Abstract
Neuroblastoma (NB) is a commonly occurring cancer among infants and young children. Recently, long non-coding RNAs (lncRNAs) have been using as prognostic biomarkers for therapeutics and interventions in various cancers. Considering the poor survival of NB, the lncRNA-based therapeutic strategies must be improved. This work proposes an overall survival time estimator called SVR-NB to identify the lncRNA signature that is associated with the overall survival of patients with NB. SVR-NB is an optimized support vector regression (SVR)-based method that uses an inheritable bi-objective combinatorial genetic algorithm for feature selection. The dataset of 231 NB patients that contains overall survival information and expression profiles of 783 lncRNAs was used to design and evaluate SVR-NB from the database of gene expression omnibus accession GSE62564. SVR-NB identified a signature of 35 lncRNAs and achieved a mean squared correlation coefficient of 0.85 and a mean absolute error of 0.56 year between the actual and estimated overall survival time using 10-fold cross-validation. Further, we ranked and characterized the 35 lncRNAs according to their contribution towards the estimation accuracy. Functional annotations and co-expression gene analysis of LOC440896, LINC00632, and IGF2-AS revealed the association of co-expressed genes in Kyoto Encyclopedia of Genes and Genomes pathways.
Introduction
Neuroblastoma (NB) is the most common cancer in children, comprising 10% of all childhood cancers1. Most cases occur in very young children under the age of one year2; hence, NB is commonly referred to as an embryonic tumour3 and is responsible for approximately 11% of cancer deaths in children. Initially, the tumour originates in tissues of the sympathetic nervous system and is thus found as lesions in the adrenal glands, pelvis or abdomen chest4. The characteristics of neoplasms are highly enigmatic because these tumours exhibit either spontaneous regression or rapid progression. The prospect of survival depends on the age at diagnosis, tumour stage, and genetic features. According to The International Neuroblastoma Staging System, NB is staged into five groups: stage 1 to 4 and 4S based on metastasis formation and lymph node involvement5,6. The treatment of NB exhibits clinical diversity; hence, the treatment response is correlated with clinical and biological factors, including cancer risk group, age, and genetic abnormalities. Children with stage 1 and stage 2 neuroblastomas can be cured with surgery alone as a primary therapy7. Infants with stage 4 neuroblastomas exhibit better prognosis in response to treatment with chemotherapy and surgery8. In contrast, patients with high-risk NB exhibit poor event-free survival after chemotherapy, whereas improved event-free survival is observed in patients with advanced-stage NB after radiotherapy and chemotherapy followed by autologous bone marrow transplantation9. Despite treatment conditions, only 40–50% of patients with NB exhibit long-term survival10. Due to the heterogeneous nature of NBs, the clinical behaviour and molecular mechanisms underlying tumour growth are largely unknown, and more efficacious therapeutics are necessary to control this cancer.
The most common genetic abnormality observed in NB is amplification of the MYCN gene in NB cells. MYCN-mediated oncogenic transformation is responsible for aggressive tumour formation and poor prognosis in NB11. Further, genetic abnormalities associated with NB include loss of heterozygosity at the distal short arm of chromosome 1, which is associated with clinical outcome12,13, hyperdiploid features14, and defects in the function of nerve growth factor (NGFR)15,16. Genome-wide studies have sought to identify protein biomarkers for improved NB therapies. For instance, pharmacodynamic biomarkers have been developed to evaluate the mechanism of PI3K/AKT/mTOR pathway signalling activity and MYCN protein expression in children with NB17. Expression of biomarkers, including X-linked inhibitor of apoptosis and vascular growth factors, regulates bone marrow metastasis in NB18. Genomic amplification of the MYCN oncogene is associated with NB tumour aggressiveness and poor prognosis in NB patients19. Germline mutations in the anaplastic lymphoma kinase gene are largely responsible for familial NB, and this germline mutation can serve as potential therapeutic target for NB20. Although advances in treatment conditions and therapeutics have improved patient prognosis, long-term survival of the high-risk group has not been considerably improved. Hence, the identification of potential targets associated with NB survival is urgently required.
Over the past several years, advancements in next-generation sequencing (NGS) and microarray technologies have increased the interest in non-coding RNAs (ncRNAs), including small non-coding RNAs, such as miRNAs, piRNAs, and snoRNAs, and long non-coding RNAs (lncRNAs), given their significant roles in specific diseases. In particular, the role of lncRNAs in evolution and genome function is a newly described phenomenon. LncRNAs are non-coding RNAs that are >200 nucleotides in length and have been implicated in pathological and biological process through post-transcriptional regulation of mRNA processing and cis regulation21. Over the last decade, several studies have identified that lncRNAs play a significant role in several biological processes22. LncRNAs are highly stable and easily detectable in body fluids23,24. Several studies have revealed the significance of lncRNAs in various cancers. For instance, specific lncRNAs are up- or down-regulated in prostate cancer; lncRNAs, such as PCGEM-1, PCAT-1, and PCA3, play critical roles in prostate cancer25,26. The lncRNA HOTAIR is up-regulated, silences genes through interactions with LSD1 and PRC2 and is also involved in protein degradation via interaction with E3 ubiquitin ligases in various cancer types, include lung, ovarian, and pancreatic cancers27–29. LncRNAs also play important roles in NB. Specifically, ncRNA possess oncogenic properties, and its overexpression is correlated with poor prognosis in NB patients30. Overexpression of NDM29 in NB cell lines is associated with chemosensitivity31. Despite of advances in RNA-sequencing technologies, functions of several lncRNAs are not yet validated. LncRNAs are emerging as crucial players in tumorigenesis by directly or indirectly acting as tumor suppressors32 or oncogenes33. Various approaches were developed to use lncRNAs as potential targets in cancer, such as post-transcriptional targeting of lncRNAs34, modulation of lncRNAs using genome-editing techniques35, and loss of lncRNA function by inhibition of RNA-protein interactions using RNA-binding small molecules36. The identification of lncRNA signature in the context of cancer provides an opportunity to explore lncRNAs as possible targets and improve our knowledge of lncRNAs association with the overall survival of NB.
Several researchers have attempted to predict NB patient survival. Oberthuer et al. predicted individual survival rates for NB patients using the automatic relevance determination (CASPAR) algorithm37. Wei et al. developed a survival predictor using an artificial neural network and identified 19 genes that predict clinical outcome in NB patients38. Gene-wide promoter methylation profiling and cox elastic net analysis were utilized to predict NB patient outcome, and the degree of methylation of retinoblastoma 1 (RB1) and teratocarcinoma-derived growth factor 1 (TDGF1) was associated with poor survival39. MicroRNA expression profiling and support vector machines (SVMs) were used to predict event-free survival in NB patients40. However, few studies exist that use lncRNAs for survival prediction in NB patients. Divya et al. utilized lncRNA expression profiles and reported that SNHG1 is highly expressed and significantly associated with poor survival in NB patients41. Another study by Divya et al. used lncRNA expression data from 493 NB patients and identified a 16-lncRNA prognostic signature that predicts event-free survival42. In addition, lncRNA expression profiling was also used in other cancer types for prediction purposes. The lncRNA signature was used to predict the overall survival in esophageal squamous cell carcinoma43. Six lncRNAs were identified which significantly correlate with the disease free survival in patients with colorectal cancer44. Zhu et al. identified a 24-lncRNA signature to predict the prognosis in gastric cancer45. Five lncRNAs were identified which significantly correlate with the prognosis of clear cell renal cell carcinoma46. Tu et al. utilized lncRNA expression profiling and a random survival forest algorithm to predict risk groups in lung cancer patients47. Zhou et al. identified four lncRNAs that were significantly associated with overall survival in multiple myeloma patients using multivariate Cox regression and stratified analysis48. Meng et al. identified four lncRNA genes using a random survival forest algorithm to predict survival in breast cancer patients49. Recently, Wang et al. identified nine immune-related lncRNA signature in patients with anaplastic gliomas50. Genome-wide analysis study on 419 patients with glioblastoma identified six lncRNAs, AC005013.5, UBE2R2-AS1, ENTPD1-AS1, RP11-89C21.2, AC073115.6, and XLOC_004803 which distinguished the high and low risk groups51. In conclusion, utilization of lncRNA expression in cancer survival prediction could aid in the understanding of the molecular mechanisms underlying cancer progression and the identification of potential biomarkers.
Accordingly, this study proposed the SVR-NB method to identify the lncRNA signature that is strongly associated with overall survival in NB patients. Different from our previous studies41,42, SVR-NB was developed based on support vector regression (SVR)52 and an inheritable bi-objective combinatorial genetic algorithm (IBCGA)53 to select a small set of lncRNAs as a signature among a large number of lncRNAs. We retrieved RNA-seq data and overall survival information of NB patients from the database of gene expression omnibus (GEO) accession GSE62564. In clinical research, the time to death is an event of interest; hence, we exclusively focused on patients who died from NB. After the filtration process, 104 patients with 104 expression profiles consisting of 783 lncRNAs and corresponding overall survival information were obtained for further analysis. SVR-NB identified 35 out of 783 lncRNAs which are strongly correlated with overall survival in NB patients. SVR-NB using 10-fold cross-validation (10-CV) achieved a mean squared correlation coefficient of 0.85 ± 0.009 and a mean absolute error of 0.56 ± 0.09 years between actual and estimated overall survival times in NB patients. We analysed the roles of identified lncRNAs in different cancers. Furthermore, functional annotation and co-regulated gene expression analyses of top ranked lncRNAs were discussed. We hope that these findings will improve multimodal therapy and survival in patients with NB.
Results and Discussion
Overall survival estimation
We utilized SVR-NB to identify the lncRNA signature that correlated with the overall survival in NB patients. We utilized 104 lncRNA expression profiles of 783 lncRNAs and the corresponding overall survival data from 104 NB patients. SVR-NB used the feature selection algorithm IBCGA to identify a small set of lncRNAs as a signature that influence overall survival of NB patients.
SVR-NB achieved a best squared correlation coefficient of 0.89 and a mean absolute error of 0.49 years between the actual and estimated overall survival time using 10-CV from 30 independent runs (Table 1). SVR-NB obtained a mean squared correlation coefficient of 0.85 ± 0.009 and a mean absolute error of 0.56 ± 0.09 years in NB patients. We measure the feature frequency score (FFS) for each of 30 independent runs of SVR-NB to select one robust feature set with the highest FFS. The obtained signature of 35 lncRNAs has the highest FFS of 7.86 indicating that each lncRNA appears 7.86 times on average in the 30 runs. The FFS values of 30 runs are given in Supplementary Fig. S1.
Table 1.
Performance of SVR-NB.
| Method | Features Selected | Squared correlation coefficient | Mean absolute error (years) |
|---|---|---|---|
| SVR-NB | 33 | 0.89 | 0.49 |
| SVR-NB(Mean) | 30.26 | 0.85 ± 0.009 | 0.56 ± 0.09 |
| SVR-NB(FFS) | 35 | 0.84 | 0.63 |
| Ridge regression | 783 | 0.62 | 0.87 |
| LASSO | 41 | 0.68 | 0.78 |
| Elastic net | 44 | 0.67 | 0.81 |
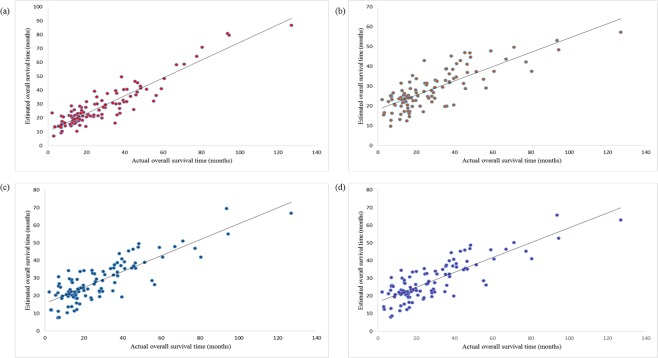
We compared the SVR-NB method with three standard linear regression methods: ridge, LASSO and elastic net regression methods. Ridge regression used all the features and obtained a squared correlation coefficient of 0.62 and a mean absolute error of 0.87 years between the actual and estimated overall survival times. LASSO identified 41 features and achieved a squared correlation coefficient and a mean absolute error of 0.68 and 0.78 years, respectively. The elastic net method identified 44 features and obtained a squared correlation coefficient and a mean absolute error of 0.67 and 0.81 years, respectively, between the actual and estimated survival time. The SVR-NB estimation performance is better than that of these three standard regression methods. The correlation plots of SVR-NB, ridge, LASSO, and elastic net are presented in Fig. 1.
Figure 1.
(a) Estimation performance of SVR-NB. (b) Estimation performance of ridge regression. (c) Estimation performance of LASSO regression. (d) Estimation performance of elastic net regression. X-axis refers to actual overall survival time, and Y-axis refers to estimated survival time.
Additionally, we used the signature of 35 lncRNAs and Naïve Bayes classifier54 to classify the 352 NB patients into high risk and low risk groups. Naïve Bayes classifier achieved a leave-one-out cross-validation accuracy, Matthews correlation coefficient, precision, recall and area under ROC curve of 86.64%, 0.73, 0.86, 0.86, and 0.94 respectively. The prediction performance of Naïve Bayes classifier was evaluated using a receiver operating curve (ROC), as shown in Supplementary Fig. S2.
SVR-NB validation
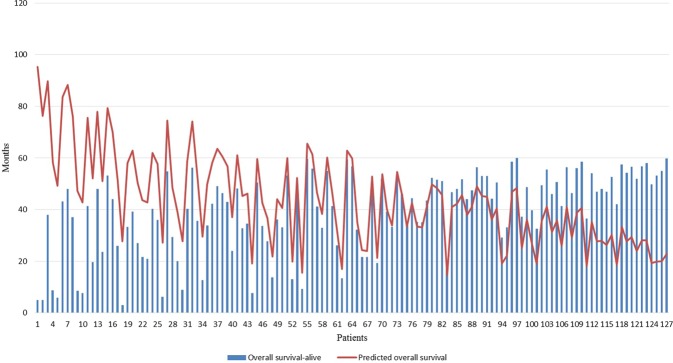
We evaluated the performance of SVR-NB in an independent test cohort of 127 patients with NB who are still living. The independent test cohort exhibits the mean overall survival time of 39.22 ± 15.42 months, whereas the predicted mean overall survival time is increased compared with the actual mean overall survival time 43.55 ± 17.58 months. The predicted mean overall survival time of 73 (50.96 ± 18.45) among the 127 patients is increased compared with the actual mean overall survival time (32.77 ± 16.18). The obtained squared correlation coefficient was 0.31 between actual overall survival time and predicted overall survival time.
The prediction error in terms of mean absolute error for the remaining 54 patients whose predicted overall survival time is smaller than the actual overall survival time, which is 1.19 years between the actual overall survival time and predicted overall survival time. Comparing to the prediction error of 0.63 years obtained for the 104 NB patients using SVR-NB (FFS), whereas the prediction error of 1.19 years is higher, due to the small sample size. However, SVR-NB would perform better by increasing the training sample size. The estimation of overall survival in the independent test cohort is presented in Fig. 2.
Figure 2.
SVR-NB validation using an independent test cohort of 127 NB patients.
Ranking of the lncRNA signature
We ranked the lncRNAs of the identified signature using main effect deference (MED) analysis55. MED analysis reveals the contribution of each lncRNA among the lncRNA signature towards estimation accuracy of the overall survival time. LncRNAs with higher MED scores indicate a greater contribution of these lncRNAs towards the estimation accuracy of overall survival time, while the lncRNAs with lower MED scores indicates the lesser contribution. The top 10 ranked lncRNAs based on the MED analysis are LOC440896, LOC729770, LINC00632, CXCR2P1, LOC643542, LOC387720, IGF2-AS, DUX4L3, HAS2-AS1, and LINC01606. We ranked all 35 lncRNAs, and their corresponding MED values are presented in Table 2. The top 10 lncRNAs and their chromosome locations are provided in Supplementary Table S1.
Table 2.
MED ranking of lncRNAs.
| Rank | Ref-Seq ID | LncRNA-Symbol | MED score |
|---|---|---|---|
| 1 | NR_015361 | LOC440896 | 2.588 |
| 2 | XR_108432 | LOC729770 | 1.694 |
| 3 | NR_028344 | LINC00632 | 1.655 |
| 4 | NR_002712 | CXCR2P1 | 1.451 |
| 5 | NR_033921 | LOC643542 | 1.388 |
| 6 | XR_109027 | LOC387720 | 1.296 |
| 7 | NR_028043 | IGF2-AS | 1.282 |
| 8 | NM_001164467 | DUX4L3 | 1.279 |
| 9 | NR_002835 | HAS2-AS1 | 0.983 |
| 10 | NR_038235 | LINC01606 | 0.981 |
| 11 | NR_030171 | MIR492 | 0.975 |
| 12 | NR_027088 | LOC284661 | 0.953 |
| 13 | NR_002145 | OR2L1P | 0.945 |
| 14 | NR_003503 | GGT8P | 0.925 |
| 15 | XR_109271 | LOC400511 | 0.857 |
| 16 | NR_027284 | LINC00602 | 0.811 |
| 17 | NR_033942 | ARHGEF34P | 0.768 |
| 18 | XM_001717149 | LOC100130503 | 0.719 |
| 19 | NR_027321 | LINC00964 | 0.649 |
| 20 | NR_002766 | MEG3 | 0.614 |
| 21 | NR_026816 | PSORS1C3 | 0.589 |
| 22 | NR_003187 | NCF1C | 0.511 |
| 23 | XR_109119 | LOC100129223 | 0.369 |
| 24 | XR_111273 | LOC100509445 | 0.300 |
| 25 | NR_033400 | CSNK1G2-AS1 | 0.290 |
| 26 | NR_029965 | MIR431 | 0.258 |
| 27 | NR_024192 | HILS1 | 0.255 |
| 28 | NR_026766 | MYCNOS | 0.236 |
| 29 | NR_038977 | LINC01239 | 0.155 |
| 30 | NR_073404 | LOC441081 | 0.125 |
| 31 | NR_037890 | DNAJB8-AS1 | 0.107 |
| 32 | NR_024119 | LINC00244 | 0.106 |
| 33 | NR_046173 | LOC254896 | 0.102 |
| 34 | XR_110545 | LOC730376 | 0.088 |
| 35 | XR_109597 | GDF5OS | 0.066 |
Significance of top ranked lncRNA in cancers
LOC440896
Uncharacterized LOC440896 alias AL353608.3 is differently expressed in various cancers. Genome-wide analysis studies on 79 small cell lung cancer patients reported that AL353608.3 is up-regulated and differently expressed in lung cell carcinoma compared with that of normal cells with a log2-fold change of 3.256. RNA-sequencing of cells derived from patients with juvenile idiopathic arthritis demonstrated that AL353608.3 was up-regulated in inflammatory cells with a log2-fold change of 5 compared with that of normal cells57. This lncRNA is actively involved in breast cancer cells, and expression of AL353608.3 is up-regulated in breast cancer cells compared with that of normal counterparts58. Additionally, AL353608.3 was down-regulated in blood platelets from patients with pancreatic adenocarcinoma with a log2-fold change of −4.2 compared with that in healthy samples59, and expression of this lncRNA expression is also involved in glioblastoma59.
LINC00632
Long intergenic non-protein coding RNA 632 (LINC00632) is implicated in several major cancers. For instance, LINC00632 expression was up-regulated in breast cancer cells with a log2-fold change of 5.2 compared with that in normal cells58,60. Up-regulation of LINC00632 was observed in prostate carcinoma cells with a log2-fold change of 4.8 compared with that in healthy cells61. Additionally, down-regulation of LINC00632 is significantly associated with different cancer types, such as non-small cell lung carcinoma59 and medulloblastoma62, and down-regulation of LINC00632 is frequently observed in glioblastoma63,64. In addition to cancer tissues, LINC00632 is highly expressed in normal brain tissue with a mean RPKM of 3.06 ± 1.5465.
LOC643542
Uncharacterized LOC643542 is highly expressed in human normal tissues and 27 other tissue types, such as fat, kidney and brain, with mean RPKM values of 0.29 ± 0.17, 0.15 ± 0.11, and 0.07 ± 0.14, respectively65. Genome-wide association studies revealed the association of LOC643542 with major depressive disorder66. A meta-analysis of 1110 major depressive disorder cases reported that LOC643542 is localized in the brain region and exhibits a higher number of single-nucleotide polymorphisms66. Genome-wide association studies further confirm the association of LOC643542 in bipolar disorder67 and hyperactivity disorder68.
IGF2-AS
RNA-sequence analysis study on breast carcinoma patients revealed that IGF2-AS is up-regulated in HER2 breast carcinoma cells with a log2-fold change of −4.1 compared with that in normal cells58. Down-regulation of IGF2-AS was also observed in amyotrophic lateral sclerosis69 and Down syndrome (trisomy 21)70 with log2-fold changes of −1.5 and −2.9, respectively, compared with those in normal cells. IGF2-AS expression was up-regulated in glioblastoma71 with a log2-fold change of 3 and in childhood brain tumour ependymoma62 with a log2-fold change of 1.3.
HAS2-AS1
HAS2-AS1 is frequently down-regulated in different cancer types. RNA sequencing of six tumour types revealed that HAS2-AS1 is down-regulated in various cancers59. HAS2-AS1 expression was down-regulated in breast carcinoma cells, pancreatic carcinoma, colorectal carcinoma, non-small cell lung carcinoma, and glioblastoma cells with log2-fold changes of −3.2, −2.6, −1.8, −1.2, and −1.2, respectively, compared with those in normal cells59. In addition, HAS2-AS1 up-regulation was also observed in glioblastoma cells with a log2-fold change of 3.571.
LINC01606
LINC01606 is implicated in various cancers. RNA-sequence analysis on LINC01606 revealed that LINC01606 is up-regulated in triple-negative breast cancer cells and HER2-positive breast carcinoma cells with log2-fold changes of 5.4 and 2.8, respectively58. Up-regulation of LINC01606 was also observed in oesophageal adenocarcinoma72. LINC01606 was down-regulated in pancreatic adenocarcinoma59 and glioma71 with log2-fold changes of −5 and −3.5, respectively. RNA-sequencing studies on different tumour types revealed down-regulation of LINC01606 in hepatobiliary carcinoma, non-small cell lung carcinoma, and colorectal carcinoma with log2-fold changes of −2.7, −2.4, and −2.1, respectively59.
Few studies reported the remaining four lncRNAs (LOC729770, CXCR2P1, LOC387720, and DUX4L3) among the top 10 ranked lncRNAs, involved in NB and other cancers. Though, these four lncRNAs LOC729770, CXCR2P1, LOC387720, and DUX4L3 have few experimental validations in NB, their contribution towards the overall survival estimation is higher ranked second, fourth, sixth, and eighth respectively. Hence, these four lncRNAs are potential biomarkers of NB survival time to be further validated. We summarize the top 10 ranked lncRNAs and their role in cancer/disorder in Supplementary Table S2.
Though there were limited number of experimental validations on lncRNAs in NB, we reported some studies to support the association between the identified lncRNAs and cancer. A study using a real-time reverse transcriptase polymerase chain reaction assay (qPCR) and western blot analysis on NB cells revealed that MYCN expression was found to be up-regulated and associated with the NB stage73. Northern blot analysis on Wilm’s tumor samples reported that IGF2-AS was found to be up-regulated in Wilms’ tumor samples compared to the healthy samples74. A qPCR and Sothern blot analysis on hepatocellular carcinoma cells revealed that IGF2-AS can significantly restrain the malignant cells and may act as gene therapeutic target75. Up-regulation of HAS2-AS1 was observed in oral squamous cell carcinoma using qPCR and western blot analysis76. LNC00964 expression was found to be down-regulated in colorectal cancer using qPCR analysis77. The qPCR and western blot analyses revealed the up-regulation of MEG3 in pancreatic ductal carcinoma78, multiple myeloma79, and ovarian cancer80.
Expression difference in amplified MYCN and non-amplified MYCN groups
The GEO database (GSE62564) included 401 patients with MYCN non-amplified disease and 92 patients with MYCN amplified disease. We measured expression levels of the top 10 ranked lncRNAs in MYCN amplified and MYCN non-amplified groups. We observed a slight difference in the expression of top ranked lncRNAs in MYCN amplified and MYCN non-amplified groups. Of the top 10 ranked lncRNAs, the mean expression of LOC440896, LOC729770, LINC00632, CXCR2P1, LOC643542, LOC387720, IGF2-AS, DUX4L3, HAS2-AS1, and LOC100507651 are 0.18 ± 0.35, 0.24 ± 0.32, 4.42 ± 3.59, 4.24 ± 4.91, 0.36 ± 0.36, 0.08 ± 0.09, 2.10 ± 5.4, 0.39 ± 1.95, 0.33 ± 0.44 and 0.12 ± 0.89, respectively, in the MYCN-amplified group and 0.11 ± 0.19, 0.32 ± 0.10, 3.59 ± 7.79, 4.91 ± 3.92, 0.36 ± 0.12, 0.09 ± 0.05, 5.47 ± 1.79, 1.95 ± 1.60, 0.44 ± 0.21 and 0.89 ± 0.10, respectively, in the MYCN-non-amplified group. Box-plot representations of lncRNA expression in the MYCN-amplified and MYCN-non-amplified group are presented in Supplementary Fig. S3.
Additionally, we performed the survival analysis of the top 10 ranked lncRNAs using Kaplan-Meir (KM) survival curves. We used median expression of the lncRNA as a threshold to classify lncRNA expression into high expression group and low expression group. The KM-survival curves were plotted for the top 10 ranked lncRNAs. The overall survival KM plots for the two groups were shown in Supplementary Fig. S4.
Six lncRNAs among the top 10 are differently expressed in various normal human tissues, such as lung, liver, ovary, brain, and other tissues. The expression levels of these six lncRNAs in different tissues are shown in Supplementary Fig. S5 using the human body map.
Functional annotations of LOC440896, IGF2-AS, and DUX4L3
We examined the functional annotations of the top 10 ranked lncRNAs using Database for Annotations Visualization and Integrated Discovery tool (DAVID)81. Each lncRNA is associated with specific functional annotations. For instance, among the top 10 ranked lncRNAs, LOC440896 is associated with the sequence feature of the putative uncharacterized protein FLJ45355. IGF2-AS is associated with the putative insulin-like growth factor2 antisense gene protein and sequence variant. DUX4L3 is associated with compositionally biased regions Ala-rich and Arg-rich and DNA binding region. The lncRNA DUX4L3 is associated with various gene-ontology terms, including nitrogen compound metabolic process (GO:0006807), biosynthetic process (GO:0009058), regulation of biological process (GO:0050789), regulation of metabolic process (GO:0019222), cellular metabolic process (GO:0044237), and biological regulation (GO:0065007).
Furthermore, the UCSC_TFBS algorithm available from DAVID was used to identify protein interactions, including transcription factors with sets of target genes. Four out of the top10 ranked lncRNAs including CXCR2P1, HAS2-AS1, DUX4L3, and LOC440896, are involved in protein interactions and have functions related to transcription factors. We summarize the functional annotations associated with the top 10 ranked lncRNAs in Table 3.
Table 3.
LncRNA and their predicted protein interactions.
| ID | Gene Name | Species | UCSC_TFBS |
|---|---|---|---|
| 3580 | C-X-C motif chemokine receptor 2 pseudogene 1 (CXCR2P1) | Homo sapiens | AP1, AP4, AREB6, ARP1, CDP, CDPCR3, CEBP, CETS1P54, CP2, E47, GATA1, GATA3, GR, GRE, HEN1, HNF1, HTF, IK3, LUN1, MYOD, MZF1, NF1, NFAT, P300, PAX4, PAX5, SEF1, SRF, TAL1ALPHAE47, TAL1BETAE47, TAL1BETAITF2, TAXCREB, TCF11, YY1 |
| 594842 | HAS2 antisense RNA 1 (HAS2-AS1) | Homo sapiens | AHR, AHRARNT, AML1, AP1, AP4, AREB6, ARNT, ATF, ATF6, BACH1, BACH2, BRACH, CART1, CDC5, CDPCR3HD, CEBP, CREB, CREBP1, CREBP1CJUN, E2F, E47, E4BP4, EGR3, EVI1, FOXJ2, FOXO3, FOXO4, FREAC3, FREAC4, FREAC7, GATA1, GFI1, GRE, HAND1E47, HEN1, HFH1, HFH3, HSF1, HSF2, HTF, IK2, IK3, LHX3, LMO2COM, LUN1, MEIS1BHOXA9, MYCMAX, MYOD, NFE2, NFKB, NFY, NKX25, NKX61, NMYC, OCT1, P300, PAX2, PAX4, PAX6, PBX1, PPARG, RFX1, S8, SOX5, SRY, STAT3, STAT5A, USF, XBP1, YY1, ZIC3 |
| 653548 | double homeobox 4 like 3 (DUX4L3) | Homo sapiens | AP2REP, AREB6, CDPCR3HD, FOXO3, FREAC4, HSF2, OCT1, P53, PAX3, PAX5, SPZ1, TCF11MAFG |
| 440896 | uncharacterized LOC440896 (LOC440896) | Homo sapiens | CEBPB, EVI1, FOXJ2, FREAC2, GATA1, IK3, ISRE, NKX25, PAX3, RP58, TCF11MAFG, TST1 |
Co-regulated gene network analysis of LOC440896, LINC00632 and IGF2-AS
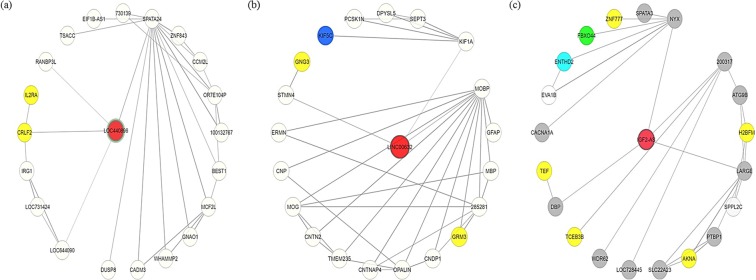
We constructed the co-regulated gene network using COXPRESdb82 to identify gene coexpression relationships among the top ranked lncRNAs. We analysed coexpressed genes and their functions for the lncRNAs LOC440896, LINC00632 and IGF2-AS. Four coexpressed genes, including cytokine receptor-like factor 2 (CRLF2), spermatogenesis associated 24 (SPATA24), uncharacterized LOC644090 (LOC644090), and RAN binding protein 3-like (RANBP3L), are directly connected to LOC440896. CRLF2 and interleukin 2 receptor alpha (IL2RA) genes are involved in the Jak-STAT signalling pathway (KEGG ID: hsa04630) and cytokine-cytokine receptor interaction (KEGG ID: hsa04060). In the co-expressed gene network, LINC00632 is directly connected to stathmin-like 4 (STMN4), myelin-associated oligodendrocyte basic (MOBP) and kinesin family member 1A (KIF1A) genes. Three genes were identified in the co-expression network of LINC00632: G protein subunit gamma 3 (GNG3) and glutamate metabotropic receptor 3 (GRM3), which are involved in the glutamatergic synapse (KEGG ID: hsa04724), and kinesin family member 5C (KIF5C), which is involved in the dopaminergic synapse (KEGG ID: hsa04728). IGF2-AS is directly connected to like-glycosyltransferase (LARGE), nyctalopin (NYX) and the D site of albumin promoter (albumin D-box) binding protein (DBP) in the co-expressed gene network. The top 100 genes coexpressed with IGF2-AS are involved five different KEGG pathways, including platelet activation (KEGG ID: hsa04611), phospholipase D signalling pathway (KEGG ID: hsa04072), Rap1 signalling pathway (KEGG ID: hsa04015), cAMP signalling pathway (hsa04024), and endocrine resistance (KEGG ID: hsa01522). The three lncRNAs and the involvement of coexpressed genes based on KEGG pathway analysis is presented in Table 4. Gene co-expression networks for LOC440896, LINC00632 and IGF2-AS are presented in Fig. 3.
Table 4.
KEGG pathway association of co-expressed genes for LOC440896, LINC00632, and IGF2-AS.
| LncRNA | Gene symbol | Gene name | Correlation with lncRNA | KEGG Pathway name (KEGG ID) |
|---|---|---|---|---|
| LOC440896 | SPATA24 | spermatogenesis associated 24 | 0.42 | ▪ Jak-STAT signaling pathway (hsa05630). ▪ Cytokine-cytokine receptor interaction (hsa04060). ▪ Chagas disease (American trypanosomiasis) (hsa05142). ▪ Tuberculosis (hsa05152). ▪ Adrenergic signaling in cardiomyocytes (hsa04261) |
| LOC644090 | uncharacterized LOC644090 | 0.33 | ||
| CRLF2 | cytokine receptor-like factor 2 | 0.29 | ||
| RANBP3L | RAN binding protein 3-like | 0.19 | ||
| LINC00632 | STMN4 | stathmin-like 4 | 0.26 | ▪ GABAergic synapse (hsa04727) ▪ Morphine addiction (hsa05032) ▪ Retrograde endocannabinoid signaling (hsa04723) ▪ Neuroactive ligand-receptor interaction (hsa04080) ▪ Nicotine addiction (hsa05033) |
| MOBP | myelin-associated oligodendrocyte basic | 0.26 | ||
| KIF1A | kinesin family member 1A | 0.24 | ||
| IGF2-AS | NYX | nyctalopin | 0.55 | ▪ Platelet activation (hsa04611) ▪ Phospholipase D signaling pathway (hsa04072) ▪ Rap 1 signaling pathway (hsa04015) ▪ cAMP signaling pathway (hsa04024) ▪ Endocrine resistance (hsa01522) |
| LARGE | like-glycosyltransferase | 0.52 | ||
| DBP | D site of albumin promoter (albumin D-box) binding protein | 0.46 |
Figure 3.
Co-expressed gene regulatory network of (a) LOC4408965, (b) LINC00632, and (c) IGF2-AS. Nodes represent lncRNAs, and edges represents coexpressed genes. The red node in the middle indicates the lncRNA. The yellow, blue, green, aqua and grey coloured nodes indicates the genes that are involved in different KEGG pathways.
Furthermore, we investigated the expression levels of these three lncRNAs in NB patients using integrated bioinformatics and wet-lab data analysis of NB data83, in which 88 human NB samples were analysed. Gene expression charts were generated using the gene expression activity chart plugin, which is available from the BioGPS gene annotation portal84. Expression charts for LOC440896, LINC00632 and IGF2-AS among 88 human NB samples are presented in Supplementary Fig. S6.
Conclusions
Recent advances in NGS data have attracted considerable attention in the exploration of the significance of ncRNAs in cancer. LncRNAs are becoming a subject of interest in cancer research due to their critical role in multiple biological processes. Recent developments in computational biology and experimental techniques have identified thousands of lncRNAs in eukaryotes. However, only few lncRNAs are characterized and experimentally validated to confirm their disease association. Hence, developing computational models to identify the lncRNAs in cancer is an important task that would aid to understand the disease at lncRNA levels, and disease diagnosis. Various computational prediction models have been developed to discover non-coding RNAs and disease association85–90. Chen et al. developed potential computational models to identify the lncRNA and disease association91,92. Identification of the lncRNA signature associated with overall survival in cancer patients using well-validated computational methods is helpful for the therapeutic strategies. LncRNAs are implicated in tumorigenesis and exhibit diverse regulatory processes in cellular process. Thus, the identification of lncRNA signature would be important in terms of disease characterization and therapy. Therefore, we attempted to identify the lncRNA signature that is associated with the overall survival of NB patients, which could aid in NB therapeutics. Accordingly, we developed a survival time estimator called SVR-NB to estimate the overall survival time and identify the lncRNA signature that is associated with overall survival in NB patients. We incorporated the feature selection algorithm IBCGA into SVR to establish the optimized SVR model. SVR-NB identified a 35-lncRNA signature that is potentially correlated with the overall survival time of NB patients. SVR-NB obtained a 10-CV squared correlation coefficient of 0.85 ± 0.009 and a mean absolute error of 0.56 ± 0.09 years between the actual and estimated overall survival times in NB patients. In addition, SVR-NB performed better than standard regression methods, including ridge, LASSO and elastic net. Although, the estimation performance of SVR-NB is promising, it has some limitations due to the small sample size. The prediction error of SVR-NB on the independent test cohort was increased when compared to that on the training dataset. Nonetheless, SVR-NB performance can be improved by increasing the number of samples.
We ranked the lncRNAs of the identified signature based on their contribution towards the survival estimation. Furthermore, we analysed the roles of the top ranked lncRNAs in cancer. Functional annotations and co-regulated gene expression of LOC440896, LINC00632 and IGF2-AS are discussed. The expression levels of these three lncRNAs in NB samples were presented using expression charts. Although some of the lncRNAs among the top 10 ranked list, such as LOC729770, CXCR2P1, LOC387720, and DUX4L3 are uncharacterized, and not involved in NB, our analysis suggests that these four lncRNAs might exhibit critical roles in NB patients’ overall survival and are promising biomarkers of NB survival time for further validation.
The development of technologies for potential identification of lncRNAs and their role in cancer are important for NB diagnostics and therapeutics. Identified lncRNAs in this study could aid in the development of lncRNA-based targeted cancer therapies in NB patients.
Materials and Methods
Dataset
We retrieved the lncRNA expression dataset of 493 NB samples from GEO accession GSE62564. The details about preprocessing and normalization of the GSE62564 dataset is described in the work41. We applied filtration to the dataset, including elimination of duplicate entries, selection of samples who died from NB, and retrieval of overall survival time by using the sample ID. We eliminated samples with the overall survival time of less than 30 days. In the lncRNA filtration process, we applied log intensity variation93 to reduce the size of candidate features from 6260 to 783 lncRNAs. After the filtration process, the training dataset consisted of 104 patients with overall survival time and 104 expression profiles of 783 lncRNAs. Another dataset of 127 patients with NB who are alive from GEO accession GSE62564 was used as an independent test cohort.
SVR-NB
This study proposed an overall survival time estimator SVR-NB based on SVR using IBCGA to identify the set of lncRNAs in NB patients. The functionality of SVR-NB is two-fold: to estimate the overall survival time and to identify significant lncRNAs strongly associated with overall survival.
The support vector machine (SVM) algorithm94, is useful in solving bioinformatics problems95,96. SVR is another version of SVM for regression. SVR has been widely applied in many biomedical fields, such as pharmaceutical research97 and cancer prognosis98. We have successfully applied an SVR incorporated with feature selection algorithm IBCGA for estimation of survival in patients with glioblastoma multiforme and lung adenocarcinoma99,100.
SVR-NB is developed based on ν-SVR for the given data points (x1, y1), … (xn, yn), where xi ∈ Rl is an NB patient input sample and, yi ∈ Rk is a target label (yi is the overall survival time). The primal problem of ν-SVR is described as follows.
| 1 |
where ; and b is a constant.
Here, 0 ≤ ν ≤ 1, and C is the regularization parameter. The ε-insensitive loss function.
To avoid the over training, we used 10-fold cross-validation (10-CV) to evaluate the performance of the model. Pearson’s correlation coefficient (CC) was used as a fitness function. Pearson’s correlation coefficient (CC) is formulated as follows:
| 2 |
where xi and yi are actual and estimated overall survival time of the ith lncRNA respectively, and and are their corresponding means. Here, N is the total number of patients with NB. We used squared correlation coefficient to evaluate the model performance.
Inheritable bi-objective combinatorial genetic algorithm
To select a minimal set of informative features from a large number of candidate features the inheritable bi-objective combinatorial genetic algorithm (IBCGA) is used. The IBCGA uses an intelligent evolutionary algorithm101 that can efficiently solve large parameter optimization problems. In this study, we propose a method for the identification of informative lncRNAs associated with NB overall survival based on the IBCGA and ν-SVR by maximizing the estimation performance in terms of correlation coefficient (CC). In this work, the LibSVM package102 was used for implementation of ν-SVR.
The encoded chromosomes and the customized IBCGA were designed as described in previous studies99,100,103. The chromosome of the IBCGA comprises 783 genes and three 4-bit genes for encoding γ, C, and ν for the ν-SVR. In this work, the parameter values are rstart = 10, rend = 50, Npop = 50, Pc = 0.8, Pm = 0.05, and Gmax = 6053.
We evaluated the prediction performance using mean absolute error (MAE):
| 3 |
where xi and yi are actual and estimated overall survival time of the ith lncRNA, respectively. Here, N is the total number of NB patients. The steps of IBCGA are as follows.
Step 1: (Initialization) Randomly generate a population of Npop individuals.
Step 2: (Evaluation) Evaluate the fitness value of all individuals using the fitness function that is the squared correlation coefficient (SCC) in terms of 10-fold cross-validation (10-CV).
Step 3: (Selection) Use a tournament selection method that selects the winner from two randomly selected individuals to generate a mating pool.
Step 4: (Crossover) Select two parents from the mating pool to perform orthogonal array crossover operation.
Step 5: (Mutation) Apply a conventional mutation operator to the randomly selected individuals in the new population. Mutation is not applied to the best individuals to prevent the best fitness value from deterioration.
Step 6: (Termination test) If the stopping condition for obtaining the solution is satisfied, output the best individual as the solution. Otherwise, go to Step 3.
Step 7: (Inheritance) If r < rend, randomly change one bit in the binary genes for each individual from 0 to 1; increase the number r by one, and go to Step 3. Otherwise, stop the algorithm.
Step 8: (Output) Obtain a set of lncRNAs from the chromosome of the best individual.
Ridge, LASSO and Elastic net
We compared three standard regression methods with SVR-NB. The Ridge regression is also called L2-penalized regression104. The Ridge regression conserves all the features to build prediction models. In the Ridge regression, the penalty term (λ) regularizes the coefficients of the predictors towards zero, if the coefficients take large values, and the optimization function is penalized. Hence, the Ridge regression shrinks the coefficients and reduces the model complexity. The least absolute shrinkage and selection operator (LASSO)105 was also employed to estimate the overall survival of NB patients. LASSO uses L1 regularization, in which some of the coefficients are neglected or regularized to zero for the evaluation of output105. Therefore, LASSO can help in the feature selection procedure. We chose λ (minimum λ) for the tuning parameter after 100 iterations of 10-CV. We used squared correlation coefficient and mean absolute error for the performance measurement.
Elastic net106 is an extension of the LASSO, in which LASSO and ridge regression are combined. The Elastic net method can be defined as follows
| 4 |
where yi is the overall survival time at observation; xi ∈ Rm is the vector of m lncRNA expression values for the i-th observation, β0 and β are regression coefficients, λ is a regularization parameter, and N is the total number of observations.
Feature frequency score (FFS)
We measure the feature frequency score for each independent run as follows:
| 5 |
where f(z) is the feature frequency for feature z that presents in the lncRNA set, nt is number of the features in the t-th signature, t = 1 …. R, and Zi is the i-th lncRNA in the t-th solution.
Supplementary information
Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma
Acknowledgements
This work was funded by Ministry of Science and Technology ROC under the contract numbers MOST 107-2221-E-009-154 –, 107-2634-F-075-001 –, 107-2218-E-009-005 –, 107-2218-E-029-001 –, and 107-2319-B-400-001 –, and was financially supported by the “Center for Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
S.Y.S. and S.Y.H. designed the system, participated in manuscript preparation, and carried out the detail study. D.S., H.C.H. and Y.C.L. participated in the data analysis and discussed the results. All authors have read and approved the final manuscript.
Data Availability
All the data used in this analysis can be found at the database of gene expression omnibus (GEO) accession GSE62564.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41553-y.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Birch JM, Blair V. The epidemiology of infant cancers. The British Journal of Cancer. Supplement. 1992;18:S2–S4. [PMC free article] [PubMed] [Google Scholar]
- 3.Kaatsch P, et al. Pediatric bone tumors in Germany from 1987 to 2011: incidence rates, time trends and survival. Acta Oncol. 2016;55:1145–1151. doi: 10.1080/0284186x.2016.1195509. [DOI] [PubMed] [Google Scholar]
- 4.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodeur GM, et al. International criteria for diagnosis, staging, and response to treatment in patients with neuroblastoma. J Clin Oncol. 1988;6:1874–1881. doi: 10.1200/jco.1988.6.12.1874. [DOI] [PubMed] [Google Scholar]
- 6.Brodeur GM, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/jco.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 7.Perez CA, et al. Biologic Variables in the Outcome of Stages I and II Neuroblastoma Treated With Surgery as Primary Therapy: A Children’s Cancer Group Study. Journal of Clinical Oncology. 2000;18:18–18. doi: 10.1200/jco.2000.18.1.18. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt ML, et al. Biologic Factors Determine Prognosis in Infants With Stage IV Neuroblastoma: A Prospective Children’s Cancer Group Study. Journal of Clinical Oncology. 2000;18:1260–1268. doi: 10.1200/jco.2000.18.6.1260. [DOI] [PubMed] [Google Scholar]
- 9.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/nejm199910143411601. [DOI] [PubMed] [Google Scholar]
- 10.Shao JB, Lu ZH, Huang WY, Lv ZB, Jiang H. A single center clinical analysis of children with neuroblastoma. Oncol Lett. 2015;10:2311–2318. doi: 10.3892/ol.2015.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 12.Maris JM, et al. Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 1995;55:4664–4669. [PubMed] [Google Scholar]
- 13.Maris JM, et al. Region-specific detection of neuroblastoma loss of heterozygosity at multiple loci simultaneously using a SNP-based tag-array platform. Genome Research. 2005;15:1168–1176. doi: 10.1101/gr.3865305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Look AT, et al. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. Journal of Clinical Oncology. 1991;9:581–591. doi: 10.1200/jco.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
- 15.Azar CG, Scavarda NJ, Reynolds CP, Brodeur GM. Multiple defects of the nerve growth factor receptor in human neuroblastomas. Cell Growth Differ. 1990;1:421–428. [PubMed] [Google Scholar]
- 16.Suzuki T, Bogenmann E, Shimada H, Stram D, Seeger RC. Lack of High-Affinity Nerve Growth Factor Receptors in Aggressive Neuroblastomas. JNCI: Journal of the National Cancer Institute. 1993;85:377–384. doi: 10.1093/jnci/85.5.377. [DOI] [PubMed] [Google Scholar]
- 17.Smith JR, et al. Novel pharmacodynamic biomarkers for MYCN protein and PI3K/AKT/mTOR pathway signaling in children with neuroblastoma. Molecular. Oncology. 2016;10:538–552. doi: 10.1016/j.molonc.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman J, Galli S, Hanafy M, Tang X, Ahmed A. Identification of novel biomarkers in neuroblastoma associated with the risk for bone marrow metastasis: a pilot study. Clinical and Translational Oncology. 2013;15:953–958. doi: 10.1007/s12094-013-1030-4. [DOI] [PubMed] [Google Scholar]
- 19.Seeger RC, et al. Association of Multiple Copies of the N-myc Oncogene with Rapid Progression of Neuroblastomas. New England Journal of Medicine. 1985;313:1111–1116. doi: 10.1056/nejm198510313131802. [DOI] [PubMed] [Google Scholar]
- 20.Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.cd-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, et al. Neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:758. doi: 10.1038/nature03016. [DOI] [PubMed] [Google Scholar]
- 23.Hessels, D. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol44, 8–15; discussion 15–16 (2003). [DOI] [PubMed]
- 24.Arita T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 25.Crea F, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warrick JI, et al. Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization–a correlative study with urine PCA3 and TMPRSS2-ERG. Mod Pathol. 2014;27:609–620. doi: 10.1038/modpathol.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhan A, et al. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, et al. Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 2013;44:57–59. [PubMed] [Google Scholar]
- 29.Nakagawa T, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 30.Yu M, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34:931–938. doi: 10.3892/ijo_00000219. [DOI] [PubMed] [Google Scholar]
- 31.Castelnuovo M, et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. The FASEB Journal. 2010;24:4033–4046. doi: 10.1096/fj.10-157032. [DOI] [PubMed] [Google Scholar]
- 32.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 33.Barsyte-Lovejoy D, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.can-06-0037. [DOI] [PubMed] [Google Scholar]
- 34.Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vickers TA, et al. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 36.Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberthuer A, et al. Subclassification and Individual Survival Time Prediction from Gene Expression Data of Neuroblastoma Patients by Using CASPAR. Clinical Cancer Research. 2008;14:6590. doi: 10.1158/1078-0432.CCR-07-4377. [DOI] [PubMed] [Google Scholar]
- 38.Wei JS, et al. Prediction of Clinical Outcome Using Gene Expression Profiling and Artificial Neural Networks for Patients with Neuroblastoma. Cancer Research. 2004;64:6883. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yáñez Y, et al. Two independent epigenetic biomarkers predict survival in neuroblastoma. Clinical Epigenetics. 2015;7:16. doi: 10.1186/s13148-015-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulte JH, et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int J Cancer. 2010;127:2374–2385. doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- 41.Sahu D, et al. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7:58022–58037. doi: 10.18632/oncotarget.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu D, Ho S-Y, Juan H-F, Huang H-C. High-risk, Expression-Based Prognostic Long Noncoding RNA Signature in Neuroblastoma. JNCI Cancer Spectrum. 2018;2:pky015–pky015. doi: 10.1093/jncics/pky015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Y, et al. A seven-lncRNA signature predicts overall survival in esophageal squamous cell carcinoma. Sci Rep. 2018;8:8823. doi: 10.1038/s41598-018-27307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y, et al. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230–2242. doi: 10.18632/oncotarget.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi D, et al. A five-long non-coding RNA signature to improve prognosis prediction of clear cell renal cell carcinoma. Oncotarget. 2017;8:58699–58708. doi: 10.18632/oncotarget.17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu Z, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for lung cancer. Oncol Rep. 2016;36:215–222. doi: 10.3892/or.2016.4817. [DOI] [PubMed] [Google Scholar]
- 48.Zhou M, et al. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. Journal of Experimental & Clinical Cancer Research. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng J, Li P, Zhang Q, Yang Z, Fu S. A four-long non-coding RNA signature in predicting breast cancer survival. Journal of Experimental & Clinical Cancer Research. 2014;33:84. doi: 10.1186/s13046-014-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136:263–271. doi: 10.1007/s11060-017-2667-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhou M, et al. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol Neurobiol. 2018;55:3684–3697. doi: 10.1007/s12035-017-0572-9. [DOI] [PubMed] [Google Scholar]
- 52.Chang CC, Lin CJ. Training nu-support vector regression: theory and algorithms. Neural Comput. 2002;14:1959–1977. doi: 10.1162/089976602760128081. [DOI] [PubMed] [Google Scholar]
- 53.Ho SY, Chen JH, Huang MH. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans Syst Man Cybern B Cybern. 2004;34:609–620. doi: 10.1109/TSMCB.2003.817090. [DOI] [PubMed] [Google Scholar]
- 54.Hall M, et al. The WEKA data mining software: an update. SIGKDD Explor. Newsl. 2009;11:10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 55.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang L, et al. Genomic Landscape Survey Identifies SRSF1 as a Key Oncodriver in Small Cell Lung Cancer. Plos Genetics. 2016;12:e1005895. doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peeters JGC, et al. Inhibition of Super-Enhancer Activity in Autoinflammatory Site-Derived T Cells Reduces Disease-Associated. Gene Expression. Cell reports. 2015;12:1986–1996. doi: 10.1016/j.celrep.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 58.Eswaran J, et al. Transcriptomic landscape of breast cancers through mRNA sequencing. Scientific Reports. 2012;2:264. doi: 10.1038/srep00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Best MG, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eswaran J, et al. RNA sequencing of cancer reveals novel splicing alterations. Scientific reports. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beaver LM, et al. Transcriptome analysis reveals a dynamic and differential transcriptional response to sulforaphane in normal and prostate cancer cells and suggests a role for Sp1 in chemoprevention. Molecular nutrition & food research. 2014;58:2001–2013. doi: 10.1002/mnfr.201400269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griesinger AM, et al. Interleukin-6/STAT3 Pathway Signaling Drives an Inflammatory Phenotype in Group A Ependymoma. Cancer immunology research. 2015;3:1165–1174. doi: 10.1158/2326-6066.cir-15-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Griesinger AM, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. Journal of immunology (Baltimore, Md.: 1950) 2013;191:4880–4888. doi: 10.4049/jimmunol.1301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Birks DK, et al. Pediatric rhabdoid tumors of kidney and brain show many differences in gene expression but share dysregulation of cell cycle and epigenetic effector genes. Pediatric blood & cancer. 2013;60:1095–1102. doi: 10.1002/pbc.24481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi J, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winham SJ, et al. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol Psychiatry. 2014;19:1010–1016. doi: 10.1038/mp.2013.159. [DOI] [PubMed] [Google Scholar]
- 68.Lasky-Su J, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147b:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 69.Sareen, D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Science translational medicine5, 208ra149, 10.1126/scitranslmed.3007529 (2013). [DOI] [PMC free article] [PubMed]
- 70.Hibaoui Y, et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO molecular medicine. 2014;6:259–277. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gill BJ, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maag JLV, et al. Novel Aberrations Uncovered in Barrett’s Esophagus and Esophageal Adenocarcinoma Using Whole Transcriptome Sequencing. Molecular cancer research: MCR. 2017;15:1558–1569. doi: 10.1158/1541-7786.mcr-17-0332. [DOI] [PubMed] [Google Scholar]
- 73.Jacobs JF, et al. Regulation of MYCN expression in human neuroblastoma cells. BMC Cancer. 2009;9:239. doi: 10.1186/1471-2407-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okutsu T, et al. Expression and imprinting status of human PEG8/IGF2AS, a paternally expressed antisense transcript from the IGF2 locus, in Wilms’ tumors. J Biochem. 2000;127:475–483. doi: 10.1093/oxfordjournals.jbchem.a022630. [DOI] [PubMed] [Google Scholar]
- 75.Yang JM, Chen WS, Liu ZP, Luo YH, Liu WW. Effects of insulin-like growth factors-IR and -IIR antisense gene transfection on the biological behaviors of SMMC-7721 human hepatoma cells. J Gastroenterol Hepatol. 2003;18:296–301. doi: 10.1046/j.1440-1746.2003.02961.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhu G, et al. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56:2210–2222. doi: 10.1002/mc.22674. [DOI] [PubMed] [Google Scholar]
- 77.Chu H, et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer. 2015;121:2044–2052. doi: 10.1002/cncr.29314. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Y, et al. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;48:670–680. doi: 10.3892/ijo.2015.3292. [DOI] [PubMed] [Google Scholar]
- 79.Zhuang W, et al. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Liu J, Xu X, Li L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother Pharmacol. 2017;79:479–487. doi: 10.1007/s00280-017-3238-4. [DOI] [PubMed] [Google Scholar]
- 81.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 82.Okamura Y, et al. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic acids research. 2015;43:D82–86. doi: 10.1093/nar/gku1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 84.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic acids research. 2016;44:D313–D316. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X, Wang L, Qu J, Guan NN, Li JQ. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 86.Chen X, et al. BNPMDA: Bipartite Network Projection for MiRNA-Disease Association prediction. Bioinformatics. 2018;34:3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 87.Chen X, Yin J, Qu J, Huang L. MDHGI: Matrix Decomposition and Heterogeneous Graph Inference for miRNA-disease association prediction. Plos Comput Biol. 2018;14:e1006418. doi: 10.1371/journal.pcbi.1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X, Huang L. LRSSLMDA: Laplacian Regularized Sparse Subspace Learning for MiRNA-Disease Association prediction. Plos Comput Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi Y, et al. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic acids research. 2017;45:D115–d118. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cui T, et al. MNDRv2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic acids research. 2018;46:D371–d374. doi: 10.1093/nar/gkx1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 93.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proceedings of the National Academy of Sciences. 2013;110:7413. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20:273–297. doi: 10.1023/a:1022627411411. [DOI] [Google Scholar]
- 95.Ng KL, Mishra SK. De novo SVM classification of precursor microRNAs from genomic pseudo hairpins using global and intrinsic folding measures. Bioinformatics. 2007;23:1321–1330. doi: 10.1093/bioinformatics/btm026. [DOI] [PubMed] [Google Scholar]
- 96.Byvatov E, Schneider G. Support vector machine applications in bioinformatics. Applied bioinformatics. 2003;2:67–77. [PubMed] [Google Scholar]
- 97.Li AP. Preclinical in vitro screening assays for drug-like properties. Drug Discov Today Technol. 2005;2:179–185. doi: 10.1016/j.ddtec.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 98.Du X, Dua S. Cancer prognosis using support vector regression in imaging modality. World Journal of Clinical Oncology. 2011;2:44–49. doi: 10.5306/wjco.v2.i1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yerukala Sathipati S, Huang H-L, Ho S-Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genomics. 2016;17:1022. doi: 10.1186/s12864-016-3321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yerukala Sathipati S, Ho S-Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Scientific Reports. 2017;7:7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shinn-Ying H, Li-Sun S, Jian-Hung C. Intelligent evolutionary algorithms for large parameter optimization problems. IEEE Transactions on Evolutionary Computation. 2004;8:522–541. doi: 10.1109/TEVC.2004.835176. [DOI] [Google Scholar]
- 102.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 103.Yerukala Sathipati S, Ho SY. Identifying a miRNA signature for predicting the stage of breast cancer. Sci Rep. 2018;8:16138. doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoerl AE, Kennard RW. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. doi: 10.1080/00401706.1970.10488634. [DOI] [Google Scholar]
- 105.Tibshirani, R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological), 267–288 (1996).
- 106.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma
Data Availability Statement
All the data used in this analysis can be found at the database of gene expression omnibus (GEO) accession GSE62564.