Abstract
Since the release of The Cancer Genome Atlas study of papillary thyroid carcinoma (PTC) in 2014, additional genomic studies of differentiated thyroid carcinoma (DTC) using massively-parallel sequencing (MPS) have been published. Recent advances in MPS technology have started to provide important insights into the molecular pathogenesis of DTC. In the genomic landscape, the most recurrently altered genes in DTC, which has a low mutational burden relative to other cancers, are BRAF, RAS, and fusion genes. Some novel driver candidates also have been identified. The frequency of these genomic alterations varies across the subtypes of DTC (classical PTC, follicular variant of PTC, and follicular thyroid carcinoma). Telomerase reverse transcriptase (TERT) promoter mutations are the alteration that makes the most important contribution to the progression of DTC. In the transcriptomic landscape, DTC can be classified according to its gene expression profile, and each subtype has a distinct mutational profile, intracellular signaling output, and clinicopathological characteristics. Herein, we review the results of genomic studies using MPS technology, and describe the types and frequencies of genomic alterations according to histological classifications of DTC and the characteristics and significance of the gene expression signatures of DTC.
Keywords: Genome; Transcriptome; Thyroid cancer, papillary; Thyroid cancer, follicular; High-throughput nucleotide sequencing
The Namgok Award is the highest scientific award of the Korean Endocrine Society, and is given to honor an individual who has made excellent contributions to progress in the field of endocrinology and metabolism. The Namgok Award is named after the pen name of Professor Hun Ki Min, who founded the Korean Endocrine Society in 1982.
Professor Young Joo Park received the Namgok Award at the Autumn Symposium of the Korean Endocrine Society in November 2018.
INTRODUCTION
According to the GLOBOCAN 2018 estimates of cancer incidence and mortality [1], the global incidence rate of thyroid carcinoma in men and women is 3.1 and 10.2 per 100,000, respectively, but mortality rates from the disease are much lower, with rates from 0.4 to 0.5 in men and women, suggesting that thyroid carcinoma generally shows indolent behavior. In particular, in Korea, the United States, and European countries, where thyroid cancer incidence has been rapidly increasing over the last few decades, mortality has steadily declined with the rise in the incidence of thyroid microcarcinomas.
The indolent behavior of thyroid carcinoma compared with other types of cancer can be explained by its genetic characteristics. A pan-cancer study using The Cancer Genome Atlas (TCGA) data showed that differentiated thyroid carcinoma (DTC) is a tumor with one of the lowest tumor mutational burdens, and usually harbors only a single driver gene alteration [2,3,4].
Meanwhile, the mutually exclusive mutation profile of thyroid carcinoma not only helps explain the indolent behavior of this cancer, but also indicates that single gene alterations, such as BRAF or RAS mutations, can induce the development or progression of cancer. Hence, it is expected that a deeper understanding of thyroid carcinoma may provide insights into tailored treatment for patients with such driver genes.
Recently, along with rapid developments of massively-parallel sequencing (MPS) technology [5], the genetic understanding of thyroid carcinoma has rapidly grown. TCGA found an association between the type of driver mutation and two distinct gene expression signatures in papillary thyroid carcinoma (PTC) [4]. Our group confirmed those results and expanded the scope of their analysis by including follicular thyroid carcinoma (FTC) as well as PTC [6].
In this review, we combine the results of genomic studies using MPS technology and describe (1) the types and frequencies of genomic alterations according to the histological phenotypes of DTC and (2) the characteristics and significance of the gene expression signatures of DTC.
CLASSIFICATION AND OVERVIEW OF FOLLICULAR CELL-DERIVED THYROID CARCINOMA
Follicular cell-derived thyroid carcinoma is classified into PTC (80% to 85%), FTC (10% to 15%), poorly-differentiated thyroid carcinoma (PDTC, <2%), and anaplastic thyroid carcinoma (ATC, <2%) [7]. The former two categories belong to DTC, and PTC accounts for most cases of DTC. PTC can be classified into various histologic subtypes, including classical PTC (cPTC) and follicular variant of PTC (FVPTC). In terms of molecular pathogenesis, the majority of DTC cases are caused by alterations of driver genes such as BRAF, RAS, or fusion genes [8]. Those alterations particularly activate the receptor tyrosine kinase (RTK)/mitogen-activated protein kinase (MAPK) pathway, a master regulator of numerous cellular processes including proliferation, differentiation, adhesion, migration, and apoptosis. DTC can progress to PDTC or ATC by additional hits on TERT, tumor suppressors, or phosphoinositide 3-kinase (PI3K) pathway genes.
GENOMIC CHARACTERISTICS OF CLASSICAL PTC
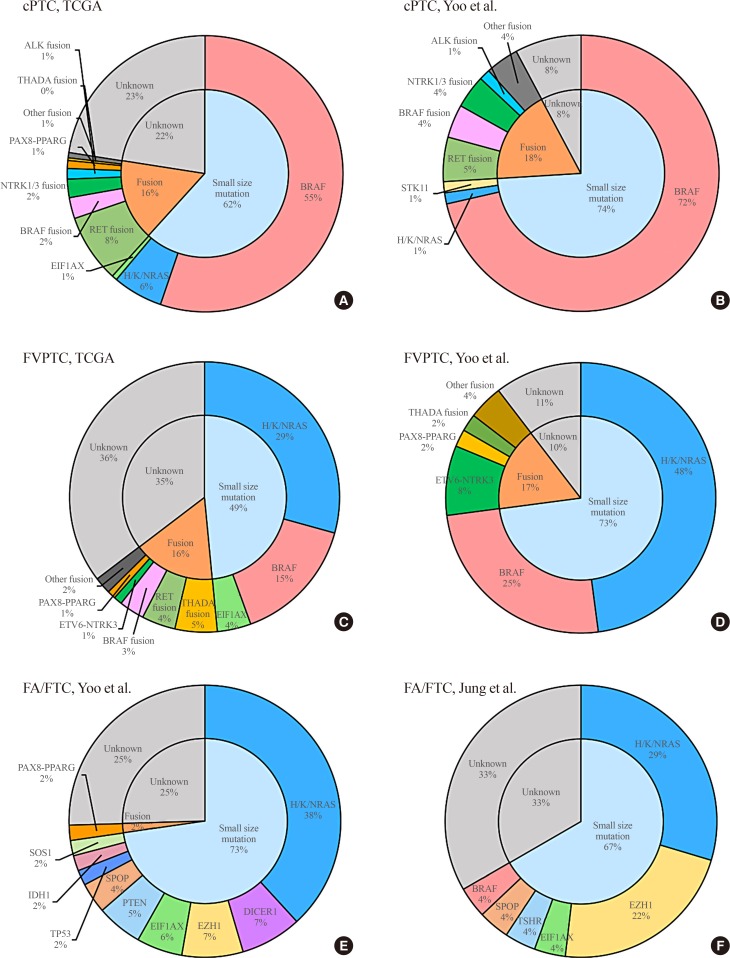
PTC is the most common type of thyroid carcinoma of follicular cell origin, and accounts for about 85% of cases of DTC [7]. It has been well documented that PTC develops through the activation of the RET/PTC-RAS-BRAF-ERK axis, which drives the oncogenic proliferation of thyroid follicular cells [9,10,11]. However, until recently, few in-depth genomic characterizations of PTC have been performed, despite its high incidence, which might be because it shows less aggressive features than any other human cancers [12]. Since the first large-scale integrative genomic analysis was accomplished by TCGA in 2014 [4], several genomic studies of DTC using MPS have been published. We summarize the major findings of these studies in Table 1, and present the frequencies of genomic alterations of DTC in several representative studies in Fig. 1.
Table 1. Summary of Major Genomic Studies of Differentiated Thyroid Carcinoma Analyzed Using the Massively-Parallel Sequencing Method.
| Study | Subtype | Country (ethnicity) | Significance |
|---|---|---|---|
| TCGA (2014) [4] | PTC (n=496) | USA (mainly Caucasian) | The first integrative genomic analysis of thyroid carcinoma |
| Illustrated the detailed mutational profile of PTC | |||
| Introduced the novel conception of a thyroid carcinoma classification (BVL and RL) based on molecular characteristics | |||
| Costa et al. (2015) [17] | PTC (n=18) | France (Caucasian) | Replicated the existence of the BVL and RL subtypes identified by TCGA in thyroid carcinoma |
| Identified a novel fusion gene rearrangement, WNK1-B4GALNT3, in PTC | |||
| Pan et al. (2016) [18] | PTC (n=402) | China (East Asian) | Identified a novel tumor suppressor, GAS8-AS1, and a driver gene, LPAR4, in PTC |
| Yoo et al. (2016) [6] | PTC (n=125) | Korea (East Asian) | The first mutational and transcriptional profiling of FA and FTC |
| FA (n=25) | Identified a third molecular subtype of thyroid carcinoma, NBNR | ||
| FTC (n=30) | Revealed the benign characteristics and the similarity between three follicular-patterned thyroid carcinomas (miFTC, FA, and EFVPTC) at the molecular level | ||
| Siraj et al. (2016) [20] | PTC (n=886) | Saudi Arabia (Middle Eastern) | Identified an association between TG alterations and tumor aggressiveness in PTC |
| Lu et al. (2017) [19] | PTC (n=138) | China (East Asian) | Identified novel fusion gene arrangements (UEVLD-RET, OSBPL9-BRAF, and SQSTM1-NTRK3) in PTC |
| Jung et al. (2016) [35] | FA (n=14) | Korea (East Asian) | Revealed the molecular similarity between FTC and FA by analysis of evolutionary age of tumors |
| FTC (n=13) |
TCGA, The Cancer Genome Atlas; PTC, papillary thyroid carcinoma; BVL, BRAFV600E-like; RL, RAS-like; FA, follicular adenoma; FTC, follicular thyroid carcinoma; NBNR, non-BRAF-non-RAS; miFTC, minimally-invasive follicular thyroid carcinoma; EFVPTC, encapsulated follicular variant of papillary thyroid carcinoma.
Fig. 1. Frequencies of driver mutations in differentiated thyroid carcinoma. Classical papillary thyroid carcinoma (cPTC) in the studies of (A) The Cancer Genome Atlas (TCGA) [4] and (B) Yoo et al. [6] (upper), follicular variant of papillary thyroid carcinoma (FVPTC) in the studies of (C) TCGA [4] and (D) Yoo et al. [6] (middle), and follicular adenoma/follicular thyroid carcinoma (FA/FTC) in the studies of (E) Yoo et al. [6] and (F) Jung et al. [35] (lower).
In the TCGA study, classical, tall cell variant, and FVPTC from 496 patients (mostly consisting of Caucasians) were analyzed with diverse types of MPS and array-based technologies. This study discovered a novel thyroid cancer driver gene, EIF1AX, as well as well-known oncogenic drivers such as BRAF, NRAS, HRAS, and KRAS. In cPTC, the frequency of BRAF, NRAS, HRAS, and KRAS mutations was 55.25%, 4.01%, 1.54%, and 0.31%, respectively. EIF1AX plays a role in recognition of the translation initiation site, and cancer-related mutations in this gene alter gene expression by increasing discrimination against AUG codons in poor contexts [13]. EIF1AX was originally reported in uveal melanoma, and in most cases showed co-mutations with GNA11 or GNAQ [14,15]. However, in PTC, EIF1AX was altered in a mutually exclusive manner with other known driver genes, and it was found in cPTC (0.62%) and FVPTC (3.03%), which are the two main types of PTC.
Furthermore, gene rearrangements including RET (8.02%), BRAF (2.47%), NTRK1/3 (2.16%), ALK (1.23%), PAX8-PPARG (0.93%), THADA (0.31%), FGFR2 (0.31%), and LTK (0.31%) were discovered in cPTC, and those alterations were also mutually exclusive with each other, in addition to mutations in BRAF, H/K/NRAS, and EIF1AX. In particular, patients who were diagnosed with PTC at a young age frequently harbored gene rearrangements, and this tendency has been confirmed by other reports [6,16].
In 2015, Costa et al. [17] described the RNA sequencing (RNA-seq)-based mutational and transcriptional profiles of 18 PTCs from French patients. They discovered somatic mutations in well-known driver genes including BRAF (16.67%), RAS (16.67%), and RET rearrangements (38.89%). Moreover, a novel fusion transcript, WNK1-B4GALNT3, was found, as well as somatic mutations in DICER1 (5.56%), MET (5.56%), and VHL (5.56%).
In 2016, our group also described the mutational profile of 77 Korean patients with cPTC based on an RNA-seq analysis [6]. In our study cohort, 71.43% of cPTC patients harbored BRAFV600E, which is a slightly higher rate than has been reported in other studies [4,17,18,19,20]. An NRAS mutation was only found in 1.3% of study subjects, whereas 18.18% of cases of cPTC had gene rearrangements in RET (5.18%), BRAF (3.9%), NTRK1/3 (3.9%), ALK (1.3%), FGFR2 (1.3%), and THADA (1.3%).
Pan et al. [18] showed a different mutational spectrum from that observed in the TCGA study in a Chinese population using 402 PTC tumors (91 and 311 tumors analyzed by whole-exome sequencing [WES] and Sanger sequencing methods, respectively). As in the TCGA cohort, BRAFV600E (59%) was the most recurrently identified driver mutation in Chinese PTC patients. However, they found that long non-coding RNA (GAS8-AS1 [9.2%]), was the second most frequently altered gene, rather than RAS (3.2%), and proposed that it plays a tumor-suppressive role in PTC. Another frequently altered gene, G protein-coupled receptor LPAR4 (2.7%), was also discovered, and the malignant transformation of PTC through LPAR4 mutation was revealed. In 2017, another Chinese PTC cohort with 138 patients was analyzed using the targeted sequencing method [19]. This study displayed a similar incidence of BRAFV600E (57.25%) as the TCGA cohort, whereas only 2.17% of tumors harbored a RAS mutation (KRAS only). Moreover, several gene rearrangements were identified, including RET (5.07%), NTRK1/3 (2.17%), and BRAF (0.72%), as well as three novel fusion gene candidates, UEVLD-RET, OSBPL9-BRAF, and SQSTM1-NTRK3.
Furthermore, Siraj et al. [20] reported the genomic characteristics of 101 PTCs from Saudi Arabian patients by WES and validated their results using 785 additional samples. They proposed that TG co-mutation is co-implicated in patients with BRAF, NRAS, or HRAS mutations. In their study, BRAF and RAS (HRAS and NRAS) were the most frequently altered genes (59.54% and 8.13%, respectively) and TG was the third most frequently mutated gene (3.05%). Patients with a TG alteration showed poorer survival than individuals without this type of mutation. Moreover, they found that advanced PTC patients frequently harbored TG mutations (12.7%).
GENOMIC CHARACTERISTICS OF FTC
FTC is the second most common type of thyroid cancer following PTC, and accounts for 10% to 30% of cases of DTC [21]. However, research on the genomic and transcriptomic characteristics of the follicular histologic type of thyroid tumors, especially FTC, is still lacking. In 2016, our group reported a comprehensive study of the transcriptional and mutational landscape of follicular-patterned thyroid tumors, including follicular adenoma (FA), minimally-invasive FTC (miFTC), and FVPTC, as well as cPTCs [6]. We analyzed miFTC in 30 Korean patients using RNA-seq. Additionally, 25 FA tumors were sequenced, as FA is a benign lesion that is difficult to differentiate from FTC based on cytologic, sonographic, or clinical features [22]. This was the first research to broaden the molecular understanding of follicular-patterned thyroid tumors, including FTC.
We identified that miFTC and FA showed a similar mutational profile, and most tumors had single nucleotide-level alterations in H/K/NRAS (38.18%), DICER1 (7.27%), EIF1AX (5.45%), EZH1 (7.27%), SPOP (3.65%), IDH1 (1.82%), and SOS1 (1.82%), rather than fusion genes. Only one PAX8-PPARG (1.82%) translocation was found in our study cohort. All mutations were mutually exclusive with each other.
This study was the first to confirm the existence of EIF1AX mutations in miFTC, as well as in PTC. Moreover, FA harbored this type of mutation, as reported in another previous study [23]. Several mutations in DICER1, which is involved in the processing of miRNA precursor hairpins and long double-stranded RNA, were found [24]. DICER1 has been reported to be associated with familial thyroid carcinoma and diverse types of cancer [25,26,27,28]. In our study, somatic DICER1 mutations were especially common in the ribonuclease IIIb domain, and those mutations associated with gene expression levels of DICER1.
After our first report regarding EZH1Q571R and EZH1Y624F mutations in follicular thyroid tumors, EZH1 was proposed to have an oncogenic impact when mutated as a second hit in autonomous thyroid adenomas [29]. This study reported that EZH1 mutations were strongly associated with alterations in cAMP pathway genes, such as TSHR, GNAS, or ADCY9. Furthermore, functional validation showed increased histone H3 trimethylation and proliferation of thyroid cells with the EZH1Q571R mutation.
An alteration of SPOP was originally reported to be found in 15% of prostate carcinomas [30,31,32], and this mutation was thought to be involved in the regulation of DAXX gene, which associates with transcriptional repressor proteins such as histone deacetylase II, core histones, and chromatin-associated proteins [33]. In our study, two FA samples harbored SPOPP94R, which is localized in the MATH domain, and the same mutation was repeatedly identified in subsequent studies [34,35]. Moreover, IDH1R132C and SOS1N233Y were found; these mutations have not been reported in thyroid carcinoma, but have been recurrently found in other types of cancer [36,37]. The aforementioned alterations of DICER1, EIF1AX, EZH1, SPOP, and IDH1 imply that several other pathways may account for a substantial proportion of FTC/FA pathogenesis, rather than the MAPK and PI3K pathways.
Regarding copy number alterations, we found that follicular-patterned thyroid tumors—miFTC, FA, and FVPTC—harbored more frequent arm-level chromosomal alterations than cPTC. In particular, chromosome 22q deletion was frequent in miFTC/FA/FVPTC or RAS-mutated tumors. Furthermore, arm-level copy number alterations were more frequent in miFTC than in FA, which might indicate that FA is a preneoplastic condition prior to miFTC.
A subsequent study from Korea described the genomic landscape of 13 FTC and 14 FA tumors [35]. Jung et al. [35] analyzed somatic mutations and copy number alterations using WES and a comparative genomic hybridization microarray, respectively. In their results, FTC and FA displayed similar genomic features, as we previously described; RAS (29.63%) and EIF1AX (7.41%) were the most frequently altered genes. Moreover, EZH1Q571R, EZH1Y642F, and SPOPR94R, which were first suggested as driver genes of FTC/FA by our group, were also found [6]. Furthermore, their analysis regarding the evolutionary age of tumors found that FA tumors are as old as FTC tumors, which suggests a similarity between FTC and FA on the molecular level.
GENOMIC CHARACTERISTICS OF FVPTC
FVPTC, a thyroid carcinoma with the nuclear features of PTC, but a follicular growth pattern rather than papillae [38], is another type of follicular-patterned thyroid tumor, in addition to FTC and FA [39]. Among all PTC variants, FVPTC is the most common subtype, and the incidence of FVPTC has increased from 20% to 30% of all cases of PTC over the last three decades in the USA [40].
In the TCGA study of PTC, 99 FVPTC patients (21.2%) were included among 496 PTC patients [4]. The mutational profile of FVPTC was characterized as RAS-dominant (29.29%), as was that of FTC/FA [6]. However, 15.15% of FVPTCs also had BRAFV600E, in contrast to FTC, which rarely harbors this mutation. Furthermore, gene rearrangements including THADA (5.05%), RET (4.04%), BRAF (3.03%), ETV6-NTRK3 (1.01%), PAX8-PPARG (1.01%), MET (1.01%), and FGFR2 (1.01%) were discovered. In FVPTC, these gene rearrangements were also mutually exclusive with BRAFV600E and RAS mutations, as in cPTC. When network-based stratification was applied to explore somatic mutation-based PTC subtypes [41], FVPTC showed a significant association with a subtype with alterations in RAS, PTEN, PPARG, and TSHR. Somatic copy number alterations were found more frequently in FVPTC than in cPTC. Moreover, an isolated deletion of chromosome 22q, a region including NF2 and CHEK2, was enriched in FVPTC compared to other subtypes of PTC, suggesting loss of these tumor suppressors (NF2 and CHEK2). This result again implies a significant association between chromosome 22q and the development of follicular-patterned thyroid carcinoma (FTC/FA and FVPTC).
In our comprehensive genomic study of follicular-patterned thyroid tumors [6], we discovered that FVPTC showed an intermediate mutational status between cPTC and miFTC. Among a total of 48 FVPTCs, RAS mutations were predominant (47.9%), followed by the BRAFV600E mutation (25.0%). Gene rearrangements, including ETV6-NTRK3 (8.33%), PAX8-PPARG (2.08%), EZR-ERBB4 (2.08%), FGFR2 (2.08%), and THADA (2.08%), were detected in 16.7% of FVPTCs. We also confirmed that FVPTC had higher percentages of arm-level copy number alterations, especially 22q deletion, which is consistent with results from previous studies [4,42].
FVPTC presents either as an encapsulated or as an infiltrative neoplasm. Recently, owing to the excellent prognosis of the non-invasive subset of encapsulated FVPTC (EFVPTC), a new entity called “non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP)” was proposed [43]. We were the first to report the integrated transcriptomic, genomic, and clinical characterization of subtypes of FVPTC [6,44], whereas previous studies, including the TCGA study, did not classify FVPTC by subtype. The mutational profiles of EFVPTC and infiltrative FVPTC were similar to those of FA/miFTC and cPTC, respectively. This is consistent with a previous study that reported that the oncogenic mutational profile of EFVPTC was very close to that of FA/FTC, while the oncogenic mutational profile of infiltrative FVPTC was in between that of FA/FTC and cPTC, although it was closer to the latter [45]. When EFVPTC was classified into two subtypes—NIFTP and invasive EFVPTC—according to capsular and vascular invasion, in NIFTP, the BRAFV600E mutation was not found, but RAS mutations were present in 64.7% as the major alterations. In contrast, in invasive EFVPTC, the BRAFV600E mutation was found in 38.5% of cases, a similar proportion to that in infiltrative FVPTC (38.9%). The proportion of RAS mutations (38.5% and 38.9%, respectively) was also similar between invasive EFVPTC and infiltrative FVPTC.
IMPACT OF TERT PROMOTER MUTATIONS IN DTC
TERT promoter mutations have been suggested as a strong prognostic biomarker in thyroid carcinoma. TERT is a core subunit of the RNA component, TERC [46]. Its upregulation reactivates telomerase, thereby maintaining the length of telomeres in human cancer cells [47]. There are two hot spots in the promoter region of TERT: chr5, 1,295,228 C>T and 1,295,250 C>T, which are localized 124 and 146 base pairs upstream of the TERT transcription start site, respectively [48,49]. The prevalence of TERT promoter mutations in PTC and FTC was reported to be approximately 12% and 18%, respectively, and was more frequent in PDTC and ATC [50]. The frequency of these mutations in DTC appears to differ among countries, and the prevalence in Korea was lower than in other countries [51].
A relationship between the progression of thyroid carcinoma and these mutations was proposed in 2013 [52], and several studies have demonstrated an association between TERT promoter mutations and aggressive clinicopathological features of PTC [50,53]. In 2014, Xing et al. [54] were the first to prove the role of the genetic duet of BRAFV600E and TERT promoter mutations in aggressive behavior and poor clinical outcomes of PTC. Remarkably, although it was reported that TERT promoter mutations in PTC without the BRAFV600E mutation did not increase the risk of recurrence or mortality after multivariate adjustment, a significant increased effect of the TERT mutations was observed when BRAFV600E was co-mutated [54,55,56].
Several studies have confirmed the synergetic effect of the coexistence of TERT promoter mutations and BRAFV600E in PTC [57], which implies the possibility that coexisting TERT promoter and RAS mutations in follicular-patterned thyroid tumors may also play a significant role in tumor aggressiveness. However, few studies have investigated the prognostic effect of the coexistence of TERT promoter and RAS mutations [55,58] Therefore, the underlying strength of their association needs to be investigated further. In an unpublished analysis, we recently discovered that all nine widely-invasive FTCs, analyzed by WES and RNA-seq, harbored various types of TERT alterations, including promoter mutations, fusion genes, and upstream translocation [59]. Although further investigation is needed, this result suggests that TERT alterations may play a significant role in the progression of FTC and tumor aggressiveness.
MOLECULAR CLASSIFICATION OF DTC
The major implication of the TCGA study was the discovery of two molecular subtypes of PTC, BRAFV600E-like (BVL) and RAS-like (RL), based on 71 gene expression signatures [4]. These two molecular subtypes represent differential regulation of mRNA/protein signaling (MAPK and PI3K signaling pathways) and thyroid differentiation, regardless of the PTC subtype. This concept of a molecular level-based classification broadened the previous understanding of thyroid carcinoma, which was based on tumor histopathology or cytology [60]. The BVL subtype was strongly associated with BRAFV600E and diverse fusion genes such as BRAF, RET, and NTRK1/3. Hence, it was usually associated with cPTC. In contrast, the RL subtype was related to mutations of RAS and EIF1AX, and fusion genes such as PAX8-PPARG, FGFR2, and THADA. The transcriptomic profile of FVPTC was frequently characterized by the RL molecular subtype. Based on the genomic findings of the TCGA study, it was suggested that a pathologic reclassification of follicular-patterned thyroid tumors is justified, meaning that FVPTC, which is currently a subtype of PTC, could be classified as FTC in the future.
The classification of thyroid carcinoma into BVL and RL subtypes according to molecular characteristics was replicated by Costa et al. [17] with differentially expressed gene analysis. They showed that the BVL (BRAF, ETV6-NTRK3, and RET fusions) and RL (HRAS, DICER1, PAX8-PPARG, and WNK1-B4GALNT3) subtypes were distinctively associated with types of altered genes.
The most noteworthy outcome of our 2016 investigation [6] was that the notion of molecular classification of thyroid carcinoma was extended. In addition to the BVL and RL subtypes established by TCGA, our analysis suggested a third molecular subtype, non-BRAF-non-RAS (NBNR), which was mainly associated with follicular-patterned thyroid tumors. In particular, alterations in DICER1, EIF1AX, EZH1, IDH1, SPOP, PAX8-PPARG, and THADA fusion represented the signature of NBNR. Thyroid tumors classified as NBNR showed low levels of MAPK signaling activation relative to the BVL and RL subtypes, whereas high thyroid differentiation and preserved metabolism were also identified. Intriguingly, most tumors with EIF1AX mutations and PAX8-PPARG, which were classified as RL in the TCGA study, were re-classified as NBNR in our analysis [4]. This result was confirmed using the TCGA cohort, as well as our Korean subjects.
The existence of the third molecular subtype, which is distinct from BVL and RL, provides further support for the relationship between the follicular histology of thyroid carcinoma and the alteration of diverse signaling pathways, rather than the MAPK or PI3K signaling pathways. Only few NBNR tumors displayed lymph node metastasis and extrathyroidal extension, unlike the BVL and RL subtypes, which demonstrates their benign and indolent nature. Recently, in benign thyroid nodules, EZH1Q571R and SPOPP94R mutations with distinctive transcriptomic features from BVL were found [34], which is consistent with our study.
CONCLUSIONS
Since the release of the TCGA study of PTC in 2014, additional genomic studies of DTC using MPS have been published. In the genomic landscape, the most recurrently altered genes in DTC, which has a low mutational burden relative to other cancers, are BRAF, RAS, and fusion genes. Some novel driver candidates also have been identified. The frequency of these genomic alterations varies across the subtypes of DTC: cPTC, FVPTC, and FTC. Moreover, TERT promotor mutations are the most important alteration that contributes to the progression of DTC, particularly when co-mutated with BRAF or RAS. In the transcriptomic landscape, DTC can be classified according to its gene expression profile as BVL, RL, or NBNR. Each subtype is associated with mutations of different genes, and the intracellular pathways related to cell proliferation or invasiveness, including the MAPK pathway, show a pattern of gradual activation from NBNR to RL to BVL, resulting in more aggressive behavior. Recent advances in MPS have provided important insights into the molecular pathogenesis of DTC beyond its histological classification.
ACKNOWLEDGMENTS
I, Young Joo Park, would like to express my sincere gratitude to emeritus professor Hun Ki Min (Namgok) for giving me the prestigious Namgok Award. I would like to thank Prof. Bo Youn Cho, Prof. Hak-Chul Jang, Prof. Kyong Soo Park, Prof. Do Joon Park, Prof. Chan Soo Shin, Prof. Ka Hee Yi and Prof. Jeong-Sun Seo at Seoul National University College of Medicine, and all the professor-mentors who guided and taught me. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning (NRF-2016R1A2B4012417).
Footnotes
CONFLICTS OF INTEREST: No potential conflict of interest relevant to this article was reported.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 3.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016;12:e1006239. doi: 10.1371/journal.pgen.1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dralle H, Machens A, Basa J, Fatourechi V, Franceschi S, Hay ID, et al. Follicular cell-derived thyroid cancer. Nat Rev Dis Primers. 2015;1:15077. doi: 10.1038/nrdp.2015.77. [DOI] [PubMed] [Google Scholar]
- 8.Gianoukakis AG, Giannelli SM, Salameh WA, McPhaul LW. Well differentiated follicular thyroid neoplasia: impact of molecular and technological advances on detection, monitoring and treatment. Mol Cell Endocrinol. 2011;332:9–20. doi: 10.1016/j.mce.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Mitsutake N, Miyagishi M, Mitsutake S, Akeno N, Mesa C, Jr, Knauf JA, et al. BRAF mediates RET/PTC-induced mitogen-activated protein kinase activation in thyroid cells: functional support for requirement of the RET/PTC-RAS-BRAF pathway in papillary thyroid carcinogenesis. Endocrinology. 2006;147:1014–1019. doi: 10.1210/en.2005-0280. [DOI] [PubMed] [Google Scholar]
- 10.Tallini G, Asa SL. RET oncogene activation in papillary thyroid carcinoma. Adv Anat Pathol. 2001;8:345–354. doi: 10.1097/00125480-200111000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Knauf JA, Ma X, Smith EP, Zhang L, Mitsutake N, Liao XH, et al. Targeted expression of BRAFV600E in thyroid cells of transgenic mice results in papillary thyroid cancers that undergo dedifferentiation. Cancer Res. 2005;65:4238–4245. doi: 10.1158/0008-5472.CAN-05-0047. [DOI] [PubMed] [Google Scholar]
- 12.Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2014. Cancer Res Treat. 2017;49:292–305. doi: 10.4143/crt.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Marcos P, Zhou F, Karunasiri C, Zhang F, Dong J, Nanda J, et al. eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast. Elife. 2017;6:e31250. doi: 10.7554/eLife.31250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang J, Cai W, Feng D, Teng H, Mao F, Jiang Y, et al. Genetic landscape of papillary thyroid carcinoma in the Chinese population. J Pathol. 2018;244:215–226. doi: 10.1002/path.5005. [DOI] [PubMed] [Google Scholar]
- 17.Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, Cacciola NA, et al. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget. 2015;6:11242–11251. doi: 10.18632/oncotarget.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan W, Zhou L, Ge M, Zhang B, Yang X, Xiong X, et al. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum Mol Genet. 2016;25:1875–1884. doi: 10.1093/hmg/ddw056. [DOI] [PubMed] [Google Scholar]
- 19.Lu Z, Zhang Y, Feng D, Sheng J, Yang W, Liu B. Targeted next generation sequencing identifies somatic mutations and gene fusions in papillary thyroid carcinoma. Oncotarget. 2017;8:45784–45792. doi: 10.18632/oncotarget.17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siraj AK, Masoodi T, Bu R, Beg S, Al-Sobhi SS, Al-Dayel F, et al. Genomic profiling of thyroid cancer reveals a role for thyroglobulin in metastasis. Am J Hum Genet. 2016;98:1170–1180. doi: 10.1016/j.ajhg.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang BH, Lo CY, Chan WF, Lam KY, Wan KY. Staging systems for follicular thyroid carcinoma: application to 171 consecutive patients treated in a tertiary referral centre. Endocr Relat Cancer. 2007;14:29–42. doi: 10.1677/erc.1.01284. [DOI] [PubMed] [Google Scholar]
- 22.McHenry CR, Phitayakorn R. Follicular adenoma and carcinoma of the thyroid gland. Oncologist. 2011;16:585–593. doi: 10.1634/theoncologist.2010-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunamurthy A, Panebianco F, J Hsiao S, Vorhauer J, Nikiforova MN, Chiosea S, et al. Prevalence and phenotypic correlations of EIF1AX mutations in thyroid nodules. Endocr Relat Cancer. 2016;23:295–301. doi: 10.1530/ERC-16-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 25.Rutter MM, Jha P, Schultz KA, Sheil A, Harris AK, Bauer AJ, et al. DICER1 mutations and differentiated thyroid carcinoma: evidence of a direct association. J Clin Endocrinol Metab. 2016;101:1–5. doi: 10.1210/jc.2015-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Kock L, Sabbaghian N, Soglio DB, Guillerman RP, Park BK, Chami R, et al. Exploring the association between DICER1 mutations and differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2014;99:E1072–E1077. doi: 10.1210/jc.2013-4206. [DOI] [PubMed] [Google Scholar]
- 27.Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz KA, Pacheco MC, Yang J, Williams GM, Messinger Y, Hill DA, et al. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: a report from the International Pleuropulmonary Blastoma Registry. Gynecol Oncol. 2011;122:246–250. doi: 10.1016/j.ygyno.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calebiro D, Grassi ES, Eszlinger M, Ronchi CL, Godbole A, Bathon K, et al. Recurrent EZH1 mutations are a second hit in autonomous thyroid adenomas. J Clin Invest. 2016;126:3383–3388. doi: 10.1172/JCI84894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115(Pt 16):3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 34.Ye L, Zhou X, Huang F, Wang W, Qi Y, Xu H, et al. The genetic landscape of benign thyroid nodules revealed by whole exome and transcriptome sequencing. Nat Commun. 2017;8:15533. doi: 10.1038/ncomms15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung SH, Kim MS, Jung CK, Park HC, Kim SY, Liu J, et al. Mutational burdens and evolutionary ages of thyroid follicular adenoma are comparable to those of follicular carcinoma. Oncotarget. 2016;7:69638–69648. doi: 10.18632/oncotarget.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosai J, Carcangiu ML, DeLellis RA. Tumors of the thyroid gland. Washington DC: Armed Forces Institute of Pathology; 1992. [Google Scholar]
- 39.Giordano TJ. Follicular cell thyroid neoplasia: insights from genomics and The Cancer Genome Atlas research network. Curr Opin Oncol. 2016;28:1–4. doi: 10.1097/CCO.0000000000000248. [DOI] [PubMed] [Google Scholar]
- 40.Lam AK, Lo CY, Lam KS. Papillary carcinoma of thyroid: a 30-yr clinicopathological review of the histological variants. Endocr Pathol. 2005;16:323–330. doi: 10.1385/ep:16:4:323. [DOI] [PubMed] [Google Scholar]
- 41.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wreesmann VB, Ghossein RA, Hezel M, Banerjee D, Shaha AR, Tuttle RM, et al. Follicular variant of papillary thyroid carcinoma: genome-wide appraisal of a controversial entity. Genes Chromosomes Cancer. 2004;40:355–364. doi: 10.1002/gcc.20049. [DOI] [PubMed] [Google Scholar]
- 43.Nikiforov YE, Seethala RR, Tallini G, Baloch ZW, Basolo F, Thompson LD, et al. Nomenclature revision for encapsulated follicular variant of papillary thyroid carcinoma: a paradigm shift to reduce overtreatment of indolent tumors. JAMA Oncol. 2016;2:1023–1029. doi: 10.1001/jamaoncol.2016.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song YS, Won JK, Yoo SK, Jung KC, Kim MJ, Kim SJ, et al. Comprehensive transcriptomic and genomic profiling of subtypes of follicular variant of papillary thyroid carcinoma. Thyroid. 2018;28:1468–1478. doi: 10.1089/thy.2018.0198. [DOI] [PubMed] [Google Scholar]
- 45.Rivera M, Ricarte-Filho J, Patel S, Tuttle M, Shaha A, Shah JP, et al. Encapsulated thyroid tumors of follicular cell origin with high grade features (high mitotic rate/tumor necrosis): a clinicopathologic and molecular study. Hum Pathol. 2010;41:172–180. doi: 10.1016/j.humpath.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheong C, Hong KU, Lee HW. Mouse models for telomere and telomerase biology. Exp Mol Med. 2003;35:141–153. doi: 10.1038/emm.2003.20. [DOI] [PubMed] [Google Scholar]
- 47.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 48.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 49.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23:R143–R155. doi: 10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song YS, Lim JA, Park YJ. Mutation profile of well-differentiated thyroid cancer in Asians. Endocrinol Metab (Seoul) 2015;30:252–262. doi: 10.3803/EnM.2015.30.3.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20:603–610. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin DT, Yu K, Lu RQ, Li X, Xu J, Lei M, et al. Clinicopathological significance of TERT promoter mutation in papillary thyroid carcinomas: a systematic review and meta-analysis. Clin Endocrinol (Oxf) 2016;85:299–305. doi: 10.1111/cen.13017. [DOI] [PubMed] [Google Scholar]
- 54.Xing M, Liu R, Liu X, Murugan AK, Zhu G, Zeiger MA, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014;32:2718–2726. doi: 10.1200/JCO.2014.55.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song YS, Lim JA, Choi H, Won JK, Moon JH, Cho SW, et al. Prognostic effects of TERT promoter mutations are enhanced by coexistence with BRAF or RAS mutations and strengthen the risk prediction by the ATA or TNM staging system in differentiated thyroid cancer patients. Cancer. 2016;122:1370–1379. doi: 10.1002/cncr.29934. [DOI] [PubMed] [Google Scholar]
- 56.Liu R, Bishop J, Zhu G, Zhang T, Ladenson PW, Xing M. Mortality risk stratification by combining BRAF V600E and TERT promoter mutations in papillary thyroid cancer: genetic duet of BRAF and TERT promoter mutations in thyroid cancer mortality. JAMA Oncol. 2017;3:202–208. doi: 10.1001/jamaoncol.2016.3288. [DOI] [PubMed] [Google Scholar]
- 57.Moon S, Song YS, Kim YA, Lim JA, Cho SW, Moon JH, et al. Effects of coexistent BRAF(V600E) and TERT promoter mutations on poor clinical outcomes in papillary thyroid cancer: a meta-analysis. Thyroid. 2017;27:651–660. doi: 10.1089/thy.2016.0350. [DOI] [PubMed] [Google Scholar]
- 58.Song YS, Lim JA, Min HS, Kim MJ, Choi HS, Cho SW, et al. Changes in the clinicopathological characteristics and genetic alterations of follicular thyroid cancer. Eur J Endocrinol. 2017;177:465–473. doi: 10.1530/EJE-17-0456. [DOI] [PubMed] [Google Scholar]
- 59.Cho AB, Yoo SK, Sohn MH, Shin JY, Kim S, Lee EK, et al. Abstract 3398: The genomic and transcriptomic analysis of nine widely invasive follicular thyroid carcinomas (wiFTC) in Korean patients. Cancer Research. 2017;77(13 Suppl):3398. [Google Scholar]
- 60.Nikiforov YE, Biddinger PW, Thompson LDR. Diagnostic pathology and molecular genetics of the thyroid. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2012. [Google Scholar]