Short abstract
Population growth, climate change, and dwindling finite resources are amongst the major challenges which are facing the planet. Requirements for food, materials, water, and energy will soon exceed capacity. Green biotechnology, fueled by recent plant synthetic biology breakthroughs, may offer solutions. This review summarizes current progress towards robust and predictable engineering of plants. I then discuss applications from the lab and field, with a focus on bioenergy, biomaterials, and medicine.
Impact statement
The plant synthetic biology field has exploded in the last five years, in part driven by techniques such as CRISPR and cheap DNA synthesis. This review summarizes the current state of research in plant synthetic biology, and how it is being applied to two topics: renewable fuels and chemicals, and medicine.
Keywords: Biofuels, gene editing, genetic engineering, plant produced vaccines, plant synthetic biology, renewable chemicals
Introduction
The human population has grown ∼9-fold since the beginning of the industrial revolution,1 requiring increased exploitation of the Earth’s natural resources. Major advances in agronomic practices and plant breeding (the Green Revolution) resulted in vastly increased crop yields but needed large inputs of irrigated water, synthetic nitrogen fertilizers, and other nutrients. These practices are energy intensive and lead to environmental problems, including salinization of soils, erosion, and nitrification of water bodies.2 Further increases in crop yields are essential to feed our future population, which is predicted to exceed 11 billion by 2100.3 Moreover, as fossil fuel reserves are depleted, the importance of agriculture for production of non-food products such as fuels, fibers, and platform chemicals will only increase. Boosting crop yields to meet these demands must be achieved (i) by maintaining or improving the productivity of land currently in cultivation to avoid further deforestation and (ii) by reducing inputs of water and nutrients.
To achieve this, a Green Revolution version 2.0 is necessary.4 In this vision, new crops are precision engineered for both optimal yield and environmental interactions. Plants are not considered in isolation but, for example, in combination with their microbiome and soil. The goals of the synthetic biology discipline: robust, predictable and rapid engineering of biology align well with this vision.5 To date, much synthetic biology research has focused on microbial engineering (with the vast bulk of that being the model organism Escherichia coli). Plant synthetic biology has necessarily been more limited and has faced different challenges, such as life-cycle length.
In this review, I will briefly define synthetic biology, discuss the history of plant engineering, and summarize the current state of the field in plant science. I will then discuss two promising applications of plant synthetic biology: production of renewable fuels and chemicals, and therapeutics. While increased yields of food crops, as well as the production of foods with increased nutritional value is clearly important, this will not be covered here due to space limitations, and because it has been recently reviewed elsewhere, e.g. Tyagi et al.6
Defining synthetic biology
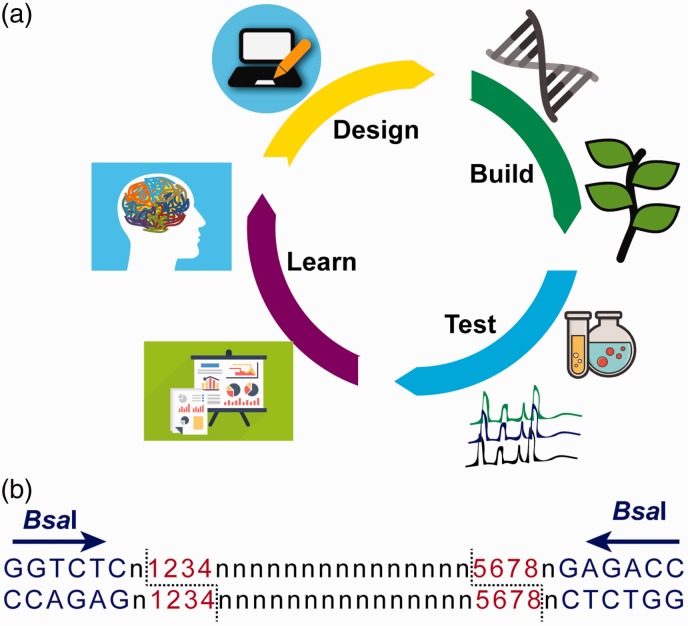
What is synthetic biology? While many definitions exist, that from the Engineering Biology Research Consortium (EBRC; www.ebrc.org) is helpful: “Synthetic biology aims to make biology easier to engineer… . It can be thought of as a biology-based toolkit that uses abstraction, standardization, and automated construction to change how we build biological systems and expand the range of possible products.” The application of engineering principles to biology requires us to be able to move from a sequenced genome to a predictable systems output, which is very challenging.7 The engineering concept of the design–build–test–learn (DBTL) cycle can underpin the application of synthetic biology to organismal engineering (Figure 1(a)).8,9 Synthetic biology aims to speed up the rate at which we move round the DBTL cycle, and, combined with systems biology, encourages us to explicitly learn from each experiment and integrate that knowledge into the next iteration. Engineers rely on interchangeable components and this concept has also been adopted by synthetic biologists. In microbial synthetic biology, this was pioneered by the BioBrick standards (https://biobricks.org/) and more recently a plant standard has been developed based on Type IIS restriction endonuclease assembly10 (Figure 1(b)).
Figure 1.
Plant synthetic biology concepts. (a) The design–build–test–learn (DBTL) cycle underpins experimental design and analysis in synthetic biology. Modified from Petzold et al.9 In the Design phase, the problem is described, the pathway selected, and the parts designed or selected. In the Build phase, the parts are synthesized, assembled, and transformed into the host organism of choice. During the Test phase, the modified organism is analyzed and data collected. In the Learn phase, the data from the Test phase are analyzed, and used to inform the next Design stage. (b) Type IIS restriction site-based DNA parts for meeting plant synthetic biology community standards.10 All DNA parts should be sequenced without a BsaI restriction site (at a minimum – BpiI and BsmBI should be avoided). Standard parts are flanked by convergent BsaI cut sites. The plasmid backbones should be free of BsaI/BpiI/BsmBI sites, and should not contain resistance genes for ampicillin/carbenicillin, or kanamycin, since these are commonly used in assembly. (A color version of this figure is available in the online journal.)
How does synthetic biology differ from conventional genetic engineering? It can be considered in some senses a continuum. Whereas genetic engineering may manipulate the expression of a single gene, synthetic biology aims to go much further – for example by the introduction of a complete metabolic pathway under tight regulatory control. While the design, synthesis and expression of the DNA parts are important, it is also the incorporation of systems data (such as transcriptomics, proteomics, metabolomics) and mathematical modeling into subsequent experiments that really sets synthetic biology apart. The eventual goal is to possess biological blueprints such that we can understand and modify an organism, as an engineer does a car. For plants, these latter goals are still very distant. However, progress has been made, and it is that which will be discussed in this review.
A brief history of plant engineering
Domestication of plants by artificial selection is central to human history.11 It began as saving seed from plants with desirable traits, and using them for subsequent plantings. It then developed into deliberate breeding, as evidenced by the work of Gregor Mendel12 and his laws of inheritance. The discovery of heterosis in 1908 (hybrid vigor), in which the offspring of certain crosses perform better than either parents, and the cytoplasmic male sterility (CMS) trait in 1933 that enables the facile production of hybrids led to great advances in yield.13 The identification of wheat cultivars with traits such as short-stature, which makes plants less prone to lodging (falling over as a result of wind-damage), led to what is now referred to as the “Green Revolution.” Norman Borlaug, an American agronomist, was awarded the Nobel Peace Prize in 1970 for using these techniques to increase wheat yields in India and Pakistan by ∼60%.
Progress in the field of genetics in the first half of 20th century suggested the promise of further yield increases. Mutagenesis (using X-rays, radiation, and chemical mutagens) was routinely employed to try to produce new commercial plant varieties. For example, much of the world’s mint oil is produced from a disease-resistant cultivar “Todd’s Mitcham” developed from a gamma irradiation breeding program started at Brookhaven National Laboratory in 1955.14 In the long-term, however, these methods have been most successful for producing new ornamental plants, such as petunias15 and cherry blossom16 (Figure 2), and the number of agriculturally important traits identified this way is relatively low, as compared to conventional breeding.
Figure 2.
Some examples of radiation plant breeding. (A) Nishina Otome cherry blossom tree (R) and the parent (L), created at RIKEN. In the absence of vernalization (exposure to winter temperatures) it is still able to flower, and will produce flowers in all four seasons. If exposed to winter temperatures, it produces three-times more flowers than the parent (B) The commercial cultivar Todd’s Mitcham mint (Mentha x piperita), which was bred at Brookhaven National Lab and has resistance to Verticillium wilt. (A) RIKEN Nishina Center for Accelerator-Based Science (B) Colonial Creek Farm (www.colonialcreekfarm.com). (A color version of this figure is available in the online journal.)
The advent of transgenic methods in the 1980s was a step-change. It allowed the introduction of new traits that may not exist within the species’ gene pool, such as resistance to a specific disease. Traditional breeding simply cannot do this. Plant science led transgenic research because of the observation that the rhizobial bacteria Agrobacterium tumefaciens can induce crown gall tumors in plants, and that this cellular proliferation was due to the presence of genes transferred from the Agrobacterium Ti (tumor inducing) plasmid and integrated into the plant genome.17 The DNA transferred (T-DNA) is flanked by defined left- and right-borders. Researchers showed that foreign genes placed between these borders could be integrated into the plant genomes, and disarmed Ti plasmids, that transferred DNA without causing tumor formation, were developed.18,19 T-DNA transformation rapidly became a common tool for plant research.20
Unfortunately, not all plants are equally susceptible to Agrobacterium transformation (including some major crop species). For example, only a few cultivars of barley can be transformed this way, and any modifications are then introduced into elite cultivars via multiple generations of back-crossing.21 Biolistic transformation (the gene gun) was developed as an alternative transformation method.22 Heavy metal particles (usually gold or tungsten) coated with DNA are fired at cells, and some of the payload integrates into the genome. This method has been used to produce many commercialized genetically modified crops, such as insect-resistant Bt corn and cotton, which expresses endotoxins derived from the soil bacterium Bacillus thuringiensis.23
However, these transgenesis techniques have limitations. For example, it is difficult to control where in the genome the transgene integrates, or the copy number of the introduced DNA. This can lead to variation in gene expression, gene-silencing, or inter-generational instability, and this unpredictably has hindered wide-spread commercialization beyond a few traits. In addition, there has been much public concern over the dangers of genetically modified organisms (GMOs), and, despite the lack of scientific evidence to support this view,24 it has limited applications of these technologies.
Gene editing
While the transgenic techniques described above have been used in early plant synthetic biology approaches (see below), the development of precise gene editing technologies that can not only precisely alter a DNA sequence, but also gene expression levels, has been fundamental to recent progress.25 Importantly, these methods do not necessarily require the introduction of foreign DNA, which can have regulatory advantages.26 Here, site-specific endonucleases are introduced into the cell. The resulting DNA double-strand break is then repaired using methods that exist in the cell: either non homologous end joining (NHEJ) or homology-directed repair (HDR).27 NHEJ is quick and dirty: efficient but error prone. HDR is slower but more precise. NHEJ usually results in non-functional genes, which is important for reverse genetics experiments, as well as some agricultural traits, e.g. powdery mildew resistance in wheat.28 HDR, by comparison, can be also be used for the introduction of foreign DNA.29
Three different endonuclease systems have been successfully deployed in plant systems: zinc fingered nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced palindromic repeats (CRISPR) – CRIPSR-Associated (Cas) proteins such as CRIPSR-Cas9. See Table 1 for a comparison of the three methods. CRISPR-based systems in particular are expanding rapidly, with new functions added via gene-fusion with the endonuclease, such as gene repression30 and, as has been shown in mammalian cells, epigenetic modification.31
Table 1.
Comparison of three gene editing techniques that have been successfully used in plant systems.
| Mechanism | Pros | Cons | |
|---|---|---|---|
| ZFN |
|
|
|
| TALEN |
|
|
|
| CRISPR-Cas9 |
|
|
|
ZFN: zinc fingered nuclease; TALEN: transcription activator-like effector nuclease;
Gene stacking and whole pathway engineering
Plant transformation is still a relatively slow and laborious process. Recent advances in which whole metabolic pathways can be introduced with a single transformation event hold great promise.32,33 COSTREL (combinatorial supertransformation of transplastomic recipient lines) takes advantage of both plasmid and nuclear transformation.33 The core metabolic pathway is transformed into the plasmid, and then these plants are then co-transformed with a cocktail of accessory protein plasmids. The resulting offspring is then screened for the abundance of the desired product. In the J-Stack method, yeast homologous recombination, combined with a toolbox of compatible synthetic DNA promoters and terminators, can be used to rapidly assemble pathways for introduction in a single transformation step, as was demonstrated both for in planta production of a potential biofuel and antibiotic.32 Both methods allow a relatively easy and rapid way to shuffle genes in a metabolic pathway, and to test different combinations. This can be essential to alleviate metabolic bottlenecks or identify potential co-factors.
Applications of plant synthetic biology
Currently, most plant synthetic biology approaches have been applied to discovery research, and this is likely to continue in the short term. Plant research can be comparatively slow, and synthetic biology provides a method to accelerate this.34 In particular, as the cost of DNA synthesis has plummeted, the use of synthetic genes (e.g. codon optimized for expression in different species) is now within reach of the average research laboratory. However, in the future, it is likely that plant synthetic biology will become increasingly important for commercial agriculture, as will be discussed below.
Biofuels and bioproducts
Almost all transportation infrastructures in the US rely on petroleum-derived hydrocarbons. Not only are fossil fuels non-renewable, but their usage contributes to climate change. Of course, petroleum is not only used for fuels. A vast range of chemicals and materials are also produced including plastics, paints, and fabrics. The reliance on petroleum has resulted in an economic, environmental, and strategic push to develop renewable alternatives.35 This includes using plant lignocellulosic biomass (i.e. the plant cell wall) as a carbon feedstock, and has coincided with, and in many cases has driven forward, advances in plant synthetic biology. This is because the economics of producing fuels and chemicals in a sustainable manner from plant biomass is challenging.
The cheapest and simplest method of producing biofuels is to convert corn starch or sugar cane sucrose into ethanol using yeast fermentation. However, this can have knock-on consequences for food prices36 and land use.35 Additionally, food crops such as corn require high inputs of water and fertilizer to produce the high yields that modern farming expects. Many fertilizers are derived from non-renewable resources, and their production contributes to greenhouse gas emissions. They also add to production costs. While agricultural lignocellulosic residues such as corn stover can be (and are) used, this alone will not be sufficient to significantly displace fossil fuels. Therefore, dedicated bioenergy crops, such as switchgrass, poplar, and biomass sorghum have been proposed.35,37
There are many barriers to the widespread implementation of dedicated biomass crops. The majority of these species have not been domesticated. Extensive improvements are required to optimize them for deconstruction and conversion, as well as making them sustainable to grow. This includes tolerance to biotic and abiotic stress, and the ability to produce high yields even in poor quality marginal soils without inputs of fertilizer (which is often non-renewable and requires fossil fuels to produce). Conventional breeding methods alone simply cannot achieve the scale of change required, at least within a reasonable timeframe. Synthetic biology has accelerated both the fundamental research required to enable predictive engineering by describing complex metabolisms, as well as the production of designed cultivars. A few examples will be highlighted here.
Lignin is a complex polyphenolic network found in some secondary plant cell walls, and it forms around 30% dry weight of biomass. While it is important for providing structural integrity to some cell types, such as the water-conducting xylem vessels, it is also challenging to break down and process as part of lignocellulosic biofuel production. The complexity arises from its chemistry. Lignin is made primarily from three monolignol units: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, which are synthesized in the cytosol, and then transported to the apoplast. Monolignol radicals are generated, and these are then incorporated by combinatorial radical coupling into the lignin polymer, forming p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits.38 The result is a highly heterogeneous polymer. Plant breeding programs have succeeded in developing lower-lignin plants, e.g. brown midrib (bm or bmr), which have up to ∼20% reduction in lignin compared to wild type.39 However, if the total lignin content is decreased much more than this, the plants will show growth defects, including lodging or stunting. The lignin biosynthetic pathway is well characterized, and synthetic biology has provided the opportunity to make “designer lignin” – lignin which is more amenable to biotechnological applications while not affecting crop yields. Multiple strategies have been implemented.
One approach has been to introduce more labile bonds into the lignin. For example, the expression of a ferulolyl-CoA:monolignol transferase (FMT) to produce ferulated monolignol subunits, which can then be incorporated into the lignin polymer to produce so-called Zip-lignin™.40 To do this, the authors searched for suitable FMT enzymes in nature, and identified an FMT from Angelica sinensis, a root used in traditional Chinese medicine which accumulates coniferyl ferulate. Expression of AsFMT in poplar indeed introduced alkali-sensitive ester bonds40 (Figure 3(a)). Poplar trees engineered in this manner show increased cell-wall digestibility under mild alkaline pre-treatment conditions.41
Figure 3.
Examples of synthetic biology strategies to engineer lignin. (a) “Zip” lignin. Introduction of ferulated monolignols into the lignin polymer results in alkali-sensitive ester bonds. (b) QsuB lignin. By reducing the amount of shikimate available to the lignin biosynthesis pathway, a lignin enriched in H-monomers is formed, and the soluble produces protocatechuate accumulates. (A color version of this figure is available in the online journal.)
An alternative approach is to specifically reduce lignin in cell types where it is not essential, e.g. fiber cells, so it only remains in cells where it is essential for plant function, e.g. xylem vessels. To precisely achieve this goal is challenging using gene silencing or gene knockouts, since it usually requires the targeting of multiple genes and results in developmental defects.42–44 A more effective method is to use a gain-of-function approach. For example, expression of a plastidial-targeted, bacterially-derived enzyme, 3-dehydroshikimate dehydratase (QsuB), under the control of the plant CINNAMATE-4-HYDROXYLASE (C4H) promoter45 (Figure 3(b)). The QsuB enzyme converts 3-dehydroshikimate into protocatechuate (PCA). This reduces the availability of an intermediate in the shikimate pathway, which in turn reduces the substrate for monolignol production, reducing total lignin synthesis. The use of the cell-specific promoter avoids altering the lignin in the xylem vessels, where it is essential to avoid vesicular collapse. As a bonus, the plant accumulates PCA, which is a promising bio-based platform chemical.46 While this initial work was performed in Arabidopsis, since the lignin and shikimate pathways are highly conserved, it is likely that this approach will work in biomass crop plants.
Plant biomass is composed of, depending on the species, ∼40% hexose (mostly derived from cellulose) and ∼30% pentose (mostly derived from hemicelluloses and pectins) sugars, with lignin as the remainder.47 In most microbes, hexose sugars are preferentially utilized as a carbon source, due to carbon catabolite repression. As a result, pentoses are only consumed once the hexose supply has been exhausted. While much research has been directed towards the development of microbes which can use both simultaneously,48 it is not yet clear that this approach will be successful in industrial production strains. An alternative approach is to optimize the biomass to match the microbial preference. In one example, the authors chose to increase the cell wall content of the pectin galactan (which is composed of the hexose galactose). Initially, they constitutively overexpressed the Golgi-localized GALACTAN SYNTHASE1 (GALS1), the enzyme responsible for galactan biosynthesis,49 achieving an increase of 50% galactan in the leaf cell wall. They have since boosted this further by co-expressing the cytosolic UDP-GLUCOSE/UDP-GALACTOSE-4-EPIMERASE2 (UGE2) gene responsible for synthesizing the GALS1 substrate, UDP-galactose.50 Finally, a gene stacking approach combined the GALS1 and UGE2 with a UDP-galactose transporter, which moves the substrate into the Golgi lumen for utilization by GALS1,51 resulting in a 50% increase in galactan in stem cell walls.
In a powerful demonstration of plant synthetic biology, this study went further, by combining the high-galactan lines with cell-specific low xylan (i.e. low pentose) and low lignin (using the QsuB strategy described above).51 The use of cell-specific promoters avoided negative growth effects while delivering plants which had ∼3 fold increase in the hexose:pentose ration and increased release (up to 73%) of fermentable sugars from the biomass.51 It remains to be seen how well this strategy will translate to crop plants, but, at least in related species like poplar which have conserved cell wall synthesis pathways, there is a high chance of success.
While the previous examples demonstrate the ability of synthetic biology to produce plant biomass which is easier to breakdown for microbial conversion, the following examples showcase the possibilities of producing platform chemicals in planta. Platform chemicals are defined as the building block molecules from which most bulk chemicals, materials, and polymers are produced from. These molecules (short-chain unsaturated hydrocarbons) are currently produced in a highly efficient manner from fossil fuels. However, a number of alternatives have been suggested as replacements which could be derived from renewable sources.52 One such molecule is the dicarboxylic acid cis,cis-muconic acid. This is a precursor for bulk chemicals that include caprolactam, adipic acid, and terephthalic acid, which in turn are precursors for plastics such as nylon and polyethylene terephthalate (PET), or can be directly incorporated into polyesters.53 Synthetic biology has been used successfully to produce muconic acid in a range of microbial systems, including Escherichia coli, Pseudomonas sp., and Sphingobium sp.54 However, large-scale microbial production requires access to a cheap carbon source, which hinders the economic competitiveness of production. An alternative is to produce muconic acid in an autotroph, such as a biomass crop, where it can be considered an added value product in addition to the biomass. This has been successfully achieved in Arabidopsis using a complex engineering strategy that required the introduction of four bacterial enzymes.55 In particular, by controlling the expression of these genes using spatially- and temporally-specific promoters, negative growth effects associated with pathway intermediates were avoided. The muconic acid produced could be extracted from senesced, mature biomass, making it compatible with existing harvesting strategies.55
Polyhydroxyalkanoates (PHAs) are a class of biodegradable, renewable plastics which include poly-3-hydroxypropionic acid (a precursor of acrylic) and poly-3-hydroxybutarate (PHB, a precursor of propylene). Some microbial species produce PHAs as a stress response in the face of nutrient limitation, where it acts as a carbon store. Engineering of microbes to produce PHAs has been relatively successful, and this is now entering limited commercial production. However, like muconic acid, there would be advantages to producing PHAs in an autotroph.56 In some species, this has been highly successful. For example, in Arabidopsis, up to ∼ 40% dry weight of PHB in leaf tissue has been reported,57 and 10.6% dry weight in Phaeodactylum tricornutum (a microalgae). It has been challenging to achieve similar levels in highly productive C4 crop plants such as sugar cane58 and switchgrass,56 where whole plant levels above 2% dry weight have not been reported, despite extensive research efforts. PHB-producing C4 plants show a strong growth phenotype, including chlorosis and stunting, and a recent systems biology study linked this to the physical presence of PHB in bundle sheath cells.59 They showed that high PHB plants are ATP starved, and hypothesized that PHB granules may shade the thylakoids, reducing photosynthetic rates.59 This suggests that future efforts to produce high-PHB C4 plants may require a careful consideration for where in the plant the PHB is produced, and at what point in the lifecycle. For example, placing the enzymes under senescence associated promoters, as has been done successfully in rice to alter biomass composition,60 may avoid such yield penalties.
Plant-produced therapeutics and vaccines
The concept of plant-based pharmaceutical production (pharming) was proposed nearly 30 years ago.61 Originally, this was conceived as the production of edible vaccines in crop plants. These would be cheap to produce, and avoid the expensive costs associated with manufacture, or supply chain issues such as refrigeration.62 However, it rapidly became clear that this goal was unachievable. The challenges of regulating dose are just too difficult to overcome. As a result, the approach has been modified, and there is now acceptance that while plant-produced vaccines and pharmaceuticals have many benefits over other expression systems, some processing of the plant material will be required prior to vaccination of the patient.
The major reason for choosing plants for the production of monoclonal antibodies (mAbs), as opposed to other industrial production systems such as bacteria, yeast, insect or mammalian cells is a combination of cost, safety, and the potential of plant systems to produce mAbs with the correct post-translational modifications.63,64 For example, mammalian production systems, which dominate the market, are expensive and challenging to scale up, whereas bacteria are unable to produce most of the correct post-translational glycosylation required for the mAb activity. Yeast, insect cells, and plants, since they are eukaryotes, can perform complex post-translational glycosylations. However, in all these systems, the exact nature of the glycosylation differs depending on the species, and all show significant differences to mammalian systems (Figure 4(a)). As a result, the concept of glycoengineering has been developed, in which the expression system is modified to produce mammalian-like glycosylated mAbs. Plants have proved to be particularly amenable to glycoengineering, and this has been accelerated by synthetic biology tools.65–67
Figure 4.
Glycoengineering of proteins in plants. (a) Simplified example structures of N-linked glycosylation in humans, plants, and yeast (b) N-linked glycosylation in ΔXF tobacco plants, which produce humanized glycans on proteins. They lack the immunogenic β1,2-xylose and core α1,3-fucose.
Perhaps the most well-known example of a plant-produced therapeutic mAb is ZMapp™, a promising treatment for Ebola virus.68,69 This consists of three mouse/human chimeric mAbs which target the surface glycoprotein of the virions, and are produced in a strain of Nicotiana benthamiana (tobacco) called ΔXF (Figure 4(b)). ΔXF plants have reduced xylosyltransferase (XT) and fucosyltransferase (FT) activity, and produce authentic human protein glycosylation.66,70 Additional plant-produced therapeutics are currently in clinical trials (clinicaltrials.gov) or have been approved. For example, Elelyso™ (taliglucerase-α), a treatment for Gaucher’s disease, is produced in carrot cell culture and was approved by the FDA in 2012. As the global market for mAbs was reported to exceed $100 billion in 2017, the expectation is that this class of plant-produced products will continue to grow.
Plants also hold promise for the commercial production of pharmaceuticals or pharmaceutical precursors. One example is the anti-malarial drug artemisinin, the main ingredient in the ACT therapies used globally (and the only current effective treatment). Artemesinin is found at relatively low abundance in the Artemisia annua plant. However, as demand for the drug increased, it became increasingly challenging to produce it from its natural source. The precursor, artemisinic acid, can be converted chemically in a low-cost process to artemisinin, and therefore it has become an important biotechnological target. The initial strategy used synthetic biology to engineer artemisinic acid production in yeast by overexpressing 14 genes, and conditionally repressing two more.71 While this approach was highly successful, producing yields in excess of 25 g/L, the costs of large-scale yeast fermentation have made this challenging to produce commercially. More recently, a similar strategy has been used to produce artemesinic acid in tobacco making use of the COSTREL gene stacking strategy discussed above.33 This has resulted in yields in excess of 120 mg/kg of biomass. The authors estimate that 200 km2 of tobacco fields would be enough to meet the current global demand of artemisinin (∼100 t).
Future work
As the precision with which plants can be engineered increases, so will the range of applications. Recently, there has been a growing interest in plants as remote sensors. Plants can be considered sustainable and self-propagating reporters of their environment, and can be surveyed remotely, e.g. by drone. Suggested purposes include environmental pollution,72 explosives,73 and radiation.74 DARPA’s new Advanced Plant Technologies (APT) research program (https://www.darpa.mil/program/advanced-plant-technologies) which was announced in 2017, will be interesting to follow in this regard. Food crops are also a major target, and engineering strategies have been developed to reduce the methane emissions of rice,75 make low-gluten wheat,76 and improve cold-storage properties of potatoes.77 This is likely to be a huge market for growth. As humans prepare for interplanetary missions, including to Mars in the near future, synthetic biology will be key to developing plants which can survive the challenges of space travel, as well as providing optimized feedstocks for microbial manufacturing efforts.78 The success of all of these approaches will be reliant on increasing the predictability of plant engineering. Some progress has been made here, as a result of the reduced cost of DNA synthesis. Groups such as Open Plant (https://www.openplant.org/) and the Joint BioEnergy Institute (https://public-registry.jbei.org/) are developing open-source registries for plant-specific DNA parts, e.g. Lao et al.79 Next steps include the development of better genetic regulators, such as synthetic riboswitches, promoters, and transcription factors, as has been done for other organisms.80,81 However, more ambitious goals such as re-designing whole plant genomes, as is being undertaken for yeast in the Sc2.0 project,82 still seem a very distant prospect. Finally, it remains to be seen how the public will view synthetic biology and gene-editing, in light of previous concerns about GMOs.
Acknowledgements
I would like to thank my colleagues at JBEI for fruitful discussions on this topic, especially Henrik Scheller, Aymerick Eudes, and Yan Liang.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) and was supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02–05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy.
References
- 1.United Nations. World population prospects. The 2015 revision, key findings and advanced tables. New York: Author, 2015 [Google Scholar]
- 2.Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. Agricultural sustainability and intensive production practices. Nature 2002; 418:671–7 [DOI] [PubMed] [Google Scholar]
- 3.Gerland P, Raftery AE, Sevcikova H, Li N, Gu D, Spoorenberg T, Alkema L, Fosdick BK, Chunn J, Lalic N, Bay G, Buettner T, Heilig GK, Wilmoth J. World population stabilization unlikely this century. Science 2014; 346:234–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pingali PL. Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci U S A 2012; 109:12302–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol 2014; 12:381–90 [DOI] [PubMed] [Google Scholar]
- 6.Tyagi A, Kumar A, Aparna SV, Mallappa RH, Grover S, Batish VK. Synthetic biology: applications in the food sector. Crit Rev Food Sci Nutr 2016; 56:1777–89 [DOI] [PubMed] [Google Scholar]
- 7.Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell 2014; 157:151–61 [DOI] [PubMed] [Google Scholar]
- 8.Liu W, Stewart CN. Plant synthetic biology. Trends Plant Sci 2015; 20:309–17 [DOI] [PubMed] [Google Scholar]
- 9.Petzold CJ, Chan LJ, Nhan M, Adams PD. Analytics for metabolic engineering. Front Bioeng Biotechnol 2015; 3:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farre G, Rogers C, Smith A, Hibberd J, Webb AA, Locke J, Schornack S, Ajioka J, Baulcombe DC, Zipfel C, Kamoun S, Jones JD, Kuhn H, Robatzek S, Van Esse HP, Sanders D, Oldroyd G, Martin C, Field R, O'Connor S, Fox S, Wulff B, Miller B, Breakspear A, Radhakrishnan G, Delaux PM, Loque D, Granell A, Tissier A, Shih P, Brutnell TP, Quick WP, Rischer H, Fraser PD, Aharoni A, Raines C, South PF, Ane JM, Hamberger BR, Langdale J, Stougaard J, Bouwmeester H, Udvardi M, Murray JA, Ntoukakis V, Schafer P, Denby K, Edwards KJ, Osbourn A, Haseloff J. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol 2015; 208:13–9 [DOI] [PubMed] [Google Scholar]
- 11.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell 2006; 127:1309–21 [DOI] [PubMed] [Google Scholar]
- 12.Mendel G. Versuche über plflanzen-hybriden. Verhandlungen des naturforschenden Vereines in Brünn. Brunn, pp.3–47 [Google Scholar]
- 13.Bohra A, Jha UC, Adhimoolam P, Bisht D, Singh NP. Cytoplasmic male sterility (cms) in hybrid breeding in field crops. Plant Cell Rep 2016; 35:967–93 [DOI] [PubMed] [Google Scholar]
- 14.van Harten AM. Mutation breeding: theory and practical applications. Cambridge: Cambridge University Press, 2007, pp.252–300. [Google Scholar]
- 15.Hase Y, Okamura M, Takeshita D, Narumi I, Tanaka A. Efficient induction of flower-color mutants by ion beam irradiation in petunia seedlings treated with high sucrose concentration. Plant Biotechnol 2010; 27:99–103 [Google Scholar]
- 16.Yamaguchi H. Mutation breeding of ornamental plants using ion beams. Breed Sci 2018; 68:71–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Larebeke N, Genetello C, Schell J, Schilperoort RA, Hermans AK, Van Montagu M, Hernalsteens JP. Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature 1975; 255:742–3 [DOI] [PubMed] [Google Scholar]
- 18.Zambryski P, Joos H, Genetello C, Leemans J, Montagu MV, Schell J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 1983; 2:2143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevan M. Binary agrobacterium vectors for plant transformation. Nucl Acids Res 1984; 12:8711–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee LY, Gelvin SB. T-DNA binary vectors and systems. Plant Physiol 2008; 146:325–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hisano H, Sato K. Genomic regions responsible for amenability to agrobacterium-mediated transformation in barley. Sci Rep 2016; 6:37505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanford JC, Wolf ED, Allen NK, inventors; Cornell Research Foundation Inc. Assignee. Method for transporting substances into living cells and tissues and apparatus therefor, US Patent, 1990. [Google Scholar]
- 23.Estruch JJ, Carozzi NB, Desai N, Duck NB, Warren GW, Koziel MG. Transgenic plants: an emerging approach to pest control. Nat Biotechnol 1997; 15:137–41 [DOI] [PubMed] [Google Scholar]
- 24.National Academies of Sciences E, Medicine. Genetically engineered crops: experiences and prospects. Washington, DC: The National Academies Press, 2016 [PubMed] [Google Scholar]
- 25.Baltes NJ, Voytas DF. Enabling plant synthetic biology through genome engineering. Trends Biotechnol 2015; 33:120–31 [DOI] [PubMed] [Google Scholar]
- 26.Surridge C. A crispr definition of genetic modification. Nat Plants 2018; 4:233. [DOI] [PubMed] [Google Scholar]
- 27.Malzahn A, Lowder L, Qi Y. Plant genome editing with TALEN and CRISPR. Cell Biosci 2017; 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 2014; 32:947–51 [DOI] [PubMed] [Google Scholar]
- 29.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009; 459:442–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y, Richardson S, Yan J, Benites VT, Cheng-Yue C, Tran T, Mortimer J, Mukhopadhyay A, Keasling JD, Scheller HV, Loque D. Endoribonuclease-based two-component repressor systems for tight gene expression control in plants. ACS Synth Biol 2017; 6:806–16 [DOI] [PubMed] [Google Scholar]
- 31.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell 2016; 167:233–47.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih PM, Vuu K, Mansoori N, Ayad L, Louie KB, Bowen BP, Northen TR, Loque D. A robust gene-stacking method utilizing yeast assembly for plant synthetic biology. Nat Commun 2016; 7:13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentes P, Zhou F, Erban A, Karcher D, Kopka J, Bock R. A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 2016; 5:e13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemhauser JL, Torii KU. Plant synthetic biology for molecular engineering of signalling and development. Nat Plants 2016; 2:16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlack RD, Wright LL, Turhollow AF, Graham RL, Stokes BJ, Erbach DC. Biomass as feedstock for a bioenergy and bioproducts industry: the technical feasibility of a billion-ton annual supply Oak Ridge, TN: Oak Ridge National Laboratory, 2005. [Google Scholar]
- 36.Chakravorty U, Hubert M-H, Nøstbakken L. Fuel versus food. Annu Rev Resour Econ 2009; 1:645–63 [Google Scholar]
- 37.Langholtz MH,BJS, Eaton LM. 2016 billion-ton report: advancing domestic resources for a thriving bioeconomy, volume 1: economic availability of feedstocks. Oak Ridge, TN: U.S. Department of Energy, 2016. [Contract No.: ORNL/TM-2016/160] [Google Scholar]
- 38.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol 2010; 153:895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Weng JK, Chapple C. Improvement of biomass through lignin modification. Plant J 2008; 54:569–81 [DOI] [PubMed] [Google Scholar]
- 40.Wilkerson CG, Mansfield SD, Lu F, Withers S, Park JY, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, Ralph J. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 2014; 344:90–3 [DOI] [PubMed] [Google Scholar]
- 41.Zhou S, Runge T, Karlen SD, Ralph J, Gonzales-Vigil E, Mansfield SD. Chemical pulping advantages of zip-lignin hybrid poplar. ChemSusChem 2017; 10:3565–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, Decker SR, Selig MJ, Sykes R, Himmel ME, Kitin P, Shevchenko O, Strauss SH. Antisense down-regulation of 4cl expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 2010; 154:874–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA. Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 2007; 68:1521–9 [DOI] [PubMed] [Google Scholar]
- 44.Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C. The arabidopsis ref8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 2002; 30:33–45 [DOI] [PubMed] [Google Scholar]
- 45.Eudes A, Sathitsuksanoh N, Baidoo EE, George A, Liang Y, Yang F, Singh S, Keasling JD, Simmons BA, Loque D. Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol J 2015; 13:1241–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, Johnson CW, Chupka G, Strathmann TJ, Pienkos PT, Beckham GT. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci U S A 2014; 111:12013–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 2008; 54:559–68 [DOI] [PubMed] [Google Scholar]
- 48.Kim JH, Block DE, Mills DA. Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl Microbiol Biotechnol 2010; 88:1077–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liwanag AJ, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MC, Clausen MH, Scheller HV. Pectin biosynthesis: GALS1 in arabidopsis thaliana is a beta-1,4-galactan beta-1,4-galactosyltransferase. Plant Cell 2012; 24:5024–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gondolf VM, Stoppel R, Ebert B, Rautengarten C, Liwanag AJ, Loque D, Scheller HV. A gene stacking approach leads to engineered plants with highly increased galactan levels in arabidopsis. BMC Plant Biol 2014; 14:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aznar A, Chalvin C, Shih PM, Maimann M, Ebert B, Birdseye DS, Loque D, Scheller HV. Gene stacking of multiple traits for high yield of fermentable sugars in plant biomass. Biotechnol Biofuels 2018; 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werpy T, Petersen G, Aden A, Bozell J, Holladay J, White J, Manheim A, Eliot D, Lasure L, Jones S. Top value added chemicals from biomass: volume I – results of screening for potential candidates from sugars and synthesis gas US Department of Energy, 2004. [Google Scholar]
- 53.Rorrer NA, Dorgan JR, Vardon DR, Martinez CR, Yang Y, Beckham GT. Renewable unsaturated polyesters from muconic acid. ACS Sustain Chem Eng 2016; 4:6867–76 [Google Scholar]
- 54.Xie NZ, Liang H, Huang RB, Xu P. Biotechnological production of muconic acid: current status and future prospects. Biotechnol Adv 2014; 32:615–22 [DOI] [PubMed] [Google Scholar]
- 55.Eudes A, Berthomieu R, Hao Z, Zhao N, Benites VT, Baidoo EEK, Loque D. Production of muconic acid in plants. Metab Eng 2018; 46:13–9 [DOI] [PubMed] [Google Scholar]
- 56.Somleva MN, Peoples OP, Snell KD. Pha bioplastics, biochemicals, and energy from crops. Plant Biotechnol J 2013; 11:233–52 [DOI] [PubMed] [Google Scholar]
- 57.Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey RN, Willmitzer L. Transgenic arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta 2000; 211:841–5 [DOI] [PubMed] [Google Scholar]
- 58.Petrasovits LA, Purnell MP, Nielsen LK, Brumbley SM. Production of polyhydroxybutyrate in sugarcane. Plant Biotechnol J 2007; 5:162–72 [DOI] [PubMed] [Google Scholar]
- 59.McQualter RB, Bellasio C, Gebbie LK, Petrasovits LA, Palfreyman RW, Hodson MP, Plan MR, Blackman DM, Brumbley SM, Nielsen LK. Systems biology and metabolic modelling unveils limitations to polyhydroxybutyrate accumulation in sugarcane leaves; lessons for c4 engineering. Plant Biotechnol J 2016; 14:567–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vega-Sanchez ME, Loque D, Lao J, Catena M, Verhertbruggen Y, Herter T, Yang F, Harholt J, Ebert B, Baidoo EE, Keasling JD, Scheller HV, Heazlewood JL, Ronald PC. Engineering temporal accumulation of a low recalcitrance polysaccharide leads to increased c6 sugar content in plant cell walls. Plant Biotechnol J 2015; 13:903–14 [DOI] [PubMed] [Google Scholar]
- 61.Hiatt A, Cafferkey R, Bowdish K. Production of antibodies in transgenic plants. Nature 1989; 342:76–8 [DOI] [PubMed] [Google Scholar]
- 62.Rybicki EP. Plant-made vaccines for humans and animals. Plant Biotechnol J 2010; 8:620–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yusibov V, Kushnir N, Streatfield SJ. Antibody production in plants and green algae. Annu Rev Plant Biol 2016; 67:669–701 [DOI] [PubMed] [Google Scholar]
- 64.Lomonossoff GP, D'Aoust MA. Plant-produced biopharmaceuticals: a case of technical developments driving clinical deployment. Science 2016; 353:1237–40 [DOI] [PubMed] [Google Scholar]
- 65.Kallolimath S, Castilho A, Strasser R, Grunwald-Gruber C, Altmann F, Strubl S, Galuska CE, Zlatina K, Galuska SP, Werner S, Thiesler H, Werneburg S, Hildebrandt H, Gerardy-Schahn R, Steinkellner H. Engineering of complex protein sialylation in plants. Proc Natl Acad Sci U S A 2016; 113:9498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loos A, Gach JS, Hackl T, Maresch D, Henkel T, Porodko A, Bui-Minh D, Sommeregger W, Wozniak-Knopp G, Forthal DN, Altmann F, Steinkellner H, Mach L. Glycan modulation and sulfoengineering of anti-hiv-1 monoclonal antibody pg9 in plants. Proc Natl Acad Sci U S A 2015; 112:12675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loos A, Steinkellner H. Plant glyco-biotechnology on the way to synthetic biology. Front Plant Sci 2014; 5:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wec AZ, Herbert AS, Murin CD, Nyakatura EK, Abelson DM, Fels JM, He S, James RM, de La Vega MA, Zhu W, Bakken RR, Goodwin E, Turner HL, Jangra RK, Zeitlin L, Qiu X, Lai JR, Walker LM, Ward AB, Dye JM, Chandran K, Bornholdt ZA. Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell 2017; 169:878–90 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davey RT, Jr., Dodd L, Proschan MA, Neaton J, Neuhaus Nordwall J, Koopmeiners JS, Beigel J, Tierney J, Lane HC, Fauci AS, Massaquoi MBF, Sahr F, Malvy D. A randomized, controlled trial of ZMapp for Ebola virus infection. N Engl J Med 2016; 375:1448–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strasser R, Stadlmann J, Schahs M, Stiegler G, Quendler H, Mach L, Glossl J, Weterings K, Pabst M, Steinkellner H. Generation of glyco-engineered nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like n-glycan structure. Plant Biotechnol J 2008; 6:392–402 [DOI] [PubMed] [Google Scholar]
- 71.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013; 496:528–32 [DOI] [PubMed] [Google Scholar]
- 72.Kovalchuk I, Kovalchuk O. Transgenic plants as sensors of environmental pollution genotoxicity. Sensors. Sensors 2008; 8:1539–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antunes MS, Morey KJ, Smith JJ, Albrecht KD, Bowen TA, Zdunek JK, Troupe JF, Cuneo MJ, Webb CT, Hellinga HW, Medford JI. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS One 2011; 6:e16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng Y, Allen S, Millwood RJ, Stewart CN. Jr, 'Fukusensor ' A genetically engineered plant for reporting DNA damage in response to gamma radiation. Plant Biotechnol J 2014;12:1329–32 [DOI] [PubMed]
- 75.Su J, Hu C, Yan X, Jin Y, Chen Z, Guan Q, Wang Y, Zhong D, Jansson C, Wang F, Schnurer A, Sun C. Expression of barley susiba2 transcription factor yields high-starch low-methane rice. Nature 2015; 523:602–6 [DOI] [PubMed] [Google Scholar]
- 76.Sanchez-Leon S, Gil-Humanes J, Ozuna CV, Gimenez MJ, Sousa C, Voytas DF, Barro F. Low-gluten, nontransgenic wheat engineered with crispr/cas9. Plant Biotechnol J 2018; 16:902–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, Tibebu R, Davison S, Ray EE, Daulhac A, Coffman A, Yabandith A, Retterath A, Haun W, Baltes NJ, Mathis L, Voytas DF, Zhang F. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 2016; 14:169–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Llorente B, Williams T, Goold H. The multiplanetary future of plant synthetic biology. Genes 2018; 9:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lao J, Oikawa A, Bromley JR, McInerney P, Suttangkakul A, Smith-Moritz AM, Plahar H, Chiu TY, Gonzalez Fernandez-Nino SM, Ebert B, Yang F, Christiansen KM, Hansen SF, Stonebloom S, Adams PD, Ronald PC, Hillson NJ, Hadi MZ, Vega-Sanchez ME, Loque D, Scheller HV, Heazlewood JL. The plant glycosyltransferase clone collection for functional genomics. Plant J 2014; 79:517–29 [DOI] [PubMed] [Google Scholar]
- 80.Auslander S, Stucheli P, Rehm C, Auslander D, Hartig JS, Fussenegger M. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat Methods 2014; 11:1154–60 [DOI] [PubMed] [Google Scholar]
- 81.Machens F, Balazadeh S, Mueller-Roeber B, Messerschmidt K. Synthetic promoters and transcription factors for heterologous protein expression in saccharomyces cerevisiae. Front Bioeng Biotechnol 2017; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CL, Chandrasegaran S, Cai Y, Boeke JD, Bader JS. Design of a synthetic yeast genome. Science 2017; 355:1040–4 [DOI] [PubMed] [Google Scholar]