Abstract
Consensus-scoring methods are commonly used with molecular docking in virtual screening campaigns to filter potential ligands for a protein target. Traditional consensus methods combine results from different docking programs by averaging the score or rank of each molecule obtained from individual programs. Unfortunately, these methods fail if one of the docking programs has poor performance, which is likely to occur due to training-set dependencies and scoring-function parameterization. In this work, we introduce a novel consensus method that overcomes these limitations. We combine the results from individual docking programs using a sum of exponential distributions as a function of the molecule rank for each program. We test the method over several benchmark systems using individual and ensembles of target structures from diverse protein families with challenging decoy/ligand datasets. The results demonstrate that the novel method outperforms the best traditional consensus strategies over a wide range of systems. Moreover, because the novel method is based on the rank rather than the score, it is independent of the score units, scales and offsets, which can hinder the combination of results from different structures or programs. Our method is simple and robust, providing a theoretical basis not only for molecular docking but also for any consensus strategy in general.
Introduction
Experimental methods for drug discovery involve high-throughput screening techniques, in which large numbers of compounds are experimentally tested and their activity is evaluated towards a biological target1. However, these procedures involve large amounts of resources and time. Computer-aided methods have emerged as a way to decrease the time and economic costs of the experimental trials by evaluating large datasets of molecules in virtual screening campaigns2–5. With these methods it is possible to filter compounds that are potentially active towards a protein target from large datasets. The impact of these in silico approaches for the discovery of new drugs has been widely documented3,5–8.
Molecular docking methods are commonly used in virtual screening campaigns of large chemical libraries2,9–12. These methods aim to find the most favourable position, orientation and conformation of each molecule upon binding to a protein target13,14, assigning a docking score to each molecule, which is an estimation of the likelihood of binding15. These high-throughput docking calculations are computationally efficient because the conformational space of the ligand is small (compared to that of the target-ligand complex) and the scoring functions are fast16. Thus, with molecular docking, it is possible to screen and rank molecules from large datasets.
Despite the success of protein-ligand docking in many virtual screening campaigns, there are several limitations. For example, the flexibility of the protein target is usually not completely taken into account17–23. To overcome this challenge, some methodologies use multiple reference target structures24–29 and merging and shrinking procedures22,30–33. Another limitation is that considerable prior knowledge of the biological system is needed, for example, for choosing the correct active site, knowing the protonation state of its amino acids, or defining the type of activity that is being searched for in the ligands16. In addition, the enrichment of the hit-list with actual ligands critically depends on the quality of the scoring functions34.
It has been found that combining results from different docking programs (i.e., consensus scoring) to obtain a final rank or score for each molecule leads to a higher success rate in virtual screening processes12,34–39. Traditional consensus methods select molecules that have the best performances for all scoring functions (i.e., the intersection between the results of the docking programs35; e.g., Fig. 1 yellow region). Alternative rank-based consensus strategies, such as ‘rank-by-rank’ and ‘rank-by-vote’40, or score-based strategies, such as ‘average of auto-scaled scores’36, ‘Z-score’41, and ‘rank-by-number’40, have become popular in recent years38,41,42 (see the Methods for a detailed description). Although these consensus methodologies have been shown to produce better results than individual scoring functions/docking programs, their implementation may be subject to biases and errors within data management40, as we show later. Consensus docking methodologies have also been used to successfully obtain the best poses of molecules in the binding pocket from multiple docking programs43–45. However, this work focuses on developing a method to extract only the best rank for each molecule from a consensus perspective.
Figure 1.
Example of the correlation between the results of two docking programs. For estrogen receptor alpha (ESR1; structure 3ERT), each point corresponds to the rank of a molecule for AutoDock Vina versus the rank of the same molecule in ICM (see Methods). Red and grey circles are ligands and decoys, respectively. There is a poor correlation between the results of docking programs, and some ligands can be ranked well by one program but poorly by another (red circles in blue region). Traditional consensus approaches only take the best molecules for all programs (yellow region), acting as a conditional “and”, whereas the novel exponential consensus ranking (ECR) strategy takes the best molecules from either program, acting as a conditional “or” (taking both the yellow and blue regions).
Here, we propose a new consensus scoring function that combines results from several docking programs using an exponential distribution for each individual rank. We tested the novel method over a wide range of datasets and found that it performed as good as or better than the top consensus strategies. Moreover, the results of the novel method are parameter-independent over a wide range. This paper is organized as follows. First, we describe the mathematical theory of the exponential consensus method. In the Results section, we show the performance of the individual docking programs in comparison to consensus strategies over different datasets, and then, we assess the performance of the consensus metrics in receptor ensemble docking. Finally, the conclusions are presented.
Theory
There are three types of docking scoring functions: force-field based, empirical and knowledge based46. The performances of these scoring functions depend on the training sets used, empirical assumptions, and parameterization protocol. It has been shown that scoring functions poorly correlate with binding free energies47; moreover, the correlation of the results between different docking programs is also poor. As an example, in Fig. 1, we show the correlation between rankings using the ICM program48 versus AutoDock Vina49 for estrogen receptor alpha (ESR1; structure 3ERT). Although both programs are designed to distinguish between ligands and decoys (red and grey circles, respectively, in Fig. 1), we find that while one program can predict a good rank for a molecule, the other program might not (red circles in blue regions Fig. 1).
Traditional consensus approaches select the intersection of the best results between docking programs, i.e., the molecules that perform well in all programs (shown by the yellow region of Fig. 1). These methods discard molecules that perform poorly in at least one program (e.g., blue regions in Fig. 1). Thus, if the results from all programs were highly correlated, then traditional consensus approaches would work well. However, because the docking programs are not optimal and have highly uncorrelated outcomes, it is necessary to propose alternative consensus strategies that will be able to select molecules that perform well in some programs but poorly in others (acting qualitatively as a conditional “or”).
For this purpose, we propose Exponential Consensus Ranking (ECR), a strategy to combine the results from several scoring functions/docking programs using an exponential distribution for each individual rank. We assign an exponential score to each molecule (i) for each scoring function (j) using the rank of the molecule () given by each individual docking program,
| 1 |
where σ is the expected value of the exponential distribution. This parameter establishes the number of molecules for each scoring function that will be considered, i.e., the threshold of the dataset to be taken into account for the consensus. The final score of each molecule i is defined as the sum of the exponential score for all of the scoring functions j
| 2 |
We note that the expected value σ can be different for each docking program. However, for the sake of simplicity, we kept this value constant. Importantly, we found that the ECR results were almost independent of σ (see the Results and Supplementary Information).
Since the score of each molecule P(i) corresponds to the summation of the ECR scores from all of the scoring functions, ECR assigns a higher score for molecules that are well ranked by several programs. However, by summing over step-like or sigmoid distributions, such as an exponential or a Gaussian function, we are able to select molecules that rank well for any of the docking programs, but it is not mandatory that the molecules rank well for all of the docking programs. The advantage of summing over sigmoid-like distributions is shown in the Supplementary Information, Table S1, for a simple case for which one program gives a very poor rank to a molecule while all the rest rank it highly. In summary, ECR acts qualitatively as a conditional “or”, e.g., by taking the molecules that fall both in the yellow and blue regions in Fig. 1 into account.
As an additional point, we also note that ECR can give the same score to many poor-performing molecules (e.g., the worst molecules can all have a score of 0), which leads to complications for correctly determining the rank of molecules that have the same score. Therefore, it is necessary to shuffle molecules that have the same score several times and calculate the average and standard deviation of the performance metrics, such as the enrichment factors. A similar issue but for all molecules (not only poor-performing ones) occurs for the RbV metric with the number of votes (see the Methods).
Results and Discussion
Consensus ranking improves enrichment using a single target structure
The performance of an array of different docking programs has been widely evaluated on several systems47,50,51. It has been found that the effectiveness of each program is system-dependent, mainly because the search algorithms used to find the correct poses and scoring functions depend on the training sets and parameterization protocols. In this work, we use six docking programs: AutoDock52, ICM48, LeDock53, rDock54, AutoDock Vina49 and Smina55. All these programs have search algorithms and scoring functions based on different approximations and parameters (see the Methods). To mimic a scenario in which a docking program has poor results, we introduce a scoring function that assigns random scores to each molecule according to a normal distribution. This Random Scoring Function (RSF) simulates the results of a docking program that fails to properly distinguish between ligands and decoys (cf. the ROC-AUC in Supplementary Table S2).
We analyse the results of the docking programs over four diverse benchmark systems: cyclin-dependent kinase 2 (CDK2), estrogen receptor α (ESR1), β2 adrenergic receptor (ADRB2), and carbonic anhydrase 2 (CAH2) with two target crystal structures each (see the Methods). In Table 1, we show the enrichment factor at 2% (EF2) (see the Methods) for each system and structure, and for each individual docking program (EF1, EF5, enrichment plots (EP) and ROC-AUC are presented in Supplementary Tables S2–S4 and Supplementary Fig. S1). In agreement with the literature, the performance of the docking programs is system-dependent and, in some cases, structure-dependent for the same system16,39,47,50. On average, ICM presented the best performance, followed by rDock, while AutoDock Vina and Smina presented worse performances. However, for the CAH2 system ICM had one of the worst performances. LeDock presented the best results for CDK2 but poor results for ESR1 and ADRB2, possibly because it was parameterized with a set of protein kinases56. No program presented the best performance over all of the structures and systems evaluated; moreover, in some cases, docking programs, such as AutoDock Vina, presented a similar performance to that of the RSF (see Table 1 for the CDK2 structure 4KD1 and ROC-AUC in Supplementary Table S2). Overall, these results highlight potential problems in over-fitting and training-set dependencies.
Table 1.
EF2 for all individual docking programs and consensus strategies using individual structures.
| System | CDK2 | ESR1 | ADRB2 | CAH2 | ||||
|---|---|---|---|---|---|---|---|---|
| Structure | 4KD1 | 1FVV | 1XP9 | 3ERT | 3PDS | 4LDO | 1BCD | 4PQ7 |
| AutoDock | 3.0 | 13.0 | 14.6 | 13.8 | 3.6 | 5.8 | 0.7 | 2.2 |
| ICM | 6.0 | 14.0 | 18.9 | 18.9 | 11.7 | 17.5 | 1.5 | 3.2 |
| LeDock | 10.0 | 14.0 | 6.7 | 9.8 | 2.2 | 3.6 | 8.1 | 7.1 |
| rDock | 4.0 | 16.0 | 16.5 | 19.7 | 6.6 | 6.8 | 5.1 | 8.8 |
| Smina | 5.0 | 6.0 | 15.4 | 15.0 | 4.4 | 1.2 | 3.4 | 5.6 |
| AutoDock Vina | 2.0 | 7.0 | 9.8 | 9.5 | 4.6 | 2.2 | 3.2 | 2.0 |
| RSF | 2.0 | 2.0 | 0.8 | 0.4 | 0.0 | 1.0 | 1.2 | 0.7 |
| AASS | 7.0 | 20.0 | 11.4 | 10.2 | 3.2 | 5.1 | 8.8 | 6.8 |
| RbN | 7.0 | 13.0 | 18.9 | 17.3 | 8.0 | 15.8 | 5.6 | 6.3 |
| Z-score | 6.0 | 19.0 | 18.1 | 20.5 | 9.2 | 10.0 | 10.5 | 9.8 |
| RbR | 7.0 | 17.0 | 17.7 | 18.5 | 9.0 | 10.0 | 9.5 | 8.1 |
| RbV | 6.8 | 19.1 | 19.9 | 21.1 | 9.2 | 9.5 | 8.3 | 9.3 |
| ECR | 6.0 | 21.0 | 21.7 | 22.8 | 11.9 | 10.4 | 8.3 | 9.0 |
RbV and ECR strategies were calculated for a threshold equal to the 5% of the dataset.
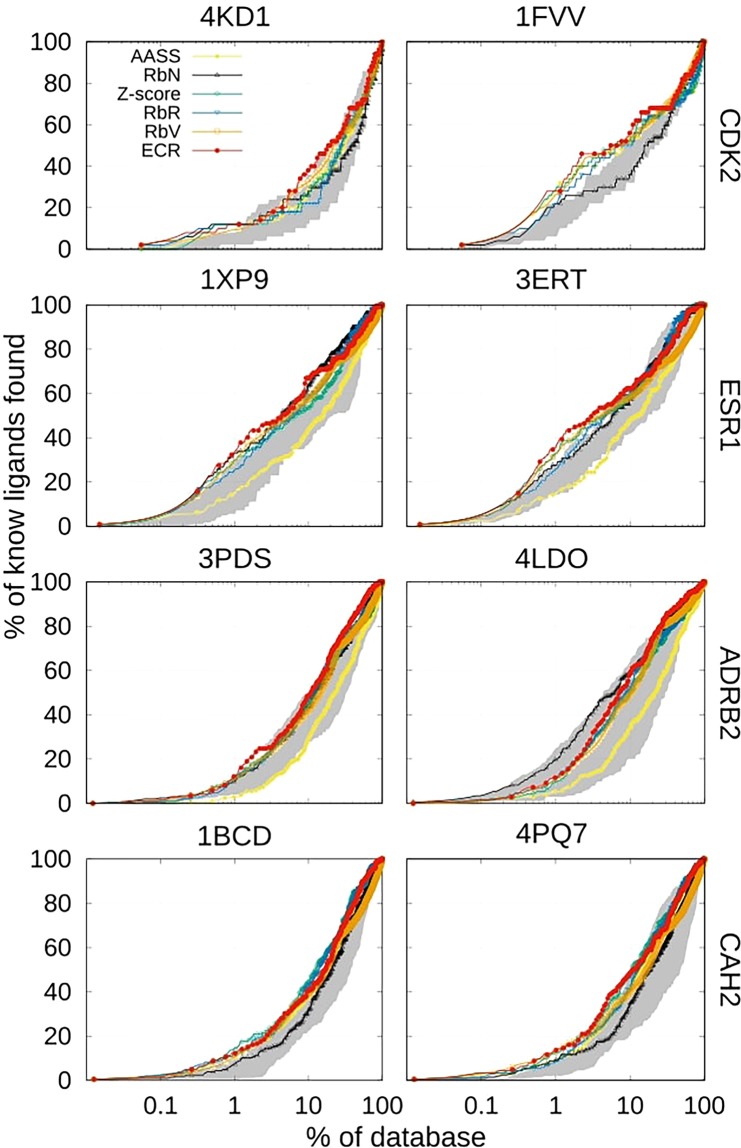
In Table 1, we also compare state-of-the-art consensus strategies [average of auto-scaled scores (AASS), rank-by-number (RbN), Z-score, rank-by-rank (RbR) and rank-by-vote (RbV), (see the Methods)] with ECR over each target structure for all of the benchmark systems. In Fig. 2, we present the EP (for the definition, see the Methods) for the consensus strategies over the benchmark systems and individual structures. The shaded regions correspond to the area between the EPs of the best and worst performances of the individual docking programs (the individual results of all the programs are presented in Supplementary Fig. S1).
Figure 2.
Enrichment plots for consensus strategies using the results obtained from each individual program using individual structures. The shaded area corresponds to the region between individual docking programs that presented the best and worst performance for each system. RbV and ECR were calculated using a threshold-parameter of 5% of the dataset.
As previously shown38,42,57, it is observed that consensus strategies improve the results obtained from individual docking programs when screening up to the top-ranked 10% of the dataset (as is usually desired). However, the performance of the score-based consensus strategies, such as RbN and AASS, is system-dependent. For example, RbN has the best performance for ADRB2 (structure 4LDO); but the worst performance for CDK2 (structure 1FVV) because RbN is biased toward programs with more negative scores (see the score distributions in Supplementary Fig. S3), which are ICM and rDock.
The system dependence of consensus strategies can be proven by generating the same table without taking the ICM results into account (see Supplementary Table S5), leading to marked deterioration of the results of RbN. On the other hand, AASS has poor performance for the ESR1, ADRB2 and CAH2 systems, despite the fact that it is not affected by the scoring scales of each program.
Rank-based consensus strategies avoid problems that occur when combining scores with different scales or offsets from individual docking programs, making them less subject to bias. This problem avoidance is demonstrated by observing that the results of the RbR, RbV and ECR consensus strategies are consistent over all of the systems. However, it is important to note that the RbV and ECR consensus strategies depend on an additional parameter that considers the percentage of the dataset that will be taken into account during the consensus protocol (see the Theory and Methods sections). The dependence of these two strategies on the threshold parameter is shown in Supplementary Table S6, where it is evident that RbV depends, to a large extent, on the threshold of the dataset that is taken into consideration in the consensus, while ECR does not.
In addition, because the score of each molecule in RbV is the number of votes received at a certain threshold (see the Methods), it is possible that a large number of molecules have the same number of votes, which can lead to large uncertainties in determining the actual rank of each molecule. A similar issue could occur in ECR, but only for molecules that have been poorly ranked by all the programs, e.g., those that have a score near zero. It is an advantage for ECR over the RbV, when only the poor-ranking molecules have uncertainties, instead of all the whole set. To estimate the error in the enrichment factors and enrichment plots due to the uncertainty in ranking molecules that have the same score, the ranking lists for these molecules were randomly shuffled 20 times. The mean and standard deviation was estimated for the EFs and EPs for the RbV and ECR strategies for molecules that had the same score. In Supplementary Fig. S4, it is observed that the RbV results are subject to large error due to the shuffling, while the ECR results are not. These results show several advantages of the ECR: (i) the stability of the results with respect to the system and structure, (ii) the ability of ECR to overcome problems stemming from different score scales and offsets, (iii) the independence of the results of ECR from the σ parameter, and (iv) the small uncertainties of ECR after shuffling molecules with the same score.
The results presented in Table 1 and Fig. 2 show that the performance of most of the strategies depends not only on the system considered but also on the target structure. In the case of CDK2, the 1FVV structure presents considerably better results than the 4KD1 structure, although both structures were prepared in the same way. Thus, minimal differences within the active site can affect the docking and consensus results, as has been shown. This result highlights the relevance of accounting for target flexibility in molecular docking using procedures such as the merging and shrinking strategy, as described below.
Consensus ranking also improves receptor ensemble docking
The merging and shrinking (MS) procedure (see the Methods) is commonly used to combine docking results from different structures of a receptor and has been widely applied to a variety of systems22,26,30–33. In this procedure, the best rank or score of an ensemble of structures is kept for each molecule. The EFs at 2% for the benchmark systems, using MS with the rank or the score, are presented for each individual docking program in Table 2 (first seven rows). We find that, in most cases, by applying this procedure, the EFs improve or remain similar to those presented by the best individual structure (see Supplementary Table S11) using either the rank or the score. In a few cases, the results are significantly worse than for the best individual structure (e.g., for rDock in CDK2 and Smina in ADRB2). Importantly, as has been previously shown in other systems22,58, the results are not as poor as those corresponding to the worst structure of each system. For example, in the case of CDK2, if the 4KD1 structure had been chosen at random, then the results would have been much worse than those obtained using MS with the two structures. Since, for a system of interest, it is difficult to know a priori what the best structure is, the use of the MS procedure provides a robust strategy to obtain the most valuable results. The results also indicate that the performance of the individual docking programs in receptor ensemble docking is system dependent. For example, LeDock performs best on the CDK2 system but severely under-performs on the ESR1 and ADRB2 systems, highlighting the necessity of applying consensus strategies.
Table 2.
EF2 for MS strategy using each individual program and consensus strategies.
| System | CDK2 | ESR1 | ADRB2 | CAH2 | ||||
|---|---|---|---|---|---|---|---|---|
| Merging and Shrinking | Score | Rank | Score | Rank | Score | Rank | Score | Rank |
| AutoDock | 12.0 | 11.0 | 14.6 | 14.6 | 5.3 | 4.9 | 1.2 | 1.2 |
| ICM | 12.0 | 14.0 | 20.5 | 20.9 | 18.0 | 18.0 | 2.0 | 2.2 |
| LeDock | 15.0 | 15.0 | 8.7 | 9.1 | 2.2 | 2.7 | 7.8 | 7.6 |
| rDock | 11.0 | 11.0 | 17.7 | 19.3 | 8.0 | 8.0 | 7.3 | 8.1 |
| Smina | 6.0 | 7.0 | 15.8 | 15.4 | 1.9 | 2.4 | 4.4 | 4.4 |
| AutoDock Vina | 7.0 | 4.0 | 9.8 | 11.0 | 3.4 | 3.4 | 2.0 | 1.4 |
| RSF | 2.0 | 1.0 | 1.2 | 1.2 | 0.5 | 0.5 | 1.5 | 1.5 |
| AASS | 18.0 | 17.3 | 4.3 | 9.0 | ||||
| RbN | 13.0 | 22.1 | 12.9 | 7.8 | ||||
| Z-score | 18.0 | 22.1 | 9.7 | 10.2 | ||||
| RbR | 13.0 | 19.3 | 8.5 | 9.5 | ||||
| RbV | 17.2 | 22.3 | 10.2 | 9.0 | ||||
| ECR | 16.0 | 23.2 | 12.9 | 10.0 | ||||
The MS procedure uses the best score or the best rank depending on the consensus strategy employed. RbV and ECR strategies were calculated for a threshold-parameter equal to 5% of the dataset.
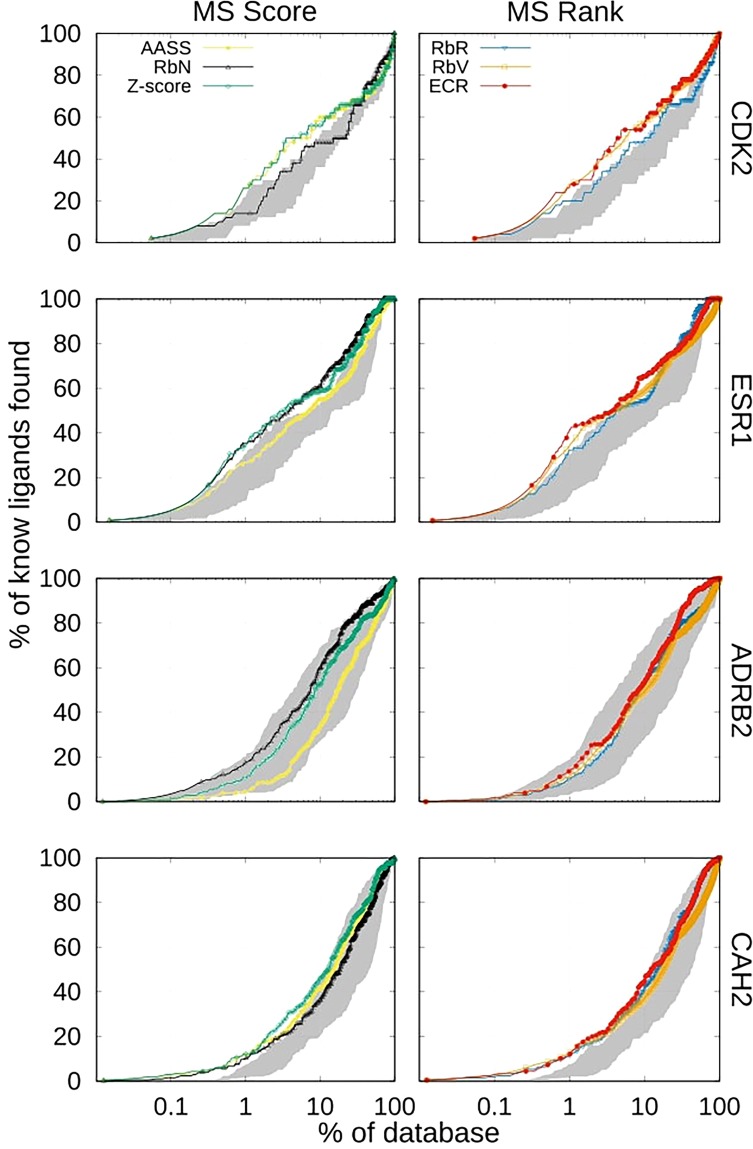
One must be careful when applying consensus strategies with the MS procedure. As shown in the Methods section, there are two sets of consensus strategies: score-based (AASS, RbN, Z-score) and ranked-based (RbR, RbV, ECR). In score-based approaches, the units, scale or offset might be different for each program and for each individual structure as well. For example, for two structures and a single docking program, it is possible that the score distributions will not overlap due to an offset difference. When performing MS using the score, only the lowest scoring molecules for the structure with the lowest offset will be used, which may be problematic because the structure with the lowest offset may not be the one with the best performance. On the other hand, when using the rank in the MS procedure, the best molecules for each structure are considered (regardless of the offset), which removes the uncertainties of the score offset and scale between structures. In agreement with these observations, score-based consensus strategies are only applied when performing MS using the score, while rank-based consensus strategies are only applied when performing MS using the rank.
In Table 2, the results of the score-based and rank-based consensus strategies are shown, including the novel ECR strategy with rank-based MS. For the CAH2 system (a case in which most docking programs perform slightly better than random), the consensus and MS strategies improve the outcome. For the ESR1 and ADRB2 systems, MS also improves the results for the consensus strategies compared to the individual structures, probably because there was not a large difference between the results of the considered structures. However, due to the different results obtained for the two structures of CDK2, MS slightly decreases the performance of the consensus strategies compared to the best structure (1FVV). Despite this result, we can conclude that MS generates more reliable results when random target structures are chosen. EF1, EF5, EPs and ROC-AUC for the individual programs and consensus scoring when applying the MS procedure are presented in Supplementary Tables S7–S9 and Supplementary Fig. S2.
Again, the dependence of the results on the system used for some of the consensus strategies is evident. For example, RbN presents the best performance for ADRB2 but the worst performance for CDK2, because for ADRB2 the ICM program presents, by far, the best results. For this particular case, as previously stated, RbN is favoured because ICM is also the program that assigns the lowest scores. However, if the ICM program is not taken into account, the RbN results worsen from 12.9 to 5.1 at EF2 for ADRB2 (see Supplementary Table S10), similar to the results obtained for a single structure. This result reaffirms the fact that consensus strategies can be biased by the individual results of some programs.
In Fig. 3, we show the EPs for MS rank-based and score-based consensus strategies. The dependency on a particular system for some of these strategies can more clearly be observed. However, rank-based consensus strategies are more stable than score-based ones, with ECR being the rank-based consensus strategy that presents the best performance overall.
Figure 3.
EPs for consensus strategies after applying MS procedure with score (left) and rank (right). The shaded area corresponds to the region of the best and worst performance for individual docking programs after applying MS for each system. RbV and ECR were calculated for a threshold-parameter of 5% of the dataset.
Analysis of the performance of ECR
In Table 3, we present the overall performance of each consensus strategy. To do so, we define a performance metric that counts how many times each consensus scoring strategy had the best EF for individual structures and the MS approach for all of the systems and structures studied. The performance metric for each strategy for the EFs at 1%, 2% and 5% and total are presented. In the cases for which two consensus strategies displayed the highest EFs, one point was given to each. The maximum number of points for individual results that a consensus strategy can reach is eight (since eight individual structures were evaluated), and for the MS results, the maximum score is three for either rank-based or score-based strategies. For example, for CDK2 and EF2 (see Table 2), one point is given to RbV (the best MS ranked-based strategy) and one point is given to both the AASS and the Z-score (the best MS score-based strategies). In this way, the success rate of each consensus strategy can be measured. The results presented in Table 3 show that ECR presents the best performance and demonstrate that ECR is the least dependent on the docking programs, structures or systems.
Table 3.
Summary of the success of the different consensus strategies.
| Consensus strategy | AASS | RbN | Z-score | RbR | RbV | ECR |
|---|---|---|---|---|---|---|
| EF1 | 3 | 4 | 6 | 1 | 2 | 7 |
| EF2 | 2 | 4 | 5 | 1 | 1 | 7 |
| EF5 | 1 | 4 | 5 | 0 | 1 | 9 |
| Total | 6 | 12 | 16 | 2 | 4 | 23 |
For each EF, a score of one (1) is given to the consensus strategy that presents the highest EF and zero (0) otherwise. In the case two or more consensus strategies have the same and highest EF, each strategy receives a point. The maximum number of points that can be obtained for a consensus method is nine (six for the individual structures and three for the MS).
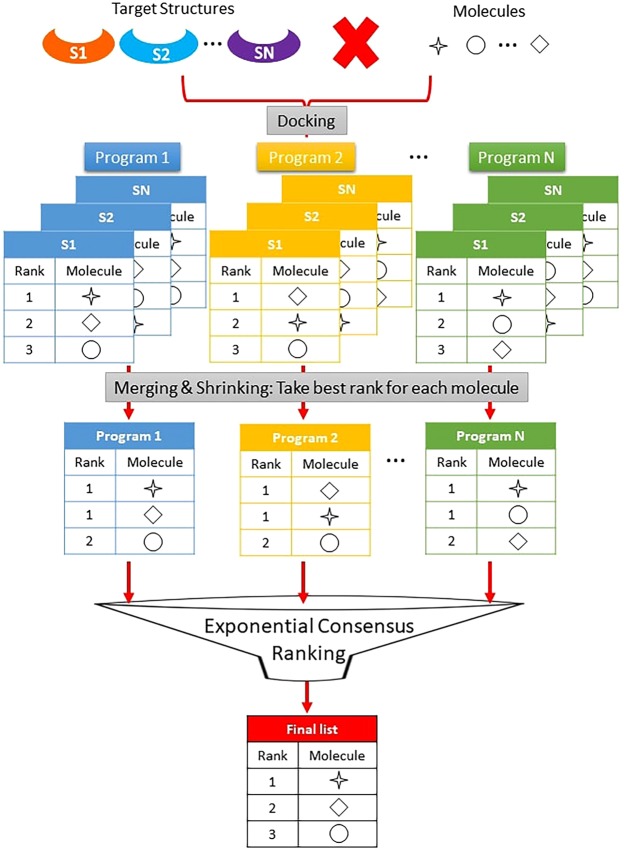
Taking these results into account, we present the protocol illustrated in Fig. 4 for docking and ensemble receptor docking. Using several structures of the same biological target, we screen the dataset with several docking programs, apply the MS protocol and use consensus scoring applying the ECR strategy. This protocol can lead to better results because it can eliminate bias towards the choice of a bad target structure or the use of a program that presents poor results for the system. The main advantages of ECR are as follows: (i) inclusion of protein target flexibility, with MS presenting more reliable results; (ii) avoidance of the problems of bias resulting from the score scales, offset or units because it is based on the rank; and (iii) ability to assign good scores to molecules that are well ranked by several docking programs, without the need to be well ranked by all of the docking programs, because it works as a conditional “or” (see Supplementary Table S1). In addition, ECR (unlike RbV) is almost independent of the threshold used and the error due to data management is negligible. In summary, these advantages allow ECR to perform better than traditional consensus strategies. Moreover, ECR requires less computational resources than other consensus strategies that have recently been proposed based on machine learning34, and it is less subject to bias due to the training-set dependencies of these methods.
Figure 4.
Proposed work flow to obtain a reliable and good performance using MS and ECR strategies. First, perform the virtual screening using different structures of the receptor and different docking programs/scoring functions. Then, for each program, apply the MS procedure maintaining the best rank of each molecule. Finally, apply ECR as consensus scoring method.
Conclusions
By performing docking-based virtual screening studies on several molecular systems and using seven scoring functions, in this work, we show that some consensus scoring methodologies avoid the system-bias effects that are typically found in individual docking programs, such as parameter training or overfitting. Moreover, we find that consensus strategies provide an overall better performance than the average of individual docking programs.
We highlight that rank-based consensus methodologies have several advantages compared to score-based strategies, stemming from scores having variable units, scales and offsets, which may significantly hinder combining results from individual structures and docking programs. Rank-based consensus metrics are not limited by score scales or offsets and may exhibit a better performance, as can be seen from the EFs, EPs and ROC-AUCs presented here.
We introduce a novel ranked-based consensus metric, Exponential Consensus Rank (ECR), which uses an exponential distribution to combine the ranks from individual docking programs. This metric has the advantage that its results are basically independent of the σ parameter, in contrast to the ‘rank-by-vote’ (RbV) methodology, which highly depends on the vote threshold. In addition, we find that ECR presents smaller errors than RbV because the assignment of the same ECR score to a molecule is less common than the assignment of the same number of votes.
Importantly, our work shows that the novel ECR metric outperforms other consensus strategies, which is independent of the σ parameter, the docking programs, or the system. Moreover, coupling our ECR metric with the merging and shrinking approach for receptor ensemble docking ensures better results with respect to the use of individual structures. These facts allow us to postulate the protocol shown in Fig. 4 to ensure the best results in a virtual screening process for docking and ensemble receptor docking.
Methods
Benchmark systems
We chose four benchmark targets to assess docking programs and consensus strategies: cyclin-dependent kinase 2 (CDK2), estrogen receptor α (ESR1), carbonic anhydrase 2 (CAH2), and β2 adrenergic receptor (ADRB2). These systems correspond to diverse protein families: protein kinases, nuclear receptors, carbonic anhydrases, and G protein-coupled receptors, respectively. To include the effect of the target’s flexibility, two crystal structures with differences near the active site were chosen for each system. We also selected different decoy/ligand datasets for each system, which contain known ligands and decoy (non-binder) molecules; to avoid artificial enrichment, the latter were chosen to ensure physicochemical similarity to the ligands but structural dissimilarity59 (see below). The diversity of these benchmark sets leads to an unbiased assessment of the protocols.
Detailed descriptions of the preparation for the structures of each system, and the molecule datasets are described below:
Cyclin-dependent kinase 2 (CDK2)
Two structures from the Protein Data Bank (PDB), codes 1FVV and 4KD1, were selected because they have acceptable crystallographic resolutions (2.8 Å and 1.7 Å, respectively), and show structural differences near the active site. The structures were protonated using Open Babel60. For this system, we used the Directory of Useful Decoys (DUD)37, which contains 72 ligands and 2074 decoys. In the case where more than one tautomer for a given molecule is present in the dataset, the docking pose with the best score for each program was chosen, resulting in only 50 ligands and 1779 decoys.
Estrogen receptor α (ESR1)
The 1XP9 and 3ERT structures (PDB codes) were selected. The NRLiSt BDB61, a dataset of ligands, decoys and targets for ESR1, was used. This dataset contains ligands with two types of activities, agonists and antagonists. We chose the dataset of antagonist compounds because both structures are bound to antagonists in the PDB. The antagonist dataset contains 133 ligands and 6555 decoys. Ligands with non-parameterized types of atoms for AutoDock were discarded, so finally, only 126 ligands remained. The protonation states of the target, ligands and decoys were preserved as they were in the dataset61.
β2 adrenergic receptor (ADRB2)
We selected the 3PDS and 4LDO PDB structures. The ligands/decoys dataset used for this system was the GPCR Decoy Database/GPCR Ligand Library (GDD/GLL)62. We chose the agonist dataset because the structures selected are in their active form63,64. This dataset has 206 ligands and 8034 decoys. The protonation state of the molecules (corresponding to pH = 7.4) was conserved.
Carbonic anhydrase 2 (CAH2)
The 1BCD and 4PQ7 structures (PDB codes) for CAH2 were selected. The structures were protonated at pH 7 using the propka3.1 server65,66 to predict the protonation states of the titrable residues. Additionally, MolProbity67 and visual inspection were used to determine the possible flips of the HIS, GLN and ASN side chains. The hydrogen atom positions were assigned using ICM, and protein and ligand polar hydrogens within 6 Å of the ligand were re-optimized using a Monte Carlo-based energy optimization68. The Directory of Useful Decoys, Enhanced (DUD-E) dataset59 was used for this enzyme. The original dataset was reduced to 7987 compounds that were randomly selected. The ligand and decoy proportions were kept as in the dataset. When more than one tautomer for a given molecule was present in the dataset, the tautomer with the best score for each program was chosen.
Docking calculations
For molecular docking the following six programs were used: AutoDock52, ICM48, LeDock53, rDock54, AutoDock Vina49 and Smina55. These programs are characterized by having different search algorithms and scoring functions, as described below.
AutoDock is the most commonly used docking program, possibly, because it is one of the oldest open-source academic programs. AutoDock has been reported to accurately predict docking poses16. AutoDock uses a Lamarckian genetic algorithm and an empirical free energy force field scoring function52.
ICM performs rigid-receptor:flexible-ligand docking, where the receptor is represented by six potential energy maps and the docked molecule is considered flexible within the energy field of the receptor and subjected to a global energy minimization protocol that consists of Monte-Carlo sampling with local energy minimization of the differentiable variables48. The lowest energy pose for each molecule is assigned an empirical score according to its fit within the binding site69.
LeDock uses a combination of simulated annealing and evolutionary optimization for the ligand pose. Physics and knowledge-based hybrid scoring schemes derived from prospective virtual screening campaigns are used53.
rDock uses a docking protocol with three stages for conformational sampling: a genetic algorithm search (GA1, GA2, GA3), followed by a low temperature Monte Carlo (MC) and a Simplex minimization stage. rDock has a scoring function constructed from the sum of several pseudo-energy scoring functions54.
AutoDock Vina employs an iterated local search global optimizer. The main advantage of this program is the speed of the calculations, which facilitates virtual screening campaigns while maintaining a good scoring power47,50.
Smina is a fork of AutoDock Vina that uses a scoring function called Vinardo70 with enhanced features based on AutoDock Vina.
The parameters of the box size remained almost the same for all programs: 20 × 20 × 20 Å for CDK2, ADRB2 and CAH2; and 25 × 25 × 25 Å for ESR1. After aligning the two structures for each system, the box centres were defined by the coordinates shown in Supplementary Table S12. For rDock, the box was automatically built using the ligand-based method54, in which the free space that can be occupied by a ligand in the binding pocket is taken as the volume of docking. For this purpose, the ligands in the structures, mentioned in the Benchmark systems sub-section, were used as references. The number of requested poses was 50 for all programs except for ICM, where the number of poses is variable and the lowest-energy pose is scored. The specific details of the search parameters for each algorithm can be found in the Supplementary Files section in the Supplementary Information. These programs were chosen based on reports from the literature44,47,50. Each program took between 1 and 25 minutes per molecule, with AutoDock being the most time consuming (see Supplementary Table S13).
In addition, we used the random scoring function (RSF), a synthetic scoring function that assigns random scores to each molecule derived from a Gaussian distribution. We used the RSF to evaluate the ability of each consensus strategy when there was a scoring function that could not differentiate between ligands and decoys in any system.
Merging and shrinking strategy
To consider the flexibility of protein targets, the merging and shrinking procedure (MS)22,58 was used. In this procedure, the docking results of the individual structures are merged, taking only the best rank (or score) for each molecule. These results are used to obtain a single rank, or score, for each molecule in receptor ensemble docking. We note that to apply the score-based consensus strategies, with MS, it is necessary to perform the MS procedure using the score. The consensus results could differ when using the rank or the score, making its selection a key issue for the virtual screening processes (see the Results section).
Consensus scoring and ranking
The results from the individual docking programs were combined using several consensus approaches. ECR was validated by comparing its results to those obtained from the consensus strategies presented below.
Rank by rank (RbR)
The molecular candidates are ranked using the average rank over all of the docking programs40. Let be the rank of molecule i for the j docking program; then, the final rank of molecule i is given by
| 3 |
where n is the total number of docking programs.
Rank by vote (RbV)
In this strategy, each molecule receives a vote if it is ranked in the top x% of the results for a certain docking program40. The final score for each molecule is given by the sum of votes obtained from all of the programs. This number can range from zero to the total number of scoring functions under consideration. All of the candidates are ranked according to their final votes. The outcome consists of the list of molecules with votes, where many molecules can have the same number of votes (e.g., for the scoring functions used in the Results section, the range is only between zero and seven). To properly estimate the performance metrics of molecules that have the same score (for ECR) or number of votes (for RbV), these molecules should be randomly shuffled several times, and the average and standard deviation of the EFs and EPs should be computed.
Rank by number (RbN)
The score of each molecule corresponds to the average score of the molecule considering all of the scoring functions,
| 4 |
where is the score of molecule i for docking program j, and n is the total number of docking programs40.
Average of auto-scaled scores (AASS)
This method is similar to RbN but it first normalizes each score between 0 and 1 and is attempt to avoid some of the issues encountered when comparing scores from different docking programs due to the score scale or offset36. For each docking program j, the score of each molecule i is scaled to a number between 0 and 1 using the minimum and maximum scores for each program ( and , respectively). The final score of each molecule is given by the average of all the normalized scores,
| 5 |
Z-score
The molecule score () is scaled using the average (μj) and standard deviation (σj) of the scores of all of the molecules for each docking program j41. The final score is the average of the scaled-score among all of the scoring functions,
| 6 |
Metric validation
Enrichment Factor (EFx%)
The EF measures the enrichment of active compounds in a molecular dataset given a specific percentage of the dataset (threshold). The EF is the ratio between ligands (hits) found using a certain threshold x% (Hitsx%) and the number of compounds at that threshold Nx% normalized by the ratio between the hits contained in the entire dataset (Hits100%) and the total number of compounds N100%,
| 7 |
Enrichment Plot (EP)
The percentage of ligands recovered as a function of the top x%-ranked compounds based on the docking and consensus scores71. The EP allows the identification of which method performs better based on the percentage of the screened dataset.
Receiver Operating Characteristics (ROC) Curve and its area under the curve (ROC-AUC)
The ROC curve indicates the ability of a program to distinguish between ligands and decoys72. The ROC curve is created by plotting the true positive rate (TPR), or sensitivity (Eq. 8), against the false positive rate (FPR), or 1 – specificity (Eq. 9), at several thresholds,
| 8 |
and
| 9 |
where TP, FP, TN and FN are the true positives, false positives, true negatives and false negatives, respectively, at a specific threshold.
An important parameter that can be obtained from ROC curves is the ROC-AUC. The ROC-AUC allows us to observe the prediction capacity of a method; in this case, it accounts for the ability of each docking program to differentiate between a ligand and a decoy. A ROC-AUC greater than 0.5 indicates that the method has a better predictive ability than a random one. The better the predictive ability of the method, the closer the ROC-AUC is to 1.
It is important to mention that EPs and ROC curves can change due to the final list sorting of RbV and ECR consensus strategies. As mentioned previously for RbV, this is because many molecules can have the same final number of votes or score. For this reason, it is necessary to calculate the error bars over the plots resulting from these scoring functions (see above and the Supplementary information).
Supplementary information
Acknowledgements
K.P.-R., I.L. and P.C. were supported by Colciencias, University of Antioquia, Colombia, and Max Planck Society, Germany. The virtual screening for the open-source docking programs were performed on the Scientific Computing Center Apolo, from Universidad EAFIT, Colombia, and in some in-house clusters. P.C. acknowledges the support for the laboratory spaces from Ruta N., Colombia. Part of this work was performed at the Instituto de Investigación en Biomedicina de Buenos Aires (IBioBA), CONICET-Partner Institute of the Max Planck Society. C.N.C. has been supported by the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (PICT 2014-3599 and 2017-3767), CONICET (PIP 2014 11220130100721). C.N.C. thanks Molsoft LLC for providing an academic license for the ICM program. The authors thank the National System of High Performance Computing (Sistemas Nacionales de Computación de Alto Rendimiento, SNCAD) and the Computational Centre of High Performance Computing (Centro de Computación de Alto Rendimiento, CeCAR) for granting use of their computational resources.
Author Contributions
K.P.-R., I.L., C.N.C. and P.C. performed the research, wrote and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Claudio N. Cavasotto, Email: CCavasotto@austral.edu.ar
Pilar Cossio, Email: pilar.cossio@biophys.mpg.de.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41594-3.
References
- 1.Phatak SS, Stephan CC, Cavasotto CN. Screenings in Drug Discovery. Expert Opinion on Drug Discovery. 2009;4:947–959. doi: 10.1517/17460440903190961. [DOI] [PubMed] [Google Scholar]
- 2.Schneider G. Automating drug discovery. Nature Reviews Drug Discovery. 2017;17:97–113. doi: 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen WL. The Many Roles of Computation in Drug Discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 4.Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nature Reviews Drug Discovery. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 5.Cavasotto CN, Orry AJ. Ligand Docking and Structure-based Virtual Screening in Drug Discovery. Current Topics in Medicinal Chemistry. 2007;7:1006–1014. doi: 10.2174/156802607780906753. [DOI] [PubMed] [Google Scholar]
- 6.Rognan, D. The impact of in silico screening in the discovery of novel and safer drug candidates. Pharmacology and Therapeutics175, 47–66, 10.1016/j.pharmthera.2017.02.034 (2017). [DOI] [PubMed]
- 7.Talele, T., Khedkar, S. & Rigby, A. Successful Applications of Computer Aided Drug Discovery: Moving Drugs from Concept to the Clinic. Current Topics in Medicinal Chemistry10, 127–141, 10.2174/156802610790232251 (2010). [DOI] [PubMed]
- 8.Jorgensen WL. Efficient drug lead discovery and optimization. Acc. Chem. Res. 2009;42:724–33. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavasotto, C. N. In silico drug discovery and design: theory, methods, challenges, and applications (CRC Press, 2015).
- 10.Ferla, S. et al. In silico screening for human norovirus antivirals reveals a novel non-nucleoside inhibitor of the viral polymerase. Scientific Reports8, 1–18, 10.1038/s41598-018-22303-y (2018). [DOI] [PMC free article] [PubMed]
- 11.Almeida TB, Carnell AJ, Barsukov IL, Berry NG. Targeting SxIP-EB1 interaction: An integrated approach to the discovery of small molecule modulators of dynamic binding sites. Scientific Reports. 2017;7:1–12. doi: 10.1038/s41598-017-15502-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang MW, Ayeni C, Breuer S, Torbett BE. Virtual screening for HIV protease inhibitors: A comparison of AutoDock 4 and Vina. PLoS One. 2010;5:1–9. doi: 10.1371/journal.pone.0011955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa, S. F. et al. Protein-ligand docking in the new millennium - A retrospective of 10 years in the field. Current medicinal chemistry20, 2296–314, http://www.ncbi.nlm.nih.gov/pubmed/23531220, 10.2174/0929867311320180002 (2013). [DOI] [PubMed]
- 14.Yuriev E, Holien J, Ramsland PA. Improvements, trends, and new ideas in molecular docking: 2012–2013 in review. Journal of Molecular Recognition. 2015;28:581–604. doi: 10.1002/jmr.2471. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S.-Y., Grinter, S. Z. & Zou, X. Scoring functions and their evaluation methods for protein–ligand docking: recent advances and future directions. Physical Chemistry Chemical Physics12, 12899, http://xlink.rsc.org/?DOI=c0cp00151a, 10.1039/c0cp00151a (2010). [DOI] [PMC free article] [PubMed]
- 16.Chen YC. Beware of docking! Trends in Pharmacological Sciences. 2015;36:78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Spyrakis F, Cavasotto CN. Open challenges in structure-based virtual screening: Receptor modeling, target flexibility consideration and active site water molecules description. Archives of Biochemistry and Biophysics. 2015;583:105–119. doi: 10.1016/j.abb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Fradera X, Cruz XD, Silva CHTP, Gelp JL. Ligand-induced changes in the binding sites of proteins. Bioinformatics. 2002;18:939–948. doi: 10.1093/bioinformatics/18.7.939. [DOI] [PubMed] [Google Scholar]
- 19.Carlson HA. Protein flexibility is an important component of structure-based drug discovery. Current Pharmaceutical Design. 2002;8:1571–1578. doi: 10.2174/1381612023394232. [DOI] [PubMed] [Google Scholar]
- 20.Cavasotto CN, Singh N. Docking and high throughput docking: successes and the challenge of protein flexibility. Current Computer-Aided Drug Design. 2008;4:221–234. doi: 10.2174/157340908785747474. [DOI] [Google Scholar]
- 21.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Science. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavasotto CN, Abagyan RA. Protein Flexibility in Ligand Docking and Virtual Screening to Protein Kinases. Journal of Molecular Biology. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Cozzini P, et al. Target flexibility: an emerging consideration in drug discovery and design. J. Med. Chem. 2008;51:6237–55. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCammon JA. Target flexibility in molecular recognition. Biochimica et Biophysica Acta - Proteins and Proteomics. 2005;1754:221–224. doi: 10.1016/j.bbapap.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 25.Tian S, et al. Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. Journal of Chemical Information and Modeling. 2014;54:2664–2679. doi: 10.1021/ci500414b. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari AM, Wei BQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. Journal of Medicinal Chemistry. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osguthorpe DJ, Sherman W, Hagler AT. Generation of Receptor Structural Ensembles for Virtual Screening Using Binding Site Shape Analysis and Clustering. Chemical Biology and Drug Design. 2012;80:182–193. doi: 10.1111/j.1747-0285.2012.01396.x. [DOI] [PubMed] [Google Scholar]
- 28.Osguthorpe DJ, Sherman W, Hagler AT. Exploring Protein Flexibility: Incorporating Structural Ensembles From Crystal Structures and Simulation into Virtual Screening Protocols. J Phys Chem B. 2013;116:6952–6959. doi: 10.1021/jp3003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leong MK, Syu RG, Ding YL, Weng CF. Prediction of N-Methyl-D-Aspartate Receptor GluN1-Ligand Binding Affinity by a Novel SVM-Pose/SVM-Score Combinatorial Ensemble Docking Scheme. Scientific Reports. 2017;7:1–15. doi: 10.1038/srep40053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. Journal of the American Chemical Society. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 31.Barril X, Morley SD. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. Journal of Medicinal Chemistry. 2005;48:4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 32.Rueda M, Bottegoni G, Abagyan R. Recipes for the Selection of Exptl Protein Conformations for Virtual Screening. Journal of Chemical Information and Modeling. 2010;50:186–193. doi: 10.1021/ci9003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovacs J, Cavasotto C, Abagyan R. Conformational sampling of protein flexibility in generalized coordinates: Application to ligand docking. J. Comp. Theor. Nanosci. 2005;2:354–361. doi: 10.1166/jctn.2005.204. [DOI] [Google Scholar]
- 34.Ericksen SS, et al. Machine Learning Consensus Scoring Improves Performance Across Targets in Structure-Based Virtual Screening. Journal of Chemical Information and Modeling. 2017;57:1579–1590. doi: 10.1021/acs.jcim.7b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charifson PS, Corkery JJ, Murcko MA, Walters WP. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. Journal of Medicinal Chemistry. 1999;42:5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 36.Oda A, Tsuchida K, Takakura T, Yamaotsu N, Hirono S. Comparison of consensus scoring strategies for evaluating computational models of protein-ligand complexes. Journal of Chemical Information and Modeling. 2006;46:380–391. doi: 10.1021/ci050283k. [DOI] [PubMed] [Google Scholar]
- 37.Huang, N., Shoichet, B. K., Irwin, J. J. & Francisco, S. Benchmarking Sets for Molecular Docking. Journal of Medicinal Chemistry 6789–6801, 10.1021/jm0608356 (2006). [DOI] [PMC free article] [PubMed]
- 38.Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative assessment of Sscoring Functions on a Diverse Test set. Journal of chemical information and modeling. 2009;49:1079–93. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 39.Kukol A, et al. Consensus virtual screening approaches to predict protein ligands. European Journal of Medicinal Chemistry. 2011;46:4661–4664. doi: 10.1016/j.ejmech.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 40.Wang R, Wang S. How Does Consensus Scoring Work for Virtual Library Screening? An Idealized Computer Experiment. Journal of Chemical Information and Computer Sciences. 2001;41:1422–1426. doi: 10.1021/ci010025x. [DOI] [PubMed] [Google Scholar]
- 41.Liu S, Fu R, Zhou LH, Chen SP. Application of consensus scoring and principal component analysis for virtual screening against β-secretase (BACE-1) PLoS One. 2012;7:e38086. doi: 10.1371/journal.pone.0038086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ece A, Sevin F. The discovery of potential cyclin A/CDK2 inhibitors: A combination of 3D QSAR pharmacophore modeling, virtual screening, and molecular docking studies. Medicinal Chemistry Research. 2013;22:5832–5843. doi: 10.1007/s00044-013-0571-y. [DOI] [Google Scholar]
- 43.Plewczynski D, Łażniewski M, Von Grotthuss M, Rychlewski L, Ginalski K. VoteDock: consensus docking method for prediction of protein–ligand interactions. Journal of Computational Chemistry. 2011;32:568–581. doi: 10.1002/jcc.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuccinardi T, Poli G, Romboli V, Giordano A, Martinelli A. Extensive consensus docking evaluation for ligand pose prediction and virtual screening studies. Journal of Chemical Information and Modeling. 2014;54:2980–2986. doi: 10.1021/ci500424n. [DOI] [PubMed] [Google Scholar]
- 45.Ren X, et al. Novel Consensus Docking Strategy to Improve Ligand Pose Prediction. Journal of Chemical Information and Modeling. 2018;58:1662–1668. doi: 10.1021/acs.jcim.8b00329. [DOI] [PubMed] [Google Scholar]
- 46.Cavasotto, C. N. Binding free energy calculation and scoring in small-molecule docking. In Physico-Chemical and Computational Approaches to Drug Discovery, 195–222 (Royal Society of Chemistry, 2012).
- 47.Wang G, Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Medicinal Chemistry. 2016;8:1707–1710. doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- 48.Abagyan R, Totrov M, Kuznetsov D. ICM - A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry. 1994;15:488–506. doi: 10.1002/jcc.540150503. [DOI] [Google Scholar]
- 49.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu W, Lucke AJ, Fairlie DP. Comparing sixteen scoring functions for predicting biological activities of ligands for protein targets. Journal of Molecular Graphics and Modelling. 2015;57:76–88. doi: 10.1016/j.jmgm.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Huang SY. Exploring the potential of global protein-protein docking: an overview and critical assessment of current programs for automatic ab initio docking. Drug Discovery Today. 2015;20:969–977. doi: 10.1016/j.drudis.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N, Zhao H. Enriching screening libraries with bioactive fragment space. Bioorganic and Medicinal Chemistry Letters. 2016;26:3594–3597. doi: 10.1016/j.bmcl.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Carmona S, et al. rDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLoS Computational Biology. 2014;10:1–8. doi: 10.1371/journal.pcbi.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. Journal of Chemical Information and Modeling. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H, Huang D. Hydrogen bonding penalty upon ligand binding. PLoS One. 2011;6:e19923. doi: 10.1371/journal.pone.0019923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park H, Eom JW, Kim YH. Consensus scoring approach to identify the inhibitors of AMP-activated protein kinase α2 with virtual screening. Journal of Chemical Information and Modeling. 2014;54:2139–2146. doi: 10.1021/ci500214e. [DOI] [PubMed] [Google Scholar]
- 58.Cavasotto, C., Orry, A. & Abagyan, R. The Challenge of Considering Receptor Flexibility in Ligand Docking and Virtual Screening. Current Computer Aided-Drug Design1, 423–440, http://www.eurekaselect.com/openurl/content.php?genre=article{&}issn=1573-4099{&}volume=1{&}issue=4{&}spage=423, 10.2174/157340905774330291 (2005).
- 59.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. Journal of Medicinal Chemistry. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Boyle NM, et al. Open Babel: An Open chemical toolbox. Journal of Cheminformatics. 2011;3:1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lagarde N, et al. NRLiSt BDB, the manually curated nuclear receptors ligands and structures benchmarking database. Journal of Medicinal Chemistry. 2014;57:3117–3125. doi: 10.1021/jm500132p. [DOI] [PubMed] [Google Scholar]
- 62.Gatica EA, Cavasotto CN. Ligand and Decoy Sets for Docking to G Protein-Coupled Receptors. Journal of Chemical Information and Modeling. 2012;52:1–6. doi: 10.1021/ci200412p. [DOI] [PubMed] [Google Scholar]
- 63.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2adrenoceptor. Nature. 2011;469:175–181. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenbaum DM, et al. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Søndergaard CR, Olsson MH, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p Kavalues. Journal of Chemical Theory and Computation. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 66.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions BT - Journal of Chemical Theory and Computation. Journal of Chemical Theory and Computation. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 67.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cavasotto CN, Aucar MG, Adler NS. Computational chemistry in drug lead discovery and design. International Journal of Quantum Chemistry. 2019;119:e25678. doi: 10.1002/qua.25678. [DOI] [Google Scholar]
- 69.Totrov M, Abagyan R. Protein-ligand docking as an energy optimization problem. New York: John Wiley and Sons; 2001. pp. 603–624. [Google Scholar]
- 70.Quiroga R, Villarreal MA. Vinardo: A scoring function based on autodock vina improves scoring, docking, and virtual screening. PLoS One. 2016;11:1–18. doi: 10.1371/journal.pone.0155183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain AN. Bias, reporting, and sharing: Computational evaluations of docking methods. Journal of Computer-Aided Molecular Design. 2008;22:201–212. doi: 10.1007/s10822-007-9151-x. [DOI] [PubMed] [Google Scholar]
- 72.Triballeau N, Acher F, Brabet I, Pin J-P, Bertrand H-O. Virtual Screening Workflow Development Guided by the “Receiver Operating Characteristic” Curve Approach. Application to High-Throughput Docking on Metabotropic Glutamate Receptor Subtype 4. Journal of Medicinal Chemistry. 2005;48:2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.