Short abstract
The traditional route to investigating biology by perturbing living systems or by individually purifying and characterizing component parts is giving way to more complex endeavors where chemists and physicists attempt to build cells from scratch. Parallel efforts are underway that either exploits extant biological parts or prebiotically plausible molecules. Both approaches help to reveal the underlying physical–chemical forces that give rise to cellular function and highlight the important role played by polymers in regulating biological chemical systems. Although the success in RNA and lipid chemistry has led to the reconstitution of specific facets of cellular life, our understanding of dynamic, dissipative networks is currently too incomplete to allow for the construction of a self-sustained, integrated protocell. However, the presence of shared chemistry points to a promising path forward.
Impact statement
Advances in the understanding of the biophysics of membranes, the nonenzymatic and enzymatic polymerization of RNA, and in the design of complex chemical reaction networks have led to a new, integrated way of viewing the shared chemistry needed to sustain life. Although a protocell capable of Darwinian evolution has yet to be built, the seemingly disparate pieces are beginning to fit together. At the very least, better cellular mimics are on the horizon that will likely teach us much about the physicochemical underpinnings of cellular life.
Keywords: Protocell, minimal cell, origin of life, RNA world, vesicles, prebiotic chemistry, peptides, protometabolism
Introduction
Recent years have brought a renewed enthusiasm to attempts at building a cell from component parts. There are laboratories that are attempting to decipher the prebiotic chemistry that led to life, to gain insight into the divide between complex chemical reactions and biological networks, and to engineer life-like technologies. The diversity in terminology used to describe the assembled chemical systems reflects the variety of approaches taken. The terms protocell, artificial cell, minimal cell, and synthetic cell are at times used to describe the same thing, and other times intended to emphasize different aspects of the assembled chemical system. For example, protocell is frequently used to describe simple cell-like units assembled from prebiotically plausible components, whereas artificial cells often indicate mimics of modern cells that consist, in part, of extant biological machinery. However, examples to the contrary are commonly encountered.
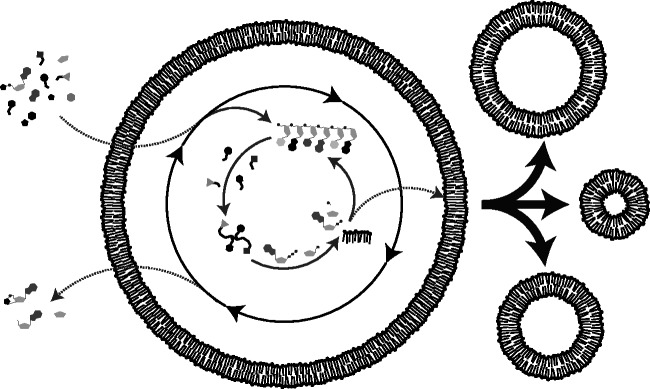
The most straightforward approach to synthesizing a model protocell (Figure 1) is to focus on developing a self-assembled, minimal unit capable of Darwinian evolution.1 As extant biology is the product of billions of years of evolution, the mechanisms exploited by living cells are quite complex in comparison to the physicochemical laws that drive the system. Therefore, in addition to being of greater prebiotic relevance, mechanisms that do not rely on biological machinery are at times easier to implement. However, the difficulty is that abiotic mechanisms proceed unregulated, which is in stark contrast to how enzymatically controlled the biological networks function.2 Moreover, the dynamics of the abiotic mechanisms that current models of protocells depend on for survival tend to decrease the robustness of the overall system. Therefore, one is often left trying to balance chemical spontaneity with robustness so that the protocell can persist through fluctuating conditions. Presumably at some point, the complexity of the system would reach a level where abiotically implausible polymers would be required to coordinate the activities inside the protocell. We have yet to determine where that boundary lies.
Figure 1.
Schematic of one conception of a hypothetical protocell. A chemical reaction network within a lipid vesicle acquires nutrients that lead to the copying of nucleic acids and the synthesis of peptides and lipids. Here, the internal chemistry of the protocell supports membrane growth by synthesizing lipid. Membrane growth then leads to the division of unstable intermediate structures through environmental shear forces. Reactive monomeric building blocks and reaction by-products, i.e. waste, passively diffuse across the membrane. Ideally, an internal protometabolic network would sustain the protocell across many generations.
Traditionally, protocells are envisioned as short-chain fatty acid vesicles containing catalytic RNA molecules. This implies that the RNA molecules are tied in some way to the maintenance of the vesicle. Although the presence of nucleic acids does affect the dynamics of some encapsulated chemical systems, as discussed below, strands of nucleic acids alone do not constitute a functioning genome any more than polymers of amino acids or sugars. Some type of supporting architecture is necessary to implement the instructions encoded within the nucleic acid polymers in a way that allows the (proto)cell to defy thermodynamic equilibrium long enough to propagate. To do so, early cell-like systems presumably made use of a rudimentary form of metabolism. It seems likely that these early protocells contained a heterogenous mixture of molecules, including RNA, peptides, metal ions, lipids, plus additional components. Although life could have begun with molecules that are no longer exploited by living cells today,3 emphasis is usually placed on molecules that have a connection to extant living systems. For example, coacervate-based and proteinosome-based cell-like systems4–7 have been constructed, but lipid-based systems have been more extensively investigated partly because lipid vesicles better resemble life as we know it.
Compartmentalization
Like all of the components of a cell, membranes are dynamic. Membranes must change shape during growth and division, mediate the uptake of nutrients and release of waste, and are integral to the maintenance of concentration gradients that are extensively used to drive thermodynamically unfavorable reactions. Many of these characteristics have been reconstituted from extant sources for the synthesis of artificial cells. For example, much of the protein division machinery has been assembled in vitro,8–12 although the robust division of lipid vesicles has yet to be demonstrated. Similarly, protein pores,13 ion pumps,14 and ATP synthase15–17 have been reconstituted in lipid vesicles.18 Importantly, some protein-mediated activities have been coupled together so that the light-dependent pumping of protons can be used to drive the synthesis of ATP.17,19 However, significant challenges remain in building in the laboratory a biological-like membrane that is responsive to the changing needs of a cell.8 This difficulty is mostly due to the fact that membranes made from biological lipids are dynamically restricted, kinetically trapped structures, which is compensated for in biology by the incorporation of up to half protein in membrane composition. Proteins are needed to modulate the properties of the membrane, and the production and degradation of the proteins themselves are regulated at a genetic level. The construction of such a complex genetic and protein-dependent system is beyond what has been reconstituted to date with cell-free synthetic biology methodologies.
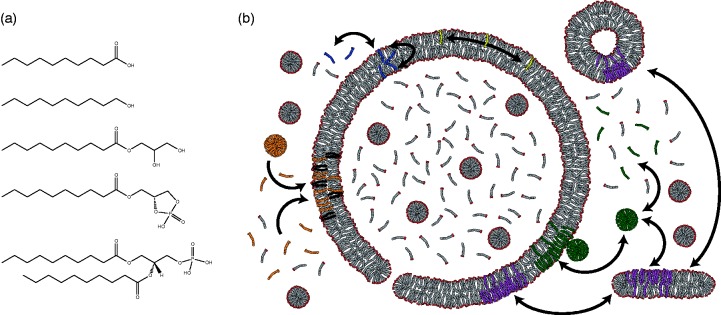
It is, therefore, perhaps unsurprising that more progress has been made with vesicles composed of model prebiotic lipids than with the phospholipids found in biological membranes (Figure 2(a)). Prebiotic lipids are typically taken to be monoacyl amphiphiles with either a carboxylate, i.e. fatty acid, or alcohol head group. Such lipids can be synthesized by spark discharge20 or Fischer–Tropsch reactions at high temperature with metal catalysts,21,22 are found in carbonaceous meteorites,23 and can form vesicles with a lipid bilayer morphologically similar to membranes made of biological phospholipids.24,25 Recent work suggests that more biologically relevant phospholipids could have formed on the prebiotic Earth;26–28 however, corroborating evidence from carbonaceous meteorites is not available. The supramolecular structures formed from fatty acids are less stable than the aggregates formed by their diacyl counterparts,29 exhibiting a much greater range of dynamics. The increased dynamics allow for activity that otherwise would require the intervention of proteins. Lipids can twist, turn, laterally diffuse, flip between leaflets, and escape into solution (Figure 2(b)). All of these phenomena are several orders of magnitude faster with fatty acids than with diacyl phospholipids.29–32 Much of the differences in lipid dynamics can be explained by considering the packing of the hydrophobic regions of the membrane. Increased packing leads to greater van der Waals interactions, and thus decreased dynamics. The dynamic nature of fatty acids allows growth,30 division,33 and the acquisition of nutrients,34–36 all in the absence of proteins. Importantly, heterogenous mixtures of monoacyl lipids, such as those that would likely arise from non-enzymatic, prebiotic synthesis, give vesicles more amenable to a protocellular life cycle.37 Heterogeneity in the length of the hydrocarbon chain promotes the self-assembly of vesicles.38,39 Moreover, heterogeneity of polar head-groups can lead to increased stability40–44 and permeability.35,36
Figure 2.
The lipids and lipid dynamics of protocellular membranes. (a) From top to bottom, the structures of model prebiotic lipids are decanoic acid, decanol, the glycerol monoester of decanoate, and cyclic-lyso-phosphodecanoic acid. The last structure is of di-decanoyl phosphatidic acid, shown for reference. (b) Above the critical aggregate concentration, fatty acids exist in equilibrium between free monomers, micelles (green), and various lipid aggregates (purple), including vesicles with bilayer membranes. Lipid monomers can be exchanged between different aggregate structures, flip between leaflets (blue), and laterally diffuse (yellow). The dynamic nature of the bilayer may lead to the formation of transient pores. Micelles can incorporate into pre-existing vesicles, leading to a net growth in volume and surface and surface area (green). Likewise, the presence of diacyl phospholipids (brown) leads to vesicle growth through the net accumulation of fatty acid (orange) by decreasing the desorption rate of fatty acids from the membrane.
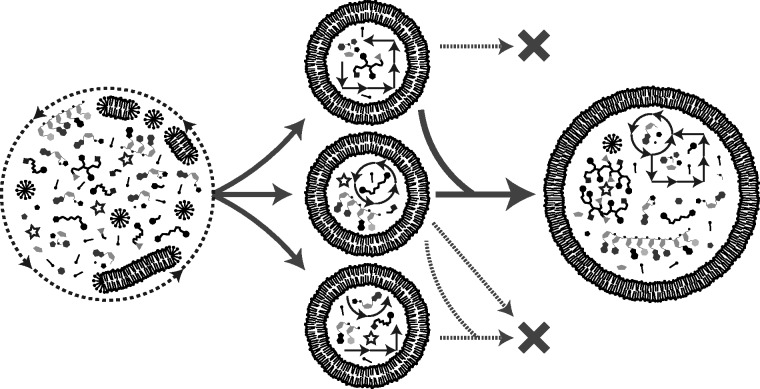
Figure 3.
Transition towards a functional protocell. A subset of complex prebiotic chemistry gave rise to protocells. Here, the process is envisaged to have proceeded in a fashion where nucleic acids were deeply integrated with peptides, and metabolic-like chemistry to generate a cell-like network from the beginning. That is, systems did not generally evolve separately and later merge. Iterative cycles of growth, fusion, and division ultimately led to competition between protocells sustained by some type of dissipative chemical (proto)metabolism.
Lipid dynamics also enable competition between fatty acid-based vesicles in a way that would be difficult to achieve with diacyl phospholipids alone. Osmotic pressure generated by plausible levels of encapsulated RNA, for example, drives vesicle growth at the expense of empty vesicles.45 Lipid exchange between the two populations of vesicles, i.e. RNA containing and empty vesicles, occurs because the energetic barrier for fatty acids to enter aqueous solution from a membrane is low.29 Therefore, the unfavorable surface area to volume ratio of osmotically swollen vesicles can be reduced through the incorporation of fatty acids that are in equilibrium between different membranes. The implication then is that the activity of an encapsulated RNA polymerase would induce growth by increasing the concentration of RNA and thus the osmotic pressure on the vesicle. Such a mechanism could also lead to the division of multilamellar vesicles under environmentally reasonable conditions, since the shape changes that arise from concentration gradients across the membranes lead to the formation of fragile thread-like structures that break apart into daughter vesicles upon agitation.33,46,47
Diacyl phospholipids are kinetically trapped in vesicle membranes29 and thus cannot contribute to the alleviation of osmotic stress in the way that fatty acids can. However, the presence of even small amounts of diacyl phospholipid significantly impacts the dynamics of fatty acids.48–50 The slower desorption rates of fatty acids from membranes containing diacyl phospholipids skew the exchange of fatty acids between different membranes.32,51–53 These altered kinetics, plus entropic effects, result in the net growth of diacyl phospholipid containing vesicles. Therefore, multiple mechanisms, including the replication of nucleic acids and the synthesis of diacyl lipids, exist that could give rise to competition between protocells.50 Since cholesterol increases the packing of membranes and decreases the permeability of membranes to solutes,34,54 it would be insightful to determine if polyaromatic hydrocarbons (PAH), which have been hypothesized to play a role in the origins of life,38,55,56 similarly impact the lipid exchange kinetics between vesicles.
Increased lipid dynamics are not without disadvantages. Fatty acid vesicles only form near the pKa of the carboxylate head group57 and are similarly sensitive to the ionic strength of the solution,58 in particular to the presence of divalent cations.59 Intuitively then, the addition of diacyl phospholipid would be expected to increase the stability of fatty acid vesicles. In fact, only 10 mol% of diacyl phospholipid decreases the critical aggregate concentration (CAC) of oleate vesicles48,50 and increases the stability of vesicles to Mg2+.48 Stability to Mg2+ is important as many chemical mechanisms, such as the polymerization of nucleic acids,49 are facilitated by the neutralizing and catalytic effects of Mg2+. Taken together, the data suggest that if a prebiotic catalyst emerged that enabled the synthesis of diacyl lipids, then vesicles containing such a catalyst would have a selective advantage due to at least two factors. Such vesicles would display increased stability to the effects of divalent cations and would be capable of preferential growth over vesicles lacking diacyl lipids, as described above. Although monoacyl glycerol esters of fatty acids increase the stability of fatty acid vesicles to Mg2+ by 2–3 fold,57,59 these monoglycerides would not be expected to mediate selective growth because of decreased acyl chain packing within the membrane.35
The presence of diacyl phospholipids could have accelerated the emergence of more complex polypeptides. The decreased permeability of diacyl phospholipids would be expected to decrease the ability of protocells to passively acquire nutrients.50 As some chemical exchange across membranes is necessary, decreased permeability would have put a selective pressure on the system to develop transport machinery, unless alternative mechanisms, such as the formation of defects and pores arising from freeze-thawing60 and macromolecular crowding,61 were available. Nevertheless, highly impermeable membranes are advantageous to a (proto)cell, since impermeability allows for the exploitation of concentration gradients to drive thermodynamically unfavorable but necessary (proto)cellular processes.62 It is also possible that diacyl phospholipids aided the emergence of soluble peptide enzymes in addition to peptides localized to the membrane. For example, fatty acids are competitive inhibitors of polymerases.63,64 In the presence of small amounts of diacyl phospholipids, however, the availability of fatty acids in solution would be greatly decreased,39,49 due to the decreased desorption rates of fatty acids from diacyl phospholipid containing membranes. Less free fatty acid could have then facilitated the emergence of enzymes with polymerase activity and thus perhaps the emergence of more complex genetic systems. In fact, model intermediate membrane compositions containing phospholipids and fatty acids can carry-out encapsulated transcription and translation while retaining the ability to acquire nucleotides and amino acids.49 Of course, there is currently insufficient data to support this speculation. It is also possible that protein enzymes emerged later, after the reliance on membranes primarily composed of fatty acids passed.
Nucleic acid and peptide polymers
The prebiotic synthesis of the building blocks of nucleic acids and peptides has been successful enough to indicate that nucleosides and amino acids were likely present on the prebiotic Earth.27,65–68 Since the early work of Miller, Urey, and Oró, the central role of cyanide-based chemistry in prebiotically producing the molecules of life has been clear. Importantly, the products of these prebiotic syntheses mirror what has been found in carbonaceous meteorites.69 Since the molecules of life tend to be photostable,70–72 and UV-light appears to be important for the prebiotic synthesis of pyrimidines73,74 and inorganic cofactors of proteins,75 UV-light seems to have played a critical role in the availability of molecules on the prebiotic Earth. Of course, factors other than photostability must have been important as well. For example, a large number of presumably photostable amino acids not found in biology have been identified in meteorites,76 suggesting that only a subset of the potential building blocks of life were selected.77,78 Nevertheless, caveats to these theories exist. Some argue that there existed an original restricted set of amino acids, for example, that were later supplemented by cellular activity.79 It is also possible that the first polymers from which modern-day polymers evolved from were chemically distinct from extant biopolymers.3,80 In other words, the original building blocks may not have resembled extant amino acids or nucleotides.
What has been more difficult to assess is how the individual building blocks could have polymerized into nucleic acids and polypeptides. Some success has been achieved in pushing these dehydration reactions forward through cycles of drying through heating followed by wetting.81,82 The presence of lipids facilitates the synthesis of RNA from mononucleotides, presumably by arranging and condensing the nucleotides between the lamellae of lipid membranes during drying.83,84 Recently, the formation of peptides was found to be facilitated by the presence of α-hydroxy acids during such dry/wet cycles.85,86 However, a significant problem emerges at temperatures above 65°C, where the hydrolysis effectively competes with polymerization,87 thereby leading to the production of short, scrambled sequences.82 Although such instability may have selected for folded sequences more resistant to degradation,81 other paths towards biological-like polymers that do not rely on heating are possible.
In biology, polymers are synthesized from activated monomers. Life exploits activation chemistry with high energy barriers so that the chemistry of the cell can be regulated by enzymes. In the absence of such catalysts, activation chemistry is needed so that the rates of polymerization in aqueous solution surpass the rates of hydrolysis. Examples of such phenomena include the carbonyl sulfide chemistry described by Ghadiri and colleagues88 for the polymerization of amino acids,88 the reactions of imidazole activated nucleotides to support the template directed synthesis of nucleic acids,89–91 and diamidophosphate-mediated chemistry, which leads to the formation of polypeptides, oligonucleotides, and phospholipids.26 Carbonyl sulfide is found in the atmosphere and is emitted from volcanoes. Prebiotically plausible routes to imidazole-activated nucleotides have been proposed via cyanogen chloride92 and methyl isocyanide93,94-assisted synthesis, and diamidophosphate is formed from the ammonolysis of metatriphosphate.95,96 Nonenzymatic polymerization reactions mediated by such activation chemistry can be enhanced by the concentrating effects of eutectic phases,97 which are also compatible with the assembly and selection of active ribozymes.98,99 It should be emphasized, however, that while it is desirable to identify conditions that favor polymerization over degradation, degradative processes are important for cellular and prebiotic chemistry.100
A central tenet of the RNA World hypothesis is that at some point, catalytic RNA molecules arose through non-enzymatic means that were capable of copying of RNA, i.e. there was once an RNA-dependent RNA polymerase ribozyme or replicase. However, the search for such a polymer has been quite difficult. After the initial report of an RNA ligase ribozyme by Bartel and Szostak in 1993,101 it took almost two decades to improve the class I RNA polymerase ribozyme102 to the point that ∼95 nucleotides could be copied, enough to generate an active hammerhead ribozyme.103 However, the processivity was not enough to synthesize another active RNA polymerase ribozyme (∼200 nucleotides in length). The successful selection of cross chiral RNA polymerase ribozymes demonstrates that RNA molecules that exploit more tertiary interactions as opposed to direct base-pairing decreases nucleotide bias104 and further suggests that at least some of the difficulties encountered in identifying more processive RNA polymerase ribozymes stem from the limitations of current selection technologies.105
Or perhaps the difficulty in developing RNA polymerase ribozymes reflects a too stringent interpretation of the RNA World. The ribosome may be a ribozyme,66,106–108 but this enzyme still depends on the presence of peptides for robust activity. In other words, if prebiotic chemistry gave rise to both RNA and peptides,27 then it is logical to assume that both worked together cooperatively. In fact, all that is needed to aminoacylate RNA, a key step in the RNA-guided biosynthesis of peptides, are short oligomers ranging between 5 and 45 nucleotides.109–112 Additionally, codon length trimer oligonucleotides facilitate both nonenzymatic113 and ribozyme-mediated copying of RNA.114 This type of reasoning led Holliger and colleagues115 to explore the influence of basic peptides on the activity of RNA polymerase ribozymes. The peptides decrease the dependence on Mg2+ down to concentrations that are compatible with fatty acid vesicles.
Compartments are thought to be important to systems of replicating RNA to protect against take over from parasitic sequences, i.e. easily copied RNA sequences that do not provide a selective advantage to the system.1,116,117 This protection does not, however, remove the ability of RNA molecules to compete against each other since efficient RNA replicators should outcompete less efficient replicators through the selective growth and division45 of the vesicle, as described above. A catalyst engaged in the synthesis of diacyl phospholipids would similarly help the catalyst-vesicle system outcompete other vesicles that lacked such a catalyst.50 However, these important mutually beneficial links between RNA and vesicles have not been adequately demonstrated experimentally, because the necessary ribozymes either have not been identified or are too inefficient. Instead, the non-enzymatic copying of oligonucleotides35,36 and the reconstitution of hammerhead ribozyme activity59 were reconstituted in fatty acid vesicles, and an RNA polymerase ribozyme copied an RNA template within phospholipid vesicles.115 While impressive, none of these systems disrupt equilibria enough to lead to vesicle growth or division. Without an intimate connection between RNA and vesicle chemistry, assembled models of protocells will continue to function as a composite of separate systems as opposed to an integrated, life-like system where the genotype directly affects a selectable phenotype.
Metabolism
Some sort of metabolic activity is needed to maintain the low entropy, out of equilibrium state of a living cell.118 To do so, biology couples thermodynamically favorable reactions to the unfavorable chemistry needed to maintain the cell. Although there is no a priori reason why life must begin with a network of chemical reactions that resembles what is found in biology today, the universality of central metabolism has often been interpreted to indicate that extant-like metabolic chemistry was present on the early Earth before there was life.119,120 However, while the plausibility of individual reactions occurring on the prebiotic Earth is high, it is much less clear if entire metabolic-like cycles could have existed.121 Nevertheless, model prebiotic versions of glycolysis,122–124 the Wood–Ljungdahl pathway,125 and the citric acid cycle126–128 have been constructed in the laboratory. In each case, activity was dependent upon the presence of metals, either in elemental, mineral, or hydrated ionic form. This dependence on metals is not surprising since extant metabolism is completely reliant on the activity of metalloproteins in which the metallocofactor itself is largely responsible for the catalysis.129
The point of metabolism is generally not to produce specific molecules. For example, ATP alone does not drive a reaction better than any other molecule in the absence of coupling chemistry. Instead, dissipative reaction networks are harnessed to maintain the far-from-equilibrium state of the cell. Early attempts at building chemical models of dissipative systems have focused on reproducing one of the hallmarks of out-of-equilibrium biological networks, such as oscillatory behavior. Proof-of-principle reactions have been demonstrated with small molecules,130 nucleic acids,131–135 and protein enzymes.136–138 More recently, a network of organic reactions dependent upon thiol and amide chemistry gave rise to oscillatory behavior in the absence of enzymatic activity.139 Nevertheless, these conceptually important papers do not describe chemistry that is directly tied to the fitness of a protocell or to a chemical reaction network. To this end, dynamic reaction networks140,141 were developed that lead to the replication or persistence of peptides or peptide-derived structures142–144 and vesicles.145–148 However, thus far, all of the developed systems are far from functioning as a metabolic network capable of supporting a cell, in part, because metabolism addresses multiple cellular needs at once. To use a real-world example, it is as if the different wheels of a car are not only spinning independently but not even touching the ground. Metabolism requires integration and must directly support the maintenance of the cell. At the very least, effort should be expended to integrate nucleic acid and peptide chemistry.149 One path forward could be to focus on the shared chemistry that is used to mediate the different dissipative systems developed thus far. Many of these dynamic networks rely on thiol chemistry. Such chemistry has long been hypothesized to have been important for the origins of life,150,151 and the photochemistry of thiols can be tied to the replication of protocells.46
Another obvious problem with nearly all of these dissipative systems is that each system is highly contrived. Prebiotically implausible conditions are used and often times precise mixing within a microfluidic device is required. This is because no molecular component is present to govern the myriad of chemical reactions. Natural living systems have enzymes that not only catalyze reactions but do so in a manner that is responsive to the needs of the cell, as recognized by Oparin nearly a century ago.2 It remains challenging to understand how a dissipative, combinatorial system could give rise to a network of polypeptides and nucleic acids that exhibit interdependent enzyme-like activity that sustains a protocell. However, the barrier to the development of the regulators of protometabolism, i.e. enzymes, may not be as high as imagined. For example, dipeptides can display catalytic activity152–156 and can selectively coordinate the types of metal ions that are important for enzymatic catalysis.75,157 Therefore, it seems likely that short, catalytically active (metallo)peptides existed on the prebiotic Earth. Since short peptides can self-assemble158–160 and bioinformatic analyses suggest that ancient peptide motifs exist within modern day proteins,161 it may be that ancient hetero-complexes of peptides displayed activity more akin to an enzyme than just a simple catalyst. Polymerization of the peptides within the aggregate into longer protein-like structures may have then improved activity.162 But again, systems need to be integrated. Without a supporting genetic system, the utility of this new enzyme-like polymer would be limited.
Conclusion
By some measures, the prospect of building a functioning model protocell seems quite close. Remarkable progress has been made in replicating strands of RNA and in developing vesicle compartments amenable to the range of tasks needed to sustain a protocell life cycle.163 Nevertheless, after much effort, such a model system is still not available. A reasonable conclusion could be that the component parts or systems constructed separately thus far do not fit together in the same way as would be the case for a mechanical machine.164 In other words, the disparate subsystems needed to maintain a cell must be deeply integrated, as would be the case for integral (proto)cellular components that have emerged and/or evolved together (Figure 3). Since biology largely relies on enzymes to manage the coordinated activity of an integrated system, it would appear that building a cell (or artificial cell) with extant biological parts would be easier than endeavors to build (proto)cells without modern enzymes. However, the artificial cell field seems no closer to achieving success. Individual protein-mediated pathways, such as those that divide vesicles8 and replicate DNA,165 have been largely reconstituted, but the disparate subsystems needed to sustain a cell are even less integrated than what has been achieved with model protocells. Instead, artificial cell research has shown more promise thus far in developing systems with biotechnological applications166–170 than in building a convincing cellular mimic. Regardless of whether the goal is to build a cell that mimics extant cellular life or to understand how life could have emerged on the prebiotic Earth, what seems to be needed is greater insight into how deeply integrated, dissipative systems can be harnessed to build truly life-like chemical systems.
Authors’ contributions
O.D.T. and S.S.M. wrote the manuscript together.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
Funding from the Simons Foundation (290358) is gratefully acknowledged.
References
- 1.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001; 85:1509–10 [DOI] [PubMed] [Google Scholar]
- 2.Oparin AI. The origin of life New York: Macmillan, 1938
- 3.Krishnamurthy R. Life’s biological chemistry: a destiny or destination starting from prebiotic chemistry? Chem A Eur J 2018;24:16708-16715 [DOI] [PubMed] [Google Scholar]
- 4.Dora Tang TY, Rohaida Che Hak C, Thompson AJ, Kuimova MK, Williams DS, Perriman AW, Mann S. Fatty acid membrane assembly on coacervate microdroplets as a step towards a hybrid protocell model. Nat Chem 2014; 6:527–33 [DOI] [PubMed] [Google Scholar]
- 5.Qiao Y, Li M, Booth R, Mann S. Predatory behaviour in synthetic protocell communities. Nat Chem 2017;9:110–119 [DOI] [PubMed]
- 6.Aumiller WM, Pir Cakmak F, Davis BW, Keating CD. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 2016; 32:10042–53 [DOI] [PubMed] [Google Scholar]
- 7.Aumiller WM, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem 2016; 8:129–37 [DOI] [PubMed] [Google Scholar]
- 8.Kretschmer S, Schwille P. Toward spatially regulated division of protocells: insights into the E. coli min system from in vitro studies. Life 2014; 4:915–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci U S A 2013; 110:11000–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terasawa H, Nishimura K, Suzuki H, Matsuura T, Yomo T. Coupling of the fusion and budding of giant phospholipid vesicles containing macromolecules. Proc Natl Acad Sci U S A 2012; 109:5942–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurihara K, Tamura M, Shohda KI, Toyota T, Suzuki K, Sugawara T. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat Chem 2011; 3:775–81 [DOI] [PubMed] [Google Scholar]
- 12.Kurihara K, Okura Y, Matsuo M, Toyota T, Suzuki K, Sugawara T. A recursive vesicle-based model protocell with a primitive model cell cycle. Nat Commun 2015; 6:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elani Y, Law RV, Ces O. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat Commun 2014; 5:1–5 [DOI] [PubMed] [Google Scholar]
- 14.Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, Thompson J, Wallace M. Droplet interface bilayers. Mol Biosyst 2008; 4:1191–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuruma Y, Suzuki T, Ono S, Yoshida M, Ueda T. Functional analysis of membranous Fo-a subunit of F1Fo-ATP synthase by in vitro protein synthesis. Biochem J 2012; 442:631–8 [DOI] [PubMed] [Google Scholar]
- 16.Kalmbach R, Chizhov I, Schumacher MC, Friedrich T, Bamberg E, Engelhard M. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. J Mol Biol 2007; 371:639–48 [DOI] [PubMed] [Google Scholar]
- 17.Kuruma Y, Ueda T. The PURE system for the cell-free synthesis of membrane proteins. Nat Protoc 2015; 10:1328–44 [DOI] [PubMed] [Google Scholar]
- 18.Sachse R, Dondapati SK, Fenz SF, Schmidt T, Kubick S. Membrane protein synthesis in cell-free systems: from bio-mimetic systems to bio-membranes. FEBS Lett 2014; 588:2774–81 [DOI] [PubMed] [Google Scholar]
- 19.Krupinski J, Hammes GG. Steady-state ATP synthesis by bacteriorhodopsin and chloroplast coupling factor co-reconstituted into asolectin vesicles. Proc Natl Acad Sci U S A 1986; 83:4233–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen GU, Lawless JG, Edelson EH. Quantification of monocarboxylic acids from a spark discharge synthesis. J Mol Evol 1981; 17:43–7 [Google Scholar]
- 21.McCollom TM, Ritter G, Simoneit BRT. Lipid synthesis under hydrothermal conditions by Fischer-Tropsch reactions. Orig Life Evol Biosph 1999; 29:153–66 [DOI] [PubMed] [Google Scholar]
- 22.Mißbach H, Schmidt BC, Duda JP, Lünsdorf NK, Goetz W, Thiel V. Assessing the diversity of lipids formed via Fischer-Tropsch-type reactions. Org Geochem 2018; 119:110–21 [Google Scholar]
- 23.Pizzarello S, Shock E. The organic composition of carbonaceous meteorites: the evolutionary story ahead of biochemistry. Cold Spring Harb Perspect Biol 2010; 2:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hargreaves WR, Deamer DW. Liposomes from ionic, single-chain amphiphiles. Biochemistry 1978; 17:3759–68 [DOI] [PubMed] [Google Scholar]
- 25.Walde P, Wick R, Fresta M, Mangone A, Luisi PL. Autopoietic self-reproduction of fatty acid vesicles. J Am Chem Soc 1994; 116:11649–54 [Google Scholar]
- 26.Gibard C, Bhowmik S, Karki M, Kim EK, Krishnamurthy R. Phosphorylation, oligomerization and self-assembly in water under potential prebiotic conditions. Nat Chem 2018; 10:212–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat Chem 2015; 7:301–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-García C, Powner MW. Selective acylation of nucleosides, nucleotides, and glycerol-3- phosphocholine in water. Synlett 2017; 28:78–83 [Google Scholar]
- 29.Luisi PL. Are micelles and vesicles chemical equilibrium systems? J Chem Educ 2001; 78:380 [Google Scholar]
- 30.Chen IA, Szostak JW. A kinetic study of the growth of fatty acid vesicles. Biophys J 2004; 87:988–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean LR, Phillips MC. Kinetics of phosphatidylcholine and lysophosphatidylcholine exchange between unilamellar vesicles. Biochemistry 1984; 23:4624–30 [DOI] [PubMed] [Google Scholar]
- 32.Nichols JW, Pagano RE. Kinetics of soluble lipid monomer diffusion between vesicles. Biochemistry 1981; 20:2783–9 [DOI] [PubMed] [Google Scholar]
- 33.Zhu TF, Szostak JW. Coupled growth and division of model protocell membranes. J Am Chem Soc 2009; 131:5705–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sacerdote MG, Szostak JW. Semipermeable lipid bilayers exhibit diastereoselectivity favoring ribose. Proc Natl Acad Sci U S A 2005; 102:6004–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansy SS, Schrum JP, Krishnamurthy M, Tobé S, Treco DA, Szostak JW. Template-directed synthesis of a genetic polymer in a model protocell. Nature 2008; 454:122–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Flaherty DK, Kamat NP, Mirza FN, Li L, Prywes N, Szostak JW. Copying of mixed-sequence RNA templates inside model protocells. J Am Chem Soc 2018; 140:5171–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanczyc MM, Monnard PA. Primordial membranes: more than simple container boundaries. Curr Opin Chem Biol 2017; 40:78–86 [DOI] [PubMed] [Google Scholar]
- 38.Cape JL, Monnard PA, Boncella JM. Prebiotically relevant mixed fatty acid vesicles support anionic solute encapsulation and photochemically catalyzed trans-membrane charge transport. Chem Sci 2011; 2:661 [Google Scholar]
- 39.Budin I, Prwyes N, Zhang N, Szostak JW. Chain-length heterogeneity allows for the assembly of fatty acid vesicles in dilute solutions. Biophys J 2014; 107:1582–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansy SS, Szostak JW. Thermostability of model protocell membranes. Proc Natl Acad Sci U S A 2008; 105:13351–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurer SE, Deamer DW, Boncella JM, Monnard PA. Chemical evolution of amphiphiles: glycerol monoacyl derivatives stabilize plausible prebiotic membranes. Astrobiology 2009; 9:979–87 [DOI] [PubMed] [Google Scholar]
- 42.Namani T, Deamer DW. Stability of model membranes in extreme environments. Orig Life Evol Biosph 2008; 38:329–41 [DOI] [PubMed] [Google Scholar]
- 43.Monnard PA, Apel CL, Kanavarioti A, Deamer DW. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: implications for a prebiotic aqueous medium. Astrobiology 2002; 2:139–52 [DOI] [PubMed] [Google Scholar]
- 44.Monnard PA, Deamer DW. Preparation of vesicles from nonphospholipid amphiphiles. Meth Enzymol 2003; 372:133–51 [DOI] [PubMed] [Google Scholar]
- 45.Chen IA, Roberts RW, Szostak JW. The emergence of competition between model protocells. Science 2004; 305:1474–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu TF, Adamala K, Zhang N, Szostak JW. Photochemically driven redox chemistry induces protocell membrane pearling and division. Proc Natl Acad Sci U S A 2012; 109:9828–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budin I, Debnath A, Szostak JW. Concentration-driven growth of model protocell membranes. J Am Chem Soc 2012; 134:20812–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalai P, Ustriyana P, Sahai N. Aqueous magnesium as an environmental selection pressure in the evolution of phospholipid membranes on early earth. Geochim Cosmochim Acta 2018; 223:216–28 [Google Scholar]
- 49.Jin L, Kamat NP, Jena S, Szostak JW. Fatty acid/phospholipid blended membranes: a potential intermediate state in protocellular evolution. Small 2018; 14:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budin I, Szostak JW. Physical effects underlying the transition from primitive to modern cell membranes. Proc Natl Acad Sci U S A 2011; 108:5249–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zakim D, Daniels C, Noy N. Rates of hydration of fatty acids bound to unilamellar vesicles of phosphatidylcholine or to albumin. Biochemistry 1985; 24:3286–92 [DOI] [PubMed] [Google Scholar]
- 52.Hamilton JA. Medium-chain fatty acid binding to albumin and transfer to phospholipid bilayers. Proc Natl Acad Sci U S A 1989; 86:2663–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massey JB, Bick DH, Pownall HJ. Spontaneous transfer of monoacyl amphiphiles between lipid and protein surfaces. Biophys J 1997; 72:1732–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean LR, Phillips MC. Mechanism of cholesterol and phosphatidylcholine exchange or transfer between unilamellar vesicles. Biochemistry 1981; 20:2893–900 [DOI] [PubMed] [Google Scholar]
- 55.Deamer DW, Weber AL. Bioenergetics and life’s origins. CHS Perspect Biol 2010; 2:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrenfreund P, Rasmussen S, Cleaves J, Chen L. Experimentally tracing the key steps in the origin of life: the aromatic world. Astrobiology 2006; 6:490–520 [DOI] [PubMed] [Google Scholar]
- 57.Apel CL, Deamer DW, Mautner MN. Self-assembled vesicles of monocarboxylic acids and alcohols: conditions for stability and for the encapsulation of biopolymers. Biochim Biophys Acta 2002; 1559:1–9 [DOI] [PubMed] [Google Scholar]
- 58.Black RA, Blosser MC, Stottrup BL, Tavakley R, Deamer DW, Keller SL. Nucleobases bind to and stabilize aggregates of a prebiotic amphiphile, providing a viable mechanism for the emergence of protocells. Proc Natl Acad Sci U S A 2013; 110:13272–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen IA, Salehi-Ashtiani K, Szostak JW. RNA catalysis in model protocell vesicles. J Am Chem Soc 2005; 127:13213–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litschel T, Ganzinger KA, Movinkel T, Heymann M, Robinson T, Mutschler H, Schwille P. Freeze-thaw cycles induce content exchange between cell-sized lipid vesicles. New J Phys 2018; 20:055008 [Google Scholar]
- 61.Su WC, Gettel DL, Chabanon M, Rangamani P, Parikh AN. Pulsatile gating of giant vesicles containing macromolecular crowding agents induced by colligative nonideality. J Am Chem Soc 2018; 140:691–9 [DOI] [PubMed] [Google Scholar]
- 62.Piedrafita G, Monnard PA, Mavelli F, Ruiz-Mirazo K. Permeability-driven selection in a semi-empirical protocell model: the roots of prebiotic systems evolution. Sci Rep 2017; 7:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizushina Y, Tanaka N, Yagi H, Kurosawa T, Onoue M, Seto H, Horie T, Aoyagi N, Yamaoka M, Matsukage A, Yoshida S, Sakaguchi K. Fatty acids selectively inhibit eukaryotic DNA polymerase activities in vitro. Biochim Biophys Acta 1996; 1308:256–62 [DOI] [PubMed] [Google Scholar]
- 64.Nakamura R, Takeuchi R, Kuramochi K, Mizushina Y, Ishimaru C, Takakusagi Y, Takemura M, Kobayashi S, Yoshida H, Sugawara F, Sakaguchi K. Chemical properties of fatty acid derivatives as inhibitors of DNA polymerases. Org Biomol Chem 2007; 5:3912. [DOI] [PubMed] [Google Scholar]
- 65.Parker ET, Cleaves HJ, Dworkin JP, Glavin DP, Callahan M, Aubrey A, Lazcano A, Bada JL. Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proc Natl Acad Sci U S A 2011; 108:5526–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 2004; 39:99–123 [DOI] [PubMed] [Google Scholar]
- 67.Oró J, Kimball AP. Synthesis of purines under possible primitive earth conditions. I. Adenine from hydrogen cyanide. Arch Biochem Biophys 1961; 94:217–27 [DOI] [PubMed] [Google Scholar]
- 68.Šponer JE, Szabla R, Góra RW, Saitta AM, Pietrucci F, Saija F, Di Mauro E, Saladino R, Ferus M, Civiš S, Sponer J. Prebiotic synthesis of nucleic acids and their building blocks at the atomic level-merging models and mechanisms from advanced computations and experiments. Phys Chem Chem Phys 2016; 18:20047–66 [DOI] [PubMed] [Google Scholar]
- 69.Callahan MP, Smith KE, Cleaves HJ, Ruzicka J, Stern JC, Glavin DP, House CH, Dworkin JP. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc Natl Acad Sci USA 2011; 108:13995–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rapf RJ, Vaida V. Sunlight as an energetic driver in the synthesis of molecules necessary for life. Phys Chem Chem Phys 2016; 18:20067–84 [DOI] [PubMed] [Google Scholar]
- 71.Szabla R, Tuna D, Góra RW, Šponer J, Sobolewski AL, Domcke W. Photochemistry of 2-aminooxazole: a hypothetical prebiotic precursor of RNA nucleotides. J Phys Chem Lett 2013; 4:2785–8 [Google Scholar]
- 72.Kolb VM, Dworkin JP, Miller SL. Alternative bases in the RNA world: the prebiotic synthesis of urazole and its ribosides. J Mol Evol 1994; 38:549–57 [DOI] [PubMed] [Google Scholar]
- 73.Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009; 459:239–42 [DOI] [PubMed] [Google Scholar]
- 74.Xu J, Tsanakopoulou M, Magnani CJ, Szabla R, Šponer JE, Šponer J, Góra RW, Sutherland JD. A prebiotically plausible synthesis of pyrimidine β-ribonucleosides and their phosphate derivatives involving photoanomerization. Nat Chem 2017; 9:303–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonfio C, Valer L, Scintilla S, Shah S, Evans DJ, Jin L, Szostak JW, Sasselov DD, Sutherland JD, Mansy SS. UV-light-driven prebiotic synthesis of iron-sulfur clusters. Nat Chem 2017; 9:1229–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burton AS, Stern JC, Elsila JE, Glavin DP, Dworkin JP. Understanding prebiotic chemistry through the analysis of extraterrestrial amino acids and nucleobases in meteorites. Chem Soc Rev 2012; 41:5459–72 [DOI] [PubMed] [Google Scholar]
- 77.Raggi L, Bada JL, Lazcano A. On the lack of evolutionary continuity between prebiotic peptides and extant enzymes. Phys Chem Chem Phys 2016; 18:20028–32 [DOI] [PubMed] [Google Scholar]
- 78.Lazcano A, Miller SL. On the origin of metabolic pathways. J Mol Evol 1999; 49:424–31 [PubMed] [Google Scholar]
- 79.Trifonov EN. The triplet code from first principles. J Biomol Struct Dyn 2004; 22:1–11 [DOI] [PubMed] [Google Scholar]
- 80.Hud NV, Cafferty BJ, Krishnamurthy R, Williams LD. The origin of RNA and “my grandfather’s axe. Chem Biol 2013; 20:466–74 [DOI] [PubMed] [Google Scholar]
- 81.Ross D, Deamer DW. Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life 2016; 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriguez-Garcia M, Surman AJ, Cooper GJT, Suárez-Marina I, Hosni Z, Lee MP, Cronin L. Formation of oligopeptides in high yield under simple programmable conditions. Nat Commun 2015; 6:8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajamani S, Vlassov A, Benner S, Coombs A, Olasagasti F, Deamer DW. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig Life Evol Biosph 2008; 38:57–74 [DOI] [PubMed] [Google Scholar]
- 84.Mungi C, Rajamani S. Characterization of RNA-like oligomers from lipid-assisted nonenzymatic synthesis: implications for origin of informational molecules on early earth. Life 2015; 5:65–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forsythe JG, Yu SS, Mamajanov I, Grover MA, Krishnamurthy R, Fernández FM, Hud NV. Ester-mediated amide bond formation driven by wet-dry cycles: a possible path to polypeptides on the prebiotic earth. Angew Chem Int Ed 2015; 54:9871–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toppozini L, Dies H, Deamer DW, Rheinstädter MC. Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: insights into assembly of nucleic acid for primitive life. PLoS One 2013; 8:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mamajanov I, Macdonald PJ, Ying J, Duncanson DM, Dowdy GR, Walker CA, Engelhart AE, Fernández FM, Grover MA, Hud NV, Schork FJ. Ester formation and hydrolysis during wet-dry cycles: generation of far-from-equilibrium polymers in a model prebiotic reaction. Macromolecules 2014; 47:1334–43 [Google Scholar]
- 88.Leman L, Orgel L, Ghadiri MR. Carbonyl sulfide-mediated prebiotic formation of peptides. Science 2004; 306:283–6 [DOI] [PubMed] [Google Scholar]
- 89.Powner MW, Sutherland JD, Szostak JW. The origins of nucleotides. Synlett 2011;14:1956-1964
- 90.Li L, Prywes N, Tam CP, O’Flaherty DK, Lelyveld VS, Izgu EC, Pal A, Szostak JW. Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J Am Chem Soc 2017; 139:1810–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weiman BJ, Lohrmann R, Orgel LE, Schneider-Bernloehr H, Sulston JE. Template-directed synthesis with adenosine-5’-phosphorimidazolide. Science 1968; 1067:13435. [DOI] [PubMed] [Google Scholar]
- 92.Yi R, Hongo Y, Fahrenbach AC. Synthesis of imidazole-activated ribonucleotides using cyanogen chloride. Chem Commun 2018; 54:511–4 [DOI] [PubMed] [Google Scholar]
- 93.Mariani A, Russell DA, Javelle T, Sutherland JD. A light-releasable potentially prebiotic nucleotide activating agent. J Am Chem Soc 2018; 140:8657–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mullen LB, Sutherland JD. Simultaneous nucleotide activation and synthesis of amino acid amides by a potentially prebiotic multi-component reaction. Angew Chem Int Ed 2007; 46:8063–6 [DOI] [PubMed] [Google Scholar]
- 95.Krishnamurthy R, Guntha S, Eschenmoser A. Regioselective alpha-phosphorylation of aldoses in aqueous solution. Angew Chem Int Ed 2000; 39:2281–5 [PubMed] [Google Scholar]
- 96.Walter F, Erich T. Zur Chemie der kondensierten phosphate und arsenate. Anorg Allg Chem 1964; 328:113–216 [Google Scholar]
- 97.Monnard PA, Ziock H. Eutectic phase in water-ice: a self-assembled environment conducive to metal-catalyzed non-enzymatic RNA polymerization. Chem Biodivers 2008; 5:1521–39 [DOI] [PubMed] [Google Scholar]
- 98.Mutschler H, Wochner A, Holliger P. Freeze-thaw cycles as drivers of complex ribozyme assembly. Nat Chem. 2015; 7:502–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Attwater J, Wochner A, Holliger P. In-ice evolution of RNA polymerase ribozyme activity. Nat Chem 2013; 5:1011–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mariani A, Sutherland JD. Non-enzymatic RNA backbone proofreading through energy-dissipative recycling. Angew Chem Int Ed 2017; 56:6563–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bartel DP, Szostak JW. Isolation of a large new ribozymes pool of random sequences. Science 1993; 261:1411–8 [DOI] [PubMed] [Google Scholar]
- 102.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 2001; 292:1319–25 [DOI] [PubMed] [Google Scholar]
- 103.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science 2011; 332:209–12 [DOI] [PubMed] [Google Scholar]
- 104.Sczepanski JT, Joyce GF. A cross-chiral RNA polymerase ribozyme. Nature 2014; 515:440–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Horning DP, Joyce GF. Amplification of RNA by an RNA polymerase ribozyme. Proc Natl Acad Sci U S A 2016; 113:9786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cech TR. The ribosome is a ribozyme. Science 2000; 289:878–9 [DOI] [PubMed] [Google Scholar]
- 107.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000; 289:905–20 [DOI] [PubMed] [Google Scholar]
- 108.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920–30 [DOI] [PubMed] [Google Scholar]
- 109.Turk RM, Chumachenko NV, Yarus M. Multiple translational products from a five-nucleotide ribozyme. Proc Natl Acad Sci U S A 2010; 107:4585–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turk RM, Illangasekare M, Yarus M. Catalyzed and spontaneous reactions on ribozyme ribose. J Am Chem Soc 2011; 133:6044–50 [DOI] [PubMed] [Google Scholar]
- 111.Suga H, Hayashi G, Terasaka N. The RNA origin of transfer RNA aminoacylation and beyond. Philos Trans R Soc Lond B 2011; 366:2959–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao H, Murakami H, Suga H, Ferré DAmaré. AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 2008; 454:358–61 [DOI] [PubMed] [Google Scholar]
- 113.Tam CP, Fahrenbach AC, Björkbom A, Prywes N, Izgu EC, Szostak JW. Downstream oligonucleotides strongly enhance the affinity of GMP to RNA primer-template complexes. J Am Chem Soc 2017; 139:571–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Attwater J, Raguram A, Morgunov AS, Gianni E, Holliger P, Avenue FC, Campus CB, Street O. Ribozyme-catalysed RNA synthesis using triplet building blocks. Elife 2018;7:e35255 [DOI] [PMC free article] [PubMed]
- 115.Tagami S, Attwater J, Holliger P. Simple peptides derived from the ribosomal core potentiate RNA polymerase ribozyme function. Nat Chem 2017; 9:325–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ichihashi N, Usui K, Kazuta Y, Sunami T, Matsuura T, Yomo T. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat. Commun 2013; 4:1–7 [DOI] [PubMed] [Google Scholar]
- 117.Tsuji G, Fujii S, Sunami T, Yomo T. Sustainable proliferation of liposomes compatible with inner RNA replication. Proc Natl Acad Sci U S A 2016;113:590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ruiz-Mirazo K, Briones C, De La Escosura A. Prebiotic systems chemistry: new perspectives for the origins of life. Chem Rev 2014; 114:285–366 [DOI] [PubMed] [Google Scholar]
- 119.Morowitz HJ, Kostelnik JD, Yang J, Cody GD. The origin of intermediary metabolism. Proc Natl Acad Sci U S A 2000; 97:7704–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith E, Morowitz HJ. Universality in intermediary metabolism. Proc Natl Acad Sci U S A 2004; 101:13168–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Orgel LE. The implausibility of metabolic cycles on the prebiotic earth. PLoS Biol 2008; 6:e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coggins AJ, Powner MW. Prebiotic synthesis of phosphoenol pyruvate by α-phosphorylation-controlled triose glycolysis. Nat Chem 2017; 2:310–317 [DOI] [PubMed] [Google Scholar]
- 123.Keller MA, Turchyn AV, Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol Syst Biol 2014; 10:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keller MA, Zylstra A, Castro C, Turchyn AV, Griffin JL, Ralser M. Conditional iron and pH-dependent activity of a non-enzymatic glycolysis and pentose phosphate pathway. Sci Adv 2016; 2:e1501235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Varma SJ, Muchowska KB, Chatelain P, Moran J. Native iron reduces CO2 to intermediates and end-products of the acetyl-CoA pathway. Nat Ecol Evol 2018; 2: 1019–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muchowska KB, Varma SJ, Chevallot-Beroux E, Lethuillier-Karl L, Li G, Moran J. Metals promote sequences of the reverse Krebs cycle. Nat Ecol Evol 2017; 1:1716–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Springsteen G, Yerabolu JR, Nelson J, Rhea CJ, Krishnamurthy R. Linked cycles of oxidative decarboxylation of glyoxylate as protometabolic analogs of the citric acid cycle. Nat Commun 2018; 9:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guzman MI, Martin ST, Guzman MI, Martin ST, Lide DR. Oxaloacetate-to-malate conversion by mineral photoelectrochemistry: implications for the viability of the reductive tricarboxylic acid cycle in prebiotic chemistry. Int J Astrobiol 2008; 7:271–8 [Google Scholar]
- 129.Belmonte L, Mansy SS. Metal catalysts and the origin of life. Elements 2016; 12:413–8 [Google Scholar]
- 130.Biosa G, Bastianoni S, Rustici M. Chemical waves. Chemistry 2006; 12:3430–7 [DOI] [PubMed] [Google Scholar]
- 131.Gines G, Zadorin AS, Galas JC, Fujii T, Estevez-Torres A, Rondelez Y. Microscopic agents programmed by DNA circuits. Nat Nanotechnol 2017; 12:351–9 [DOI] [PubMed] [Google Scholar]
- 132.Krishnan Y, Simmel FC. Nucleic acid based molecular devices. Angew Chem Int Ed Engl 2011; 50:3124–56 [DOI] [PubMed] [Google Scholar]
- 133.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992; 355:850–2 [DOI] [PubMed] [Google Scholar]
- 134.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science 2006; 314:1585–9 [DOI] [PubMed] [Google Scholar]
- 135.Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N. Spontaneous network formation among cooperative RNA replicators. Nature 2012; 491:72–7 [DOI] [PubMed] [Google Scholar]
- 136.Semenov SN, Wong ASY, Van Der Made RM, Postma SGJ, Groen J, Van Roekel HWH, De Greef TFA, Huck WTS. Rational design of functional and tunable oscillating enzymatic networks. Nat Chem 2015; 7:160–5 [DOI] [PubMed] [Google Scholar]
- 137.Pogodaev AA, Wong ASY, Huck WTS. Photochemical control over oscillations in chemical reaction networks. J Am Chem Soc 2017; 139:15296–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Postma SGJ, Te Brinke D, Vialshin IN, Wong ASY, Huck WTS. A trypsin-based bistable switch. Tetrahedron 2017; 73:4896–900 [Google Scholar]
- 139.Semenov SN, Kraft LJ, Ainla A, Zhao M, Baghbanzadeh M, Campbell VE, Kang K, Fox JM, Whitesides GM. Autocatalytic, bistable, oscillatory networks of biologically relevant organic reactions. Nature 2016; 537:656–60 [DOI] [PubMed] [Google Scholar]
- 140.Kauffman SA. Autocatalytic sets of proteins. J Theor Biol 1986; 119:1–24 [DOI] [PubMed] [Google Scholar]
- 141.Hordijk W, Steel M, Kauffman S. The structure of autocatalytic sets: evolvability, enablement, and emergence. Acta Biotheor 2012; 60:379–92 [DOI] [PubMed] [Google Scholar]
- 142.Carnall JMA, Waudby CA, Belenguer AM, Stuart MCA, Peyralans JJP, Otto S. Mechanosensitive self-replication driven by self-organization. Science 2010; 327:1502–6 [DOI] [PubMed] [Google Scholar]
- 143.Omosun TO, Hsieh MC, Childers WS, Das D, Mehta AK, Anthony NR, Pan T, Grover MA, Berland KM, Lynn DG. Catalytic diversity in self-propagating peptide assemblies. Nat Chem 2017; 9:805–9 [DOI] [PubMed] [Google Scholar]
- 144.Nanda J, Rubinov B, Ivnitski D, Mukherjee R, Shtelman E, Motro Y, Miller Y, Wagner N, Cohen-Luria R, Ashkenasy G. Emergence of native peptide sequences in prebiotic replication networks. Nat Commun 2017; 8:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maiti S, Fortunati I, Ferrante C, Scrimin P, Prins LJ. Dissipative self-assembly of vesicular nanoreactors. Nat Chem 2016; 8:725–31 [DOI] [PubMed] [Google Scholar]
- 146.Bhattacharya A, Brea RJ, Devaraj NK. De novo vesicle formation and growth: an integrative approach to artificial cells. Chem Sci 2017;8:7912–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zepik HH, Blöchliger E, Luisi PL. A chemical model of homeostasis. Angew Chem Int Ed 2001; 40:199–202 [DOI] [PubMed] [Google Scholar]
- 148.Bissette AJ, Odell B, Fletcher SP. Physical autocatalysis driven by a bond-forming thiol-ene reaction. Nat Commun 2014; 5:1–8 [DOI] [PubMed] [Google Scholar]
- 149.Taran O, Chen C, Tolulope O, Hsieh M, Rha A, Goodwin JT, Mehta AK, Grover MA, Lynn DG. Expanding the informational chemistries of life: peptide/RNA networks. Philos Trans A Math Phys Eng Sci 2017; 375:20160356 [DOI] [PubMed] [Google Scholar]
- 150.De Duve C. Vital dust: the origin and evolution of life on earth New York: Basic Books, 1995.
- 151.Goldford JE, Hartman H, Smith TF, Segrè D. Remnants of an ancient metabolism without phosphate. Cell 2017; 168:1126–34.e9 [DOI] [PubMed] [Google Scholar]
- 152.Chen YH, Sung PH, Sung K. Synthesis of proline-derived dipeptides and their catalytic enantioselective direct aldol reactions: catalyst, solvent, additive and temperature effects. Amino Acids 2010; 38:839–45 [DOI] [PubMed] [Google Scholar]
- 153.Imanishi Y, Tanihara M, Higashimura T. Histidine-containing cyclic dipeptides as catalysts in the hydrolysis of carbonic acid p-nitrophenyl esters. Biopolymers 1977; 16:2203–15 [DOI] [PubMed] [Google Scholar]
- 154.Luppi G, Cozzi PG, Monari M, Kaptein B, Broxterman QB, Tomasini C. Dipeptide-catalyzed asymmetric aldol condensation of acetone with (N-alkylated) isatins. J Org Chem 2005; 70:7418–21 [DOI] [PubMed] [Google Scholar]
- 155.Wieczorek RD, Dörr M, Chotera A, Luisi PL, Monnard PA. Formation of RNA phosphodiester bond by histidine-containing dipeptides. Chem Bio Chem 2013; 14:217–23 [DOI] [PubMed] [Google Scholar]
- 156.Gorlero M, Wieczorek R, Adamala K, Giorgi A, Schininà ME, Stano P, Luisi PL. Ser-His catalyses the formation of peptides and PNAs. FEBS Lett 2009; 583:153–6 [DOI] [PubMed] [Google Scholar]
- 157.Belmonte L, Rossetto D, Forlin M, Scintilla S, Bonfio C, Mansy SS. Cysteine containing dipeptides show a metal specificity that matches the composition of seawater. Phys Chem Chem Phys 2016; 18:20104–8 [DOI] [PubMed] [Google Scholar]
- 158.Kumar M, Ing NL, Narang V, Wijerathne NK, Hochbaum AI, Ulijn RV. Amino-acid-encoded biocatalytic self-assembly enables the formation of transient conducting nanostructures. Nat Chem 2018; 10:696–703 [DOI] [PubMed] [Google Scholar]
- 159.Mondal S, Varenik M, Bloch DN, Atsmon-Raz Y, Jacoby G, Adler-Abramovich L, Shimon LJW, Beck R, Miller Y, Regev O, Gazid E. A minimal length rigid helical peptide motif allows rational design of modular surfactants. Nat Commun 2017; 8:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Frederix Pwjm Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Exploring the sequence space for (tri-)peptide self-assembly to design and discover new hydrogels. Nat Chem 2015; 7:30–7 [DOI] [PubMed] [Google Scholar]
- 161.Alva V, Söding J, Lupas AN. A vocabulary of ancient peptides at the origin of folded proteins. Elife 2015; 4:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scintilla S, Bonfio C, Belmonte L, Forlin M, Rossetto D, Li J, Cowan JA, Galliani A, Arnesano F, Assfalg M, Mansy SS. Duplications of an iron–sulphur tripeptide leads to the formation of a protoferredoxin. Chem Commun 2016; 52:13456–9 [DOI] [PubMed] [Google Scholar]
- 163.Blain JC, Szostak JW. Progress Toward Synthetic Cells. Annu Rev Biochem 2014; 83:615–40 [DOI] [PubMed] [Google Scholar]
- 164.Mansy SS, Pohflepp S. Living machines. In: Ginsberg AD, Calvert J, Schyfter P, Elfick A and Endy D (eds) Synthetic aesthetics. Cambridge, MA: The MIT Press, 2014, pp.233–244
- 165.Nies P, Van Westerlaken I, Blanken D, Salas M, Mencía M. Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat Commun 2018; 9:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hindley JW, Elani Y, McGilvery CM, Ali S, Bevan CL, Law RV, Ces O. Light-triggered enzymatic reactions in nested vesicle reactors. Nat Commun 2018; 9:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lentini R, Martín NY, Forlin M, Belmonte L, Fontana J, Cornella M, Martini L, Tamburini S, Bentley WE, Jousson O, Mansy SS. Two-way chemical communication between artificial and natural cells. ACS Cent Sci 2017; 3:117–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Van Hest JCM. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc Chem Res 2017; 50:769–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Booth MJ, Schild VR, Graham AD, Olof SN, Bayley H. Light-activated communication in synthetic tissues. Sci Adv 2016; 2:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Langton MJ, Scriven LM, Williams NH, Hunter CA. Triggered release from lipid bilayer vesicles by an artificial transmembrane signal transduction system. J Am Chem Soc 2017; 139:15768–73 [DOI] [PubMed] [Google Scholar]