain target genes. In our study, the data clearly indicated that higher E2F expression was significantly correlated with poor prognosis in patients treated with 5FU adjuvant treatment. Thus, we recommend that 5FU-based adjuvant therapy should be avoided in GC patients, who have higher E2F expression.
Conclusion
In the current study the prognostic values of mRNA expression of E2F family members in human GC was analysed using the KM Plotter. Our result demonstrated that three family members of E2F (E2F1, E2F3, E2F4) mRNA expressions were significantly associated with unfavourable OS in GC patients. However, increased expressions of E2F2, E2F5, EF6 and E2F7 were significantly associated with favourable OS, especially for higher clinical stages in GC patients. In addition to our investigation, we have observed that high expressions of E2F2, E2F5, E2F6 and E2F7 suggested favourable prognosis under HER2 negative status and E2F2 and E2F5 also suggested favourable prognosis under HER2 positive status. In terms of treatment strategies in GC patients, our results showed that treatment with 5FU-based adjuvant was significant with unfavourable prognosis in E2F family members. According to our results, the prognostic values of E2F mRNA expression were significantly favourable, except for E2F1, E2F3 and E2F4 in GC patients. These results provided a better insight into the prognostic functions of E2F mRNA genes in GC. Although the results should be further verified in clinical trials, our findings may be a favourable prognostic predictor in the development of newer therapeutic drugs in the treatment of GC.
Abbreviations
- 5FU
5-Fluorouracil
- CI
confidence interval
- DP
dimerisation protein
- E2F1
E2F transcription factor 1
- E2F2
E2F transcription factor 2
- E2F3
E2F transcription factor 3
- E2F4
E2F transcription factor 4
- E2F5
E2F transcription factor 5
- E2F6
E2F transcription factor 6
- E2F7
E2F transcription factor 7
- GAC
gastric adenocarcinoma
- GC
gastric cancer
- GSE
Genet Sel Evol
- HCC
hepatic cellular carcinoma
- HER2
human epidermal growth factor receptor-2
- HR
hazard ratio
- KM Plotter
Kaplan–Meier Plotter
- OS
overall survival
- Rb
retinoblastoma
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province [grant number Y17H100013].
Author contribution
T.M. and B.-C.C. conceived and designed the present study. F.N., Y.Y. and T.M. performed the experiments and analysed the data. T.M. and X.F. wrote the manuscript. F.N. and Y.Y. revised the manuscript.
References
- 1.Ferlay J., Shin H.R., Bray F.. et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2.Lauren P. (1965) The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 64, 31–49 10.1111/apm.1965.64.1.31 [DOI] [PubMed] [Google Scholar]
- 3.Yang W., Raufi A. and Klempner S.J. (2014) Targeted therapy for gastric cancer: Molecular pathways and ongoing investigations. Biochim. Biophys. Acta 22, 00048–00041 [DOI] [PubMed] [Google Scholar]
- 4.Oba K. et al. (2013) Role of chemotherapy for advanced/recurrent gastric cancer: an individual-patient-data meta-analysis. Eur. J. Cancer 49, 1565–1577 10.1016/j.ejca.2012.12.016 [DOI] [PubMed] [Google Scholar]
- 5.Trimarchi J.M. and Lees J.A (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- 6.Chen H.Z., Tsai S.Y. and Leone G. (2009) Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attwooll C., LazzeriniDenchi E. and Helin K. (2004) The E2F family: specific functions and overlapping interests. EMBO J. 23, 4709–4716 10.1038/sj.emboj.7600481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trimarchi J.M. and Lees J.A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- 9.Li J. et al. (2008) Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell 14, 62–75 10.1016/j.devcel.2007.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giangrande P.H., Zhu W., Schlisio S., Sun X., Mori S., Gaubatz S.. et al. (2004) A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 18, 2941–2951 10.1101/gad.1239304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A.. et al. (2002) E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16, 245–256 10.1101/gad.949802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giangrande P.H., Zhu W., Schlisio S., Sun X., Mori S., Gaubatz S.. et al. (2004) A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 18, 2941–2951 10.1101/gad.1239304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa H., Ishiguro K., Gaubatz S., Livingston D.M. and Nakatani Y. (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 10.1126/science.1069861 [DOI] [PubMed] [Google Scholar]
- 14.Nevins JR. (2001) The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10, 699–703 10.1093/hmg/10.7.699 [DOI] [PubMed] [Google Scholar]
- 15.Xie Y., Wang C., Li L., Ma Y., Yin Y. and Xiao Q. (2009) Overexpression of E2F-1 inhibits progression of gastric cancer in vitro. Cell Biol. Int. 33, 640–649 10.1016/j.cellbi.2009.02.015 [DOI] [PubMed] [Google Scholar]
- 16.Szász A.M., Lánczky A., Nagy Á.. et al. (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H.K., Choi I.J., Kim C.G., Kim H.S., Oshima A., Yamada Y.. et al. (2012) Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. Pharmacogenomics J. 12, 119–127 10.1038/tpj.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster S., Gretschel S., Jons T., Yashiro M. and Kemmner W. (2011) THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Mod. Pathol. 24, 1390–1403 10.1038/modpathol.2011.99 [DOI] [PubMed] [Google Scholar]
- 19.Busuttil R.A., George J., Tothill R.W., Ioculano K., Kowalczyk A., Mitchell C.. et al. (2014) A signature predicting poor prognosis in gastric and ovarian cancer represents a coordinated macrophage and stromal response. Clin. Cancer Res. 20, 2761–2772 10.1158/1078-0432.CCR-13-3049 [DOI] [PubMed] [Google Scholar]
- 20.Seo J., Kee H.J., Choi H.J., Lee J.E., Park S.Y., Lee S.H.. et al. (2018) Inhibition of Wntless/GPR177 suppresses gastric tumorigenesis. BMB Rep. 51, 255–260 10.5483/BMBRep.2018.51.5.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Gao X., Han L., Yu J. and Li H. (2018) Identification of hub genes with prognostic values in gastric cancer by bioinformatics analysis. World J. Surg. Oncol. 16, 114 10.1186/s12957-018-1409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J. and Li Q. (2010) SzallasiZ.An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 23.Gyorffy B., Lánczky A. and Szállási Z. (2012) Implementing an online tool for genomewide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 19, 197–208 10.1530/ERC-11-0329 [DOI] [PubMed] [Google Scholar]
- 24.Gyorffy B., Surowiak P., Budczies J. and Lánczky A. (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 8, e82241 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Park J.-S., Wei Y., Rajurkar M., Cotton J.L., Fan Q.. et al. (2013) TRIB2 acts downstream of Wnt/TCF in liver cancer cells to regulate YAP and C/EBPα function. Mol. Cell 51, 211–225 10.1016/j.molcel.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xanthoulis A. and Tiniakos D.G. (2013) E2F transcription factors and digestive system malignancies: how much do we know? World J. Gastroenterol. 19, 3189–3198 10.3748/wjg.v19.i21.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebihara Y., Miyamoto M., Shichinohe T., Kawarada Y., Cho Y., Fukunaga A.. et al. (2004) Over-expression of E2F-1 in esophageal squamous cell carcinoma correlates with tumor progression. Dis. Esophagus 17, 150–154 10.1111/j.1442-2050.2004.00393.x [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.L., Uen Y.H., Li C.F., Horng K.C., Chen L.R., Wu W.R.. et al. (2013) The E2F transcription Factor 1 transactives Stathmin 1 in hepatocellular carcinoma. Ann. Surg. Oncol. 20, 4041–4054 [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki K., Yajima T., Nagao T., Shinkawa H., Kondo F., Hanami K.. et al. (2003) Expression of transcription factor E2F-1 in pancreatic ductal carcinoma: an immunohistochemical study. Pathol. Res. Pract. 199, 23–28 10.1078/0344-0338-00348 [DOI] [PubMed] [Google Scholar]
- 30.Huang C.L. I, Liu D., Nakano J., Yokomise H., Ueno M., Kadota K.. et al. (2007) E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non-small-cell lung cancer. Clin. Cancer Res. 13, 6938–6946 10.1158/1078-0432.CCR-07-1539 [DOI] [PubMed] [Google Scholar]
- 31.Han S., Park K., Bae B.N., Kim K.H., Kim H.J., Kim Y.D.. et al. (2003) E2F1 expression is related with the poor survival of lymph node-positive breast cancer patients treated with fluorouracil, doxorubicin and cyclophosphamide. Breast Cancer Res. Treat. 82, 11–16 10.1023/B:BREA.0000003843.53726.63 [DOI] [PubMed] [Google Scholar]
- 32.Atienza C., Elliott M.J., Dong Y.B., Yang H.L., Stilwell A., Liu T.J.. et al. (2000) Adenovirus-mediated E2F-1 gene transfer induces an apoptotic response in human gastric carcinoma cells that is enhanced by cyclin dependent kinase inhibitors. Int. J. Mol. Med. 6, 55–63 [DOI] [PubMed] [Google Scholar]
- 33.Xie Y., Yin Y., Li L., Ma Y. and Xiao Q. (2009) Short interfering RNA directed against the E2F-1 gene suppressing gastric cancer progression in vitro. Oncol. Rep. 21, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 34.Xu T.-P., Wang Y.-F., Xiong W.-L., Ma P., Wang W.-Y., Chen W.-M.. et al. (2017) E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 8, e2837 10.1038/cddis.2017.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimer D., Sadr S., Wiedemair A., Goebel G., Concin N., Hofstetter G.. et al. (2006) Expression of the E2F family of transcription factors and its clinical relevance in ovarian cancer. Ann. N.Y. Acad. Sci. 1091, 270–281 [DOI] [PubMed] [Google Scholar]
- 36.Reimer D., Sadr S., Wiedemair A., Stadlmann S., Concin N., Hofstetter G.. et al. (2007) Clinical relevance of E2F family members in ovarian cancer–an evaluation in a training set of 77 patients. Clin. Cancer Res. 13, 144–151 10.1158/1078-0432.CCR-06-0780 [DOI] [PubMed] [Google Scholar]
- 37.Suzuki D.E., Nakahata A.M. and Okamoto O.K. (2014) Knockdown of E2F2 inhibits tumorigenicity, but preserves stemness of human embryonic stem cells. Stem Cells Dev. 23, 1266–1274 10.1089/scd.2013.0592 [DOI] [PubMed] [Google Scholar]
- 38.Xanthoulis A., Kotsinas A., Tiniakos D., Fiska A., Tentes A.A., Kyroudi A.. et al. (2014) The relationship between E2F family members and tumor growth in colorectal adenocarcinomas: a comparative immunohistochemical study of 100 cases. Appl. Immunohistochem. Mol. Morphol. 22, 471–477 10.1097/PAI.0b013e3182598198 [DOI] [PubMed] [Google Scholar]
- 39.Olsson A.Y., Feber A., Edwards S., TePoele R., Giddings I., Merson S.. et al. (2007) Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 26, 1028–1037 10.1038/sj.onc.1209854 [DOI] [PubMed] [Google Scholar]
- 40.Zeng X., Yin F., Liu X., Xu J., Xu Y., Huang J.. et al. (2014) Upregulation of E2F transcription factor 3 is associated with poor prognosis in hepatocellular carcinoma. Oncol. Rep. 31, 1139–1146 10.3892/or.2014.2968 [DOI] [PubMed] [Google Scholar]
- 41.Crosby M.E. and Almasan A. (2004) Opposing roles of E2Fs in cell proliferation and death. Cancer Biol. Ther. 3, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DuPree E.L., Mazumder S. and Almasan A. (2004) Genotoxic stress induces expression of E2F4, leading to its association with p130 in prostate carcinoma cells. Cancer Res. 64, 4390–4393 10.1158/0008-5472.CAN-03-3695 [DOI] [PubMed] [Google Scholar]
- 43.Ma Y., Freeman S.N. and Cress W.D. (2004) E2F4 deficiency promotes drug-induced apoptosis. Cancer Biol. Ther. 3, 1262–1269 [DOI] [PubMed] [Google Scholar]
- 44.Khaleel S.S, Andrews E.H, Ung M., DiRenzo J. and Cheng C. (2014) E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast Cancer Res. 16, 486 10.1186/s13058-014-0486-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farman F.U., Haq F., Muhammad N., Ali N., Rahman H. and Saeed M. (2018) Aberrant promoter methylation status is associated with upregulation of the E2F4 gene in breast cancer. Oncol. Lett. 15, 8461–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Sturgis E.M., Zhu L., Cao X., Wei Q., Zhang H.. et al. (2017) E2F transcription factor 2 variants as predictive biomarkers for recurrence risk in patients with squamous cell carcinoma of the oropharynx. Mol. Carcinog. 56, 1335–1343 10.1002/mc.22595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang D.Z., Wang Y.P., Liu J., Hui X.B., Wang X.D., Chen X.. et al. (2018) MicroRNA-129-3p suppresses tumor growth by targeting E2F5 in glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 22, 1044–1050 [DOI] [PubMed] [Google Scholar]
- 48.Li S.L., Sui Y., Sun J., Jiang T.Q. and Dong G. (2018) Identification of tumor suppressive role of microRNA-132 and its target gene in tumorigenesis of prostate cancer. Int. J. Mol. Med. 41, 2429–2433 [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Zhu X., Zhu X., Wu Y., Liu Y., Yao B.. et al. (2017) MiR-613 suppresses retinoblastoma cell proliferation, invasion, and tumor formation by targeting E2F5. Tumour Biol. 39 (3), 10.1177/1010428317691674 [DOI] [PubMed] [Google Scholar]
- 50.Sun C.C., Zhou Q., Hu W., Li S.J., Zhang F., Chen Z.L.. et al. (2018) Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging (Albany N.Y.) 10, 973–987 10.18632/aging.101441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang C., Dai C.Y., Mei Z., Jiang M.J., Gu D.N., Huang Q.. et al. (2018) microRNA-193a stimulates pancreatic cancer cell repopulation and metastasis through modulating TGF-β2/TGF-βRIII signalings. J. Exp. Clin. Cancer Res. 37, 25 10.1186/s13046-018-0697-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang H., Liu P., Yang L., Xie X., Ye F., Wu M.. et al. (2014) miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol. Cancer Ther. 13, 3185–3197 10.1158/1535-7163.MCT-14-0243 [DOI] [PubMed] [Google Scholar]
- 53.Zhang W., Zeng Z., Zhou Y., Xiong W., Fan S., Xiao L.. et al. (2009) Identification of aberrant cell cycle regulation in Epstein-Barr virus-associated nasopharyngeal carcinoma by cDNA microarray and gene set enrichment analysis. Acta Biochim. Biophys. Sin. (Shanghai) 41, 414–428 10.1093/abbs/gmp025 [DOI] [PubMed] [Google Scholar]
- 54.Ye Y.Y., Mei J.W., Xiang S.S., Li H.F., Ma Q., Song X.L.. et al. (2018) MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 9, 410 10.1038/s41419-018-0444-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin W., Wang B., Ding M., Huo Y., Hu H., Cai R.. et al. (2016) Elevated E2F7 expression predicts poor prognosis in human patients with gliomas. J. Clin. Neurosci. 33, 187–193 10.1016/j.jocn.2016.04.019 [DOI] [PubMed] [Google Scholar]
- 56.Hazar-Rethinam M., de Long L.M., Gannon O.M., Boros S., Vargas A.C., Dzienis M.. et al. (2015) RacGAP1 is a novel downstream effector of E2F7-dependent resistance to doxorubicin and is prognostic for overall survival in squamous cell carcinoma. Mol. Cancer Ther. 14, 1939–1950 10.1158/1535-7163.MCT-15-0076 [DOI] [PubMed] [Google Scholar]
- 57.Chu J., Zhu Y., Liu Y., Sun L., Lv X., Wu Y.. et al. (2015) E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget. 6, 31944–31957 10.18632/oncotarget.5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gravalos C. and Jimeno A. (2008) HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann. Oncol. 19, 1523–1529 10.1093/annonc/mdn169 [DOI] [PubMed] [Google Scholar]
- 59.Andrechek E.R. (2015) HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene 34, 217–225 10.1038/onc.2013.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leelakanok N., Geary S. and Salem A. (2018) Fabrication and use of poly(d,l-lactide-co-glycolide)-based formulations designed for modified release of 5-fluorouracil. J. Pharm. Sci. 107, 513–528 10.1016/j.xphs.2017.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu B., Gu D., Zhang X., Liu B. and Xie J. (2017) The role of GLI2-ABCG2 signaling axis for 5Fu resistance in gastric cancer. J. Genet. Genomics 44, 375–383 10.1016/j.jgg.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]