Abstract
Most of the redox proteomics strategies are focused on the identification and relative quantification of cysteine oxidation without considering the variation in the total levels of the proteins. However, protein synthesis and protein degradation also belong to the regulatory mechanisms of the cells, being therefore important to consider the changes in total protein levels in PTMs-focused analyses, such as cysteine redox characterization. Therefore, a novel integrative approach combining the SWATH-MS method with differential alkylation using a combination of commonly available alkylating reagents (oxSWATH) is presented, by which it is possible to integrate the information regarding relative cysteine oxidation with the analysis of the total protein levels in a cost-effective high-throughput approach.
The proposed method was tested using a redox-regulated protein and further applied to a comparative analysis of secretomes obtained from cells cultured under control or oxidative stress conditions to strengthen the importance of considering the overall proteome changes. Using the OxSWATH method it was possible to determine both the relative proportion of reduced and reversible oxidized oxoforms, as well as the total levels of each oxoform by taking into consideration the total levels of the protein. Therefore, using OxSWATH the comparative analyses can be performed at two different levels by considering the relative proportion or the total levels at both peptide and protein level. Moreover, since samples are acquired in SWATH-MS mode, besides the redox centered analysis, a generic differential protein expression analysis can also be performed, allowing a truly comprehensive evaluation of proteomics changes upon the oxidative stimulus.
Data are available via ProteomeXchange and SWATHAtlas with the identifiers PXD006802, PXD006802, and PASS01210.
Abbreviations: Acry, acrylamide; cysTMT, cysteine tandem mass tag; DMEM, Dulbecco's modified Eagle medium; FA, Formic acid; FBS, fetal bovine serum; FDR, False Discovery Rate; IAM, iodoacetamide; ICAT, isotope coded affinity tag; IDA, information-dependent acquisition; IS, internal standard; PBS, phosphate buffered saline; PTMs, Post Translational Modifications; ROS, Reactive oxygen species; TCA, Trichloroacetic acid; WT, wild-type; XIC, extracted-ion chromatogram
Highlights
-
•
Determination of redox changes considering protein total levels.
-
•
Integrative redoxomics and common differential proteomics in a single analysis.
-
•
Differential alkylation strategy using commonly available alkylating agents.
-
•
First untargeted label-free quantitative method to study cysteine oxidation.
Graphical abstract
1. Introduction
Reactive oxygen species (ROS) are a common by-product of the metabolic active cells, being associated with deleterious effects and disease development but also with a physiological role as second messengers in the regulation of several pathways [1]. ROS-dependent signaling involves the reversible oxidation and reduction of some specific amino acids, with crucial reactive cysteine residues being the most frequent target [2], [3]. In fact, changes in the redox states of protein cysteine residues serve as regulatory switches between essential redox signaling and the chronic effects of oxidative stress [4] being therefore tightly regulated by ferredoxin-thioredoxin and glutathione-glutaredoxin systems responsible for a reversible regulation of the oxidized cysteines [3], [5].
At physiological conditions, the cysteine residues exist as thiolate anion (Cys-S-), an unprotonated form of the cysteine thiol (Cys-SH) which is highly susceptible to oxidation [6]. When subjected to mild oxidative stress, this residue is easily oxidized to one of the many different redox forms of the cysteine, including disulphide bonds (S–S), S-glutathionylation (S-SG), S-nitrosylation (SNO) and S-sulfenylation (SOH) which corresponds to reversible oxidized cysteine oxoforms that can be reduced again to the thiol form (SH) [7]. However, if subjected to higher levels of oxidative stress and the sulfenic moieties are not reduced back they can continue to be oxidized to higher oxidation states such as sulfinic (–SO2) and sulfonic (–SO3) acids, which are considered “over-oxidized” modifications that are usually irreversible at normal intracellular condition by chemical or enzymatic means [3], [8], [9]. The cysteine oxoforms associated with low levels of oxidation are required for cellular functions, helping cells to adapt to moderate levels of oxidative stress, while cysteines’ overoxidation leads to permanent protein dysfunction and damage, which can be associated with a disease state or even cell death [9], [10].
To identify and quantify cysteine redox modifications on the proteome scale, several approaches have been developed, mostly using thiol-specific reagents and isotope tags, which can be divided in two main groups: gel-based and gel-free approaches [5], [8]. While, two-dimensional gel electrophoresis combined with fluorescent dye labeling (usually cyanine dyes [11]) can allow the inspection of spot patterns related to redox but also protein-level changes, this method lacks in multiplexing capability and most importantly in the capacity to identify the modified cysteines [5], [11]. On the other hand, the common gel-free technology using isotope tagging, such as isotope coded affinity tag (ICAT [12]) and cysteine tandem mass tag (cysTMT [13]) are often associated with the enrichment of cysteine-containing peptides being useful for the identification of the potential redox-sensitive cysteine residues and for the quantification of the redox changes but missing the capacity to determine the changes in the total levels of the proteins [5]. Only more recently, two new approaches were proposed to simultaneously analyze the cysteine redox changes and total protein levels changes in the same experiment, the cysTMTRAQ [5] and the GELSILOX [14]. cysTMTRAQ combines in the same experiment two types of isobaric tags, cysTMT for redox changes and iTRAQ for total levels changes, considerably increasing the cost of this type of experiments [5]. On the other hand, the GELSILOX approach is based on the use of 18O enzymatic labeling of peptides, a labor-intensive and time-consuming procedure, which may be affected by several factor that can influence the labeling efficiency and which is limited to the direct comparison of two experimental conditions [14], [15].
Among the several strategies for identifying and quantifying the redox state of cysteines, special attention has been given to the method commonly referred as differential alkylation due to its intrinsic capacity to preserve the thiol oxidation state observed at physiological conditions, but most importantly by being able to differentiate the different cysteine oxoforms associated with reversible oxidation [2], [8], [16]. Regardless of the target oxoform, the principles behind this approach can be summarized into three steps. The initial step consists in blocking the SH groups (reduced cysteines) with an alkylating reagent, then modification-specific or general reductants capable to reduce all reversible oxoforms are added to the sample and finally the nascent thiols are blocked with a distinct alkylating reagent, usually with the isotopically modified version of the first alkylating agent [2]. The value of this approach is that the labile oxidation-modified cysteines are replaced by highly stable alkylated forms which can then be analyzed by mass spectrometry approaches resulting in a comprehensive characterization of redox proteomes including the analysis of various cysteine oxoforms, the estimation of their relative abundances, and identification of the modified site [8], [17].
As stated above, mass spectrometry strategies are essential to identify and quantify the peptide/protein being modified. While the quantification in these methods is performed at the intact peptide level, the fragmentation profile is essential to obtain the identification of the peptides by comparing the acquired spectra with the ones generated in-silico. On the other hand, the gold standard method for MS quantification – Multiple Reaction Monitoring (MRM) – is based on the quantification of the fragments of molecules instead of the intact molecules themselves, which results in an increase in the specificity of the method; however, this method required a prior knowledge of the molecules to be quantified and an exhaustive method development. Considering all of these aspects, in the last years, a special attention has been given to the SWATH-MS approach due to its capacity to obtain a “complete record of the fragmentation profiles” of all the peptides of a sample organized into individualized spectra of a selected small group of intact molecules (generically called “SWATH-windows”). Thus allowing to isolate the fragmentation spectrum of very similar peptides within different SWATH windows and to obtain the quantification of those peptides at the fragmentation levels without interference, as performed in MRM. In addition to that, the SWATH-MS pipeline comprises an informed/targeted data analysis (extraction of quantitative data) limited to the peptides confidently identified in a previous conventional proteomics approach, ensuring that potentially all the proteins/peptides that have been identified could be quantified [18], [19]. These characteristics of the SWATH-MS make it an appealing strategy to quantify PTMs, in particular for the study of cysteine modifications using differential alkylation that relies on the tagging of the cysteines with different reagents to induce a mass shift between reduced and oxidized peptides.
Considering the aspects presented above, a new integrative approach combining the label-free SWATH-MS acquisition method with a differential alkylation pipeline using commonly available alkylating reagents (OxSWATH) is proposed in the present work to integrate the information regarding relative cysteine oxidation with a generic comparative analysis of protein levels. The proposed method was firstly tested using a single protein with a well define regulation by oxidative stress, and further extended to the evaluation of the oxidative state of a large set of proteins secreted from cells exposed to an oxidative stress condition caused by acute stimulation with hydrogen peroxide.
2. Experimental procedures
2.1. Chemicals and reagents
All reagents used in cell culture were cell culture-tested. The Dulbecco's modified Eagle medium (DMEM) with Glutamax™ and low glucose (1 g/L), fetal bovine serum (FBS), trypsin 0.05% solution in phosphate buffered saline (PBS), Fungizone® Antimycotic (amphotericin B), penicillin-streptomycin solution (Pen-Strep) solution, and Dulbecco′s phosphate buffered saline (DPBS) (10×) were obtained from Invitrogen. The hydrogen peroxide (H2O2) used in oxidative stress stimulation was obtained from Sigma-Aldrich.
The reagents used in the differential alkylation procedure have different origins: the iodoacetamide was obtained from Sigma-Aldrich, the 40% acrylamide/bis-acrylamide solution (37.5:1) from BioRad and the dithiothreitol (DTT) from GE Healthcare Life Sciences. For in gel digestion, the precast polyacrylamide gels “4–20% TGX Stain-Free Gel” and all the buffers, including the Laemmli buffer, used in the electrophoresis were obtained from Bio-Rad, and the Trypsin Modified Sequencing Grade used in protein digestion, was obtained from Roche Diagnostics.
The reagents used in MS analysis were all high-quality chemical or reagents (ACS Reagent Chemicals & Lab Grades). Formic acid (FA) was obtained from AMRESCO and water, methanol and acetonitrile (ACN) from Fisher. Ortho-phosphoric acid and ammonium sulfate were from MERK and ammonium bicarbonate from Fluka.
2.2. Experimental design and statistical rationale
Two types of experiments were performed in this study: i) an initial validation of the method by an in-vitro assay using a redox-regulated recombinant protein subjected to direct oxidation with H2O2; ii) application of the method in a biological context by assessing the oxidative status of the proteins secreted under control and oxidative stress conditions. For the in-vitro study, two recombinant forms of the protein DJ-1 were used: the [WT]DJ-1 and [C106DD]DJ-1, for the wild-type (non-oxidized) and a constitutive C106 oxidized form of the protein, respectively. Both recombinant proteins were analyzed under reduced condition (control condition) and after a direct oxidation by H2O2 (positive control for cysteine oxidation). Each in-vitro reaction was performed in a total of four replicates (each reaction was performed with a different aliquots of the recombinant protein). Cells’ secretomes were collected from HeLa cells under control and oxidative stress conditions (induced by stimulation with H2O2) in a total of four biological replicates each. Each secretome (also known as conditioned medium) was split into three parts to be subjected to the three reactions of alkylation (see subheading “Differential alkylation” for details). A recombinant protein (MBP-GFP) was added to the media after collection, to be used as internal standard [20].
Different statistical approaches were applied depending on the data being analyzed. The comparison tests applied were mainly parametric methods (Student's t-test and ANOVA) with statistically significant differences being considered for p-values below 0.05. No multiple comparisons correction was applied. Data normality was assessed by different methods, including Q-Q plots and Shapiro-Wilk test, and homogeneity of variances was evaluated by Levene's test. Outliers’ detection was performed by the ROUT method.
Both experimental design and the respective data analysis will be detailed in the following subsections.
2.3. Recombinant DJ-1 production
2.3.1. DNA constructs
The synthetic DNA coding for the human protein DJ-1 with the codons optimized for E. coli expression was chemically synthetized from GeneArt® Gene Synthesis (Invitrogen) and amplified by PCR to include the restriction sites for NheI and XhoI at 5′- and 3′-ends, respectively using the forward primer 5’-GCTAGCAAACGTGCACTGGTTATTCTG-3’ and the reverse primer 5’-CTCGAGTCAATCTTTCAGAACCAGCGGTG-3’. The purified product was cloned into pGEM®-T-Easy plasmid (Promega) and subcloned to pSKB-3 expression vector (DJ-1_pSKB-3) after digestion of both with NheI and XhoI (New England Biolabs, Inc.), in frame with a N-terminal hexahistidine-tag and a TEV (tobaco etch virus) recognition site, for the expression of a recombinant DJ-1 with a TEV-cleavable His-tag. The constitutively C106-oxidized mutant of DJ-1 was generated by replacing the cysteine 106 residue by two aspartic acid residues (C106DD) using the Quick-Change Site-Directed Mutagenesis kit (Stratagene) and the primers 5′-CGCAAAGGTCTGATTGCAGCAATTGATGATGCAGGTCCGACCGCACTGC-3′ (forward) and 5′-GCAGTGCGGTCGGACCTGCATCATCAATTGCTGCAATCAGACCTTTGCG-3′ (reverse) (mutation underlined). All positive clones were selected by restriction analysis and confirmed by DNA sequencing.
2.3.2. Expression and purification
The DJ-1_pSKB-3 constructions (for both WT and C106DD mutant) were transformed into competent E. coli BL21star (DE3) strain and transformed cells were allowed to grow at 37 °C in LB supplemented with 50 μg/mL kanamycin until an optical density at 600 nm of 0.5 was reached, after which the protein expression was induced for 16 h by the addition of IPTG to a final concentration of 1 mM. After protein expression, the pellet of cells was resuspended in 20 mM sodium phosphate, 500 mM NaCl, 10 mM Imidazole, pH 7.2 and disrupted. The cellular extract was clarified by centrifugation, the supernatant was filtered through 0.2 µm syringe filters and the protein was applied to a 5 mL HisTrap HP column (GE Healthcare) pre-equilibrated in the same buffer. Protein elution was obtained by stepwise increasing of imidazole concentration up to 500 mM (50, 100, 300 and 500 mM) and the fraction containing the higher amount of the protein of interest was further purified by size-exclusion chromatography with a HiLoad 26/600 Superdex 200 prep grade column (GE Healthcare Life Sciences) and eluted using PBS (8 mM K2HPO4, 2 mM NaH2PO4·H2O, 150 mM NaCl).
2.4. Conditioned medium collection
HeLa cells were seeded at 12 × 103 cell/cm2 in 148 cm2 plates (Corning) in a total of 4 plates per replicate. After 48 h in culture (37 °C with 5% of CO2/95% air and 95% humidity) the culture medium (DMEM medium with 10% FBS) was discarded and cells were washed twice with warm PBS to remove the remaining FBS. Then, the culture medium was changed to DMEM without FBS (control condition) or to a solution of 1 mM of H2O2 in DMEM without FBS (stress condition) for 40 min, after which the medium was changed again to DMEM without FBS. The conditioned media were collected 24 h after the stimulation. The collected media were centrifuged at 290g, for 5 min at 4 °C to remove cell debris, and then split into three parts for differential alkylation. A recombinant protein (MBP-GFP) was added to the media after collection, to be used as internal standard in the quantitative analysis [21]. Each condition was performed in a total of four replicates.
2.5. Differential alkylation
2.5.1. In-vitro assay with the recombinant DJ-1 proteins
Ten micrograms of each recombinant protein ([WT]DJ-1 and [C106DD]DJ-1) were used per reaction (Supplementary Fig. S1-A). The first alkylation steps were performed using iodoacetamide (IAM) at a final concentration of 66 mM (R1 and R2) and acrylamide (Acry) at 6% (v/v) (R3), and their concentrations were triplicated in the second steps (R3 in the case of IAM, and R1 and R2 for Acry) to compensate for the excess of DTT. DTT was added to a final concentration of 11 mM and the reduction and alkylation steps were performed using ultrasonication for 10 min. Ultrasonication was performed in a 750 W Ultrasonic processor with cuphorn using 20% intensity and pulses ON/OFF of 1 s each. The reagents from the first alkylation were removed using cut-off filters of 5 kDa (Vivaspin500, Sartorius) followed by a washing step with 0.5 M TEAB, and this step was repeated before the protein digestion to remove the excess of reducing and alkylating reagents. To promote the in-vitro oxidation of the recombinant proteins prior to the differential alkylation, they were incubated with 1 mM of hydrogen peroxide for 30 min at room temperature. Each reaction was performed in a total of four replicates and all samples were subjected to liquid digestion. In addition, more reactions were performed using a single alkylating reagent, with or without reduction of the samples prior to alkylation. These reactions were only used for peptide identification.
2.5.2. Conditioned media
The conditioned medium of each sample was split into three parts, one of them to be immediately subjected to the alkylation step using iodoacetamide (to block the reduced cysteines, Fig. 1A – R1) while the other two parts were subjected to reduction with DTT prior to the alkylation step (R2 and R3). All the reactions, except the alkylation with iodoacetamide in R1, were performed after the concentration of the media to 600 µL using the cut-off filters of 5 kDa (Vivaspin20, Sartorius). Prior to media concentration, acetonitrile was added to all samples to a final concentration of 20% (v/v) [2] to promote some protein denaturation, and iodoacetamide was added to the R1 part at a final concentration of 66 mM. After the concentration of the medium, cysteines were reduced by the addition of DTT to a final concentration of 11 mM and alkylated with 66 mM of IAM (R2), 6% (v/v) of Acrylamide in R3 and 12% (v/v) of acrylamide in R1. All the reactions were performed in an ultrasonic bath (USC 1200 THD & THD/HF) at maximum power for 45 min.
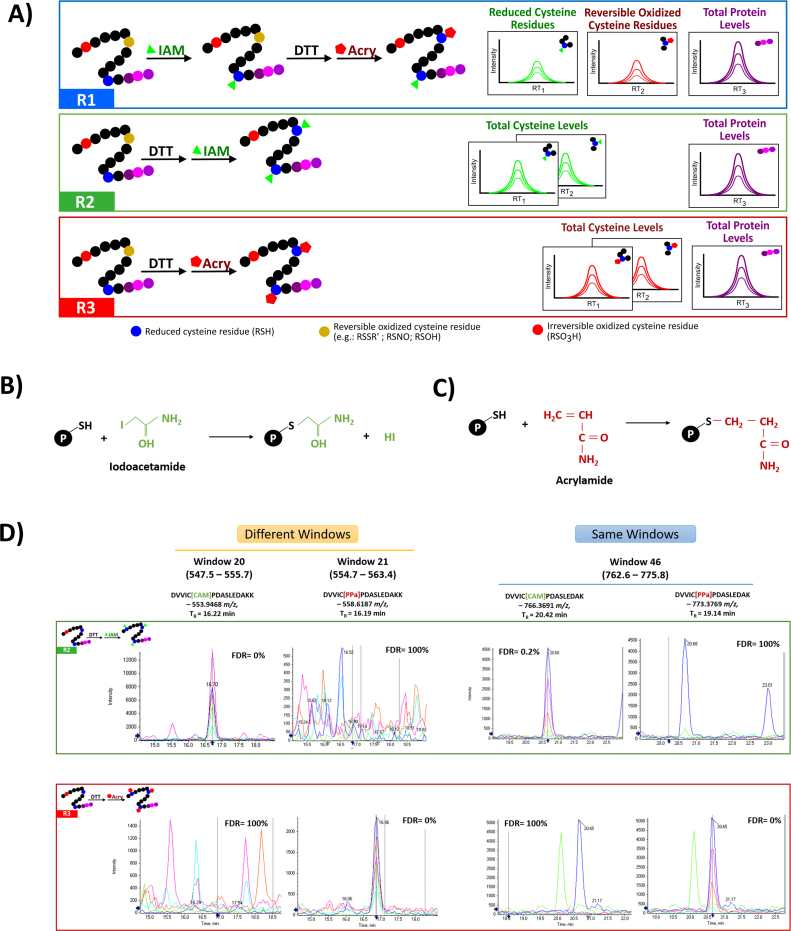
Fig. 1.
Differential alkylation combined with SWATH-MS acquisition: oxSWATH. A) Schematic representation of the experimental workflow used to evaluate the proposed label free differential alkylation method. In this method, the differential alkylation (R1) is performed by using two widely used alkylating agents, iodoacetamide (IAM) and acrylamide (Acry), to block the reduced and the reversible oxidized cysteines, respectively. This reaction is compared with two additional reactions where sample is completely reduced prior to alkylation (R2 and R3 with IAM and acrylamide, respectively) to access the total level of the cysteine peptides. Both cysteine containing peptides and peptides without cysteines are monitoring by SWATH-MS, to measure the changes in cysteine-redox state and total protein levels, respectively. B) Iodoacetamide reaction with free thiols. C) Acrylamide reaction with free thiols. D) Detection of pairs of cysteine-alkylated peptides acquired in the different and in the same SWATH-MS window. The selectivity of the method is verified through the analysis of representative LC-MS/MS chromatograms of two pairs of DJ-1 peptides DVVIC[IAA]PDASLEDAKK/DVVIC[Acryl]PDASLEDAKK (acquired in different windows – see Supplementary Table S2) and DVVIC[IAA]PDASLEDAK/DVVIC[Acryl]PDASLEDAK (acquired in the same window) in the R2 (samples only alkylated with IAA) and R3 (samples only alkylated with Acryl). For each peak group is presented the FDR value obtained using SWATH™ processing plug-in for PeakView. Retention time (TR) was adjusted for each sample and the XICs were obtained in a window of 4 min centered at the aligned retention time. See Supplementary Fig. S4 for detailed information regarding the areas integrated in the pair of peptides acquired in the same window.
After the differential alkylation, the concentrated conditioned media were precipitated using Trichloroacetic acid (TCA) – Acetone. Briefly, TCA was added to each sample to a final concentration of 20% (v/v), followed by an incubation at − 80 °C and centrifugation at 20,000g for 20 min. Protein pellets were washed and solubilized with ice-cold (−20 °C) acetone, followed by a centrifugation at 20,000g for 20 min [22]. The washed pellets were re-suspended in 2× Laemmli buffer, aided by ultrasonication and denaturation at 95 °C for 5 min [23].
2.6. Protein digestion
Recombinant DJ-1 samples were subjected to liquid digestion as described in Anjo et al. [24], without the alkylation step. The digestions were performed with trypsin in a 1:50 ratio, overnight at 37 °C.
The secretomes were subjected to in gel digestion using the short GeLC approach [24]. Seventy microliters of each sample were used for SWATH-MS analysis, and 20 µL of each replicate were combined to create representative pools of each group of samples to be used for protein identification. A total of 6 pools (two experimental conditions from 3 different alkylation reactions) were made and subjected to the short-GeLC approach. The peptides of each digested pooled sample were separated by high pH Reverse Phase Chromatography as presented in Silva et al. [21] and the 28 collected fractions throughout the chromatographic run were joined into 6 samples per pool, in order to increase the number of proteins and peptides identified (in particular cysteine containing peptides) [25].
2.7. Mass spectrometry analysis by SWATH mode
2.7.1. Acquisition
Samples were analyzed on an AB Sciex® 5600 TripleTOF in two modes: information-dependent acquisition (IDA) for protein identification and library generation, and SWATH acquisition for quantitative analysis. Peptide separation was performed using liquid chromatography (nanoLC Ultra 2D, Eksigent®) on a MicroLC ChromXP™ C18CL reverse phase column (300 µm × 15 cm, 3 µm, 120 Å, Eksigent®) at 5 µL/min with a multistep gradient: 0–2 min linear gradient from 5% to 10%, 2–45 min linear gradient from 10% to 30%, and 45–46 min to 35% of acetonitrile in 0.1% FA. Peptides were eluted into the mass spectrometer using an electrospray ionization source (DuoSpray™ Source, AB Sciex®) with a 50 µm internal diameter (ID) stainless steel emitter (AB Sciex®).
Information dependent acquisition (IDA) experiments were performed in the single alkylated samples in the case of the recombinant DJ-1, and for each fraction of the representative pooled samples of the conditioned media. The mass spectrometer was set to scanning full spectra (350–1250 m/z) for 250 ms, followed by up to 100 MS/MS scans (100–1500 m/z from a dynamic accumulation time – minimum 30 ms for precursor above the intensity threshold of 1000 – in order to maintain a cycle time of 3.3 s). Candidate ions with a charge state between + 2 and + 5 and counts above a minimum threshold of 10 counts per second were isolated for fragmentation and one MS/MS spectra was collected before adding those ions to the exclusion list for 25 s (mass spectrometer operated by Analyst® TF 1.7, AB Sciex®). Rolling collision was used with a collision energy spread of 5.
For SWATH-MS based experiments, the mass spectrometer was operated in a looped product ion mode with the same chromatographic conditions used as in the IDA run described above. The SWATH-MS setup was designed specifically for the samples to be analyzed (Supplementary Tables S1 and S2 for recombinant DJ-1 and conditioned media experiments, respectively). A set of 45 or 60 windows of variable width (for recombinant DJ-1 or conditioned medium, respectively) was constructed covering the precursor mass range of 350–1250 m/z. A 200 ms survey scan (350–1250 m/z) was acquired at the beginning of each cycle for instrument calibration and SWATH MS/MS spectra were collected from 100 to 1500 m/z for 70 or 50 ms (for recombinant DJ-1 or conditioned medium, respectively) resulting in a cycle time of 3.25 s, which is compatible with the acquisition of at least 8 points per chromatographic peak. The collision energy for each window was determined according to the calculation for a charge +2 ion centered upon the window with a collision energy spread of 15.
2.7.2. Protein identification and library generation
Protein identification was performed in ProteinPilot™ software (v5.0, AB Sciex®) using the Paragon™ Algorithm (5.0.0.0, 4767, AB Sciex®). Recombinant DJ-1 proteins were searched against the complete database from SwissProt (released at February 2015, composed by 547,351 entries) and conditioned medium was searched against a database composed by the Homo sapiens database from SwissProt (released at February 2015, composed by 20,200 entries) and the sequences of the recombinant proteins used as internal standard (IS). The searches were performed with the following parameters: trypsin digestion; and iodoacetamide or acrylamide as cysteine alkylating reagent, for R2 and R3 samples, respectively; and a special focus option for gel-based approaches was used in the analysis of the conditioned medium. In the conditioned media experiments, the files acquired from control and stress conditions were combined into a single search per reaction of alkylation (R2 or R3). Mass tolerances and exceptions to cleavage rules like missed or semi-specific cleavages were defined automatically according with predefined probabilities. Briefly, probabilities of 0.75 and 0.00001 were set for missed cleavage and non-specific cleavages, respectively, and the mass tolerances were set in 0.05 ± 0.0011 Da and 0.1 ± 0.01 Da for precursor and fragment ions, respectively. An independent False Discovery Rate (FDR) analysis, using the target-decoy approach provided by ProteinPilot™, was used to assess the quality of the identifications. Positive identifications were considered when identified proteins and peptides reached a 5% local FDR.
A specific library of precursor masses and fragment ions was obtained from each identification file using the SWATH™ processing plug-in for PeakView™ (v2.0.01, AB Sciex®). The libraries were exported as text files to be manually adapted to the redoxomics analysis. For each library, the peptides containing alkylated cysteines were isolated and further combined with the peptides from the internal standard, this step generates cysteine specific libraries (one for the iodoacetamide alkylated peptides and one for acrylamide alkylated peptides). In addition, another library is created using only peptides without cysteines, to be used to determine the total levels of the proteins. In total, three libraries are used per redoxomics assay.
2.7.3. SWATH data file processing
Peptides were selected automatically from the library using the criteria described in Anjo et al. [24]. Up to 15 peptides were chosen per protein for the determination of the protein total levels, and all the peptides containing cysteines were used in the case of the cysteine specific libraries. SWATH quantitation was attempted for all proteins in the library files that were identified below 5% local FDR from ProteinPilot™ search. For each peptides, up to 5 target fragment ions (corresponding to a peak group) were automatically selected and scored [24]. Peak group confidence threshold was determined based on a FDR analysis using the target-decoy approach and the peptide that met the 1% FDR threshold in all the replicates (in the case of IS), or at least three replicates were retained for posterior analysis. The peak areas of the target fragment ions of those peptides were extracted across the experiment using a 4 min extracted-ion chromatogram (XIC) window adjusted in order to accommodate the entire chromatographic peaks. The retention time was adjusted to each sample using the IS peptides.
Total protein levels were estimated by summing all the transitions from all the peptides [24] without cysteine of a given protein, and for each protein the levels of its cysteine alkylated peptides were determined by summing all the transitions of that group of peptides. Peptides alkylated with iodoacetamide and acrylamide were analyzed in separate.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [26] via the PRIDE [27] partner repository with the dataset identifier PXD006802 and PXD006803 (for conditioned media and recombinant DJ-1 data, respectively, and it is also available as a public resource in the SWATHAtlas database (www.SWATHAtlas.org) with the deposit no. PASS01210.
2.8. Western blot
Conditioned media were spiked with the same amount of IS solution prior to culture medium concentration. The samples were denatured by boiling at 95 °C for 5 min, and the entire volume was loaded per lane and separated on 12.5% SDS-polyacrylamide gels using a mini-PROTEAN® Tetra Electrophoresis System (Bio-Rad). Proteins were transferred to low fluorescence polyvinylidene fluoride (PVDF) membranes (TBT RTA TRANSFER KIT, Bio-Rad) using a Trans-Blot Turbo Transfer System (BioRad) during 30 min at a constant voltage of 25 V (with the current limited to 1 A). Following transfer, the membranes were blocked for 1 h at room temperature (RT) with 5% (w/v) skimmed milk powder in PBS-Tween 20 (PBS-T) [0.1% (v/v)]. The membrane was incubated sequentially with anti-DJ-1 (1:1000; ADI-KAM-SA100-E, Enzo Life Sciences, Inc.) and anti-GFP (1:1000; SICGEN – Research and Development in Biotechnology Ltd.) overnight at 4 °C followed by 1 h at RT, prepared in the blocking solution. Primary antibodies were removed, and membranes were extensively washed with PBS-T (3 times, 15 min under agitation each time). Blots were then incubated for 1 h at RT with the respective secondary antibodies conjugated with alkaline phosphatase (anti-mouse and anti-goat for DJ-1 and GFP, respectively) in 5% (w/v) skimmed milk powder dissolved in PBS-T followed by extensive washes as above. The membrane was firstly incubated with the DJ-1 specific antibody followed by re-probing of the membrane with antibody against a GFP specific antibody.
Protein-immunoreactive bands were developed using the “Enhanced Chemifluorescence (ECF) detection system” (GE Healthcare) and visualized in a Molecular Imager FX System (Bio-Rad). The adjusted volumes (total intensities in a given area with local background subtraction) for each band were obtained using the Image Lab software (version 5.1, Bio-Rad). DJ-1 levels were normalized for GFP levels (used as internal standard) and the differences in the secretion of DJ-1 were evaluated by a Student's t-test performed in GraphPad Prism (version 6.01).
2.9. Data analysis
2.9.1. Differential proteomics analysis of the secretome
Differential secretome analysis was performed using the results from the total levels of the proteins. Protein total levels were normalized for the values of the internal standard, and the normalized values of the three reactions (R1, R2 and R3) were combined into a single value per replicate. The proteins altered between the two conditions were identified by an ANOVA test performed in InfernoRDN (version 1.1.5581.33355) using the log10 transformed values, and statistically significant differences were considered for p-values < 0.05. Data normality was assessed by the analysis of Q–Q plots [28] obtained in InfernoRDN [29].
2.9.2. Determination of the reduced and reversible oxidized fraction of the proteins
The iodoacetamide alkylated peptides (from R1 and R2) are used to determine the reduced fraction of a give protein, and the acrylamide alkylated peptides (R1 and R3) are used to determine the reversible oxidized levels of the proteins (Supplementary Fig. S2). The levels of the cysteine alkylated peptides are firstly normalized to the total levels of the correspondent protein within the same reaction of alkylation, i.e., the values calculated from the R1 are compared with the total protein levels in R1, and so forth. The normalized values are used to determine the relative proportion of the reduced fraction by calculating the ratio between the IAM-alkylated peptides in R1 and the IAM-alkylated peptides in R2 (correspondent to the total amount of free available cysteines). To determine the levels of the reversible oxidized fraction, a similar ratio is performed but using the Acry-alkylated peptides from R1 and the Acry-alkylated peptides from R3. The combination of these two values should be closer to one, therefore values smaller than one may be an indication of irreversible oxidations. To determine the total levels of each oxoform (reduced and reversible oxidized fraction expressed in procedure defined unit, p.d.u), the relative proportions obtained in each replicate are multiplied by the correspondent protein total levels (summed peptides’ intensity) in R1 reaction (the reaction common to the two types of alkylating agents). See Supplementary Fig. S2 for a schematic representation of these calculations.
2.9.3. Comparative analysis of the oxidative state of DJ-1
The reduced and oxidized fractions were compared between the experimental conditions ([WT]DJ-1 vs [C106DD]DJ-1 and Control vs Stress conditions) using student's t-tests with statistical significance considered for p-values below 0.05. Parametric assumptions (data normality and homogeneity of variance) were tested using Shapiro-Wilk Test and Levene's Test, respectively. All the tests were performed in IBM® SPSS® Statistics version 22. Outliers’ detection was performed in GraphPad Prism (version 6.01) using the ROUT method.
2.9.4. Differential redoxomics analysis
Comparative redoxomics analysis of the secretome was performed using the results from the reduced and reversible oxidized fractions, for both the relative and the total proportions. In the case of the total levels, data was further normalized using the internal standard to accommodate potential sample processing errors. The proteins altered between the two experimental conditions (control vs stress) were identified as indicated above for the differential proteome analysis of the secretome. The analysis was repeated for each type of results related to the oxidative state of the secreted proteins.
2.9.5. Functional analysis
Protein domain enrichment analysis was performed with FunRich (version 3.1.3) using Protein domain database, and statistically analyzed with hypergeometric test using FunRich human genome database as the background [30]. A Bonferroni corrected p-value < 0.05 indicates the sub-groups of protein domains that are significantly enriched in the sample against the background database (9952 entries). Reactome (version 57) [31] (available at http://www.reactome.org/) was used to perform pathway enrichment analysis of the total list of altered proteins (considering their secretion levels or alteration in redox state) in the present study. Pathways were considered enriched for FDR analysis below 5%.
2.9.6. Row-clustered heat maps
Heat maps were done using GPRroX (version 1.1.15) using the standardized levels of the quantified proteins within all the replicates of the two reactions.
3. Results
3.1. Evaluation of the SWATH-MS approach combined with differential alkylation (oxSWATH) for redoxomics studies
Differential alkylation has been widely applied to evaluate the redox state of proteins usually associated with the addition of isotopically labeled alkylated agents to be able to perform quantitative comparisons [8]. In the present work, a modification of the usual pipeline is presented which consist in the use of a combination of two common alkylating agents instead of isotopically labeled species (Fig. 1A). In addition, this method is combined with the analysis of samples by SWATH-MS allowing the quantification of the cysteine alkylated peptides along with other peptides that are used to access the differences in the total levels of the proteins.
Considering that SWATH-MS quantification is obtained at the transition level, it is extremely important to guarantee that the peptides alkylated with the two different agents (see Figs. 1B and 1C for the modifications caused by the two reagents used in the present work) have a mass shift sufficient to clearly differentiate the two peptides and their fragments, either by separating the two peptides into different SWATH windows or by inducing a clear mass shift even at the fragmentation level that results into two largely different MS/MS spectra (Fig. 1D and Supplementary Figs. S3 and S4). Despite of the recent developments that allow to create SWATH windows with widths as smaller as 2–3 Da [32], in this pipeline a combination of two alkylating agents which mass shift induced in the peptides differs in 14 Da were used: iodoacetamide (which induce a mass shift of +57.0214 Da) and acrylamide (+71.0371 Da). This difference is compatible with the majority of the windows created in a SWATH setup with variable windows (Supplementary Table S1 and S2) ensuring that each peptide is acquire without the interference of the other ionic species. Besides that, the scoring algorithm used to integrate the peak groups also reveals to be able to even differentiate pairs of peptides that are acquired within the same window (Supplementary Fig. S7).
Since, two different alkylating agents were used to block the reduced (IAM) and the reversible oxidized (Acry) cysteines, the alkylated peptides (different ion species) cannot be directly compared as it is performed in the common pipeline of differential alkylation using isotopically labeled reagent. Therefore, an additional reaction in which the sample is reduced prior to the first alkylation step is required to determine the total levels of cysteine peptides with exception of those subjected to irreversible oxidation (Supplementary Fig. S7). Considering that two different alkylation reagents were used, this reaction must be performed in duplicate to address the total levels using each reagent (Fig. 1A, R2 for IAM and R3 for Acry) and thus allowing the comparison of the results of the two alkylating agents by their normalizations to R2 or R3 condition. In this sense, the R1/R2 ratio using the IAM alkylated peptides corresponds to the reduced fraction of the protein of interest, while the R1/R3 ratio using the Acry alkylated peptides will refer to the reversible oxidized levels of that protein. Although in most of the scenarios the use of only one type of the results (reduced or oxidized fractions) can be enough to perform comparative analysis or to infer the other fraction of the data (as can be seen in Supplementary Fig. S7 that clearly demonstrate the complementarity between the results from the two fractions), the use of both data is extremely valuable in particular when the total levels of the proteins are considered, and to infer potential irreversible oxidations. Therefore, the proposed pipeline comprises the parallel analysis of the sample using the three reactions presented above, its acquisition in SWATH-MS mode and a data analysis that comprises an initial normalization to the total levels of the protein in study (obtained from a set of peptides without cysteines) followed by the determination of the reduced and oxidized fraction by R1/R2 and R1/R3 ratios, respectively.
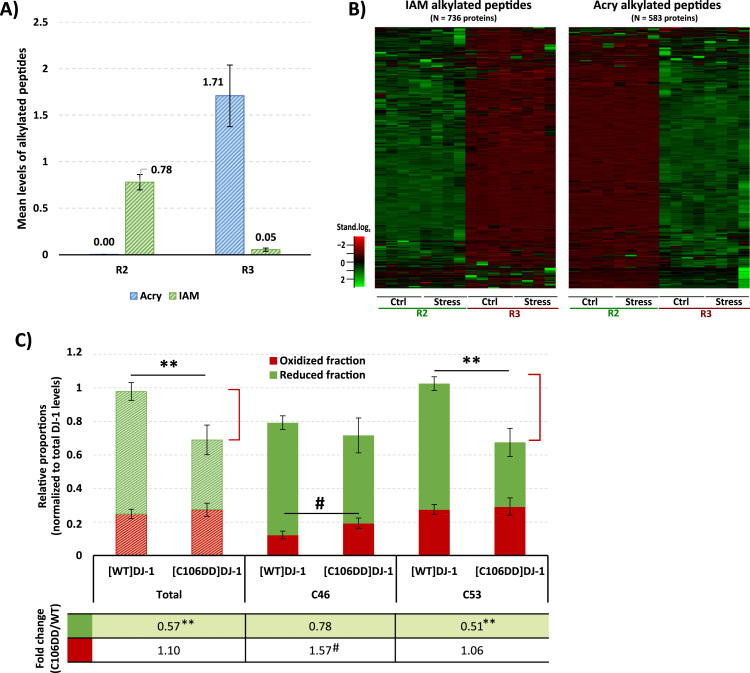
To test the proposed method, a well-known redox sensitive protein was used: the protein DJ-1 [33], [34]. The protein was produced and purified both as a wild-type (WT) form and a C106DD mutant, which mimics a constitutive form of DJ-1 oxidized on cysteine 106 [35]. Besides the cysteine 106, this protein contains two additional cysteines at the positions 46 and 53, which may be responsible for the modulation of the oxidation of the central cysteine 106 [36]. An additional step was performed (Supplementary Fig. S1A, dashed lines) to evaluate the degree of alkylation and the capacity to differentiate the peptides alkylated with IAM from those alkylated with Acry. For that, the samples from R2 and R3 were further subjected to reduction and alkylation with the opposite reagent. Considering that in R2 and R3 all the cysteines are reduced and blocked in the first part of the reaction, no signal would be expected from the second alkylating agent. Also, by being able to acquire the differently alkylated peptides in distinct SWATH-MS windows no signal interference by the peptides alkylated with the other alkylated agent is expected. The results (Fig. 2B) show that the mean levels of [WT]DJ-1 IAM-alkylated peptides are higher in R2, while the Acry-alkylated peptides are higher in the R3 reaction and a very small, almost negligible, values are observed in the second step of alkylation (R2 for Acryl and R3 for IAM). The same is observed if individual cysteines of DJ-1 (C43 and C53) were considered (Supplementary Fig. S1B and C). These results confirm that both reactions of alkylation have a good yield, which is particularly high using IAM justifying its used as the first alkylating reagent in R1. Moreover, no signal interference is observable even if a larger set of different proteins were considered (Fig. 2B) meaning that, there is also no interference associated with the potential acquisition of the same peptide with different alkylations in the same SWATH window (Fig. 1D and Supplementary Fig. S4 for details). Altogether, these results confirm that the proposed method fulfil the required conditions to be applied with success in the analysis of the redox state of proteins.
Fig. 2.
In-vitro evaluation of the capacity of the oxSWATH approach to access the changes in cysteine redox state. A) To confirm the yield of alkylation, the proposed method was applied to the redox sensitive protein DJ-1 by subjecting equal amounts of this protein to the three reactions, and an additional step of reduction and alkylation using the opposite alkylating agent (Acry and IAA, for R2 and R3, respectively, as indicated in Supplementary Fig. S1A by the dotted lines) was performed in R2 and R3. The yield of alkylation can be inferred by the mean alkylation levels of acrylamide (Acry, blue bar) and iodoacetamide (IAM, green bar) in R2 and R3 reactions where the cysteines were reduced prior to the first alkylation step. The results are presented as the mean ± SD of four independent replicates. B) The capacity to differentiate the two types of alkylated peptides, was evaluated by comparing the combined levels of the IAM-alkylated peptides or Acry-alkylated peptides of thousands of secreted proteins in the R2 and R3 reaction. IAM was only used in R2 and acrylamide was only used in R3. The row-clustered heat maps represent the standardized levels of the quantified proteins within all the replicates of the two reactions. C) oxSWATH applied to the comparative redox characterization of DJ-1 WT and the mutant C106DD forms (correspondent to the constitutive C106-oxidized form) subjected to direct oxidation via hydrogen peroxide. For each protein, the relative proportion of reversible oxidized (red bar) and reduced (green bar) fractions is presented, and potential irreversible oxidations are indicated by the red bracket. The analysis was performed for each individual cysteine (C46 and C53) and combining the results from the two monitored cysteine residues (Total). The table below the graphic presents the fold change between the C106DD and the WT form of the DJ-1 protein for each fraction. Data correspond to the mean ± SD of four independent experiments in the case of the WT oxidized fraction, and three replicates in the case of C106DD and in the reduced fraction of the WT experiments. **ρ < 0.01, for significant differences between the reduced fractions of the proteins and # ρ < 0.05, for significant differences between the oxidized fractions of the proteins using independent Student's t-test.
This approach was then tested in an in-vitro assay in which the [WT]DJ-1 and the [C106DD]DJ-1 were oxidized by the direct action of hydrogen peroxide. To that, the recombinant proteins were incubated with a concentration of hydrogen peroxide known to cause some degree of DJ-1 oxidation not associated with an irreversible oxidation state [37]. Separate data analysis was performed for the two groups of peptides obtained in DJ-1 analysis (Supplementary Table S3): i) the group containing the peptides with cysteine residues, those peptides were used to determine the reduced and reversible oxidized fractions of the protein; and, ii) the group of peptides without cysteines which was used to determine the total levels of the recombinant proteins. From the cysteine peptides, those alkylated with iodoacetamide were used to determine the reduced fraction of DJ-1, while the peptides alkylated with acrylamide were used to address the reversible oxidized fraction of the protein. The levels of the alkylated peptides were adjusted to the total levels of recombinant DJ-1, and the normalized values were then used to calculate the relative proportions of oxidized and reduced fractions of each recombinant protein (Fig. 2C, with the reduced fraction in green and the oxidized fraction in red). The analysis was performed for each individual cysteine (C46 and C53) and for the total oxidative state by combining the results from the two cysteine residues monitored. From the results obtained (Fig. 2C), it is possible to observe that in general there is an overall decrease in the levels of the reduced C106DD compared with the WT protein when subjected to the same oxidative stress. The major contribution for this decrease comes from the large reduction in the C53 reduced fraction which may be associated to a potential irreversible oxidation of that cysteine (indicated by the red line), since no difference is observed in the reversible oxidized fraction (red bars). On the other hand, it is observed an increased in the oxidized fraction of C46 without a meaningful difference in the reduced fraction of the two forms of the protein. These results may be justified by the localization of these two cysteine residues: while C46 is partially buried within the DJ-1 dimmer, C53 is localized at the surface of the protein being therefore more susceptible to be oxidized [36]. The differences in the oxidative state of the WT protein and the C106DD mutant may be associated with the inaccessibility to transfer the ROS to the cysteine 106 of the mutant protein (which is substituted by two aspartic acids), therefore affecting in higher extent the other two cysteines that form the DJ-1 redox center (which is evident by the increase in the reversible oxidized form of the C46 and the induction of irreversible oxidation in C53).
Taking into consideration the results observed for the two recombinant forms of DJ-1, it is possible to confirm that this method can be applied to evaluate and characterize the redox state of ROS-sensitive proteins, being also capable to indirectly indicate potential irreversible oxidations, which can be confirmed (quantified) depending on the capacity to identify those modified forms (sulfinic and sulfonic acid) in the regular experiment for protein identification.
3.2. Application of the oxSWATH-MS in a comparative proteomics analysis – the example of a secretome analysis
The method proposed in the present work was applied in the differential proteome analysis of secretomes obtained from cells cultured under control or stress conditions, in which a large proteome variation is expected. To promote the oxidative stress, cells were stimulated with an acute concentration of hydrogen peroxide for a short period, and then were left to recovery for 24 h (Fig. 3A). By removing the oxidative stimulus, it is possible to focus the analysis on the regulation caused by endogenous ROS and not by the direct oxidative action of the hydrogen peroxide, corresponding therefore to a more physiological condition. However, considering that there is a gap of 24 h between the stimulation and the collection of the conditioned medium (also known as secretome), a large impact in the oxidative state of the secreted proteins is not expected.
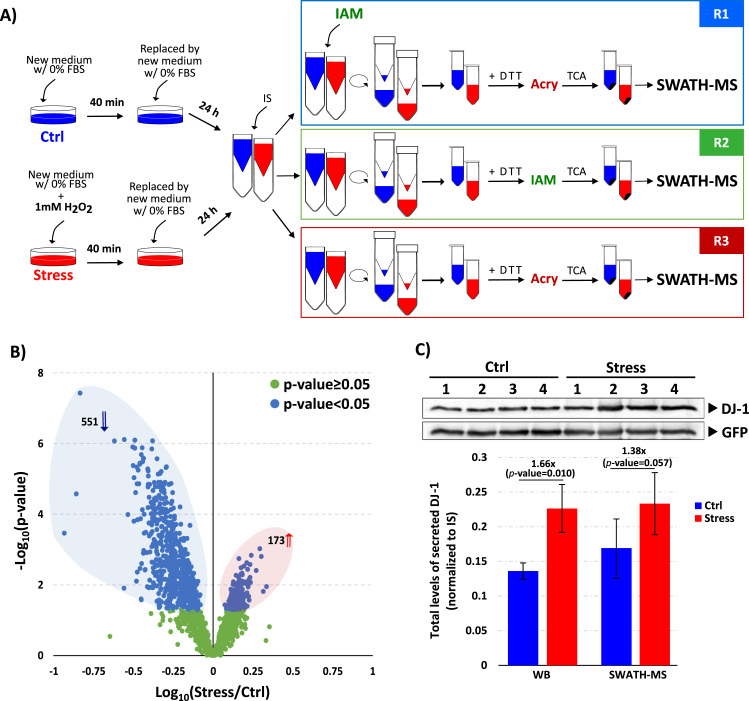
Fig. 3.
OxSWATH applied to differential secretome analysis. A) Adaptation of the proposed differential alkylation approach to the analysis of proteins secreted by cells under different oxidative stress conditions. Cells were subjected to 1 mM of hydrogen peroxide stimulus for 40 min, after which the medium was replaced by new medium without the stimulus. The culture medium (secretome) was collected 24 h after the stimulus. The secretome obtained from cells not exposed to the hydrogen peroxide was used as control, and an internal standard was added to the medium after the collection. The collected medium was split and processed in parallel to perform the three reactions. The alkylation with IAM in the R1, was performed before the concentration of the medium, and 20% acetonitrile was added to the medium to induce protein denaturation to promote the access of IAM to the cysteines not available in the folded proteins. Sample reduction and the subsequent alkylation were performed in the concentrated media, after which the proteins were precipitated, and the entire content was analyzed by short GeLC-SWATH-MS. B) Volcano plot representing the differential total proteome analysis of the secretome between the two conditions (ctrl and stress). The total levels of the 1691 secreted proteins were obtained by summing all the peptides without cysteines. The protein levels were normalized to the values of the IS in each sample after which the results of the three reactions were combined into a single result per replicate. Statistical analysis was performed by Student's t-test and statistical significance was considered for p-values < 0.05 (blue dots). A total of 551 proteins were significantly decreased in the oxidative stress condition (blue arrow) and 173 proteins were significantly increased (red arrow). C) Evaluation of secreted levels of total DJ-1 in control and stress conditioned medium. Data correspond to the mean ± SD of four independent experiments.
After the collection of the medium, an internal standard was added to monitor sample loss during processing, and the medium was split into three parts to be subjected to the three reactions described above (Fig. 3A). In the case of the R1 the first alkylation step was immediately performed after the collection of the medium, to avoid the oxidation of the free thiols by the contact with air and to take advantage of the concentration step to remove the first alkylating agent before the second step of reaction. Additionally, to promote some protein denaturation, and thus facilitate the access to all the cysteines, 20% of an organic solvent (in this case acetonitrile) was added to the samples [2]. The remaining reactions were performed after the concentration of the medium, allowing a drastic reduction in the amount of reagent required for the reactions. After the differential alkylation procedure, the secreted proteins were precipitated with TCA/acetone and the entire samples were subjected to the short GeLC-SWATH approach [24].
As previously mentioned, by acquiring the data on SWATH-MS mode besides the quantification of the alkylated peptides, it is also possible to acquire information from peptides not containing cysteines, which will be used to calculate the total levels of the proteins in the experimental conditions being tested. Therefore, with the present method the information obtained from the sample is not limited to the redox sensitive proteins, being also possible to perform a common differential proteome analysis of the samples at the same time of a redoxomics analysis. In this sense, taking into consideration the total levels of the secreted proteins (normalized to the IS to accommodate for sample's processing error) a differential characterization of the secreted proteins under control or stress condition allowed to highlight 724 altered proteins from a total of 1691 quantified proteins (Supplementary Table S4), with 551 proteins being more secreted in the control conditions than the stress condition, and 173 proteins being more secreted in the stress conditions (Fig. 3B). Additionally, it is possible to observe that, in some cases the fold changes are higher than 30–50%, pointing to the importance of considering the total levels of the analyzed proteins when comparative redox characterization of samples is performed.
Taking into consideration the regulation of the protein DJ-1, used in the in-vitro assay, which is known to be secreted at different levels depending on the condition [38], it is possible to verify (by SWATH-MS and WB) that DJ-1 secreted levels are in fact modulated by the oxidative stress, being more secreted under the stress condition than in the control condition (Fig. 3C). This evidence allows the use of the protein DJ-1 as a model to validate the importance of taking into consideration the total levels of the protein to perform a truly comprehensive redoxomics characterization of the proteome (Fig. 4).
Fig. 4.
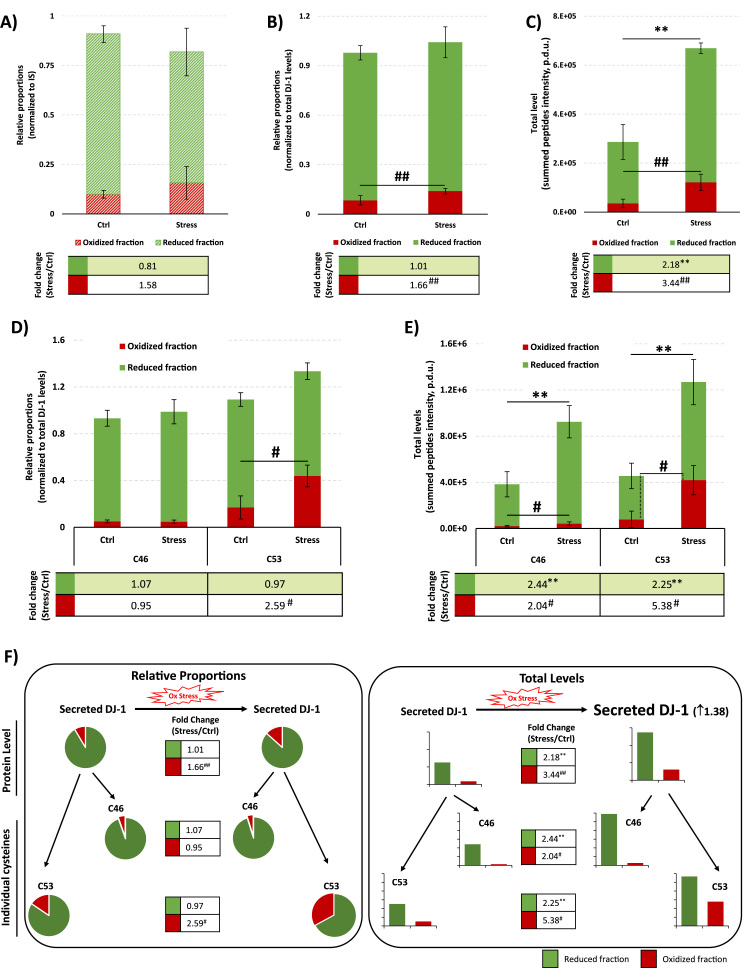
Application of OxSWATH for a comparative redox characterization of the secreted DJ-1. A) Comparative redox characterization of the secreted DJ-1 without considering the total levels of the protein in each condition. The values were normalized to the IS to adjust for sample processing deviations. Data are presented as the relative fractions of oxidized (red bar) and reduced (green bar) secreted DJ-1 from each pool. Data correspond to the mean ± SD of four independent experiments in the case of the oxidized fraction and three replicates in the reduced fraction of the stress experiments B to E) Comparative redox characterization of the secreted DJ-1 considering the levels of DJ-1 in each conditioned medium instead of the IS standard levels. Data can be presented as the relative proportions of oxidized (red bar) and reduced (green bar) fractions of the secreted DJ-1 in each condition (B and D), as performed in a common differential alkylation approach, or as the total levels of each DJ-1 oxoform (reduced or reversible oxidized DJ-1) in each condition (C and E) by taking into account the difference in the amount of DJ-1 secreted in the two conditions, i.e. by multiplying the calculated relative proportions by the total levels of DJ-1 secreted in each condition (Fig. 3C and Supplementary Fig. S2). The analysis was performed considering all the cysteine residues monitored in the experiment (B and C) and for each individual DJ-1 cysteine (C46 and C53 in D and E). The tables below the graphics present the fold change of each fraction between the protein secreted in stress and control conditions. Data correspond to the mean ± SD of four independent experiments in the case of the oxidized fraction in the combination of all cysteines and in the case of C46, and three replicates in the remaining cases. **ρ < 0.01, for significant differences between the reduced fractions of the proteins and #ρ < 0.05 or ## ρ < 0.01 for significant differences between the oxidized fractions of the proteins using independent Student's t-test. F) Schematic representation of the characterization of the oxidative state of secreted DJ-1 under control and oxidative stress conditions (summary of the results presented from B to E). p.d.u. stands for procedure defined unit.
To demonstrate the impact of considering the differences in the total levels of the targeted protein, the relative proportions of the oxidized and reduced fraction of DJ-1 were firstly determined using only the IS to account for sample processing errors which can have an important impact in label free quantifications (Fig. 4A). These results were compared with a similar analysis but taking into consideration the levels of DJ-1 in the control and stress conditions (Fig. 4B). A similar result was observed with both approaches, however some important differences can be detected: i) by omitting the total levels of the protein, as performed in most of the methods, it is observable (Fig. 4A) a tendency for a decrease in the reduced fraction of the protein under stress conditions which is not observed when the total amount of DJ-1 is considered (Fig. 4B); and ii) although an increase in the oxidized fraction was observed in both analysis under stress conditions, the normalization of the values to the total levels of DJ-1 results in a more accurate quantification and in an improvement of the reproducibility of the measurements, which are reflected in a gain of statistic power sufficient to considered the observed difference as statistically meaningful (Fig. 4B). Therefore, although leading to very similar results, the normalization to the total levels of the protein results in the inclusion of the protein DJ-1 in the list of the proteins regulated by oxidative stress, which is in line with the proposed identification of DJ-1 as a redox sensor [39], while following a generally used strategy this regulation would be lost.
Additionally, by being able to monitor the total levels of the proteins, it is possible to calculate the total levels of the reduced and oxidized fractions of DJ-1. With this type of analysis, the real difference in the levels of each fraction of the protein can be evaluated (Supplementary Fig. S2). Considering the results for the protein DJ-1 (Fig. 4C), this analysis revealed that the differences observed in the total levels of the protein (Fig. 3C) are a result of an increase in approximately 2× the levels of the pool of reduced DJ-1, and an increase of more than 3× in the pool of oxidized protein.
Considering the mechanism of redox regulation of the protein DJ-1 and the results obtained in the analysis performed for the recombinant protein, the analysis of the redox state of the secreted protein can be performed for each individual cysteine (Fig. 4C–E) but also the total level by combining the results of all the cysteines of the protein, demonstrating that the SWATH-MS approach has sufficient power to characterize the redox state of the proteins even at their physiological levels. Considering the results obtained, C53 is again the most affected cysteine residue by the oxidative stress, presenting not only a larger increase (more than 5×) in the total levels of the oxidized protein, but also a large increase (2.59×) in the proportion of the oxidized protein, which reach more than 40% of the pool of the DJ-1 secreted under oxidative stress.
With this set of results, it is clear the importance of considering the total levels of the proteins to access the real alterations in their redox state, being the capacity to provide this type of results one of the major advantages of the present method over the usual approaches. Moreover, it also demonstrates that different types of complementary information that can be obtained from the same experiment including i) a common differential proteome analysis, and ii) the characterization of the reduced and oxidized fractions of individual proteins, which can be performed considering each cysteine individually or the combination of all the cysteines (Supplementary Fig. S9 and Supplementary Tables S5 and S6), and presented as relative proportions or total levels. All the data obtained can be further combined to get a more comprehensive characterization of the samples.
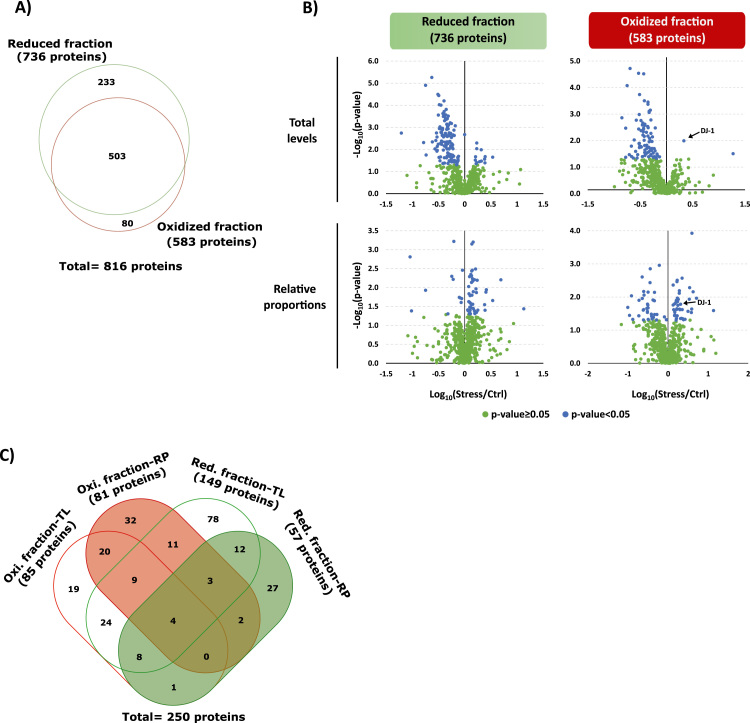
Most importantly, the experimental setup proposed in this work can be used to simultaneously address the differential secretome analysis (Fig. 3B) and an untargeted redoxomics analysis of the same dataset (Fig. 5), in this case the secretome obtained from cells subjected to control and oxidative stress conditions. For the untargeted redoxomics analysis, proteins with cysteine alkylated peptides (with both iodoacetamide or acrylamide) were considered and subjected to the normalization against the respective protein total levels. A total of 2140 iodoacetamide-alkylated peptides (Supplementary Table S7) and 1603 acrylamide-alkylated peptides (Supplementary Table S8) (values corresponding to unique peptides, i.e., no repetition of the same peptide was considered) were quantified in the assay without any type of cysteine enrichment. Those peptides can cover up until 2671 and 1977 cysteine residues (note that some of the peptides can correspond to the same residue, and more than one residue could be also present in the same peptide), from the 736 proteins quantified in the reduced fraction (by considering the iodoacetamide-alkylated peptides) or the 583 proteins quantified in the oxidized fraction (Acry-alkylated peptides), respectively. Considering the complete list of the proteins quantified in this study (the 1691 proteins – Fig. 3B) which contains a total of 18935 cysteine residues, it was observable that for many proteins it was not possible to cover any of their cysteine residues (Supplementary Fig. S10) which is largely associated with the lower sequence coverage achieved for those proteins. Nevertheless, this tendency is also observable in a similar work, where no cysteine-enrichment is performed (Supplementary Fig. S10C) [14]. Besides that, it was also possible to observed that in general, for most of the proteins quantified, it was possible to achieve up to 20–30% coverage of the cysteine residues per proteins, which is better that the observable in a similar approach (Supplementary Fig. S10C) and is very close to the coverage obtained in a strategy with cysteine-enrichment (Supplementary Fig. S11) [40]. As expected, a lower number of cysteine residues were quantified and covered when compared with an approach that is based on a cysteine-enrichment strategy, nevertheless in the present work it was possible to quantify some of the cysteine residues of the proteins that were studied by this approach (Supplementary Figs. S11 and S12). Moreover, near 50% of the proteins evaluated in the redox-centered analysis of this study were already identified in an exhaustive study of cysteine residues response to oxidative stimuli, which involved the enrichment of the peptides containing cysteines [40] (Supplementary Fig. S12).
Fig. 5.
oxSWATH applied to the differential redoxomics analysis of the secreted proteins under control and stress condition. A) Venn diagrams comparing the proteins quantified based on the IAM-alkylated peptides (reduced fraction in green) or based on Acry-alkylated peptides (oxidized fraction in red). B) Volcano plot representing the differential proteome analysis of the secretomes considering the reduced and reversible oxidized fractions of the secreted proteins. The analysis was performed at different levels either considering the total levels of reduced and reversible oxidized forms of the proteins or the relative proportions of each oxoform. All the values were normalized for the total protein levels obtained from the analysis of peptides without cysteines. Statistical analysis was performed by Student's t-test and statistical significance was considered for p-values < 0.05 (blue dots). C) Venn diagrams comparing all the proteins considered altered in one of the four comparisons performed in the study. The combination of these four results reveals that at least 250 secreted proteins are modulated by oxidative stress. Abbreviations: Oxi, oxidized; Red, reduced; RP, relative proportions; TL, total levels.
By considering the levels of the iodoacetamide-alkylated peptides, the proteins were compared based on their reduced fraction (736 proteins) while by using the peptides alkylated with acrylamide it was evaluated the oxidized fraction of the protein (583 proteins). The combination of the two results leads to an increase (of at least 10%) in the number of proteins subjected to the analysis, increasing therefore the sample coverage (Fig. 5A), and corresponding to another evidence of the advantage of performing a combined analysis using the results from the two alkylating agents. The results obtained in this work from the protein DJ-1 and from a group of another 23 selected proteins (Supplementary Fig. S9 and Supplementary Tables S5 and S6) and the results obtained in other works such as the results presented for the proteins p53 and PTP1B [41], reveal that evaluation of the redox status at the protein level may be a good reflection of the regulation of the individual cysteines, at least for some type of redox-sensitive proteins/regulations. In this sense the screening analysis was performed at the protein level instead of the cysteine-peptide level, in order to select a small group of proteins for which a deeper evaluation at the cysteine-level can be performed.
With that in mind, the comparative analysis between control and oxidative stress samples was performed using the relative proportions and the total levels of both the reduced and oxidized fractions (Fig. 5B, Supplementary Fig. S8 and Supplementary Tables S9–S12). From these results, it is clear that in general there are no major differences between the redox state of the quantified proteins with no statistically meaningful alterations being observed for near 70% of the proteins. Moreover, an overall tendency to a generalized reduced state of the protein is also observable even at the stress conditions (Supplementary Fig. S8). Nevertheless, it is still possible to identify proteins significantly altered considering their oxidative state. Bearing in mind the results from the relative proportions it is possible to observe a generalized tendency to small differences (fold change values closer to 0) with only few proteins being considered altered. However, when the protein amount is considered in the analysis, and the comparisons are performed with the total levels of reduced and reversible oxidized oxoforms of the proteins, large differences are detected between the two secretomes, as can be observed by the example of the protein DJ-1 (indicated by the arrows in Fig. 5B).
Overall, these results reflect the importance of being able to determine the effective levels of each fraction, which will be a better indicator of the real fold changes. However, the analysis of the relative proportions should not be discarded since it is the indicator in the shifts of the oxidative state within a pool of a specific protein. In this sense, the results should be combined to achieve a better characterization of each sample, and to identify the most representative set of proteins differentially regulated by the oxidative stress. This is particularly evident in the analysis of secreted proteins (Fig. 5C) where the combination of the proteins considered altered with statistical evidence (with p-values below 0.05) in the four-independent analysis (total of 250 proteins) results in an increase in at least 68% when compared to the larger individual group (containing 149 proteins). Further analyses of the set of proteins identified as potential redox altered proteins should be performed to recognize not only those that are already known to be regulated by oxidative stress, as the protein DJ-1 (Fig. 5B), but also for other interesting proteins to better understand their regulation under oxidative stress stimuli, including the individual characterization of the monitored cysteines.
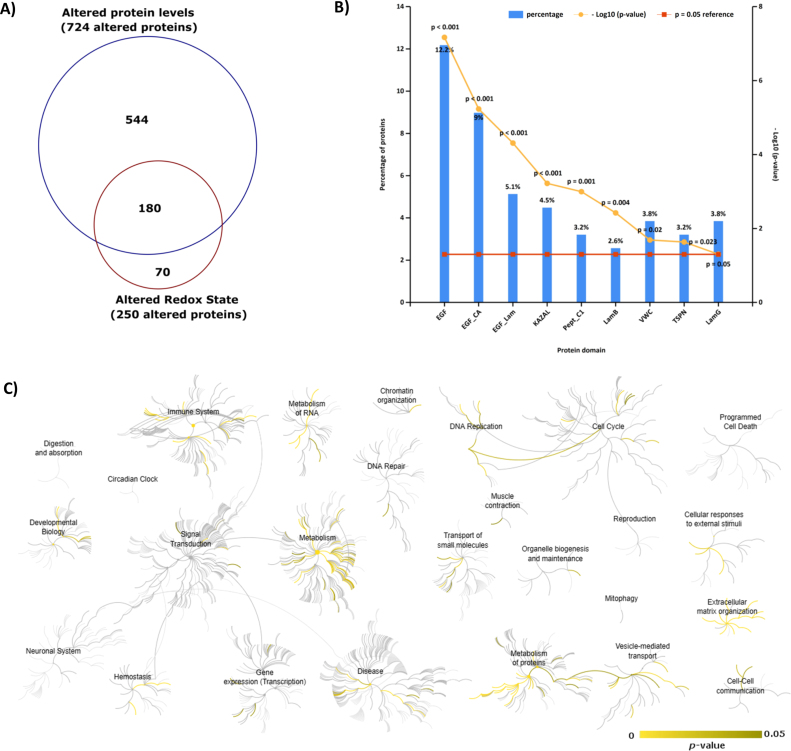
As stated above, one of the major vantages of the oxSWATH approach is its capacity to address in a single experiment a common differential proteomics analysis (Fig. 3) with a cysteine redox-centered analysis (Fig. 5), allowing a truly comprehensive evaluation of proteomics changes upon an oxidative stimulus (Fig. 6). This allows a functional characterization of the changes by either a redox-centered perspective such as the identification of common protein groups/protein domains (Fig. 6B and Supplementary Table S13) among the redox altered proteins or a generic evaluation of the main pathways altered (Fig. 6C and Supplementary Table S14) by taking into consideration all the proteins being modulated by the stimuli (including the ones differentially secreted and those with different cysteine redox state). In the present study, it was possible to identify from the 250 proteins with altered redox-state (Fig. 5B and Supplementary Fig. S13), three interesting groups of proteins having protein domains characterized by the presence of cysteines in their core (EGF_CA and KAZAL) including the group of cysteine proteases (PepT_C1), which has cysteine as their catalytic residue. In addition, by combining all the proteins altered in the study (Fig. 6C) it is possible to verify that mechanisms associated with the extracellular milieu such as those involved in Extracellular matrix organization, Vesicle-mediated transport and Cellular responses to external stimuli are altered among the secretomes from control and oxidative stress conditions, indicating a potential reorganization of the extracellular space as well as an active response to stimuli.
Fig. 6.
Functional characterization of the secretome alterations induced as response to oxidative stress. A) Venn diagrams combining the proteins altered in the differential proteomics analysis (Fig. 3) with those altered in the redox-centered analysis (Fig. 5). B) Protein domain enrichment analysis of the 250 proteins with altered redox status (see Supplementary Table S13 for the list of proteins presented in each domain). C) Pathways analysis of all the 794 proteins found statistically altered in the two analyses (FDR analysis below 5%; see Supplementary Table S14 for the list of proteins belonging to each pathway).
4. Discussion
In this work, we present a new integrative approach that combines the untargeted label-free SWATH-MS acquisition method with differential alkylation, allowing to integrate the information regarding relative cysteine oxidation of thousands of cysteine residues with a generic differential proteomics analysis. Thus, the present method reveals to be an innovative approach when compared with the common proteomics screenings focused on post translational modifications (PTMs), including cysteine redox centered analysis, which are usually focused only on the PTMs’ changes lacking the evaluation of the relative levels of the modified and non-modified proteins.
The importance of considering the total levels of the proteins to access the real alterations in their redox state was clearly demonstrated in the set of results presented, being the capacity to provide this type of results one of the major advantages of the present method over the usual redox centered approaches where enrichment of the cysteine-containing peptides is commonly performed.
The use of different alkylating agents allows the independent untargeted analysis of the reduced and reversible oxidative forms of the proteins as relative proportions or total levels, which can be further combined to get a more comprehensive characterization of the samples. Furthermore, different types of complementary information can be obtained from a single experiment including i) the common differential redoxomics analysis used to identified altered proteins, and ii) the deeper characterization of the oxidative state (reduced and oxidized fractions) of individual targeted proteins, which can be performed considering each cysteine individually or for the combination of all the cysteines and presented as relative proportions or total levels.
Moreover, the approach presented can be easily adapted to the researcher needs, for instances different pairs of alkylating reagents can be used depending on their compatibility, i.e., their capacity to induce mass differences easily distinguishable in a SWATH-MS method. By using non-isotopically labeled alkylating reagents, which are commonly available in most of the proteomics laboratories, the costs associated with this type of analysis are very reduced when compared with the usual labeling strategies. On the other hand, stable isotope labeled variants could be an interesting alternative to reduce the amount of sample required for the analysis (by reducing the analysis to a single reaction). Nevertheless, the usual small mass difference observed between the labeling alternatives, makes it harder to combine with SWATH-MS. An interesting alternative, can be the use of monobromobimane as the first alkylating agent, which is known to induce a mass shift of 190 Da [42] been therefore easily paired with another alkylating reagent for the proteomics analysis, and may also allow to perform a parallel analysis of the low molecular weight thiols (including disulphides and glutathione disulphide) by using the flow-through and the supernatant of protein precipitation as previously described [43], [44], and thus complementing the results from the proteomics evaluation with an additional analysis of the global cellular thiol-state. Different reducing agents can be also used beside the ones that promote a complete reduction of the reversible modification (such as the DTT or TCEP), including reagents that promote a specific reduction of a particular type of cysteine modifications (such as ascorbate, m-arsenite and glutaredoxin which are responsible for the reduction of s-nitrosylations, s-sulfenylation and s-glutathionylation, respectively) [45]. Finally, cysteine exposure can also be adjustable by promoting different degrees of protein denaturation, by using organic solvents or some detergents [44]; nevertheless, a prior study indicates that the cysteines more affected by different oxidants are the ones more exposed [40] which may indicate that for most of the redox-centered analysis it is not necessary to induce a complete denaturation of the proteins.
Additional advantages of the present approach includes the fact that there is no limits in the number of samples to be compared, and the possibility to, not only quantify non-cysteine alkylated peptides but also quantify other modified peptides, such as cysteine irreversible modification (sulfinic and sulfonic acid) or other type of PTM that can be important in the cellular context being studied, depending on the capacity to identify those modified forms in the regular experiment for protein identification. Notably, the method presented proves to have enough power to characterize individual residues even at physiological levels without any type of enrichment, and the results obtained also indicate that this method can be applicable to challenging samples such as membrane-enriched samples (it was possible to quantify some cysteine residues from membrane proteins – Supplementary Fig. S14 – possibly located at extracellular vesicles). This may be important for subcellular characterization of some redox relevant organelles such as mitochondrial, nucleus and reticulum [46], which may be achieved by performing the isolation of the specific organelle prior to the differential alkylation.
In summary, the presented pipeline allows to integrate the information regarding relative cysteine oxidation with the changes in the protein total levels, being able to present both the relative proportions and total levels of reduced and reversible oxidized fractions of the analyzed proteins. Additionally, it is also capable of indicating indirectly the presence of potential irreversible oxidations by combining the relative proportions of the reduced and reversible oxidized fractions, which can be directly quantified if the peptides with those modification were also identified in the study. The untargeted redoxomics analysis performed by combining all these different types of data improve the identification of the proteins modulated by oxidative stress, for which a detailed characterization of the oxidative state can be performed considering each cysteine individually or the combination of all the cysteines. Finally, this redox centered analysis can be further complemented with a simultaneous generic differential protein expression analysis of the same dataset, leading therefore to a more comprehensive evaluation of proteome changes upon oxidative stimulus.
Acknowledgments
This work was financed by the European Regional Development Fund (ERDF) through the COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation and Portuguese national funds via FCT – Fundação para a Ciência e a Tecnologia, I.P., under projects: PTDC/NEU-NMC/0205/2012, POCI-01-0145-FEDER-007440 (ref.; UID/NEU/04539/2013), POCI-01-0145-FEDER-016428 (ref.: SAICTPAC/0010/2015), POCI-01-0145-FEDER-016795 (ref.: PTDC/NEU-SCC/7051/2014), POCI-01-0145-FEDER-029311 (ref.: PTDC/BTM-TEC/29311/2017), POCI-01-0145-FEDER-30943 (ref.: PTDC/MEC-PSQ/30943/2017) and PTDC/MED-NEU/27946/2017; and by The National Mass Spectrometry Network (RNEM) under the contract POCI-01-0145-FEDER-402-022125 (ref.: ROTEIRO/0028/2013). S.I.A, was supported by Ph.D. fellowship SFRH/BD/81495/2011, co-financed by the European Social Fund (ESF) through the POCH – Programa Operacional do Capital Humano and national funds via FCT.
Acknowledgments
Author contribution
S.I.A. performed the experiments and wrote the paper. M.N.P., L.R.L., L.S. and P.C. performed the recombinant protein production. S.I.A., M.G., and B.M. were involved in the design of the experiments and discussion of the results, and all the authors were involved in the revision of the manuscript.
The authors have declared no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101130.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Holmstrom K., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 2.Schilling B., Yoo C.B., Collins C.J., Gibson B.W. Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 2004;236:117–127. [Google Scholar]
- 3.Miki H., Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J. Biochem. 2012;151:255–261. doi: 10.1093/jb/mvs006. [DOI] [PubMed] [Google Scholar]
- 4.Jones D.P., Go Y.-M. Mapping the cysteine proteome: analysis of redox-sensing thiols. Curr. Opin. Chem. Biol. 2011;15:103–112. doi: 10.1016/j.cbpa.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Parker J., Balmant K., Zhu F., Zhu N., Chen S. cysTMTRAQ—an integrative method for unbiased thiol-based redox proteomics. Mol. Cell. Proteom. 2015;14:237–242. doi: 10.1074/mcp.O114.041772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel T. From sulfenylation to sulfhydration: what a thiolate needs to tolerate. Sci. Signal. 2012;5 doi: 10.1126/scisignal.2002943. (pe10-pe10) [DOI] [PubMed] [Google Scholar]
- 7.Wojdyla K., Rogowska-Wrzesinska A. Differential alkylation-based redox proteomics – lessons learnt. Redox Biol. 2015;6:240–252. doi: 10.1016/j.redox.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojdyla K., Rogowska-Wrzesinska A. Differential alkylation-based redox proteomics – lessons learnt. Redox Biol. 2015;6:240–252. doi: 10.1016/j.redox.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Held J.M., Gibson B.W. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol. Cell. Proteom. 2012;11 doi: 10.1074/mcp.R111.013037. (R111.013037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurd T.R., James A.M., Lilley K.S., Murphy M.P. Measuring redox changes to mitochondrial protein thiols with redox difference gel electrophoresis (Redox‐Dige) Methods Enzymol. 2009;456:343–361. doi: 10.1016/S0076-6879(08)04419-4. [DOI] [PubMed] [Google Scholar]
- 12.Fu C., Hu J., Liu T., Ago T., Sadoshima J., Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J. Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker J., Zhu N., Zhu M., Chen S. Profiling thiol redox proteome using isotope tagging mass spectrometry. JoVE (J. Vis. Exp.) 2012 doi: 10.3791/3766. (e3766-e3766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Acedo P., Nunez E., Gomez F.J., Moreno M., Ramos E., Izquierdo-Alvarez A., Miro-Casas E., Mesa R., Rodriguez P., Martinez-Ruiz A., Dorado D.G., Lamas S., Vazquez J. A novel strategy for global analysis of the dynamic thiol redox proteome. Mol. Cell. Proteom.: MCP. 2012;11:800–813. doi: 10.1074/mcp.M111.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capelo J.L., Carreira R.J., Fernandes L., Lodeiro C., Santos H.M., Simal-Gandara J. Latest developments in sample treatment for 18O-isotopic labeling for proteomics mass spectrometry-based approaches: a critical review. Talanta. 2010;80:1476–1486. doi: 10.1016/j.talanta.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 16.Held J.M., Danielson S.R., Behring J.B., Atsriku C., Britton D.J., Puckett R.L., Schilling B., Campisi J., Benz C.C., Gibson B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteom. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen-Heininger Y.M., Mossman B.T., Heintz N.H., Forman H.J., Kalyanaraman B., Finkel T., Stamler J.S., Rhee S.G., van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anjo S.I., Santa C., Manadas B. SWATH‐MS as a tool for biomarker discovery‐from basic research to clinical applications. Proteomics. 2017 doi: 10.1002/pmic.201600278. [DOI] [PubMed] [Google Scholar]
- 19.Santa C., Anjo S.I., Mendes V.M., Manadas B. Neuroproteomics—LC-MS quantitative approaches. Recent Adv. Proteom. Res. 2015 [Google Scholar]
- 20.Anjo S.I., Simões I., Castanheira P., Grãos M., Manadas B. A generic normalization method for proper quantification in untargeted proteomics screening. bioRxiv. 2018 [Google Scholar]
- 21.Silva C., Santa C., Anjo S.I., Manadas B. A reference library of peripheral blood mononuclear cells for SWATH-MS analysis. Proteom. Clin. Appl. 2016 doi: 10.1002/prca.201600070. [DOI] [PubMed] [Google Scholar]
- 22.Manadas B.J., Vougas K., Fountoulakis M., Duarte C.B. Sample sonication after trichloroacetic acid precipitation increases protein recovery from cultured hippocampal neurons, and improves resolution and reproducibility in two-dimensional gel electrophoresis. Electrophoresis. 2006;27:1825–1831. doi: 10.1002/elps.200500757. [DOI] [PubMed] [Google Scholar]
- 23.Santa C., Anjo S.I., Manadas B. Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach. Proteomics. 2016 doi: 10.1002/pmic.201600024. [DOI] [PubMed] [Google Scholar]
- 24.Anjo S.I., Santa C., Manadas B. Short GeLC-SWATH: a fast and reliable quantitative approach for proteomic screenings. Proteomics. 2015;15:757–762. doi: 10.1002/pmic.201400221. [DOI] [PubMed] [Google Scholar]
- 25.Manadas B., English J.A., Wynne K.J., Cotter D.R., Dunn M.J. Comparative analysis of OFFGel, strong cation exchange with pH gradient, and RP at high pH for first-dimensional separation of peptides from a membrane-enriched protein fraction. Proteomics. 2009;9:5194–5198. doi: 10.1002/pmic.200900349. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Riverol Y., Xu Q.W., Wang R., Uszkoreit J., Griss J., Sanchez A., Reisinger F., Csordas A., Ternent T., Del-Toro N., Dianes J.A., Eisenacher M., Hermjakob H., Vizcaino J.A. PRIDE inspector toolsuite: moving toward a universal visualization tool for proteomics data standard formats and quality assessment of ProteomeXchange datasets. Mol. Cell. Proteom.: MCP. 2016;15:305–317. doi: 10.1074/mcp.O115.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oveland E., Muth T., Rapp E., Martens L., Berven F.S., Barsnes H. Viewing the proteome: how to visualize proteomics data? Proteomics. 2015;15:1341–1355. doi: 10.1002/pmic.201400412. [DOI] [PubMed] [Google Scholar]
- 29.Polpitiya A.D., Qian W.J., Jaitly N., Petyuk V.A., Adkins J.N., Camp D.G., 2nd, Anderson G.A., Smith R.D. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., Bacic A., Hill A.F., Stroud D.A., Ryan M.T., Agbinya J.I., Mariadason J.M., Burgess A.W., Mathivanan S. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 31.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R., Jassal B., Jupe S., Matthews L., May B., Palatnik S., Rothfels K., Shamovsky V., Song H., Williams M., Birney E., Hermjakob H., Stein L., D'Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Bilbao A., Bruderer T., Luban J., Strambio-De-Castillia C., Lisacek F., Hopfgartner G., Varesio E. The use of variable Q1 isolation windows improves selectivity in LC-SWATH-MS acquisition. J. Proteome Res. 2015;14:4359–4371. doi: 10.1021/acs.jproteome.5b00543. [DOI] [PubMed] [Google Scholar]
- 33.Aleyasin H., Rousseaux M.W., Marcogliese P.C., Hewitt S.J., Irrcher I., Joselin A.P., Parsanejad M., Kim R.H., Rizzu P., Callaghan S.M., Slack R.S., Mak T.W., Park D.S. DJ-1 protects the nigrostriatal axis from the neurotoxin MPTP by modulation of the AKT pathway. Proc. Natl. Acad. Sci. USA. 2010;107:3186–3191. doi: 10.1073/pnas.0914876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinumi T., Kimata J., Taira T., Ariga H., Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 35.Wilson M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox Signal. 2011;15:111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waak J., Weber S.S., Gorner K., Schall C., Ichijo H., Stehle T., Kahle P.J. Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J. Biol. Chem. 2009;284:14245–14257. doi: 10.1074/jbc.M806902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madian A.G., Hindupur J., Hulleman J.D., Diaz-Maldonado N., Mishra V.R., Guigard E., Kay C.M., Rochet J.-C., Regnier F.E. Effect of single amino acid substitution on oxidative modifications of the Parkinson's disease-related protein, DJ-1. Mol. Cell. Proteom. 2012;11 doi: 10.1074/mcp.M111.010892. (M111.010892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagida T., Tsushima J., Kitamura Y., Yanagisawa D., Takata K., Shibaike T., Yamamoto A., Taniguchi T., Yasui H., Taira T., Morikawa S., Inubushi T., Tooyama I., Ariga H. Oxidative stress induction of DJ-1 protein in reactive astrocytes scavenges free radicals and reduces cell injury. Oxid. Med. Cell. Longev. 2009;2:36–42. doi: 10.4161/oxim.2.1.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox Signal. 2011;15:111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu L., Liu K., Sun M., Tian C., Sun R., Morales Betanzos C., Tallman K.A., Porter N.A., Yang Y., Guo D., Liebler D.C., Yang J. Systematic and quantitative assessment of hydrogen peroxide reactivity with cysteines across human proteomes. Mol. Cell. Proteom. 2017;16:1815–1828. doi: 10.1074/mcp.RA117.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Held J.M., Danielson S.R., Behring J.B., Atsriku C., Britton D.J., Puckett R.L., Schilling B., Campisi J., Benz C.C., Gibson B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteom. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dias G.M., López M.L., Ferreira A.T.S., Chapeaurouge D.A., Rodrigues A., Perales J., Retamal C.A. Thiol-disulfide proteins of stallion epididymal spermatozoa. Anim. Reprod. Sci. 2014;145:29–39. doi: 10.1016/j.anireprosci.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Rossi R., Giustarini D., Milzani A., Dalle-Donne I. Cysteinylation and homocysteinylation of plasma protein thiols during ageing of healthy human beings. J. Cell. Mol. Med. 2009;13:3131–3140. doi: 10.1111/j.1582-4934.2008.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen R.E., Roth D., Winther J.R. Quantifying the global cellular thiol–disulfide status. Proc. Natl. Acad. Sci. USA. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray C.I., Van Eyk J.E. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ. Cardiovasc. Genet. 2012;5 doi: 10.1161/CIRCGENETICS.111.961425. (591-591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.A. Scirè, L. Cianfruglia, C. Minnelli, D. Bartolini, P. Torquato, G. Principato, F. Galli, T. Armeni, Glutathione compartmentalization and its role in glutathionylation and other regulatory processes of cellular pathways, BioFactors. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material