Short abstract
Phospholipid membranes are necessary for the compartmentalization of chemistries within biological cells and for initiation and propagation of cell signaling. The morphological and chemical complexities of cellular membranes represent a challenge for dissecting the biochemical processes occurring at these interfaces. Therefore, investigations of the biological events occurring at the membrane require suitable models to reproduce the intricacy of this surface. Solid-supported lipid bilayers (SLBs) are simplified physical replicas of biological membranes that allow for bottom-up reconstruction of the molecular mechanisms occurring at cellular interfaces. In this brief review, we introduce how the properties of SLBs can be tuned to mimic biological membranes, highlighting the engineering approaches for creating spatially resolved patterns of lipid bilayers and supported membranes with curved geometries. Additionally, we present how SLBs have been employed to reconstitute molecular mechanisms involved in intercellular signaling and more recently, membrane trafficking.
Impact statement
Artificial membranes with complex topography aid the understanding of biological processes where membrane geometry plays a key regulatory role. In this review, we highlight how emerging material and engineering technologies have been employed to create minimal models of cell signaling pathways, in vitro. These artificial systems allow life scientists to answer ever more challenging questions with regards to mechanisms in cellular biology. In vitro reconstitution of biology is an area that draws on the expertise and collaboration between biophysicists, material scientists and biologists and has recently generated a number of high impact results, some of which are also discussed in this review.
Keywords: Solid-supported lipid bilayers, membrane patterning, membrane curvature, synthetic biology, in vitro reconstitution
Introduction
Phospholipid membranes allow for compartmentalization of chemical signals within a biological cell. They separate the extracellular milieu from the cytoplasmic interior, defining a cell’s boundaries and delimit all the organelles within its cytosol. They also protect the genome from the cytosolic environment and attack from nucleases. Biological membranes are dynamic structures that permit bidirectional transport of cargo, cellular communication and signaling. Therefore, they are more than just a diffusion barrier and participate in cellular signaling, serving as scaffolds and surfaces for protein complexes.1
The phospholipid bilayer has been known as a functional two-dimensional fluid since the 1970s2 with the first bilayer structure based on amphiphilic phospholipids presented in 1925.3 Today, our understanding of molecular processes at the biological membrane has been built upon studies performed on models of the cell membrane. These include structures such as liposomes, giant unilamellar vesicles (GUVs), monolayers and polymer-supported lipid bilayers4,5 (Figure 1).
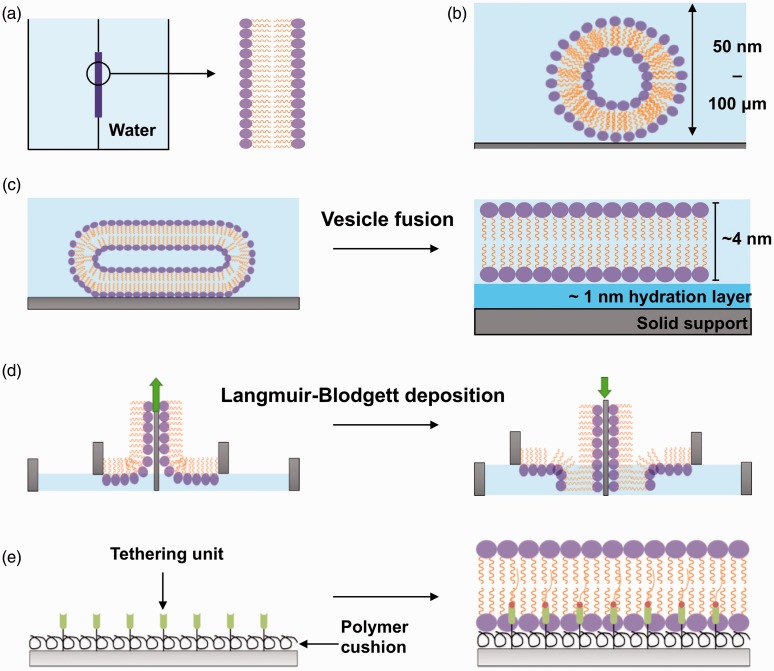
Figure 1.
In vitro approaches used to model the biological membrane. (a) Black lipid membranes are formed when dried down lipids are hydrated with an aqueous medium over an aperture between two aqueous phases. (b) Liposomes (50 nm) or giant uni-lamellar vesicles (100 µm) are formed when dried down lipids are hydrated in an aqueous medium. (c) SLB formation by the vesicle fusion method. Liposomes are incubated onto a hydrophilic surface where they fuse, deform, and spontaneously rupture to form a uniform, continuous, fluid lipid bilayer. (d) SLB formation with the Langmuir-Blodgett technique. Lipid monolayers are formed at the air–water interface when lipids are added to an aqueous medium. This monolayer is transferred onto a hydrophilic solid support twice to form a lipid bilayer. The green arrow indicates the direction the substrate (grey) is moving to pick up another monolayer of lipids. (e) Polymer-supported/tethered lipid bilayers are formed on top of a polymer cushion hydrogel and also tethering units such as nickel NTA for anchoring of proteins. (A color version of this figure is available in the online journal.)
The planar lipid bilayers or solid-supported lipid bilayers (SLBs) are model systems constructed via self-assembly of lipids on a solid support. They can be engineered to exhibit similar fluid properties of crowded natural cell membranes, displaying lateral diffusion coefficients between 1 μm2 s−1 (similar to cell membranes) or slightly faster diffusion rates up to 3–4 μm2 s−1, depending on the physical properties of the support interface.6 The lateral diffusion of lipids in SLBs is also influenced by the liquid-crystalline properties of the bilayer that change with lipid composition.7 Lipid diffusion is confined to the plane of the solid support where the fluidity of lipid leaflets is sustained by a ∼1 nm layer of water parting the bilayer from the substrate.8
SLBs have been utilized to investigate membrane organization,9 protein self-assembly,10 membrane remodeling11,12 and deduce mechanisms for spatial and temporal initiation of cell signaling at the plasma membrane.13–15 The major impact from investigating biochemical processes occurring at the membrane may be in the field of drug development, given that around 50% of drugs target transmembrane proteins.16
Importantly, SLBs can be designed to be more than just a flat lipid bilayer. Solid support and membrane bilayer morphology can be easily altered to accurately mimic a system of interest. Specifically, the spatial organization and mechanical deformation of supported membranes can be manipulated by patterning the underlying substrate with modern micro and nano-fabrication techniques (Table 1).
Table 1.
Outline of the techniques for patterning SLBs, their resolutions, and disadvantages/advantages.
| Technique | Patterning process | Lateralresolution | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Micro-contact printing | Form of soft lithography that fabricates structures using elastomeric stamps, molds and conformable photomasks. | 500 nm |
|
|
40,46 |
| Photo-lithography | Uses light to transfer a pattern from a photo-mask to a light-sensitive chemical (photoresist), on a substrate. | <100 nm |
|
|
60 |
| Scanning probe lithography | Set of nano-lithographic methods to pattern materials using scanning probes. | 10 nm |
|
|
42,57,61 |
| Electron beam lithography | A beam of electrons is scanned over a surface covered with an electron-sensitive resist, removing regions of exposed or non-exposed resist to draw a pattern. | <10 nm |
|
|
53,62 |
| Multi-photon lithography(direct laser writing) | Structuring is achieved by direct fabrication using near infrared femtosecond lasers to induce polymerization at the focal point. | 100 nm |
|
|
45,58,59 |
In this mini review, we introduce the technologies to engineer spatially resolved flat lipid bilayers and membranes containing patterns of curvature. We discuss how these artificial systems facilitate the study of the biological pathways controlled by the interplay between cellular membranes and membrane-interacting proteins.
Towards building a realistic model of the cell membrane
Cellular membranes exhibit heterogeneity in lipid composition and display a variety of shapes and morphologies, which are necessary for specialization of function. For example, asymmetry in cholesterol lateral composition at the plasma membrane plays a role in the compartmentalization of protein complexes and in activating and regulating cell signaling via membrane trafficking.17,18 The function of membrane-bound organelles is linked to their complex morphologies. An exquisite example is the continuous membrane network of the endoplasmic reticulum (ER), with its closely packed membrane sheets. The ER membrane architecture maximizes the surface areas for protein synthesis, whereas the tubules in the peripheral are used for contacting other membrane organelles.19 Some organelles change morphology progressively to acquire diverse functions, such as endosomes ‘maturing’ into multivesicular bodies during cargo trafficking to the lysosomes.20
The earliest model system of a cell membrane was the black lipid membrane, developed by Mueller in the 1960s,21 but it was not very versatile (Figure 1(a)). Today, liposomes and GUVs (Figure 1(b)) offer the possibility to study the role of lipid composition and membrane mechanics in the context of protein–membrane interactions.4 Generally, GUVs are more appropriate for studying how a wide range of membrane physical parameters affect the interaction between membranes and other molecules. Nonetheless, SLBs offer specific advantages for quantifying kinetic processes at the molecular level since they are amenable to coupling with surface-sensitive analysis techniques, such as surface plasmon resonance and quartz microbalance. Low- and high-resolution imaging techniques, such as atomic force microscopy (AFM), fluorescence recovery after photobleaching, and total internal reflection microscopy (TIRF) permit image acquisition of membrane topography, quantification of membrane fluidity, and localization of protein assemblies at the supported membrane.22
There is a choice of methods available to produce SLBs, depending on the extent of surface coverage required and the type of solid support used. The direct vesicle fusion23 method is performed in aqueous buffers and involves liposome interaction, deformation, and spontaneous rupture into a single lipid bilayer, coating uniformly the solid substrate, usually silica or mica (Figure 1(c)). It is the most straightforward and commonly used approach to form SLBs, although it is less suitable to form membranes containing a high proportion of cholesterol, which impacts negatively on the efficiency of vesicle rupture.24 In the solvent-assisted lipid bilayer method, the bilayer is formed following gradual exchange of a water-miscible organic solvent with an aqueous buffer solution, over lipids deposited onto the solid support.25 The method is suitable to form bilayers onto a wide range of surfaces (e.g. silicon dioxide and gold26), since it does not require an interaction between lipid vesicles and the surface. In the Langmuir-Blodgett technique,27,28 a lipid monolayer formed at an air–water interface is transferred to a hydrophilic surface. This technique can be used to produce asymmetric lipid bilayers, which mimic natural cell membranes more closely and are not accessible with the vesicle fusion method (Figure 1(d)). Recently, spin coating has also been established as a facile method to deposit membranes onto a variety of solid supports.29,30
A major difference between reconstituted flat lipid bilayers and biological membranes is the lack of the bulk aqueous phase of the cytosol underneath the lipid leaflet, which confers fluidity to cellular membranes. The small hydration layer, spacing the membrane from the solid support, does not accurately mimic the properties of the cellular cytosol. Therefore, in SLBs, the solid support and the lipid bilayer are not fully decoupled, and the nature of the surface can have major effects on the properties of the membrane. For instance, stable lipid bilayers can form around highly curved substrates, such as 100 nm diameter silica nanoparticles,31 by means of electrostatic interactions between the lipid vesicles and the silica support. Moreover, supported lipid bilayers can adapt to changes induced by substrate plasticity,32,33 a behavior that can be exploited to control the mechanical properties of the membrane.
In biological membranes, cytoskeletal scaffolds disengage the lipid leaflets from the bulk phase of the cytosol. Synthetic materials that exhibit mechanical properties similar to those of the cytoskeleton can be sandwiched between the bilayer and the solid support. For example, polymer brushes34 or generic polymer nanofibers35 can be used to recapitulate the function of actin networks and provide the reconstituted lipid bilayer with fluidity close to that observed in biological membranes. These polymer-supported SLBs can also be spatially patterned and incorporate proteins of interest36,37 (Figure 1(e)). Conversely, SLBs have also been used also to study the assembly of cytoskeletal proteins, such as the polymerization of an actin scaffold triggered from vesicles containing ponticulin, an initiator of actin self-assembly.38
Engineering of spatially patterned bilayers to investigate cell signaling
SLBs can be engineered as spatially organized patterns of lipid bilayers with specific compositions. Spatial patterning can be achieved with methods where either the membrane is locally confined by the substrate topography (e.g. microcontact printing)39–41 or parts of the membrane are removed after lipid bilayer deposition (e.g. photolithographic techniques).42–45 Therefore, solid supports can be engineered to introduce ad hoc morphological features to study the biological process of interest. An overview of methods for patterning a solid support is presented in Table 1 and summarized below.
Microcontact printing is a fairly versatile approach to form patterned membranes. It is based on a form of soft lithography as it utilizes soft elastomers, typically polydimethylsiloxane (PDMS).39 The PDMS surface is patterned and used as a contact stamp to transfer a thin layer of material onto a substrate. Its main limitations include stamp deformation or stretching and compressing of the stamp, introducing unwanted features into the pattern. During transfer, the ink can laterally spread into unwanted regions not replicating the desired pattern.46 Polymer “lift-off” techniques are a form of microcontact printing where a thin layer of a sacrificial material (aluminum or a polymer) is patterned onto glass. Once an SLB has formed, the material is etched away removing lipids in the patterned regions47–49 (Figure 2(a)).
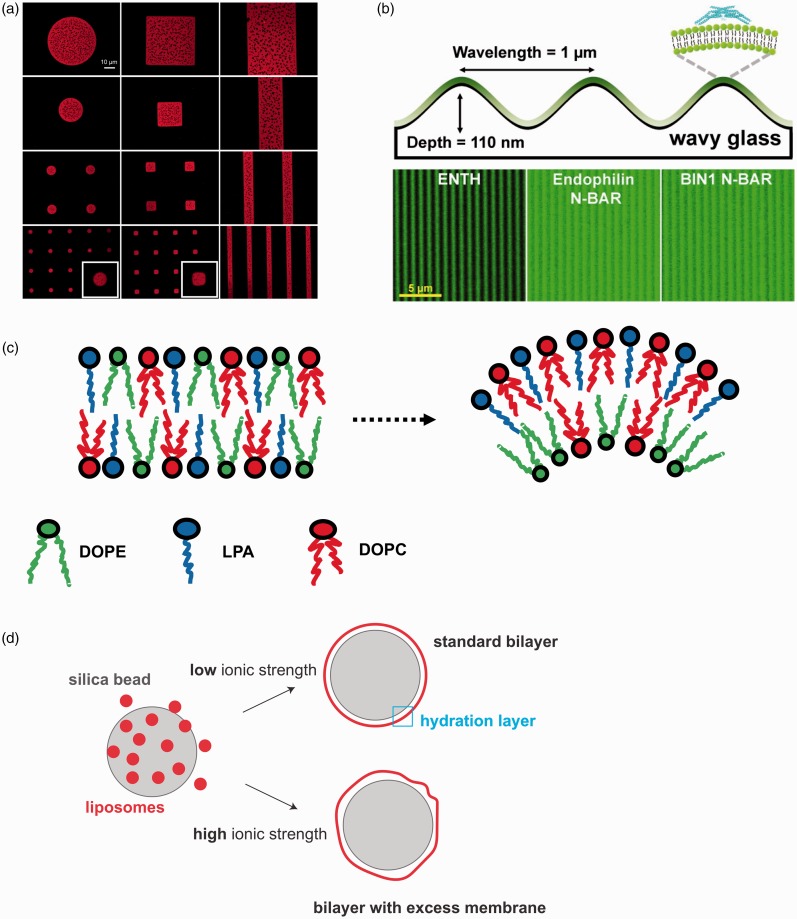
Figure 2.
Examples of spatial and curved patterns of lipid bilayers on solid support. (a) High quality, fully mobile SLBs forming well-defined patterned arrays of phase-segregating DOPC:DSPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)) SLBs through the polymer stencil lift-off technique. DHPE-LR (red) (Lissamine rhodamine-labeled 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) partitions preferentially with DOPC (Lα phase), rendering the DSPC (Lβ phase) domains dark. The insets are magnified images of the small circular and square patterns. Adapted from Zhou and Mornal-Mirabal47 with permission from the publisher Copyright (2016) American Chemical Society. (b) Curvature-sensing proteins incubated on fluid wavy membranes and visualized via confocal fluorescence microscopy. Partitioning of ENTH-GFP, N-BAR-Alexa Fluor 488 and BIN1 N-BAR-Alexa Fluor 488 into positivecurvature regions. Adapted from Hsieh et al.50 with permission from the publisher. Copyright (2012) American Chemical Society. (c) Spontaneous lipid sorting occurs for specific lipid mixtures. For instance, lysophosphatidic acid (LPA) inverted conical shape matches best positive curvature regions when in combinations with cylindrical lipids such as DOPC and both tend to distribute in the outer membrane leaflet. If present, DOPE (DOPE is 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine) prefers to occupy the inner membrane leaflet.51 (d) SUPER templates are formed by exploiting the swelling of membranes in high ionic strength buffers. Fusion of liposomes on size silica nanoparticles (∼5 nm) in high ionic strength solution generates bilayers with excess membranes.52 (A color version of this figure is available in the online journal.)
Currently, the techniques that can fabricate membrane arrays with the highest resolution are electron beam53 and scanning probe lithography,42 which can reproduce features down to 10 nm in size (Table 1). Scanning probe lithography uses an AFM tip to pattern substrates, meaning that features with size beyond the diffraction limit are accessible via this technique. Other types of scanning probe lithography either involve removal of material from the surface of the substrate (thermal scanning probe lithography)43 or use diffusion at the tip of the scanning probe to coat patterns onto substrates, such as alkanethiol arrays on gold substrates (dip-pen lithography).44,54 Patterned bilayers can be formed by removal of lipids from a membrane using UV/ozone photolithography55,56 or using atomic force microscopy.57 Multi-photon lithography exploits the chemical changes occurring in substrates that can simultaneously absorb two or more photons, such as hydrogels, natural polymers, and even proteins.58 The critical size of the features generated with this technique can be as small as 100 nm, since the chemical transformations occur in a region with a volume of few attoliters.59
Patterned SLBs provide an advanced platform for the investigation of complex biological processes at the molecular level. For instance, they can be used to study the molecular mechanisms of endocytosis,63 cell–cell interactions,64 and communication.65,66 Lipid membranes can be spatially organized with micrometer size diffusion barriers and employed to study molecular events occurring within confined geometries, such as the molecular details of T-cell activation.67 Similarly, patterned arrays of lipid bilayers68 can be used as sensor arrays for cell adhesion.13,69 Physical barriers for lipid diffusion can be made using masks created from materials that prevent membrane formation (e.g. metals, some polymers). These barriers are usually taller than the average height of a lipid bilayer (∼4 nm), but recently, it has been shown that graphene70 offers the possibility to create diffusion barriers as thin as one-atom.
The role of spatial organization of transmembrane receptors in intercellular signaling has been extensively studied using patterned SLBs.71 At the plasma membrane, internal and external inputs can alter the spatial organization of cell surface receptors. For instance, intercellular signaling requires close contact between transmembrane receptors to allow direct communication between neighboring cells. Reproducing, in vitro, an intercellular junction is a complex task. However, the combination of solid-state nanolithography with supported lipid bilayers allow to study the relationship between spatial organization and transmembrane receptor activity.72 For example, Greene et al.73 used electron beam lithography to create physical barriers to lipid bilayer lateral mobility creating membrane corrals containing the tyrosine kinase receptor Ephrin A1 (EphA1). This system was used to guide breast cancer cells to the EphA1-patterned membranes via the interaction with the cognate receptor Ephrin A2 (EphA2), thereby creating a semi-synthetic junction. This spatial setup of Ephrin receptors revealed the relevant molecules involved in the endocytosis of EphA2, an important step in the regulation of the EphA2 receptor signaling pathway.73
Similar experimental approaches have been applied to the investigation of events involved in the mechanical regulation of cell surface proteins. For instance, patterned SLBs have been employed to study the relationship between the organization of epidermal growth factor receptor (EGFR) and its phosphorylation levels74 and for the understanding of Ras signaling at a single-molecule level.75
Engineering curved membranes to understand the mechanisms of membrane remodeling
Membrane geometry and cellular signaling are intimately connected,76 particularly in biological processes where macromolecular assembly at the membrane is regulated by curvature.1,77,78 Membrane shape and lipid composition are features of membrane organelles, which are not only evolutionarily conserved but are also finely controlled to maintain spatial segregation of molecules during membrane trafficking.79,80 Over a specific threshold (∼0.8 µm−1), curvature appears to regulate the spatial organization of lipid phases composed of cholesterol81 and the segregation of lipids such as hexadecanoic acid82 or cardiolipin.83 Importantly, local membrane curvature also controls the segregation of curvature-sensing proteins50 (Figure 2(b)). For instance, lipidated proteins appear to be selectively recruited by regions of specific membrane curvature.84 The correlation between substrate curvature and lipid organization underlines the existence of feedback mechanisms that regulate the interaction between the membrane and protein assemblies.85 Therefore, methods to introduce regions of curvature in model membranes are highly desirable to study events where lipid sorting and membrane deformation are crucial mechanistic steps for recruiting protein complexes,86 such as membrane trafficking.
Curved supported membranes are formed via direct fusion of lipid vesicles onto the pre-patterned solid support. Localized regions of membrane curvature can be introduced by means of grooves, edges, or other non-flat features at the surface of the SLB solid support. For example, patterns of curvature can be produced by manufacturing cylindrical concavities with a focused-ion beam, on a chromium-coated coverslip onto which the lipid bilayers are assembled.87 Alternatively, nanopits88 and nanoparticles89 can be used to guide the formation of regions of local curvature in lipid bilayers. It is worth noting that there is a physical limit to the curvature achievable for a phospholipid membrane on a solid support. This was exemplified by the study of Roiter et al.,90 which explored the behavior of membranes deposited on surfaces decorated with nanoparticles. They observed that lipid bilayers form continuously on silica nanoparticles smaller than 1.2 nm or larger than 22 nm, while pore formation in the membrane around the particle was likely within this size range.
Non-flat membranes facilitate the investigation of processes where an asymmetric segregation of lipids occurs laterally or between the leaflets of the membrane at very high curvatures (e.g. tens of nm).91 Coupling between lipid structures and curvature preference is a consequence of the interactions between different lipid shapes, which prefer to arrange as locally curved bilayers.51,81 Lipid sorting can be a key signal to drive transmembrane protein clustering92 and cell signaling.93,94 For instance, lysophosphatidic acid displays an inverted cone shape that confers its tendency to populate regions of membranes with positive curvature51,95,96 (Figure 2(c)).
Complex membrane topographies containing curves and ridges97 can enable the investigation of how protein complexes remodel phospholipid membranes. Membrane remodeling proteins display curved structures, which are used for sensing curvature and for inducing further membrane bending.1,86,98 Special types of SLBs have been recently developed to specifically follow membrane remodeling reactions. The SUPported bilayers with Excess membrane Reservoir52,99 are assembled on micron-scale silica beads in such a manner that a loosely fitted membrane is deposited on the solid support (Figure 2(d)). The excess membrane allows for membrane deformation to occur and be monitored by fluorescence and electron microscopy. These supported membranes have been successfully used to reveal the physical basis of membrane remodeling promoted by protein crowding11 or performed by endocytic100–103 and autophagic proteins.104 Similarly, the supported membrane tubes (SMrT)105 mimic the membrane tethers pulled by proteins106,107 and can be used to study the impact of local curvature on protein–membrane interactions.
A unique membrane remodeling protein complex has recently received attention in the membrane biophysics community, the Endosomal Sorting Complex Required for Transport (ESCRT). ESCRT function is vital for key cellular processes, such as multivesicular body biogenesis, cytokinesis, neuron pruning, plasma and nuclear membrane repair, and viral budding108,109 (Figure 3(a)). In all these processes, the topology of membrane deformation by ESCRT is the same: the budding occurs away from the cytosol,110 in contrast, for example, to bud formation by dynamin in endocytosis.103,111
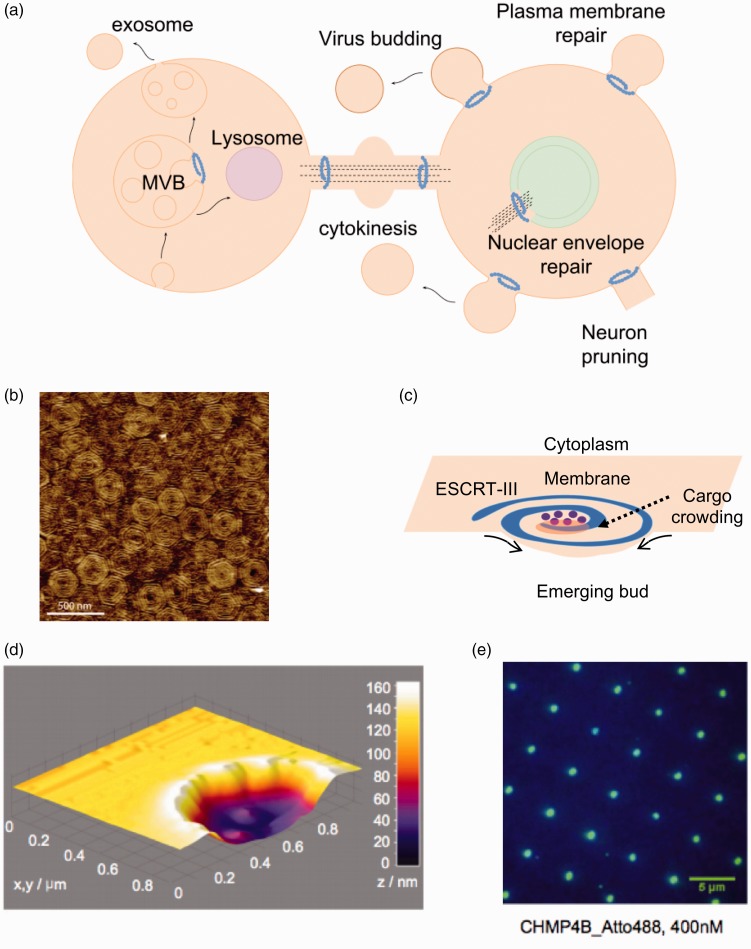
Figure 3.
Membrane remodeling activity by the ESCRT complex. (a) Brief overview of some of the cellular functions of the ESCRT complex in mammalian cells. In all processes, ESCRT-III remodel membranes away from the cytosol. The core remodeling machinery (ESCRT-III) is indicated as a blue spiral. ESCRT-III organizes as circular filaments to stabilize the neck of membranes buds. (b) Atomic force microscopy topographic image of the center of a Snf7 patch assembled on GUV membrane. Image reprinted from Chiaruttini et al.115 with permission from Elsevier. (c) The spiral structure acts as a spring collecting elastic energy which upon release buckles the membrane. (d) 3D surface plot of the atomic force microscopy profile showing lipid bilayer formation on top of the 100-nm-deep invaginated template for negative curvature, which approximates the shape of a nascent HIV-1 bud produced by focused ion beam. Image reproduced from Lee et al.87 with permission from Proceedings of the National Academy of Sciences USA. (e) Total internal reflection fluorescence (TIRF) image after 20-min incubation with 400 nM of the human ESCRT-III core subunit CHMP4B-Atto488 (DiD fluorescein used as a blue marker for lipids). Image reproduced from Lee et al.87 with permission from Proceedings of the National Academy of Sciences USA. (A color version of this figure is available in the online journal.)
Given its unique activity, the biophysics and biochemistry of budding and scission reactions by the ESCRT complex have been the focus of intense investigations. The assembly of ESCRT proteins into an active complex has been reproduced using GUVs112,113 and SLBs, which allowed to dissect the biophysical basis of membrane remodeling. The main scission machinery of ESCRT, namely ESCRT-III, has been observed to assemble into concentric circle-like structures on lipid bilayers114,115 (Figure 3(b)). These studies suggested a potential mechanism for membrane deformation that involves flat “spirals” of ESCRT-III acting like springs. ESCRT-III spirals undertake lateral compression on the membrane and incorporate elastic energy, which leads to membrane deformation upon energy release (Figure 3(c)).115 In another study, SLBs were used to decipher how the growth of ESCRT-III circular structures is self-regulated and how ESCRT-mediated membrane remodeling is coupled to ATP hydrolysis.116
ESCRT proteins assemble and stabilize the neck of a budding membrane; therefore, membrane curvature plays a role in protein recruitment and assembly at these locations.108 Lee et al.87 created a SLB with features resembling membrane bud necks, by assembling a lipid bilayer on a substrate with 100 nm deep concavities, creating regions of negative curvature (Figure 3(d)). TIRF microscopy was used to monitor the assembly of the human ESCRT-III complex at the membrane, which revealed a clustering of the proteins specifically at the invaginations of the membrane, demonstrating ESCRT activity at membrane bud necks87 (Figure 3(e)).
Perspective and conclusions
We have presented a brief overview of the SLB technology and reviewed the fabrication of spatial and curved patterns for in vitro reconstitutions of biological processes at the membrane.
Flat synthetic membranes are a powerful tool to investigate how the biophysics of lipid–lipid interactions contributes to the assembly of complex biological structures.117–119 Advances in the engineering of structural features in solid supports is allowing the generation of ever more complex spatial or curvature patterned bilayers, thus creating more realistic mimics of cellular membranes. SLBs with rough surfaces facilitate the systematic dissection of the interplay between curvature and lipid sorting and crucially, how these physical properties are coupled and controlled in a complex biological process. Lipid phases can be coupled with nanometer-scale curvature patterns,120 providing a strategy for organizing a controlled segregation of biomolecules and lipids.
A yet unmet challenge in the SLB field is the in vitro reconstruction of more complex biological membranes, such as the nuclear envelope. The nuclear envelope is a double membrane,121 in which transmembrane proteins couple the inner membrane to chromatin and the outer membrane relays signals from the cytoskeleton.122 Such an in vitro model would allow the study of the molecular basis of epigenetic mechanisms and nuclear envelope membrane repair in physiological and diseased contexts.123 We are still far from being able to design a fit-for purpose model of the nuclear envelope although SLBs composed of multiple bilayers have been recently developed.48,124 Therefore, it appears that the field is ripe for moving towards increasing complexity with the destination of yet more complex models to interrogate biological mechanisms and/or reconstruct cell biology from the bottom-up.
Authors’ contributions
All authors contributed equally to the writing and editing of this article.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
BC and FC thank the EPSRC for a studentship to Sarina Chand under the DTA scheme (EP/L505055/1). BC and PB thank the EPSRC (grants EP/M027821/1 and EP/M027929/1) for funding.
References
- 1.Beales PA, Ciani B, Cleasby AJ. Nature’s lessons in design: nanomachines to scaffold, remodel and shape membrane compartments. Phys Chem Chem Phys 2015; 17:15489–507 [DOI] [PubMed] [Google Scholar]
- 2.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science 1972; 175:720–31 [DOI] [PubMed] [Google Scholar]
- 3.Gorter E, Grendel F. On bimolecular layers of lipoids on the chromocytes of the blood. J Exp Med 1925; 41:439–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagny TJ, Bassereau P. Bioinspired membrane-based systems for a physical approach of cell organization and dynamics: usefulness and limitations. Interface Focus 2015; 5:20150038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loose M, Schwille P. Biomimetic membrane systems to study cellular organization. J Struct Biol 2009; 168:143–51 [DOI] [PubMed] [Google Scholar]
- 6.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science 1997; 275:651–3 [DOI] [PubMed] [Google Scholar]
- 7.Chapman D. Phase transitions and fluidity characteristics of lipids and cell membranes. Q Rev Biophys 1975; 8:185–235 [DOI] [PubMed] [Google Scholar]
- 8.Zwang TJ, Fletcher WR, Lane TJ, Johal MS. Quantification of the layer of hydration of a supported lipid bilayer. Langmuir 2010; 26:4598–601 [DOI] [PubMed] [Google Scholar]
- 9.Morigaki K, Tanimoto Y. Evolution and development of model membranes for physicochemical and functional studies of the membrane lateral heterogeneity. Biochim Biophys Acta 2018; 1860:2012–7 [DOI] [PubMed] [Google Scholar]
- 10.Yip CM, Darabie AA, McLaurin JA. Aβ42-peptide assembly on lipid bilayers. J Mol Biol 2002; 318:97–107 [DOI] [PubMed] [Google Scholar]
- 11.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein crowding. Nat Cell Biol 2012; 14:944–9 [DOI] [PubMed] [Google Scholar]
- 12.Marklew CJ, Booth A, Beales PA, Ciani B. Membrane remodelling by a lipidated endosomal sorting complex required for transport-III chimera, in vitro. Interface Focus 2018; 8:20180035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groves JT, And LKM, Bertozzi CR, Mahal L, Bertozzi CR, And LKM, Bertozzi CR, Mahal L, Bertozzi CR, And LKM, Bertozzi CR. Control of cell adhesion and growth with micropatterned supported lipid membranes. Langmuir 2001; 17:5129–33 [Google Scholar]
- 14.DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys J 2008; 94:3286–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mossman KD, Campi G, Groves JT, Dustin ML. Immunology altered TCR signaling from geometrically repatterned immunological synapses. Science 2005; 310:1191–3 [DOI] [PubMed] [Google Scholar]
- 16.Yin H, Flynn AD. Drugging membrane protein interactions. Annu Rev Biomed Eng 2016; 18:51–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010; 327:46–50 [DOI] [PubMed] [Google Scholar]
- 18.Ewers H, Helenius A. Lipid-mediated endocytosis. Cold Spring Harb Perspect Biol 2011; 3:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Hu J. Shaping the endoplasmic reticulum into a social network. Trends Cell Biol 2016; 26:934–43 [DOI] [PubMed] [Google Scholar]
- 20.Woodman PG, Futter CE. Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol 2008; 20:408–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller P, Rudin DO, Ti Tien H, Wescott WC. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature 1962; 194:979–80 [DOI] [PubMed] [Google Scholar]
- 22.Schwille P, Diez S. Synthetic biology of minimal systems. Crit Rev Biochem Mol Biol 2009; 44:223–42 [DOI] [PubMed] [Google Scholar]
- 23.Kalb E, Frey S, Tamm LK. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. BBA – Biomembr 1992; 1103:307–16 [DOI] [PubMed] [Google Scholar]
- 24.Hardy GJ, Nayak R, Zauscher S. Model cell membranes: techniques to form complex biomimetic supported lipid bilayers via vesicle fusion. Curr Opin Colloid Interface Sci 2013; 18:448–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabaei SR, Jackman JA, Kim SO, Zhdanov VP, Cho NJ. Solvent-assisted lipid self-assembly at hydrophilic surfaces: factors influencing the formation of supported membranes. Langmuir 2015; 31:3125–34 [DOI] [PubMed] [Google Scholar]
- 26.Tabaei SR, Choi JH, Haw Zan G, Zhdanov VP, Cho NJ. Solvent-assisted lipid bilayer formation on silicon dioxide and gold. Langmuir 2014; 30:10363–73 [DOI] [PubMed] [Google Scholar]
- 27.Tamm LK, Mcconnell HM. Supported phospholipid bilayers. Biophys J 1985; 47:105–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blodgett KB. Films built by depositing successive monomolecular layers on a solid surface. J Am Chem Soc 1935; 57:1007–22 [Google Scholar]
- 29.Simonsen AC, Bagatolli LA. Structure of spin-coated lipid films and domain formation in supported membranes formed by hydration. Langmuir 2004; 20: 9720–9728 [DOI] [PubMed] [Google Scholar]
- 30.Boye TL, Jeppesen JC, Maeda K, Pezeshkian W, Solovyeva V, Nylandsted J, Simonsen AC. Annexins induce curvature on free-edge membranes displaying distinct morphologies. Sci Rep 2018; 8:10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mornet S, Lambert O, Duguet E, Brisson A. The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy. Nano Lett 2005; 5:281–5 [DOI] [PubMed] [Google Scholar]
- 32.Stubbington L, Arroyo M, Staykova M. Sticking and sliding of lipid bilayers on deformable substrates. Soft Matter 2017; 13:181–6 [DOI] [PubMed] [Google Scholar]
- 33.Stubbington L, Arroyo M, Staykova M. Sticking and sliding of lipid bilayers on deformable substrates. Soft Matter 201. 7;13:181–6 [DOI] [PubMed] [Google Scholar]
- 34.Blakeston AC, Alswieleh AM, Heath GR, Roth JS, Bao P, Cheng N, Armes SP, Leggett GJ, Bushby RJ, Evans SD. New poly(amino acid methacrylate) brush supports the formation of well-defined lipid membranes. Langmuir 2015; 31:3668–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi F, Xu J, Smith AM, Parikh AN, Lavan DA. Nanofiber-supported phospholipid bilayers. Soft Matter 2009; 5:5037–41 [Google Scholar]
- 36.Johnson A, Madsen J, Chapman P, Alswieleh A, Al-Jaf O, Bao P, Hurley CR, Cartron ML, Evans SD, Hobbs JK, Hunter CN, Armes SP, Leggett GJ. Micrometre and nanometre scale patterning of binary polymer brushes, supported lipid bilayers and proteins. Chem Sci 2017; 8:4517–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia S, Cartron M, Morby J, Bryant DA, Hunter CN, Leggett GJ. Fabrication of nanometer- and micrometer-scale protein structures by site-specific immobilization of histidine-tagged proteins to aminosiloxane films with photoremovable protein-resistant protecting groups. Langmuir 2016; 32:1818–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson B, Colyer J, Bushby RJ, Evans SD. Self-assembly of actin scaffolds at ponticulin-containing supported phospholipid bilayers. Biophys J 2006; 90:L21–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilbur JL, Kim E, Xia Y, Whitesides GM. Lithographic molding: a convenient route to structures with sub‐micrometer dimensions. Adv Mater 1995; 7:649–52 [Google Scholar]
- 40.Wilbur JL, Kumar A, Biebuyck HA, Kim E, Whitesides GM. Microcontact printing of monolayers : applications in microfabrication. Nanotechnology 1996; 7:452–7 [Google Scholar]
- 41.Lenz P, Ajo-Franklin CM, Boxer SG. Patterned supported lipid bilayers and monolayers on poly(dimethylsiloxane). Langmuir 2004; 20:11092–9 [DOI] [PubMed] [Google Scholar]
- 42.Krämer S, Fuierer RR, Gorman CB. Scanning probe lithography using self-assembled monolayers. Chem Rev 2003; 103:4367–418 [DOI] [PubMed] [Google Scholar]
- 43.Hua Y, Saxena S, Lee JC, King WP, Henderson CL. Direct three-dimensional nanoscale thermal lithography at high speeds using heated atomic-force microscope cantilevers. In: Lercel MJ (ed.) International society for optics and photonics, p 65171-L.
- 44.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nat Nanotechnol 2007; 2:145–55 [DOI] [PubMed] [Google Scholar]
- 45.Callis PR. Two-photon-induced fluorescence. Annu Rev Phys Chem 1997; 48:271–97 [DOI] [PubMed] [Google Scholar]
- 46.Perl A, Reinhoudt DN, Huskens J. Microcontact printing: limitations and achievements. Adv Mater 2009; 21:2257–68 [Google Scholar]
- 47.Zhu Y, Moran-Mirabal J. Micropatterning of phase-segregated supported lipid bilayers and binary lipid phases through polymer stencil lift-off. Langmuir 2016; 34:11021–8 [DOI] [PubMed] [Google Scholar]
- 48.Zhu Y, Negmi A, Moran-Mirabal J. Multi-stacked supported lipid bilayer micropatterning through polymer stencil lift-off. Membranes (Basel) 2015; 5:385–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson BL, Groves JT. Hybrid protein-lipid patterns from aluminum templates. Langmuir 2007; 23:2052–7 [DOI] [PubMed] [Google Scholar]
- 50.Hsieh WT, Hsu CJ, Capraro BR, Wu T, Chen CM, Yang S, Baumgart T. Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir 2012; 28:12838–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callan-Jones A, Sorre B, Bassereau P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb Perspect Biol 2011; 3:A004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann S, Pucadyil TJ, Schmid SL. Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir. Nat Protoc 2013; 8:213–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y. Nanofabrication by electron beam lithography and its applications: a review. Microelectron Eng 2015; 135:57–72 [Google Scholar]
- 54.Ginger DS, Zhang H, Mirkin CA. The evolution of dip-pen nanolithography. Angew Chem Int Ed Engl 2004; 43:30–45 [DOI] [PubMed] [Google Scholar]
- 55.Yu CH, Parikh AN, Groves JT. Direct patterning of membrane-derivatized colloids using in-situ UV-ozone photolithography. Adv Mater 2005; 17:1477–80 [Google Scholar]
- 56.Shi J, Chen J, Cremer PS. Sub-100 nm patterning of supported bilayers by nanoshaving lithography. J Am Chem Soc 2008; 130:2718–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson BL, Groves JT. Scanning probe lithography on fluid lipid membranes supported bilayer deposition. J Am Chem Soc 2004; 126:13878–9 [DOI] [PubMed] [Google Scholar]
- 58.Selimis A, Mironov V, Farsari M. Direct laser writing: principles and materials for scaffold 3D printing. Microelectron Eng 2014; 132:83–9 [Google Scholar]
- 59.Li L, Fourkas JT. Multiphoton polymerization. Mater Today 2007; 10:30–7 [Google Scholar]
- 60.Wallraff GM, Hinsberg WD. Lithographic imaging techniques for the formation of nanoscopic features. Chem Rev 1999; 99:1801–22 [DOI] [PubMed] [Google Scholar]
- 61.Garcia R, Knoll AW, Riedo E. Advanced scanning probe lithography. Nat Nanotechnol 2014; 9:577–87 [DOI] [PubMed] [Google Scholar]
- 62.Crivello JV, Reichmanis E. . Photopolymer materials and processes for advanced technologies. Chem Mater 2014; 26:553–48 [Google Scholar]
- 63.Zhao W, Hanson L, Lou HY, Akamatsu M, Chowdary PD, Santoro F, Marks JR, Grassart A, Drubin DG, Cui Y, Cui B. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat Nanotechnol 2017; 12:750–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moulick RG, Panaitov G, Choi SE, Mayer D, Offenhäusser A. Patterning artificial lipid bilayer on nanostructured surfaces. Int J Nanomed 2018; 13:55–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McConnell HM, Watts TH, Weis RM, Brian AA. Supported planar membranes in studies of cell-cell recognition in the immune system. Biochim Biophys Acta 1986; 864:95–106 [DOI] [PubMed] [Google Scholar]
- 66.Jung H, Robison AD, Cremer PS. Detecting protein-ligand binding on supported bilayers by local pH modulation. J Am Chem Soc 2009; 131:1006–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watts TH, Gaub HE, Mcconnell HM. T-cell-mediated association of peptide antigen and major histocompatibility complex protein detected by energy transfer in an evanescent wave-field. Nature 1986; 320:179–81 [DOI] [PubMed] [Google Scholar]
- 68.Cremer PS, Yang T. Creating spatially addressed arrays of planar supported fluid phospholipid membranes. J Am Chem Soc 1999; 121:8130–1 [Google Scholar]
- 69.Biswas KH, Hartman KL, Zaidel-Bar R, Groves JT. Sustained α-catenin activation at E-cadherin junctions in the absence of mechanical force. Biophys J 2016; 111:1044–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W, Chung JK, Lee YK, Groves JT. Graphene-templated supported lipid bilayer nanochannels. Nano Lett 2016; 16:5022–6 [DOI] [PubMed] [Google Scholar]
- 71.Manz BN, Groves JT. Spatial organization and signal transduction at intercellular junctions. Nat Rev Mol Cell Biol 2010; 11:342–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nair PM, Salaita K, Petit RS, Groves JT. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat Protoc 2011; 6:523–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Greene AC, Lord SJ, Tian A, Rhodes C, Kai H, Groves JT. Spatial organization of EPHA2 at the cell-cell interface modulates trans-endocytosis of ephrina1. Biophys J 2014; 106:2196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stabley D, Retterer S, Marshall S, Salaita K. Manipulating the lateral diffusion of surface-anchored EGF demonstrates that receptor clustering modulates phosphorylation levels. Integr Biol 2013; 5:659–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee YK, Low-Nam ST, Chung JK, Hansen SD, Lam HYM, Alvarez S, Groves JT. Mechanism of SOS PR-domain autoinhibition revealed by single-molecule assays on native protein from lysate. Nat Commun 2017; 8:15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmick M, Bastiaens PIH. The interdependence of membrane shape and cellular signal processing. Cell 2014; 156:1132–8 [DOI] [PubMed] [Google Scholar]
- 77.Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 2007; 447:461–4 [DOI] [PubMed] [Google Scholar]
- 78.Arumugam S, Bassereau P. Membrane nanodomains: contribution of curvature and interaction with proteins and cytoskeleton. Essays Biochem 2015; 57:109–19 [DOI] [PubMed] [Google Scholar]
- 79.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 2009; 25:329–54 [DOI] [PubMed] [Google Scholar]
- 80.Heald R, Cohen-Fix O. Morphology and function of membrane-bound organelles. Curr Opin Cell Biol 2014; 26:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parthasarathy R, Yu C, Groves JT. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir 2006; 22:5095–9 [DOI] [PubMed] [Google Scholar]
- 82.Cheney PP, Weisgerber AW, Feuerbach AM, Knowles MK. Single lipid molecule dynamics on supported lipid bilayers with membrane curvature. Membranes (Basel) 2017; 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boyd KJ, Alder NN, May ER. Buckling under pressure: curvature-based lipid segregation and stability modulation in cardiolipin-containing bilayers. Langmuir 2017; 33:6937–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Larsen JB, Kennard C, Pedersen SL, Jensen KJ, Uline MJ, Hatzakis NS, Stamou D. Membrane curvature and lipid composition synergize to regulate N-Ras anchor recruitment. Biophys J 2017; 113:1269–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vanni S, Hirose H, Barelli H, Antonny B, Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun 2014; 5:4916. [DOI] [PubMed] [Google Scholar]
- 86.McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci 2015; 128:1065–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee I-H, Kai H, Carlson L-A, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci USA 2015; 112:15892–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samiee KT, Moran-Mirabal JM, Cheung YK, Craighead HG. Zero mode waveguides for single-molecule spectroscopy on lipid membranes. Biophys J 2006; 90:3288–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Black JC, Cheney PP, Campbell T, Knowles MK. Membrane curvature based lipid sorting using a nanoparticle patterned substrate. Soft Matter 2014; 10:2016–23 [DOI] [PubMed] [Google Scholar]
- 90.Roiter Y, Ornatska M, Rammohan AR, Balakrishnan J, Heine DR, Minko S. Interaction of lipid membrane with nanostructured surfaces. Langmuir 2009; 25:6287–99 [DOI] [PubMed] [Google Scholar]
- 91.Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J 2006; 91:487–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol 2013; 202:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Head BP, Patel HH, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: Impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta – Biomembr 2014; 1838:532–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Foster DA. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol Metab 2013; 24:272–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kooijman EE, Chupin V, de Kruijff B, Burger KNJ. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003; 4:162–74 [DOI] [PubMed] [Google Scholar]
- 96.Kooijman EE, Chupin V, Fuller NL, Kozlov MM, De Kruijff B, Burger KNJ, Rand PR. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 2005; 44:2097–102 [DOI] [PubMed] [Google Scholar]
- 97.Hoopes MI, Faller R, Longo ML. Lipid domain depletion at small localized bends imposed by a step geometry. Langmuir 2011; 27:2783–8 [DOI] [PubMed] [Google Scholar]
- 98.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 2006; 7:9–19 [DOI] [PubMed] [Google Scholar]
- 99.Pucadyil TJ, Schmid SL. Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission. Biophys J 2010; 99:517–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Johnson A, Hanna M, Audhya A. Eps15 membrane-binding and -bending activity acts redundantly with Fcho1 during clathrin-mediated endocytosis. MBoC 2016; 27:2675–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dar S, Pucadyil TJ. The pleckstrin-homology domain of dynamin is dispensable for membrane constriction and fission. Mol Biol Cell 2017; 28:152–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mattila JP, Shnyrova AV, Sundborger AC, Hortelano ER, Fuhrmans M, Neumann S, Müller M, Hinshaw JE, Schmid SL, Frolov VA. A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature 2015; 524:109–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roux A, Koster G, Lenz M, Sorre B, Manneville J-B, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci 2010; 107:4141–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hervás JH, Landajuela A, Antón Z, Shnyrova AV, Goñi FM, Alonso A. Human ATG3 binding to lipid bilayers: role of lipid geometry, and electric charge. Sci Rep 2017; 7:15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dar S, Kamerkar SC, Pucadyil TJ. Use of the supported membrane tube assay system for real-time analysis of membrane fission reactions. Nat Protoc 2017; 12:390–400 [DOI] [PubMed] [Google Scholar]
- 106.Roux A, Cappello G, Cartaud J, Prost J, Goud B, Bassereau P. A minimal system allowing tubulation with molecular motors pulling on giant liposomes. Proc Natl Acad Sci 2002; 99:5394–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leduc C, Campàs O, Zeldovich KB, Roux A, Jolimaitre P, Bourel-Bonnet L, Goud B, Joanny J-F, Bassereau P, Prost J. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci USA 2004; 101:17096–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hurley JH. ESCRTs are everywhere. Embo J 2015; 34:2398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Olmos Y, Carlton JG. The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol 2016; 38:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol 2010; 11:556–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Daumke O, Roux A, Haucke V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell 2014; 156:882–92 [DOI] [PubMed] [Google Scholar]
- 112.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010; 464:864–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature 2009; 458:172–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 2008; 180:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 2015; 163:866–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mierzwa BE, Chiaruttini N, Redondo-Morata L, Moser Von Filseck J, König J, Larios J, Poser I, Müller-Reichert T, Scheuring S, Roux A, Gerlich DW. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 2017; 19:787–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miles P, Cassidy P, Donlon L, Yarkoni O, Frankel D. In vitro assembly of a viral envelope. Soft Matter 2015; 11:7722–7 [DOI] [PubMed] [Google Scholar]
- 118.Miles P, Frankel D. Lipid directed assembly of the HIV capsid protein. Soft Matter 2014; 10:9562–7 [DOI] [PubMed] [Google Scholar]
- 119.Boura E, Ivanov V, Carlson LA, Mizuuchi K, Hurley JH. Endosomal sorting complex required for transport (ESCRT) complexes induce phase-separated microdomains in supported lipid bilayers. J Biol Chem 2012; 287:28144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ogunyankin MO, Longo ML. Compositional sorting dynamics in coexisting lipid bilayer phases with variations in underlying e-beam formed curvature pattern. Analyst 2013; 138:3719–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Torbati M, Lele TP, Agrawal A. Ultradonut topology of the nuclear envelope. Proc Natl Acad Sci USA 2016; 113:11094–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aureille J, Jma Belaadi N, Guilluy C, Lechler T, Salas RC. Mechanotransduction via the nuclear envelope: a distant reflection of the cell surface. Curr Opin Cell Biol 2017; 44: 59–67 [DOI] [PubMed] [Google Scholar]
- 123.Campsteijn C, Vietri M, Stenmark H. Novel ESCRT functions in cell biology: spiraling out of control? Curr Opin Cell Biol 2016; 41:1–8 [DOI] [PubMed] [Google Scholar]
- 124.Heath GR, Li M, Polignano IL, Richens JL, Catucci G, O’Shea P, Sadeghi SJ, Gilardi G, Butt JN, Jeuken LJCC. Layer-by-layer assembly of supported lipid bilayer poly-l-lysine multilayers. Biomacromolecules 2015; 17:324–35 [DOI] [PubMed] [Google Scholar]