Abstract
Mammalian cells are organized into different compartments that separate and facilitate physiological processes by providing specialized local environments and allowing different, otherwise incompatible biological processes to be carried out simultaneously. Proteins are targeted to these subcellular locations where they fulfill specialized, compartment-specific functions. Spatial proteomics aims to localize and quantify proteins within subcellular structures.
Introduction
Mammalian cells are organized into different compartments that separate and facilitate physiological processes by providing specialized local environments and allowing different, otherwise incompatible biological processes to be carried out simultaneously. Proteins are targeted to these subcellular locations where they fulfill specialized, compartment-specific functions. Spatial proteomics aims to localize and quantify proteins within subcellular structures to provide three important biological insights: First, placing a protein in a specific location within the cell provides a hypothesis about what function the protein might have. For example, proteins localized to the mitochondria could have roles in energy production or apoptosis. Second, it can indicate a specific state of the cell or provide potential hypotheses about a new function of a protein if the protein is found in different subcellular locations simultaneously or upon perturbation. Third, determining the localization of proteins is important to understand the functions of organelles and compartments. Most importantly though, spatial proteomics of the non-perturbed state also provides a baseline for detecting aberrant localization of proteins, which is an important cause for a number of different human diseases.
Protein biology and subcellular localization
Because spatial proteomics requires the enrichment of proteins prior to identification, results are fundamentally limited with regards to several basic aspects in subcellular biology of proteins. One aspect is that proteins can be present at different subcellular locations simultaneously. For example, actin prominently assembles the actin cytoskeleton in the cytoplasm. While β-actin has been also observed in nuclear preparations by mass spectrometry, it was usually discarded as non-nuclear contaminant, which it well may have been in cases. Recently, elegant genetic and imaging studies now show that nuclear actin plays a role in the repair of DNA double strand breaks and in transcriptional activation in the nucleus [38-41].
Another problem complicating spatial proteomics is the fact that proteins can shuttle between different subcellular localizations and their function can change accordingly. The tumor suppressor protein p53 and the Rel-family transcription factor NF-κB are two well studied example proteins that shuttle from the cytosol to the nucleus [20,21]. In both cases, not only the subcellular location of the two proteins change but also their activity and protein-protein interactions. When p53 is located in the cytosol in healthy cells, it is usually in an inactive state and bound to MDM2, which degrades p53 [22-24]. In the event of cell stress, p53 is released from MDM2, translocated into the nucleus, and acts as transcription factor to induce apoptosis [25]. The same is true for NF-κB, which is inactive in the cytosol, but is activated by induced nuclear import [26]. Spatial proteomic approaches to study diseases such as cancer and inflammatory diseases thus need to account for such dynamic protein location and specific cellular circumstances as well as protein modifications and mutations, which may determine the cellular “zipcode” of a protein.
Furthermore, protein location may not be the same during the entire life-cycle of a protein. For example, during protein biogenesis, membrane proteins will be cotranslationally inserted into the ER before journeying in vesicles through the ERGIC (ER-Golgi-intermediate compartment), the Golgi, and in different vesicles to their target destinations at the plasma membrane (either apical or basolateral). Thus, individual protein molecules change location and protein-protein interaction partners several times before arriving at their final cellular destination. Other aspects of changing location and function include protein turnover, receptor recycling and degradation as well as protein localization change depending on stimulus. Thus, a protein may be found in several different cellular compartments at the same time. Furthermore, proteins may change their location depending on the cell cycle phase, circadian rhythm or upon various stressors and may become concentrated in exceptional subcellular compartments that constitute itself through liquid-liquid phase transitions.
Such details of localization need to be considered when designing spatial proteomics approaches and validation experiments. Several newer approaches try to address these challenges and improve the confidence in protein localization studies to complement more traditional approaches.
Methods in spatial proteomics
Several methods are available to monitor protein localization in cells and tissues, the most prominent being imaging methods. A recent large scale study used immunofluorescence to localize 12,073 proteins to 33 subcellular structures comprising 13 human organelles [1], (http://www.proteinatlas.org). Other large-scale studies successfully localized several hundreds of proteins that were directly labeled with fluorophores [2,3]. Still, the function and localization of approximately 50% of the human proteome has not been characterized in detail and is annotated solely based on structural or sequence features of the protein or by inference from other species, some of them quite distant from human.
Combining proteomic technologies with biochemical purification of subcellular compartments enabled enormous synergies and opportunities to characterize the content of organelles and, thus spatial proteomics was developed as a global method to characterize the subcellular location of proteins in an unbiased way to provide functional context [4-6]. Classical spatial proteomics approaches include the purification of organelles by subcellular fractionation. Such approaches have produced some clinically relevant results. The MitoCarta studies, which aim to characterize the mouse and human mitochondrial proteome, found several proteins in mitochondria that were not previously annotated as mitochondrial, but that play a role in the activity and assembly of respiratory chain complex I. Mutation of one of the newly localized proteins, C8orf38, was subsequently found to be causative for Inherited Complex I deficiency [7,8]. Spatial proteomics studies have also helped to detect developmental differences in protein location. McClatchy et al., for example, quantitated synaptosomal and mitochondrial proteins at different developmental time points in different locations of the rat brain and found surprising differences in protein expression and protein abundances [9]. Amin et al., used the INTACT (isolation of nuclei tagged in specific cell types) method to enrich the nuclei from heart cells of Xenopus [10]. This strategy identified 12 human protein orthologs whose genes have been identified to be important in congenital heart disease and thus important in heart development. In addition, spatial proteomics has helped to define the proteome of the post synaptic density (PSD) of neurons. Cell biological techniques for enriching PSD are quite well established and thus there have been a number of studies to identify the proteins present in PSD [11,12], many of which are involved in neurodegenerative and psychological diseases. The proteome of large, stable structures such as chromatin [13,14] or cilia have also been performed and led to the identification proteins linked to ciliopathies [15].
Despite the success of subcellular fractionation approaches, several difficulties remain, such as the co-purification of different organelles with similar biophysical properties or the co-purification of cytoplasmic localized proteins that associate with or are in close proximity to the purified organelle. Other organelles and subcellular structures also do not lend themselves as easily to purification as nuclei and mitochondria or synapses. For example, more filamentous structures such as the Endoplasmic Reticulum (ER) that are intricately connected to other compartments, are generally “shredded” during isolation, generating microsomes, which are very difficult to separate from other cellular vesicles.
A strategy called Protein Correlation Profiling (PCP) was developed to better assess the enrichment of organelles using standard enrichment methods like density centrifugation [16]. The method follows the “least common denominator” approach and uses mass spectrometry to identify proteins of which the relative abundance maximally correlates with the band of interest in the density gradient that contains a marker protein for the organelle of interest. A similar strategy was used to assign proteins to the nuclear envelope by using subtractive analysis based on spectral counting [17]. As the nuclear envelope is contiguous with the ER, sorting out which proteins belong to the ER or the nuclear envelope is difficult. Therefore, Schirmer et al., first isolated the ER enriched microsomal fraction and then enriched the nuclear envelope using several different strategies. The proteins were then identified and quantified using mass spectrometry and this information was used to subtract away from the nuclear envelope those proteins likely to be from the ER. Interestingly, this strategy identified 67 ORFs as novel nuclear envelope localized proteins, of which 23 mapped to genomic regions associated with a variety of dystrophies. The concepts of the PCP method were subsequently further advanced by incorporating more accurate and robust quantification by labeling proteins in density gradient bands with isotope tags (LOPIT), which offers the ability to multiplex up to 11 fractions for simultaneous analysis of peptide abundances [18,19]. Protein abundance is therefore co-localized to the enrichment distribution with well-annotated marker proteins in a manner similar to PCP. However, the approach can only determine proteins that are significantly enriched in one compartment over the remainder and thus any protein that co-localizes to different compartments will be missed in this protein centric quantification approach.
Interactome studies for spatial proteomics
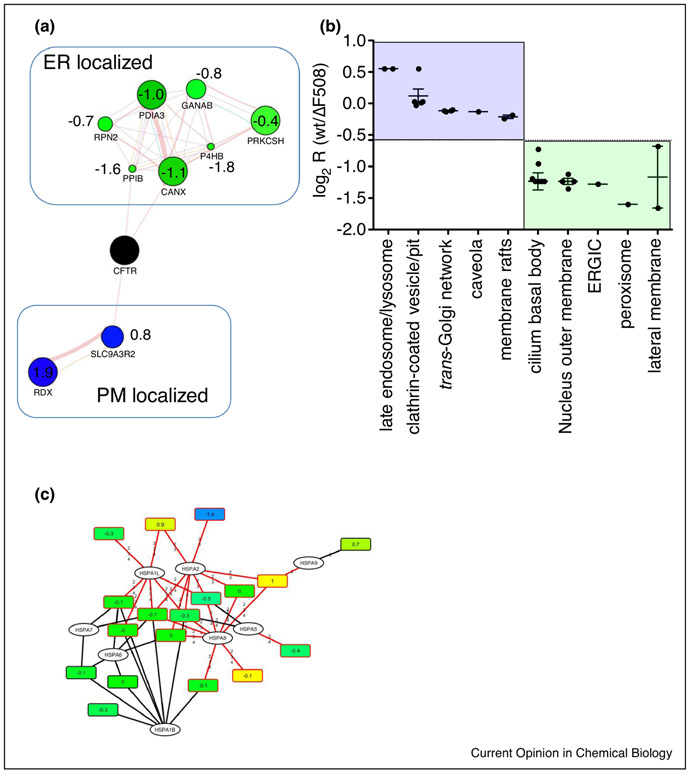
Interactome studies can provide insights into the life-cycle and the subcellular locations of proteins. For example, a protein known to localize to a specific compartment can be used to enrich for interacting proteins in the same cellular compartment. A very large-scale study of the human interactome has been conducted and is known as Bioplex [27,28]. The Bioplex interactome has designated several uncharacterized proteins as members of known complexes with specific functions and localizations, and led to subcellular localization predictions for a few other hundred proteins based on their first and second degree interactors. Through this “guilt-by-association” strategy, several proteins were discovered that are in complexes associated with disease. The interactome of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR), shed light onto the complicated life-cycle of membrane proteins and revealed protein interaction partners at different subcellular locations [29]. Interactome approaches are also useful to better understand defects associated with aberrant protein localization, often based on misfolding of the respective protein. Comparison with the interactome of the misfolded ΔF508 CFTR variant, which causes Cystic Fibrosis in more than 90% of CF patients, revealed for example different cellular location as well as differences in trafficking of the wt and misfolded mutant ΔF508 CFTR proteins, using well-annotated first or second-degree interactors in a strategy similar to “guilt-by-association”. The results reflected the well-established fact that the misfolded ΔF508 CFTR mutant is mainly localized to the ER and early secretory pathway, whereas wt CFTR is also localized to the plasma membrane (Figure 2A). GO subcellular location enrichment analysis revealed similar results – proteins interacting mainly with wt CFTR were enriched in plasma membrane associated locations, whereas proteins interacting with ΔF508 CFTR were enriched in the ER-Golgi intermediate compartment (ERGIC) or the lateral plasma membrane. This latter analysis did not reveal as clear a picture as the first strategy though - mainly because of ambiguous annotation of proteins such as kinectin, which is involved in vesicle mediated transport, but is annotated as localized to the cilium basal body (Figure 2B). It is clear that by looking at the interactome by these different approaches, different interactions occurring in different compartments of the cell can be discovered and that this information can be used to gain clinical insight. CF-therapies for example have aimed to correct the misfolding defect and move the ΔF508 CFTR protein from the ER to the plasma membrane. Since protein isoforms as well as protein homologues often share significant sequence similarity, but may be localized in different cellular locations and have different functions, smart software that can differentiate between these proteoforms in quantitative bottom-up proteomics experiments is also needed to address differential subcellular location in health and disease states.
Figure 2:
A. Quantitative analysis of CFTR Co-IPs with subcellular marker proteins in a guilt-by-association approach reveals different subcellular locations for wt CFTR and the Cystic Fibrosis causing ΔF508 CFTR mutant. Blue nodes, enhanced interaction with wt CFTR; green nodes, reduced interaction with wt CFTR. Values represent log2 ratios (wt/ΔF508 CFTR). B. GO analysis using Uniprot subcellular location also reveals different subcellular locations for wt and ΔF508 CFTR, with interactors in subcellular locations associated with the plasma membrane enriched for wt CFTR. C. Bipartite protein-peptide network shows enhanced interaction of ΔF508 CFTR with HSPA2, a chaperone which initiates proteolytic processing of misfolded proteins.
New software such as ProteinClusterQuant (PCQ) can address this issue by assembling bipartite peptide-protein networks and using the quantitative peptide information to tease out the regulation of proteoforms [30]. If used to quantify different heat shock proteins, some of which are located in the ER (such as endoplasmin and HSPA5) and some of which are located in the cytoplasm, more detailed insight into the quantity and interaction of a protein at different subcellular locations can be gained. An analysis of the heat shock proteoform quantities within the CFTR interactome dataset for example revealed increased interaction of the ΔF508 CFTR mutant with HSPA2, which is implicated in the activation of proteolysis of misfolded proteins - a result, which would likely have been missed by conventional quantitative analysis, as HSPA2 shares peptides with other heat shock proteins that were differentially regulated (see Figure 2C).
Two new methods, APEX and BioID, localize proteins within close proximity (~150 nm) to epitope tagged proteins and can be used to determine the subcellular location of neighboring proteins [31-35]. In both cases, a protein of known subcellular localization is genetically fused with a protein designed to chemically label nearby proteins. The appended protein moiety is either an engineered ascorbate peroxidase (APEX) or the BirA protein which is a promiscuous bacterial biotin ligase (BioID). In APEX, phenoxyl free radicals are generated from hydrogen peroxide and biotin phenol. The phenoxyl radical labels proteins that are within the diffusion limit of its free radical state in cells. Free radicals are rapidly quenched in aqueous solution and do not diffuse far and this turn proximity labeling with APEX in a fast and location-sensitive method. The biotinylation reaction by the biotin ligase BirA is slower and requires high intracellular concentrations of biotin for a prolonged period of time. A molecular engineered version of BirA labels within 10 min and has been named TurboID (in press at Nature Biotech). Recent studies have therefore used APEX to map proteins in the intermembrane space of human mitochondria and characterized the proteome of the synaptic cleft of neurons with APEX [34,36] whereas BioID mapped proteins in subcellular structures like cilia [37]. Carefully crafted experimental setups mitigate issues like background noise due to endogenously biotinylated proteins as well as a potential mis-localization of the tagged proteins due to the inevitable change in amino acid sequence. The studies cited verified and complemented newly identified subcellular localizations of proteins with orthogonal approaches such as immunofluorescence in conjunction with high-resolution confocal or super resolution microscopy.
The role of bioinformatics in spatial proteomics
Predicting subcellular localizations based on amino acid sequence motifs was introduced more than three decades ago with the study of signal peptides [42]. One of the first methods to predict multiple localizations was PSORT [43] which has been extended over the last years to support the prediction of sequences from a larger variety of organisms [44,45]. While some methods focus on predicting the presence of a protein in one single, specific subcellular compartment ([46], [47]) others can predict protein localization in a few [48] or in a wide range of localizations [49-52]. Most software use different sources of information, such as the sequence composition (the amino acid distribution and their physicochemical properties or sequence motifs annotations [53-58]), phylogenetic information, and gene ontology annotations [49,50,59,60]. Other tools combine different sources of information or multiple predictor algorithms [61,62].
Other computational tools use the available MS–based information to obtain more confident insight on the localization of proteins. One of these tools is pRoloc (Bioconductor R package, www.bioconductor.org)[63], which uses quantitative mass-spectrometry data to create organelle-specific profiles using known organelle markers and matches these to the distribution profiles of the unknown proteins. This software implements several classification methods previously used in the field such as: partial least-square discriminant analysis [64], SVMs [65], random forest [66], neural networks [67] and naïve Bayes [68], as well as novel algorithms such as PerTurbo [69].
More integrative and comprehensive tools may include additional annotations and information such as protein structure and protein pathways. In this regard, it is important to consider the nature of the already known annotations to infer the localization of other proteins. Protein localization annotations, such as gene ontology (GO) terms, can be weighted according to their discriminative power in a training dataset [60] or by origin and metadata like whether the localization is in silico or experimentally derived or whether the localization was described in a particular tissue or for a specific cell type. Databases should integrate this metadata so that it can be used properly by external tools.
While databases like UniprotKB often correctly annotate proteins with multiple cellular localizations, and the origin of the annotation is usually present, additional information upon protein life cycle, enzymatic activity, or even experimental design is missing which makes it difficult to filter and interpret spatially resolved annotations of proteins. Linking spatial information to different protein-annotated features such as pathways, functions or 3D conformations will aid to resolve and better understand ambiguities in spatial localization. For example to spatially resolve the initial event of protein misfolding in sporadic neurodegenerative diseases is of high importance but challenging. The self-templating ability of amyloid-b peptide and tau protein that are implicated in Alzheimer disease amplifies protein aggregation at one cellular location but might obscure the location of initial protein misfolding. In sporadic Alzheimer disease the underlying conjuncture for a misfolding of both proteins remains unclear because both protein aggregates are observed as temporally as well as potentially spatially separated.
Summary statement
Taken together, if designed and validated carefully and complemented by appropriate bioinformatics solutions, spatial proteomics is an invaluable tool to identify aberrant location of proteins in human diseases, which is often the first step to understand underlying molecular mechanisms of disease to enable new therapeutic avenues.
Figure 1:
Different approaches used in spatial proteomics reveal different aspects of subcellular organization.
Acknowledgments.
The authors would like to acknowledge funding from the NIH: P41 GM103533, R01 MH067880, 1R56AG057459, R01 EY011261, U01-EY027261, 1R33CA212973, R01 HL131697, 5U01 DA040709, R01 MH100175.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. : A subcellular map of the human proteome. Science 2017, 356.The most comprehensive map of the subcellular human proteome using flourescence labeled antibodies.
- 2.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al. : Subcellular localization of the yeast proteome. Genes Dev 2002, 16:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK: Global analysis of protein localization in budding yeast. Nature 2003, 425:686–691. [DOI] [PubMed] [Google Scholar]
- 4.Bell AW, Nilsson T, Kearney RE, Bergeron JJ: The protein microscope: incorporating mass spectrometry into cell biology. Nat Methods 2007, 4:783–784. [DOI] [PubMed] [Google Scholar]
- 5.Brunet S, Thibault P, Gagnon E, Kearney P, Bergeron JJ, Desjardins M: Organelle proteomics: looking at less to see more. Trends Cell Biol 2003, 13:629–638. [DOI] [PubMed] [Google Scholar]
- 6.Yates JR, Gilchrist A, Howell KE, Bergeron JJM: Proteomics of organelles and large cellular structures. Nature Reviews Molecular Cell Biology 2005, 6:702–714. [DOI] [PubMed] [Google Scholar]
- 7.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. : A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo SE, Clauser KR, Mootha VK: MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016, 44:D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClatchy D, Liao L, Park SK, Lee JH, Yates JR: Dynamics of Subcellular Proteomes During Brain Development. J Proteome Res 2012.A quantitative analysis of proteins localized to the synaptosome and mitochondria across developmental timepoints of brain development.
- 10.Amin NM, Greco TM, Kuchenbrod LM, Rigney MM, Chung MI, Wallingford JB, Cristea IM, Conlon FL: Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (INTACT). Development 2014, 141:962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorokina O, Sorokin A, Armstrong JD: Towards a quantitative model of the post-synaptic proteome. Mol Biosyst 2011, 7:2813–2823. [DOI] [PubMed] [Google Scholar]
- 12.Lowenthal MS, Markey SP, Dosemeci A: Quantitative mass spectrometry measurements reveal stoichiometry of principal postsynaptic density proteins. J Proteome Res 2015, 14:2528–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sureka R, Wadhwa R, Thakur SS, Pathak RU, Mishra RK: Comparison of Nuclear Matrix and Mitotic Chromosome Scaffold proteins in Drosophila S2 cells - Transmission of hallmarks of nuclear organization through mitosis. Mol Cell Proteomics 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A: Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 2014, 16:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa H, Thompson J, Yates JR, Marshall WF: Proteomic analysis of mammalian primary cilia. Curr Biol 2012, 22:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M: Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003, 426:570–574.An approach establishing the use of abundance correlation between organelle marker proteins and resident proteins.
- 17.Schirmer EC, Florens L, Guan TL, Yates JR, Gerace L: Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003, 301:1380–1382. [DOI] [PubMed] [Google Scholar]
- 18.Mulvey CM, Breckels LM, Geladaki A, Britovsek NK, Nightingale DJH, Christoforou A, Elzek M, Deery MJ, Gatto L, Lilley KS: Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat Protoc 2017, 12:1110–1135.The use of quantitative isobaric tagging to assess proteins resident in organelles.
- 19.Lilley KS, Dunkley TP: Determination of genuine residents of plant endomembrane organelles using isotope tagging and multivariate statistics. Methods Mol Biol 2008, 432:373–387. [DOI] [PubMed] [Google Scholar]
- 20.Levine AJ: p53, the cellular gatekeeper for growth and division. Cell 1997, 88:323–331. [DOI] [PubMed] [Google Scholar]
- 21.Verma IM: Nuclear factor (NF)-kappaB proteins: therapeutic targets. Ann Rheum Dis 2004, 63 Suppl 2:ii57–ii61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momand J, Zambetti GP, Olson DC, George D, Levine AJ: The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992, 69:1237–1245. [DOI] [PubMed] [Google Scholar]
- 23.Kubbutat MH, Jones SN, Vousden KH: Regulation of p53 stability by Mdm2. Nature 1997, 387:299–303. [DOI] [PubMed] [Google Scholar]
- 24.Haupt Y, Maya R, Kazaz A, Oren M: Mdm2 promotes the rapid degradation of p53. Nature 1997, 387:296–299. [DOI] [PubMed] [Google Scholar]
- 25.Lane DP: Cancer. p53, guardian of the genome. Nature 1992, 358:15–16. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S, May MJ, Kopp EB: NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998, 16:225–260. [DOI] [PubMed] [Google Scholar]
- 27.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. : The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162:425–440.A large scale protein-protein interaction study that could be useful to assess protein localizations.
- 28.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. : Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pankow S, Bamberger C, Calzolari D, Martinez-Bartolome S, Lavallee-Adam M, Balch WE, Yates JR 3rd: F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature 2015.A protein-protein interaction study of CFTR that tracks the biogenysis of CFTR through the secretory system that assesses protein localization.
- 30.Bamberger C, Martinez-Bartolome S, Montgomery M, Pankow S, Hulleman JD, Kelly JW, Yates JR 3rd: Deducing the presence of proteins and proteoforms in quantitative proteomics. Nat Commun 2018, 9:2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ: Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 2014, 111:E2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DI, Roux KJ: Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 2016, 26:804–817.A method to assess protein localization through a biotin based proximity assay.
- 33.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY: Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 2012, 30:1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY: Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell 2014, 55:332–341.A method to assess protein localization using a proximity assay using a peroxidase to create a phenoxy-biotin freee radical to label proteins.
- 35.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY: Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 2015, 12:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. : Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 2016, 166:1295–1307 e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. : A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 2015, 163:1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falahzadeh K, Banaei-Esfahani A, Shahhoseini M: The potential roles of actin in the nucleus. Cell J 2015, 17:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baarlink C, Wang H, Grosse R: Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013, 340:864–867. [DOI] [PubMed] [Google Scholar]
- 40.Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J: Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 2018, 559:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caridi CP, D'Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, et al. : Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Heijne G: A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 1986, 14:4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakai K, Kanehisa M: Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 1991, 11:95–110. [DOI] [PubMed] [Google Scholar]
- 44.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, et al. : PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardy JL, Spencer C, Wang K, Ester M, Tusnady GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, et al. : PSORT-B: Improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res 2003, 31:3613–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cokol M, Nair R, Rost B: Finding nuclear localization signals. EMBO Rep 2000, 1:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savojardo C, Martelli PL, Fariselli P, Casadio R: TPpred2: improving the prediction of mitochondrial targeting peptide cleavage sites by exploiting sequence motifs. Bioinformatics 2014, 30:2973–2974. [DOI] [PubMed] [Google Scholar]
- 48.Emanuelsson O, Nielsen H, Brunak S, von Heijne G: Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 2000, 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- 49.Blum T, Briesemeister S, Kohlbacher O: MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 2009, 10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briesemeister S, Blum T, Brady S, Lam Y, Kohlbacher O, Shatkay H: SherLoc2: a high-accuracy hybrid method for predicting subcellular localization of proteins. J Proteome Res 2009, 8:5363–5366. [DOI] [PubMed] [Google Scholar]
- 51.Goldberg T, Hamp T, Rost B: LocTree2 predicts localization for all domains of life. Bioinformatics 2012, 28:i458–i465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu CS, Chen YC, Lu CH, Hwang JK: Prediction of protein subcellular localization. Proteins 2006, 64:643–651. [DOI] [PubMed] [Google Scholar]
- 53.Almagro Armenteros JJ, Sonderby CK, Sonderby SK, Nielsen H, Winther O: DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33:3387–3395. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Duan X: Prediction of protein subcellular localization with oversampling approach and Chou's general PseAAC. J Theor Biol 2018, 437:239–250. [DOI] [PubMed] [Google Scholar]
- 55.Wang LY, Wang D, Chen YH: Prediction of protein subcellular multisite localization using a new feature extraction method. Genet Mol Res 2016, 15. [DOI] [PubMed] [Google Scholar]
- 56.Arango-Argoty GA, Jaramillo-Garzon JA, Rothlisberger S, Castellanos-Dominguez CG: Prediction of protein subcellular localization based on variable-length motifs detection and dissimilarity based classification. Conf Proc IEEE Eng Med Biol Soc 2011, 2011:945–948. [DOI] [PubMed] [Google Scholar]
- 57.Shi R, Xu C: Prediction of rat protein subcellular localization with pseudo amino acid composition based on multiple sequential features. Protein Pept Lett 2011, 18:625–633. [DOI] [PubMed] [Google Scholar]
- 58.Huang WL, Tung CW, Huang HL, Hwang SF, Ho SY: ProLoc: prediction of protein subnuclear localization using SVM with automatic selection from physicochemical composition features. Biosystems 2007, 90:573–581. [DOI] [PubMed] [Google Scholar]
- 59.Zhou H, Yang Y, Shen HB: Hum-mPLoc 3.0: prediction enhancement of human protein subcellular localization through modeling the hidden correlations of gene ontology and functional domain features. Bioinformatics 2017, 33:843–853. [DOI] [PubMed] [Google Scholar]
- 60.Chi SM, Nam D: WegoLoc: accurate prediction of protein subcellular localization using weighted Gene Ontology terms. Bioinformatics 2012, 28:1028–1030. [DOI] [PubMed] [Google Scholar]
- 61.Shen YQ, Burger G: 'Unite and conquer': enhanced prediction of protein subcellular localization by integrating multiple specialized tools. BMC Bioinformatics 2007, 8:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salvatore M, Warholm P, Shu N, Basile W, Elofsson A: SubCons: a new ensemble method for improved human subcellular localization predictions. Bioinformatics 2017, 33:2464–2470. [DOI] [PubMed] [Google Scholar]
- 63.Gatto L, Breckels LM, Wieczorek S, Burger T, Lilley KS: Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics 2014, 30:1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. : Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A 2006, 103:6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trotter MW, Sadowski PG, Dunkley TP, Groen AJ, Lilley KS: Improved sub-cellular resolution via simultaneous analysis of organelle proteomics data across varied experimental conditions. Proteomics 2010, 10:4213–4219. [DOI] [PubMed] [Google Scholar]
- 66.Ohta S, Bukowski-Wills JC, Sanchez-Pulido L, Alves Fde L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, et al. : The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 2010, 142:810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugiere S, Hippler M, Ferro M, Bruley C, Peltier G, et al. : PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 2012, 29:3625–3639. [DOI] [PubMed] [Google Scholar]
- 68.Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TP, Munro S, Lilley KS, Dupree P: Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 2012, 160:1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Courty N, Burger T, Laurent J: Perturbo: a new classification algorithm based on the spectrum perturbations of the laplace-beltrami operator In European Conference ECML/PKDD; Athens, Edited by Gunopulos D, Hofmann T, Malerba D, Vazirgiannis M: Springer: 2011:359–374. [Google Scholar]