Abstract
Parasitic infections induce host immune responses that eliminate the invading parasites. However, parasites have evolved to develop many strategies to evade host immune attacks and survive in a hostile environment. The complement system acts as the first line of immune defense to eliminate the invading parasites by forming the membrane attack complex (MAC) and promoting an inflammatory reaction on the surface of invading parasites. To date, the complement activation pathway has been precisely delineated; however, the manner in which parasites escape complement attack, as a survival strategy in the host, is not well understood. Increasing evidence has shown that parasites develop sophisticated strategies to escape complement-mediated killing, including (i) recruitment of host complement regulatory proteins on the surface of the parasites to inhibit complement activation; (ii) expression of orthologs of host RCA to inhibit complement activation; and (iii) expression of parasite-encoded proteins, specifically targeting different complement components, to inhibit complement function and formation of the MAC. In this review, we compiled information regarding parasitic abilities to escape host complement attack as a survival strategy in the hostile environment of the host and the mechanisms underlying complement evasion. Effective escape of host complement attack is a crucial step for the survival of parasites within the host. Therefore, those proteins expressed by parasites and involved in the regulation of the complement system have become important targets for the development of drugs and vaccines against parasitic infections.
Keywords: complement system, immune evasion, parasites, complement activation pathways, complement regulatory proteins
Introduction
Parasites are pathogens that live in or on hosts, from which they obtain benefits for their growth, development, and propagation, and thereby cause inevitable harm (Poulin, 2006; Auld and Tinsley, 2015). There are three main classes of parasites that can cause disease in humans, including parasitic protozoa, helminths, and ectoparasites. Parasitic protozoans are single-celled organisms that parasitize the intestinal tract and other tissues, thereby causing various diseases (De Bona et al., 2018). Helminths are multicellular eukaryotic worms, including nematodes, trematodes, and cestodes, which infect billions of people worldwide (Scholte et al., 2018). In addition, severe disease could also be caused by ectoparasites, like Sarcoptes scabiei, which burrows into the skin to induce intense itching and the condition known as scabies (Arlian and Morgan, 2017).
Parasitic infections are the most common infections in humans, affecting billions of people worldwide and causing deadly diseases (such as malaria) and often neglected tropical diseases (Hotez et al., 2008). Parasitic infections induce the host immune response, which occurs in an attempt to kill and clear the invasion (Motran et al., 2017; Silva-Barrios and Stager, 2017). The host complement system serves as the first line of defense against parasitic invasion. Complement is a major part of innate immunity that is activated by a robust and efficient proteolytic cascade that eventually results in the opsonization and lysis of many invading pathogens. It is also connected to adaptive immunity and generates inflammatory responses through the production of proinflammatory molecules (Holers, 2014). In addition to its action as an innate and adaptive immunity enhancer in the host’s defense against infection, the complement system plays an alternative role in cell homeostasis by promoting tissue regeneration, morphogenesis, and the coagulation cascade (Kimura et al., 2003; Ricklin et al., 2010). The complement system consists of more than 50 components, including plasma proteins and membrane-bound proteins, some of which serve as PRM that trigger complement activation, and others that act as regulatory proteins that downregulate complement activation to prevent self-damage or autoimmune inflammation (Ricklin et al., 2016). The orchestrated balance between the efficient destruction of pathogens and prevention of unnecessary complement activation in the host tissue is tightly regulated by complement regulatory mechanisms (Ricklin et al., 2016).
To survive within the host, the parasite must overcome the host’s immune response. As a survival strategy, parasites have developed sophisticated mechanisms to escape and defend against complement attack. Parasites express various proteins to effectively play these roles (Morais et al., 2018; Song et al., 2018). Increasing evidence reveals that parasites escape complement attack by using several approaches, including expression of proteins to capture host regulatory proteins; expression of proteins that are homologous to host regulators and interfere with the functions of the host complement system (Jozsi, 2017); and expression of proteins that specifically bind to host complement components and interfere with the final formation of the MAC by inhibiting the classical, lectin, or alternative activation pathways (Braschi and Wilson, 2006; Zhao et al., 2017; Mendes-Sousa et al., 2018; Verma et al., 2018). In this review, we compiled pertinent information regarding parasitic abilities to escape host complement attack as a survival strategy in the hostile environment, and the mechanisms underlying complement evasion.
Complement System and Activation
The complement system in human consists of about 50 molecules that are soluble plasma proteins produced mainly by the liver or membrane-tethered proteins expressed on cell surface, including effector molecules, receptors and regulators (Merle et al., 2015a; Schmidt et al., 2016). Under normal circumstances, complement components remain inactive pro-enzymes or zymogens. On apoptotic cells and the pathogens lacking specific regulators of complement, the complement activation occurs, and the proteases become enzymatically active, resulting in a rapid and efficient cascade (Merle et al., 2015a). Three possible pathways exist for complement activation: the classical pathway, the alternative pathway, and the lectin pathway. Although the initiation steps of the three pathways differ, they converge at the terminal pathway resulting in the lysis of the apoptotic cells and the pathogens (Merle et al., 2015a). The activation cascades are shown in Figure 1.
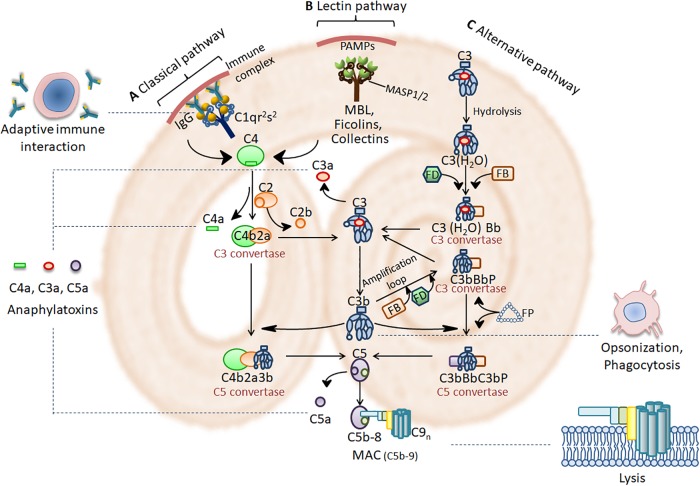
FIGURE 1.
Complement activation cascades and functions. (A) Immune complex (IC) activates the classical pathway through activating C1 complex (C1qr2s2). (B) PRMs such as MBL, ficolins and collectins, found in complexes with serine proteases (MASPs), bind to pathogen-associated molecular patterns (PAMPs) on the pathogen surface to activate the lectin pathway. Activation of the classical and lectin pathway leads to cleavage of C4 and C2 to form a C3 convertase (C4b2a). (C) The alternative pathway is initiated spontaneously by hydrolyzing C3 into C3(H2O) with factors FB, FD and FP. This leads to the formation of C3 convertases of the alternative pathway [C3(H2O)Bb or C3bBb]. Complement activation then comes to a core stage that C3 convertase cleave C3 into the anaphylatoxin C3a and the opsonin C3b. C3b then participates in the formation of the classical and lectin pathway C5 convertase (C4b2a3b) and the alternative pathway C5 convertase (C3bBbC3b). C5 convertase cleave C5 into the anaphylatoxin C5a and C5b. Afterwards, C5b assembles with C6, C7, C8, and multiple C9 molecules on the target surface to form MAC (C5b-9). MAC is a 10-nm aperture inserting into the target membrane, which results in the lysis of invading pathogens. The anaphylatoxins C3a and C5a bind to their corresponding receptors, C3aR and C5aR, to mediate inflammation. C3b triggers opsonization which facilitate phagocytic removal of the target. Complement modulates a variety of immune activities and acts as a linker between the native and the adaptive immune response such as augmentation of antibody response and enhancement of immunologic memory.
Triggering the Classical Pathway
The classical pathway is antibody-dependent and initiated by C1 complex, when binding of C1q to antibody IgM or IgG complexed with antigens (Duncan and Winter, 1988; Gaboriaud et al., 2014; Pednekar et al., 2016). Such binding events cause C1q conformational changes, which lead to the activation of C1r and C1s. Consequently, activated C1s splits C4 and C2, which then form C3 convertase (C4b2a) (Law and Dodds, 1997).
In addition to antigen-antibody immune complexes, C1q can bind directly with certain proteins on the surface of pathogens like bacterial cell walls, or polyanionic structures such as the lipoteichoic acid (Prellner, 1981). The binding of phosphatidylserine of the apoptotic cells, and other agents such as pentraxins (C-reactive protein and pentraxin 3) to C1q also initiates the classical pathway (McGrath et al., 2006; Biro et al., 2007).
Initiation of the Lectin Pathway
The lectin pathway is activated by microbial sugars instead of immune complexes. It is stimulated when MBL, ficolins (M-ficolin, H-ficolin, and L-ficolin), or collectins (collectin-10 and collectin-11) bind to mannose residues on the surface of the pathogen (Bohlson et al., 2007; Garred et al., 2016). Once bound with glycans, the MASPs become activated from inactive zymogens. A conformational change occurs in MASP-1, following which MASP-2 is cleaved (Garred et al., 2016). Activated MASP-2 then cleaves C4 and C2 to form C3 convertase (C4b2a) (Heja et al., 2012), which continues the cascade as the classical pathway until the formation of the MAC on the membrane of the pathogen. As the lectin pathway can be activated in the absence of antibody, it plays an important role in early infection even before the antibody is generated.
Initiation of the Alternative Pathway
The alternative pathway is activated by the spontaneous hydrolysis of C3 to from C3(H2O). Physiologically, the alternative pathway maintains constitutive activation at low-levels in plasma by a hydrolytic process known as tick-over. This spontaneous hydrolysis of a labile thioester bond converts C3 to a bioactive form C3(H2O) (Merle et al., 2015a). This results in a dramatic change in structure that exposes a binding site for FB (Merle et al., 2015a). FB binds to both C3b and C3(H2O), which can be cleaved into Ba and Bb by a plasma protease, FD. At this point, C3bBb and C3(H2O)Bb are formed as the C3 convertase of the alternative pathway. As C3bBb and C3(H2O)Bb are unstable, properdin (factor P, FP) binds with them to stabilize the alternative pathway C3 convertase (Kemper and Hourcade, 2008).
Generation of C3 Convertase, C5 Convertase and Formation of Activation of the MAC
All three pathways converge at the formation of C3 convertase, which promotes the cleavage of C3 into C3a and C3b. When a large quantity of C3b is generated, C5 convertase is formed by the binding of C3b to C4b2a or C3bBb (C4b2a3b or C3bBbC3b), which marks the activation of the terminal pathway (Schmidt et al., 2016). C5 is cleaved into the potent anaphylatoxin C5a and the bigger split fragment C5b by C5 convertases (Rawal and Pangburn, 2001). Large quantities of C3a and a limited quantity of C5a are released into the fluid phase, and then act on endothelial cells and mast cells (Dias Da Silva and Lepow, 1967; Kohl, 2001; Schraufstatter et al., 2002) as anaphylatoxins because of their ability to induce a shock-like syndrome similar to an allergic reaction (Coulthard and Woodruff, 2015; Hawksworth et al., 2017). The cascade of reactions leading to the assembly of the MAC on the membrane of the pathogen is initiated by C5b. This cascade starts with the binding of C5b to the monomer C6 to form the C5b6 complex, followed by further binding to the monomer C7 to expose the hydrophobic site on C7. The conformationally altered C7 is inserted into the membrane to bind C8 onto itself. Subsequently, C8β binds C5b67, and C8α-γ is inserted into the membrane, leading to the polymerization and insertion of 10–16 molecules of C9 into the membrane to form a pore (Parker and Sodetz, 2002). This 10-nm aperture is formed by the MAC, which disrupts the integrity of the lipid bilayer and allows the transfer of solutes and water across the membrane, resulting in osmotic lysis of the pathogen (Ricklin et al., 2010).
Regulation of Complement Activation
Activation of the complement system is tightly regulated to prevent uncontrolled amplification of the effects that may cause inflammation or autoimmune diseases (Zipfel and Skerka, 2009; Liszewski and Atkinson, 2015). The first method of regulation is hydrolysis. The complement-activated components that are not bound to the surface of the pathogen, such as C3b and C4b, are rapidly inactivated by hydrolysis. The second method of regulation of complement activation is through a variety of CRPs including RCAs. RCA proteins are the family of RCA gene cluster including complement receptor 1 (CR1 and CD35), membrane cofactor protein (MCP and CD46), decay accelerating factor (DAF and CD55), C4BP, and FH and its alternative splice product Factor H-like 1 (FHL-1) (Schmidt et al., 2016). Other CRPs include C1-INH, FI, CPN1, S-protein, clusterin (SP40/40), CRIg, C8BP, and protectin (CD59) (Noris and Remuzzi, 2013). They target different components to modulate and balance complement activation.
C1 Inhibitor
To inhibit the initiation stage, C1-INH, a plasma serine protease inhibitor, binds to the active enzymes C1r/s through covalent bond and dissociates it from C1q. C1-INH also binds through covalent bond to MASP-1 and MASP-2 and makes them dissociated from MBL. This leads to limit activation of the classical pathway and lectin pathway (Jiang et al., 2001).
C3 and C5 Convertase Inhibitors
Negative regulatory proteins that are also present in the plasma and/or on the cell membrane inhibit C3 and C5 convertases (Zipfel and Skerka, 2009). The C4BP has a high binding affinity to C4b and the C4b binding leads to the displacement of C2a from C3 convertase (C4b2a) (Schmidt et al., 2016). FI, an active serine protease in plasma, prevents the formation of C3 convertase and inactivates C4b2a and C3bBb through binding to C3b and C4b with the help of the cofactors FH, C4BP, MCP, and CR1. C3b is cleaved by FI and MCP first into membrane-bound iC3b and C3f, after which C3f is released into the fluid phase. The iC3b is then further cleaved by FI and CR1 into C3dg bound on the surface, and C3c released into the fluid phase. In the alternative pathway, C3bBb is permanently inactivated. Similarly, C4b is inactivated by FI and cleaved into C4c and C4d, thereby inhibiting the formation of C4b2a.
Conformational change in C3b exposes an extended surface in C3b that allows complement regulators FH, DAF, MCP, and CR1 to bind, resulting in the accelerated decay of the alternative pathway C3-convertase and inactivation of C3b (Alcorlo et al., 2015). FH is capable of competing with FB to bind with C3b (Zipfel and Skerka, 2009). DAF binds to C4b and dissociates C2a and Bb from C3 convertases (Fujita et al., 1987). It also displaces Bb from C3bBb to inhibit the formation of C3bBb or C3bBbC3b (Dho et al., 2018). In addition, CRIg can bind with remnant fragments of C3b (C3b, iC3b, and C3c) on the cell membrane to inhibit the alternative pathway (Zipfel and Skerka, 2009).
MAC Inhibitors
At the stage of MAC formation, several inhibitory proteins prevent the insertion of the MAC into the lipid bilayer. The S-protein, also known as vitronectin, binds with C5b67, C5b-8, and C5b-9 to inhibit MAC insertion. The SP40/40 not only binds directly to the C5b-9 complex but serves as a cofactor of the S-protein. Both C8BP and CD59 inhibit the binding of C9 to C5b-8, and C9 polymerization (Zipfel and Skerka, 2009).
Thus, regulation of complement activation at various stages of all three pathways spontaneously maintains homeostasis. Otherwise, any breaks in this balance could lead to autoimmune syndromes, such as atypical hemolytic uremic syndrome, with clinical features of damaged red blood cells and platelets, and kidney inflammation due to malfunction of the complement system (Yoshida et al., 2018).
Biological and Alternative Functions of Complement Activation
Complement Is Activated to Eliminate Pathogens by MAC or Opsonization
When complement is activated, it contributes to the defense against pathogens, which is not only evolved as part of the innate immune system but remains a link to the adaptive immune system. Complement destroys pathogens directly via the MAC. Once complement activation has been initiated, all three pathways lead to the formation of C3 convertase, which initiates a cascade of enzymatic reactions that eventually lead to the formation of the MAC on the surface of pathogens, resulting in osmotic lysis of invading bacteria (especially Gram-negative bacteria), viruses, and parasites (Merle et al., 2015b).
Moreover, complement fragment C5a recruits neutrophils, monocytes, and macrophages to clear pathogens through a process known as opsonization (Ricklin et al., 2010; Martin and Blom, 2016). The C3b and C4b bind with CR1 expressed on phagocytes and erythrocytes to enhance IgG-mediated phagocytosis or clear soluble immune complexes (Freeley et al., 2016). In addition, iC3b can be specifically recognized by CR3 and CR4 on monocytes and neutrophils (Wang et al., 2015; Lubbers et al., 2017). This causes the phagocytes to release toxic reactive oxygen compounds and microbicidal components, such as lysozyme and proteases that kill pathogens (Martin and Blom, 2016).
Alternative Functions of Activated Complement Components
Besides the participation of complement in innate immune defense, some of the original complement proteins, or intermediate components generated during activation, have many other immunity-enhancing functions. In addition to initiating the classical activation of complement, C1q has many unconventional functions. It is noteworthy that apart from its role as part of the complement system, C1q recognizes its receptors expressed on phagocytes, such as neutrophils and macrophages, and stimulates these phagocytes to release reactive oxygen species, such as H2O2 and superoxide, to attack pathogens. Furthermore, C1q plays a role in modulating dendritic cell (DC) maturation (Son et al., 2012), B cell tolerogenic capacity, and IgM or IgG production (Young et al., 1991). Moreover, C1q is reportedly involved in the clearance of apoptotic cells via CD91, which is a multi-protein-binding scavenger receptor complex containing CRT (Ogden et al., 2001).
Anaphylatoxins C3a and C5a stimulate DCs to express the major histocompatibility complex II (MHC-II) and CD86 via C3a and C5a receptors, thereby promoting their maturation as APCs (Peng et al., 2008, 2009). Locally produced C3a and C5a also interact with their receptors on T lymphocytes involved in maintaining T cell proliferation and differentiation (Strainic et al., 2008). The interaction between C3d and complement receptor CR2 on B cells during antigen-induced activation promotes B cell activation and facilitates the transformation of naïve B cells into antibody-producing effector and memory B cells (van den Elsen and Isenman, 2011; Donius et al., 2013). These alternative functions of complement components bridge the innate immune response with the adaptive immune response to coordinate the elimination of invading pathogens. The functions of the complement system are shown in Figure 1.
Complement Evasion by Parasites
Over millions of years of evolution, parasites, including parasitic protozoans, helminths and ectoparasites, have developed sophisticated mechanisms as strategies of survival to escape immune attack in the host. The first strategy developed was the inhibition of complement activation as the initial step to evade host immune clearance, particularly at the early stages of invasion. Complement plays an important role in the defense against parasitic infections, which relies on its direct lysis on invading parasites through the formation of the MAC, and acts as a bridge to adaptive immune responses. Parasites have evolved to develop several mechanisms that inhibit or interrupt the functions of host complement as the first steps to escape host immune attack.
Recruitment of Host Regulatory Proteins to Inhibit Complement Activation
The C1-INH is a soluble regulator of complement activation that negatively regulates the classical and lectin pathways by inhibiting C1r, C1s, MASP-1, and MASP-2, the activating proteases of the complement cascade. The intracellular protozoa, Plasmodium falciparum, which causes the deadliest malaria in humans (Rich et al., 2009), could recruit and utilize C1-INH to inhibit complement activation (Mejia et al., 2016). Merozoites, the invasive form of blood-stage malarial parasites, actively recruit C1-INH to their surface when exposed to human serum. A member of the merozoite surface protein 3 family, PfMSP3.1, worked as a direct interactive partner to bind with C1-INH, to inhibit C1s, MASP1, and MASP2 (Kennedy and Wijeyewickrema, 2017).
Host complement FH, a CRP, is the main soluble inhibitor of the alternative pathway against the formation of C3-convertase. It achieves this inhibition by supporting the conversion of C3b to iC3b (Zipfel and Skerka, 2009). Echinococcus granulosus, a cestode that causes cystic echinococcosis in humans, expresses a protein that sequestrates FH on the hydatid cyst wall to inhibit complement C3b deposition (Diaz et al., 1997; Breijo et al., 2008). The binding molecule remained unknown until further research showed that it was InsP6, a major component of the acellular laminated layer (LL) of the hydatid cyst wall, that was bound with FH to inhibit the alternative pathway (Irigoin et al., 2008). Mosquito midgut epithelial cells also express two proteins (40 and 100 kDa) as receptors that captured FH to inhibit the deposition of C3b and impair activation of the alternative complement pathway (Khattab et al., 2015).
Decay-accelerating factor (DAF; CD55), another human complement regulatory factor, causes the accelerated decay of C3 and C5 convertases by associating with C4b and C3b deposited on the cell membrane (Fujita et al., 1987; Heine et al., 2003). Schistosoma mansoni is a blood fluke that causes intestinal schistosomiasis. When incubated with normal human erythrocytes, but not with DAF-deficient erythrocytes, S. mansoni became resistant to complement lysis in vitro (Horta and Ramalho-Pinto, 1991). Further study showed that S. mansoni acquired DAF from host erythrocytes via the expression of a GPI anchor on the surface of the worm (Ramalho-Pinto et al., 1992). The ability of the trypsin-treated S. mansoni worm to acquire DAF was reduced (Ramalho-Pinto et al., 1992). Treatment with GPI-specific phospholipase D (GPI-PLD) facilitated the binding of DAF to the surface of the schistosomula (Carvalho et al., 1994).
As a membrane-bound inhibitor of the cytolytic MAC, CD59 reduces C9 polymerization on the cell surface by binding to C8α and C9 (Venneker and Asghar, 1992). The N-linked glycosylation of CD59 is related to its complement-inhibitory activity (Ninomiya et al., 1992). P. falciparum is able to acquire the intrinsic host factor, CD59, to restrict complement attack on the infected erythrocyte (Wiesner et al., 1997). Furthermore, P. falciparum expressed mannosyltransferase (PfPIG-M), which is involved in GPI synthesis, and thereafter increased the levels of the GPI-anchored protein, CD59, on the cells, indicating that the GPI anchor is involved in the capture of CD59 on the surface of P. falciparum-infected erythrocytes (Kim and Hong, 2007). Trichomonas vaginalis, an anaerobic flagellated protozoan parasite, also acquired CD59 from different host cells, including red blood cells, during infection, to protect the parasite from being lysed by host complement (Ibanez-Escribano et al., 2015).
Expression of Homologous Proteins With Host Regulators of Complement Activation
To avoid complement-mediated lysis, some parasites express a variety of CRPs on their surfaces. As more parasite genomes have been deciphered, parasitic helminths have been shown to express many mammalian-like receptors for host growth factors, cytokines, or hormones to regulate the growth, development, signaling, and reproduction of the parasite (Hu et al., 2003). More results suggest that parasites modulate host anti-parasite immune responses by expressing host immunological inhibitors and receptors of some immunological components (Ramirez-Toloza and Ferreira, 2017). The antigenic similarity between host- and parasite-expressed proteins may mask the host’s immune system to recognize invading parasites, and thus protect the parasite from elimination (Abu-Shakra et al., 1999).
Parasites express orthologs of host complement components or regulators to modulate or inhibit the functions of the host complement. S. japonicum, a blood fluke that causes Asian schistosomiasis, expresses protein (Schistosome CRIT) that share similarities with host complement C2 receptor inhibitor trispanning (CRIT) (Inal, 2005). This expression is indicative of their involvement in host complement activation or regulation. Schistosome CRIT is located on the surface tegument of the Schistosoma parasite and enables it to bind C2 via its extracellular domain. It subsequently inhibits the binding of C2 to C4b, to interfere with the formation of C3 convertase (C4b2a). The CRIT is an example of molecular mimicry, as it reportedly binds C2 with a domain that is homologous to one region of human C4b. Both the classical and lectin complement pathways are interrupted when C2 is hijacked (Cestari Idos et al., 2009). The C2 binding site of schistosome CRIT is located at an 11-amino acid sequence at the C-terminus of the first extracellular domain, which is involved in the inhibition of the classical complement pathway and reduction of immune complex-mediated inflammation (Inal et al., 2003). Trypanosoma cruzi, an intracellular protozoan parasite that causes Chagas disease, also expresses CRIT on the surface of trypomastigotes to inhibit C2-associated complement activation (Ramirez-Toloza and Ferreira, 2017).
In addition to the C2 receptor inhibitor, adult schistosomes express the C3 receptor on their surface tegument. During complement activation, C3 binds to the worm’s surface through the C3 receptor and stimulates the replacement of the outer tegument, which is shed during complement attack (Silva et al., 1993). Through this expression of the C3 receptor and shedding of the C3–C3 receptor, parasites are able to consume C3 from serum, and consequently become non-activators of the alternative pathway (Marikovsky et al., 1986; Rasmussen and Kemp, 1987).
In addition to its recruitment of host DAF on its surface to avoid host complement lysis, the T. cruzi trypomastigote also expresses DAF (T-DAF) on the surface of its virulent forms to inhibit complement activation by blocking C3, similar to mammalian DAF (Joiner et al., 1988; Kipnis et al., 1988; Tambourgi et al., 1993). Further studies have demonstrated that T. cruzi expressed a 160 kDa (GP160) complement regulatory glycoprotein on the surface of trypomastigotes (Norris et al., 1989). The gp160 gene was verified to share significant DNA sequence homologous with the human DAF gene (Norris et al., 1991). GP160 can inhibit the formation of the alternative and classical C3 convertase as it is a member of the C3/C4 binding family of complement regulators. This prevents the activation and amplification of the complement cascade on the parasite’s surface (Norris and Schrimpf, 1994; Norris et al., 1997).
An earlier study described a schistosome complement inhibitor, a 94-kD protein of S. mansoni (SCIP-1), expressed on the surface of S. mansoni larvae and adults, which was found to be functionally and antigenically related to human CD59. It binds to human C8 and C9, and inhibits the assembly of C5b-9 (Parizade et al., 1994). In addition, other CD59 homologs have been identified in the schistosome genome displaying the consensus CCXXXCN sequence at the C terminus (Wilson and Coulson, 2009) and in the membrane fraction of the live schistosome tegument (Castro-Borges et al., 2011). CD59 homologs (FhCD59-1,2,3) have also been found on the surface tegument of the trematode, Fasciola hepatica, a liver fluke (Shi et al., 2014). FhCD59-2 showed a phylogenetic relationship with SmCD59-2 on the surface tegument of S. mansoni (Shi et al., 2014). However, analogs of mammalian cell-expressed recombinant schistosome CD59 showed no inhibition of complement activity in vitro, which possibly differs from the action of native proteins expressed in the tegument (Farias et al., 2013). And both the location and potential function of the CD59-like proteins in F. hepatica even require further biochemical analyses to elucidate.
Expression of Proteins to Inhibit Host Complement Activation
In addition to their expression of parasite-encoded regulators, which mimic host complement regulators, to inhibit complement activation, parasites also express or secrete a variety of proteins that directly bind to some complement components to inhibit their activation by targeting various stages (Figure 2).
FIGURE 2.
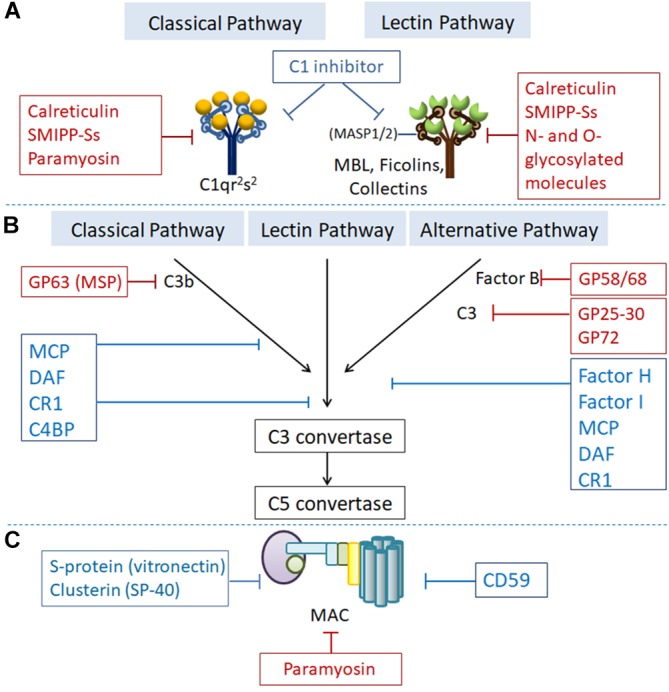
Regulation of complement activation by RCAs or parasite-expressed proteins targeting different complement components at the different steps of complement activation. Proteins showed in the blue boxes are human complement regulatory proteins. Proteins showed in the red boxes are parasites-generated to inhibit complement activation. (A) At the initiation step, C1 inhibitor binds to the active enzymes C1r/s and MASP-2 and dissociates them from C1q and MBL, respectively. Calreticulin, paramyosin, SMIPP-Ss, and N- and O-glycosylated molecules expressed by parasitic protozoa or helminths bind to C1q, MBL and ficolins to inhibit initiation of the classical and lectin pathway. (B) C3 convertase (C3bBb) undergoes an accelerated decay mediated by complement receptor 1 (CR1), decay accelerating factor (DAF), C4b-binding protein (C4BP) or factor H (FH). C3b is inactivated to iC3b by factor I (FI) with cofactor CR1, FH, C4BP or membrane cofactor protein (MCP). GP63 expressed by Leishmania binds to C3 leading to proteolysis of the active component C3b to form the inactive iC3b, thus preventing the formation of C3-convertase. GP58/68 expressed by T. cruzi binds with FB and interferes the formation of C3 convertase to inhibit alternative pathway. (C) Under normal conditions, CD59, S-protein (vitronectin), and Clusterin (SP-40) prevent the formation of the MAC. Paramyosin generated by helminth like T. spiralis bind with C8 and C9 to inhibit the formation of MAC.
Expression of Molecules That Inhibit C1q and MBL
The initiator of the classical complement pathway, C1q, plays an important role both in complement activation and in the activation of some immune cells by binding to the C1q receptor on their surfaces. CRT is a calcium-binding protein with a broad range of functions associated with immunomodulation (Malhotra et al., 1990). It contains the globular N-domain, proline-rich P-domain, and acidic C-domain, which can bind to C1q, resulting in the inhibition of C1q-dependent complement activity (Boelt et al., 2016). Studies have shown that CRTs are expressed on the surfaces of several parasites, such as Trypanosoma carassii, Necator americanus, Trichinella spiralis, and Brugia malayi, where it acts as an inhibitor of C1q, to facilitate the survival of the parasites in the hosts (Pritchard et al., 1999; Oladiran and Belosevic, 2010; Yadav et al., 2014; Zhao et al., 2017).
The tissue-dwelling nematode, T. spiralis, which causes serious trichinellosis in humans, expresses CRT (Ts-CRT) that can bind to C1q to inhibit the activation of the classical complement pathway. This results in the failure of MAC generation on the parasites, thereby conferring protection from the attack by the activated complement (Zhao et al., 2017). Binding of Ts-CRT to C1q also inhibited C1q involved pathogen-clearance functions, such as reduced recruitment of immune effect cells (neutrophils, eosinophils, and macrophages) to the site of parasitic infection and reduced the release of reactive oxygen intermediates (ROIs) and reactive nitrogen intermediates (RNIs) (Zhao et al., 2017).
The CRT expressed by B. malayi, a nematode that causes lymphatic filariasis in humans, could bind to human C1q. Bm-CRT inhibited C1q-dependent lysis of immunoglobulin-sensitized red blood cells (Yadav et al., 2017). Molecular docking has identified that interactions between Bm-CRT and C1q occurred at the site of the C1qB chain (IgG/M and CRP binding sites on C1q), and Bm-CRT conserved and non-conserved regions of the N/P-domain, in which 37 amino acids were involved in the interaction (Yadav et al., 2014, 2017). Mapping of the C1q binding site in the Hc-CRT secreted by H. contortus has shown that the N-terminal sequences of GKYYDDAKRD and AKFPKKFT were involved in the binding of C1q and suppression of complement-mediated cell lysis (Naresha et al., 2009).
Taenia solium, a tapeworm in the intestine of pigs that causes cysticercosis in humans, secreted CRT bound to C1 and induced a dose-dependent inhibition of C1-initiated complement activation (Laclette et al., 1992). More studies have shown that the S. scabiei secreted proteolytically inactive serine protease paralogs (SMIPP-Ss) that are bound both to C1q to inhibit the classical pathway, and to FP resulting in assembly failure of the alternative pathway convertases and thereby inhibiting the alternative pathway (Bergstrom et al., 2009).
Haemonchus contortus, a gastrointestinal nematode that parasitizes domestic animals, expresses GAPDH, one of the components of H. contortus ES products. The parasite enzyme trapped C1q and inhibited the complement-mediated lysis of sensitized sheep erythrocytes. The C1q binding site of Hc-GAPDH was mapped to the N-terminal (Vedamurthy et al., 2015).
Trypanosoma cruzi utilizes N- and O-glycosylated molecules on the surface of trypomastigotes and amastigotes to bind with L- and, H-ficolins, and MBL (Kahn et al., 1996). This results in the failure of MASP-2-induced C2 and C4 cleavage (Cestari and Ramirez, 2010; Ribeiro et al., 2015). Similarly, CRT of T. cruzi could bind to the collagenous portion of L-ficolin to inhibit the activation of the lectin pathway (Sosoniuk et al., 2014). Tc-CRT also bound MBL thus inactivating the lectin pathway (Lidani et al., 2017; Sosoniuk-Roche et al., 2017). Two scabies mite proteins, SMIPP-S D1 and I1, were also shown to be capable of binding with MBL to inhibit activation of the MBL-initiated lectin pathway (Reynolds et al., 2014).
One more important C1-INH is the helminth-secreted Pmy, which plays an important role in immunomodulatory functions by evading complement-mediated lysis. T. spiralis and S. mansoni express Pmy on their surfaces, which is also able to bind to C1q to inhibit C1q-initiated complement activation (Deng et al., 2007; Zhao et al., 2014).
Inhibition Occurs at the C3 and C5 Convertase Steps
Formation of the C3 and C5 convertases is a critical step for complement activation. Inhibitors bind to C3 and C5 convertases and interfere with their enzymatic reactions, thereby inhibiting the final formation of the MAC. Inhibition of C3 and C5 convertases also inhibits the release of anaphylatoxin C5a and the C5a-mediated recruitment of neutrophils.
During T. cruzi infection in mammals, the infective trypomastigote becomes resistant to lysis induced by the alternative complement pathway. The majority of C3 on the trypomastigote is in the inactive form, iC3b, which fails to form C5 convertase (Joiner et al., 1986). T. cruzi express the C3 acceptor in the infective trypomastigote stage and is a molecule of 25–30 kDa, whereas the C3 acceptor in the epimastigote stage is GP72 to inhibit the formation of alternative pathway C3 convertase (Joiner et al., 1986). A glycoprotein expressed on the surface of the trypomastigote, with molecular weight of 58/68 kDa (Gp 58/68), is another CRP that inhibits the formation of cell-bound and fluid phase alternative pathway C3 convertase, possibly through its interaction with FB rather than C3b (Fischer et al., 1988). Further investigation with recombinant protein demonstrated that this protein binds to human complements C3b and C4b to inhibit the activation of the complement cascade (Beucher et al., 2003).
Hc-GAPDH, which inhibits the C1q-initiated classical complement pathway, also could act as a C3 binding protein (C3BP) to inhibit complement activation, as measured by the hemolytic assay and MAC formation (Sahoo et al., 2013). The binding of C3BP to C3 through its N-terminal domain resulted in the inhibition of complement activation (Vedamurthy et al., 2015).
The GP63 expressed by Leishmania, an intracellular protozoa that causes leishmaniasis, is the most abundant surface glycoprotein on Leishmania promastigotes (Yao et al., 2003) that binds to C3 with high affinity, leading to proteolysis of the active component C3b to form the inactive iC3b, thus preventing the formation of C3-convertase (Yao, 2010).
MAC Formation Is Impeded
The terminal complement pathway is the final cytolytic step in the complement cascade, which results in formation of the MAC, a lytic assembly of C5b, C6, C7, C8, and multiple molecules of C9. In addition to the expression of host CD59 homologs on their surfaces to inhibit formation of the MAC and directly restrict complement-mediated lysis, parasites also express several other molecules that interfere with MAC formation as a final measure of protection against complement attack.
Trichinella spiralis expressed paramyosin (Ts-Pmy) on the surface of larvae and adult worms acts as an important immunomodulatory protein by not only binding to C1q to inhibit the initiating step of classical complement activation (Zhang et al., 2011; Sun et al., 2015), but binding to C8, C9 to interfere with MAC formation, and thus protecting the parasite from complement-mediated killing (Hao et al., 2014). Mapping of the C8/9 complement binding site has been narrowed down to 14 amino acid residues at the C-terminus (866Val-879Met) of Ts-Pmy, which inhibited C9 polymerization and complement-mediated lysis of rabbit erythrocytes (Zhao et al., 2014). The similar C8/C9 binding sites can also be found in S. mansoni-Pmy, located within the amino acid sequence of 744Asp–866Met at the C-terminus (Deng et al., 2007). The identification of complement binding sites on CRPs is important to gain a better understanding of their inhibitory mechanisms, as it relates to the design of drugs and vaccines against parasitic infections.
The host CRPs used by parasites and the parasite-generated CRPs described in this review are listed in Table 1.
Table 1.
A selection of host complement regulatory proteins utilized by parasite or parasite-produced complement regulatory proteins described in this review.
| Recruitment of host regulatory proteins to inhibit complement activation | ||||
|---|---|---|---|---|
| Proteins | Species | Ligands | Effects | Reference |
| PfMSP3.1 | Plasmodium falciparum | C1-INH | To recruit and utilize C1-INH to inactivate C1s and MASP-2 | Kennedy and Wijeyewickrema, 2017 |
| InsP6 | Echinococcus granulosus | FH | To recruit and utilize FH to inactivate the conversion of C3b to iC3b | Irigoin et al., 2008 |
| Proteins (40 and 100 kDa) | Mosquito midgut epithelial cells | FH | To capture FH to inhibit the deposition of C3b and impair activation of the alternative complement pathway | Khattab et al., 2015 |
| A GPI anchor on the surface of the worm | Schistosoma mansoni | DAF | To acquire DAF from host erythrocytes to form a complex with C4b and C3b deposited on the cell membrane causing accelerated decay of C3 and C5 convertases | Ramalho-Pinto et al., 1992 |
| PfPIG-M | Plasmodium falciparum | CD59 | To acquire the intrinsic CD59 to reduce C9 polymerization on the cell surface by binding to C8α and C9 | Kim and Hong, 2007 |
| Unknown | Trichomonas vaginalis | CD59 | To acquire CD59 from different host cells, including red blood cells to reduce C9 polymerization | Ibanez-Escribano et al., 2015 |
| Expression of homologous proteins with host regulators of complement activation | ||||
| Proteins | Species | Ligands | Effects | Reference |
| Complement Receptor Inhibitor Trispanning (CRIT) | Schistosoma japonicum Trypanosoma cruzi | C2 | To be homologous with host CRIT; To hijack C2 to interrupt the classical and lectin complement pathways | Cestari Idos et al., 2009; Ramirez-Toloza and Ferreira, 2017 |
| Schistosoma mansoni C3 receptor | Schistosom mansoni | C3 | To be homologous with host C3 receptor; To consume C3 from serum, and consequently become non-activators of the alternative pathway | Silva et al., 1993 |
| T-DAF (GP160) | Trypanosoma cruzi | C3 C4 | To be homologous with host DAF; To inhibit complement activation by blocking C3 and C4 | Norris et al., 1991, 1997; Tambourgi et al., 1993; Norris, 1998 |
| SCIP-1 | Schistosom mansoni | C8 C9 | To be homologous with host CD59; To bind to human C8 and C9 and inhibit the assembly of C5b-9 | Parizade et al., 1994 |
| Expression of proteins to inhibit host complement activation | ||||
| Proteins | Species | Ligands | Effects | Reference |
| Calreticulin | Trichinella spiralis Trypanosoma carassii Necator americanus Brugia malayi Trypanosoma cruzi | C1q L-Ficolin MBL | To inhibit activation of the classical and lectin complement pathway | Pritchard et al., 1999; Oladiran and Belosevic, 2010; Sosoniuk et al., 2014; Yadav et al., 2014; Sosoniuk-Roche et al., 2017; Zhao et al., 2017 |
| Paramyosin | Trichinella spiralis Schistosom mansoni | C1q C8 C9 | To inhibit activation of the classical complement pathway and the formation of MAC | Deng et al., 2007; Zhang et al., 2011; Zhao et al., 2014 |
| SMIPP-Ss | Sarcoptes scabiei | C1q FP | To inhibit activation of the classical and alternative pathway | Bergstrom et al., 2009 |
| SMIPP-S D1 and I1 | Sarcoptes scabiei | MBL | To inhibit activation of the lectin complement pathway | Reynolds et al., 2014 |
| N- and O-glycosylated molecules | Trypanosoma cruzi | L-Ficolin H-Ficolin | To inhibit activation of the lectin complement pathway | Kahn et al., 1996 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Haemonchus contortus | C1q C3 | To inhibit haemolysis and MAC formation | Sahoo et al., 2013; Vedamurthy et al., 2015 |
| GP58/68 | Trypanosoma cruzi | FB | To bind with FB and inhibit alternative pathway | Fischer et al., 1988 |
| GP63 | Leishmania mexicana | C3b | To form iC3b | Yao et al., 2003 |
| Protein (25–30 kDa) (trypomastigote) GP72 (epimastigote) | Trypanosoma cruzi | C3 | To inhibit the formation of the C3 convertase in the alternative pathway | Joiner et al., 1986 |
Parasite-Generated Complement Regulatory Proteins as Vaccine and Drug Targets
Parasite-generated complement inhibitory proteins are actively involved in the inhibition of host complement activation and are important for the survival of parasites within the host. Therefore, these proteins have emerged as prime targets for the development of drugs and vaccines against infections, or regulation of disease progression. Immunization with these proteins may reduce the defensive ability of the parasites against host complement attack, thereby rendering them more susceptible to the host’s immune defense, and ultimately leading to their expulsion from the host. Immunization with C4BP-fused MSP119 induced protective immunity in BALB/c mice against the otherwise lethal malarial parasitic challenge of Plasmodium yoelii, possibly through protection of the parasite from complement lysis (Ogun et al., 2008). Vaccination with complement regulator CRIT ed1 synthetic peptide conferred protection against the challenge of Schistosoma japonicum in mice through the inhibition of complement activity both in vitro and in vivo (Ma et al., 2017). T. cruzi expresses a complement regulatory protein (Tc-CRP) as a major antigen that induces the production of lytic antibodies during T. cruzi infections, making it a potential target for vaccine development (Henrique et al., 2016).
As an important CRP, Pmy has become another target of interest for the development of vaccines against various helminth infections. Mice immunized with native or recombinant Pmy of S. mansoni show a significant reduction in worm burden when challenged with S. mansoni cercariae (Pearce et al., 1988). The monoclonal antibody, 9G3, which targets the complement binding site of Ts-Pmy, could partially block its complement inhibitory activity, thereby increasing complement-mediated killing of larvae (Hao et al., 2014). Mice immunized with recombinant protein (Yang et al., 2010), epitope peptides (Wei et al., 2011; Gu et al., 2013, 2017), or the DNA (Wang et al., 2016, 2017) of Ts-Pmy were conferred with significant protection against the challenge of T. spiralis muscle larvae. Thus, Ts-Pmy has become the leading vaccine candidate for the control of trichinellosis. The RNAi-mediated silencing of Pmy expression in T. spiralis was also verified to reduce viability and infectivity of treated infective larvae (Chen et al., 2012).
Conclusion
The complement system plays a major role in combating the establishment of invading pathogens, and acts as the first line of host defense against the invading parasites. Parasites have developed sophisticated mechanisms as survival strategies to defend against complement attack including recruitment of host regulatory proteins and expression of proteins either homologs to host regulators or directly blocking activated complement molecules to evade or inhibit complement activation. In this review, we analyzed a considerable number of proteins that are expressed or recruited by parasites and involved in the regulation of the host complement system as a strategy to escape host complement-mediated killing. Effective evasion of host complement attack is a crucial step for the survival of parasites within the host. Therefore, parasite-expressed CRPs have now become important targets for the development of drugs and vaccines against parasitic infections.
Author Contributions
SS, XS, and XZ conceived the concept for this review article. SS and XS wrote the manuscript. SS constructed the table and figures. BZ, XZ, and YC read, edited, and reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- APC
antigen presenting cells
- C1-INH
C1 inhibitor
- C4BP
C4b-binding protein
- C8BP
C8-binding protein
- CPN1
carboxypeptidase N
- CRIg
complement receptor of the immunoglobulin family
- CRIT
complement receptor inhibitor trispanning
- CRP
complement regulatory protein
- CRT
calreticulin
- DAF
decay-accelerating factor
- FB
factor B
- FD
factor D
- FH
factor H
- FI
factor I
- FP
properdin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- InsP6
myo-inositol hexakisphosphate
- MAC
membrane attack complex
- MASP
MBL-associated serine protease
- MBL
mannose-binding lectin
- MCP
membrane-cofactor protein
- MSP
major surface protease
- MSP1
merozoite surface protein 1
- PAMPs
pathogen-associated molecular patterns
- PfMSP3.1
Plasmodium falciparum merozoite surface protein 3 family
- PfPIG-M
P. falciparum mannosyltransferase
- Pmy
paramyosin
- PRM
pattern recognition molecule
- RCA
regulators of complement activation
- SCIP-1
schistosome complement inhibitor-1
- SMIPP-S
scabies mite proteolytically inactive serine protease paralog
- SP40/40
S-protein
- TOR
trispanning orphan receptor
Footnotes
Funding. This work was funded by the National Natural Science Foundation of China (81672042).
References
- Abu-Shakra M., Buskila D., Shoenfeld Y. (1999). Molecular mimicry between host and pathogen: examples from parasites and implication. Immunol. Lett. 67 147–152. 10.1016/S0165-2478(99)00004-8 [DOI] [PubMed] [Google Scholar]
- Alcorlo M., Lopez-Perrote A., Delgado S., Yebenes H., Subias M., Rodriguez-Gallego C., et al. (2015). Structural insights on complement activation. FEBS J. 282 3883–3891. 10.1111/febs.13399 [DOI] [PubMed] [Google Scholar]
- Arlian L. G., Morgan M. S. (2017). A review of Sarcoptes scabiei: past, present and future. Parasit. Vectors 10:297. 10.1186/s13071-017-2234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld S. K., Tinsley M. C. (2015). The evolutionary ecology of complex lifecycle parasites: linking phenomena with mechanisms. Heredity 114 125–132. 10.1038/hdy.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom F. C., Reynolds S., Johnstone M., Pike R. N., Buckle A. M., Kemp D. J., et al. (2009). Scabies mite inactivated serine protease paralogs inhibit the human complement system. J. Immunol. 182 7809–7817. 10.4049/jimmunol.0804205 [DOI] [PubMed] [Google Scholar]
- Beucher M., Meira W. S., Zegarra V., Galvao L. M., Chiari E., Norris K. A. (2003). Expression and purification of functional, recombinant Trypanosoma cruzi complement regulatory protein. Protein Expr. Purif. 27 19–26. 10.1016/S1046-5928(02)00562-4 [DOI] [PubMed] [Google Scholar]
- Biro A., Rovo Z., Papp D., Cervenak L., Varga L., Fust G., et al. (2007). Studies on the interactions between C-reactive protein and complement proteins. Immunology 121 40–50. 10.1111/j.1365-2567.2007.02535.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelt S. G., Norn C., Rasmussen M. I., Andre I., Ciplys E., Slibinskas R., et al. (2016). Mapping the Ca(2+) induced structural change in calreticulin. J. Proteomics 142 138–148. 10.1016/j.jprot.2016.05.015 [DOI] [PubMed] [Google Scholar]
- Bohlson S. S., Fraser D. A., Tenner A. J. (2007). Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol. 44 33–43. 10.1016/j.molimm.2006.06.021 [DOI] [PubMed] [Google Scholar]
- Braschi S., Wilson R. A. (2006). Proteins exposed at the adult schistosome surface revealed by biotinylation. Mol. Cell. Proteomics 5 347–356. 10.1074/mcp.M500287-MCP200 [DOI] [PubMed] [Google Scholar]
- Breijo M., Anesetti G., Martinez L., Sim R. B., Ferreira A. M. (2008). Echinococcus granulosus: the establishment of the metacestode is associated with control of complement-mediated early inflammation. Exp. Parasitol. 118 188–196. 10.1016/j.exppara.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Carvalho E. M., Metz C. N., Davitz M. A., Ramalho-Pinto F. J. (1994). Binding of GPI-PLD-treated DAF to the surface of Schistosoma mansoni schistosomula. Braz. J. Med. Biol. Res. 27 457–462. [PubMed] [Google Scholar]
- Castro-Borges W., Dowle A., Curwen R. S., Thomas-Oates J., Wilson R. A. (2011). Enzymatic shaving of the tegument surface of live schistosomes for proteomic analysis: a rational approach to select vaccine candidates. PLoS Negl. Trop. Dis. 5:e993. 10.1371/journal.pntd.0000993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I., Ramirez M. I. (2010). Inefficient complement system clearance of Trypanosoma cruzi metacyclic trypomastigotes enables resistant strains to invade eukaryotic cells. PLoS One 5:e9721. 10.1371/journal.pone.0009721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari Idos S., Krarup A., Sim R. B., Inal J. M., Ramirez M. I. (2009). Role of early lectin pathway activation in the complement-mediated killing of Trypanosoma cruzi. Mol. Immunol. 47 426–437. 10.1016/j.molimm.2009.08.030 [DOI] [PubMed] [Google Scholar]
- Chen X., Yang Y., Yang J., Zhang Z., Zhu X. (2012). RNAi-mediated silencing of paramyosin expression in Trichinella spiralis results in impaired viability of the parasite. PLoS One 7:e49913. 10.1371/journal.pone.0049913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard L. G., Woodruff T. M. (2015). Is the complement activation product C3a a proinflammatory molecule? Re-evaluating the evidence and the myth. J. Immunol. 194 3542–3548. 10.4049/jimmunol.1403068 [DOI] [PubMed] [Google Scholar]
- De Bona E., Lidani K. C. F., Bavia L., Omidian Z., Gremski L. H., Sandri T. L., et al. (2018). Autoimmunity in chronic chagas disease: a road of multiple pathways to cardiomyopathy? Front. Immunol. 9:1842. 10.3389/fimmu.2018.01842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Gold D., LoVerde P. T., Fishelson Z. (2007). Mapping of the complement C9 binding domain in paramyosin of the blood fluke Schistosoma mansoni. Int. J. Parasitol. 37 67–75. 10.1016/j.ijpara.2006.09.011 [DOI] [PubMed] [Google Scholar]
- Dho S. H., Lim J. C., Kim L. K. (2018). Beyond the role of CD55 as a complement component. Immune Netw. 18:e11. 10.4110/in.2018.18.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Da Silva W., Lepow I. H. (1967). Complement as a mediator of inflammation. II. Biological properties of anaphylatoxin prepared with purified components of human complement. J. Exp. Med. 125 921–946. 10.1084/jem.125.5.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Ferreira A., Sim R. B. (1997). Complement evasion by Echinococcus granulosus: sequestration of host factor H in the hydatid cyst wall. J. Immunol. 158 3779–3786. [PubMed] [Google Scholar]
- Donius L. R., Handy J. M., Weis J. J., Weis J. H. (2013). Optimal germinal center B cell activation and T-dependent antibody responses require expression of the mouse complement receptor Cr1. J. Immunol. 191 434–447. 10.4049/jimmunol.1203176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. R., Winter G. (1988). The binding site for C1q on IgG. Nature 332 738–740. 10.1038/332738a0 [DOI] [PubMed] [Google Scholar]
- Farias L. P., Krautz-Peterson G., Tararam C. A., Araujo-Montoya B. O., Fraga T. R., Rofatto H. K., et al. (2013). On the three-finger protein domain fold and CD59-like proteins in Schistosoma mansoni. PLoS Negl. Trop. Dis. 7:e2482. 10.1371/journal.pntd.0002482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E., Ouaissi M. A., Velge P., Cornette J., Kazatchkine M. D. (1988). gp 58/68, a parasite component that contributes to the escape of the trypomastigote form of T. cruzi from damage by the human alternative complement pathway. Immunology 65 299–303. [PMC free article] [PubMed] [Google Scholar]
- Freeley S., Kemper C., Le Friec G. (2016). The “ins and outs” of complement-driven immune responses. Immunol. Rev. 274 16–32. 10.1111/imr.12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Inoue T., Ogawa K., Iida K., Tamura N. (1987). The mechanism of action of decay-accelerating factor (DAF). DAF inhibits the assembly of C3 convertases by dissociating C2a and Bb. J. Exp. Med. 166 1221–1228. 10.1084/jem.166.5.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C., Ling W. L., Thielens N. M., Bally I., Rossi V. (2014). Deciphering the fine details of c1 assembly and activation mechanisms: “mission impossible”? Front. Immunol. 5:565. 10.3389/fimmu.2014.00565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garred P., Genster N., Pilely K., Bayarri-Olmos R., Rosbjerg A., Ma Y. J., et al. (2016). A journey through the lectin pathway of complement-MBL and beyond. Immunol. Rev. 274 74–97. 10.1111/imr.12468 [DOI] [PubMed] [Google Scholar]
- Gu Y., Sun X., Li B., Huang J., Zhan B., Zhu X. (2017). Vaccination with a paramyosin-based multi-epitope vaccine elicits significant protective immunity against Trichinella spiralis infection in mice. Front. Microbiol. 8:1475. 10.3389/fmicb.2017.01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Wei J., Yang J., Huang J., Yang X., Zhu X. (2013). Protective immunity against Trichinella spiralis infection induced by a multi-epitope vaccine in a murine model. PLoS One 8:e77238. 10.1371/journal.pone.0077238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Zhao X., Yang J., Gu Y., Sun R., Zhu X. (2014). Monoclonal antibody targeting complement C9 binding domain of Trichinella spiralis paramyosin impairs the viability of Trichinella infective larvae in the presence of complement. Parasit. Vectors 7:313. 10.1186/1756-3305-7-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth O. A., Li X. X., Coulthard L. G., Wolvetang E. J., Woodruff T. M. (2017). New concepts on the therapeutic control of complement anaphylatoxin receptors. Mol. Immunol. 89 36–43. 10.1016/j.molimm.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Heine H., El-Samalouti V. T., Notzel C., Pfeiffer A., Lentschat A., Kusumoto S., et al. (2003). CD55/decay accelerating factor is part of the lipopolysaccharide-induced receptor complex. Eur. J. Immunol. 33 1399–1408. 10.1002/eji.200323381 [DOI] [PubMed] [Google Scholar]
- Heja D., Kocsis A., Dobo J., Szilagyi K., Szasz R., Zavodszky P., et al. (2012). Revised mechanism of complement lectin-pathway activation revealing the role of serine protease MASP-1 as the exclusive activator of MASP-2. Proc. Natl. Acad. Sci. U.S.A. 109 10498–10503. 10.1073/pnas.1202588109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique P. M., Marques T., da Silva M. V., Nascentes G. A. N., de Oliveira C. F., Rodrigues V., et al. (2016). Correlation between the virulence of T. cruzi strains, complement regulatory protein expression levels, and the ability to elicit lytic antibody production. Exp. Parasitol. 170 66–72. 10.1016/j.exppara.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Holers V. M. (2014). Complement and its receptors: new insights into human disease. Annu. Rev. Immunol. 32 433–459. 10.1146/annurev-immunol-032713-120154 [DOI] [PubMed] [Google Scholar]
- Horta M. F., Ramalho-Pinto F. J. (1991). Role of human decay-accelerating factor in the evasion of Schistosoma mansoni from the complement-mediated killing in vitro. J. Exp. Med. 174 1399–1406. 10.1084/jem.174.6.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P. J., Brindley P. J., Bethony J. M., King C. H., Pearce E. J., Jacobson J. (2008). Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 118 1311–1321. 10.1172/jci34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yan Q., Shen D. K., Liu F., Zhu Z. D., Song H. D., et al. (2003). Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat. Genet. 35 139–147. 10.1038/ng1236 [DOI] [PubMed] [Google Scholar]
- Ibanez-Escribano A., Nogal-Ruiz J. J., Perez-Serrano J., Gomez-Barrio A., Escario J. A., Alderete J. F. (2015). Sequestration of host-CD59 as potential immune evasion strategy of Trichomonas vaginalis. Acta Trop. 149 1–7. 10.1016/j.actatropica.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Inal J. M. (2005). Complement C2 receptor inhibitor trispanning: from man to schistosome. Springer Semin. Immunopathol. 27 320–331. 10.1007/s00281-005-0009-9 [DOI] [PubMed] [Google Scholar]
- Inal J. M., Schneider B., Armanini M., Schifferli J. A. (2003). A peptide derived from the parasite receptor, complement C2 receptor inhibitor trispanning, suppresses immune complex-mediated inflammation in mice. J. Immunol. 170 4310–4317. 10.4049/jimmunol.170.8.4310 [DOI] [PubMed] [Google Scholar]
- Irigoin F., Laich A., Ferreira A. M., Fernandez C., Sim R. B., Diaz A. (2008). Resistance of the Echinococcus granulosus cyst wall to complement activation: analysis of the role of InsP6 deposits. Parasite Immunol. 30 354–364. 10.1111/j.1365-3024.2008.01034.x [DOI] [PubMed] [Google Scholar]
- Jiang H., Wagner E., Zhang H., Frank M. M. (2001). Complement 1 inhibitor is a regulator of the alternative complement pathway. J. Exp. Med. 194 1609–1616. 10.1084/jem.194.11.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner K., Sher A., Gaither T., Hammer C. (1986). Evasion of alternative complement pathway by Trypanosoma cruzi results from inefficient binding of factor B. Proc. Natl. Acad. Sci. U.S.A. 83 6593–6597. 10.1073/pnas.83.17.6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner K. A., daSilva W. D., Rimoldi M. T., Hammer C. H., Sher A., Kipnis T. L. (1988). Biochemical characterization of a factor produced by trypomastigotes of Trypanosoma cruzi that accelerates the decay of complement C3 convertases. J. Biol. Chem. 263 11327–11335. [PubMed] [Google Scholar]
- Jozsi M. (2017). Factor H family proteins in complement evasion of microorganisms. Front. Immunol. 8:571. 10.3389/fimmu.2017.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn S. J., Wleklinski M., Ezekowitz R. A., Coder D., Aruffo A., Farr A. (1996). The major surface glycoprotein of Trypanosoma cruzi amastigotes are ligands of the human serum mannose-binding protein. Infect. Immun. 64 2649–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C., Hourcade D. E. (2008). Properdin: new roles in pattern recognition and target clearance. Mol. Immunol. 45 4048–4056. 10.1016/j.molimm.2008.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. T., Wijeyewickrema L. C. (2017). Recruitment of human C1 esterase inhibitor controls complement activation on blood stage Plasmodium falciparum merozoites. J. Immunol. 198 4728–4737. 10.4049/jimmunol.1700067 [DOI] [PubMed] [Google Scholar]
- Khattab A., Barroso M., Miettinen T., Meri S. (2015). Anopheles midgut epithelium evades human complement activity by capturing factor H from the blood meal. PLoS Negl. Trop. Dis. 9:e0003513. 10.1371/journal.pntd.0003513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. U., Hong Y. (2007). Functional analysis of the first mannosyltransferase (PIG-M) involved in glycosylphosphatidylinositol synthesis in Plasmodium falciparum. Mol. Cells 24 294–300. [PubMed] [Google Scholar]
- Kimura Y., Madhavan M., Call M. K., Santiago W., Tsonis P. A., Lambris J. D., et al. (2003). Expression of complement 3 and complement 5 in newt limb and lens regeneration. J. Immunol. 170 2331–2339. 10.4049/jimmunol.170.5.2331 [DOI] [PubMed] [Google Scholar]
- Kipnis T. L., Joiner K. A., da Silva W. D., Rimoldi M. T., Hammer C. H., Sher A. (1988). Identification of membrane components of Trypanosoma cruzi modulators of complement system. Mem. Inst. Oswaldo Cruz. 83(Suppl. 1), 571–575. 10.1590/S0074-02761988000500069 [DOI] [PubMed] [Google Scholar]
- Kohl J. (2001). Anaphylatoxins and infectious and non-infectious inflammatory diseases. Mol. Immunol. 38 175–187. 10.1016/S0161-5890(01)00041-4 [DOI] [PubMed] [Google Scholar]
- Laclette J. P., Shoemaker C. B., Richter D., Arcos L., Pante N., Cohen C., et al. (1992). Paramyosin inhibits complement C1. J. Immunol. 148 124–128. [PubMed] [Google Scholar]
- Law S. K., Dodds A. W. (1997). The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6 263–274. 10.1002/pro.5560060201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidani K. C. F., Bavia L., Ambrosio A. R., de Messias-Reason I. J. (2017). The complement system: a prey of Trypanosoma cruzi. Front. Microbiol. 8:607. 10.3389/fmicb.2017.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski M. K., Atkinson J. P. (2015). Complement regulators in human disease: lessons from modern genetics. J. Intern. Med. 277 294–305. 10.1111/joim.12338 [DOI] [PubMed] [Google Scholar]
- Lubbers R., van Essen M. F., van Kooten C., Trouw L. A. (2017). Production of complement components by cells of the immune system. Clin. Exp. Immunol. 188 183–194. 10.1111/cei.12952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Zai J., Han Y., Hong Y., Zhang M., Cao X., et al. (2017). Characterization of Schistosoma japonicum tetraspanning orphan receptor and its role in binding to complement C2 and immunoprotection against murine schistosomiasis. Parasit. Vectors 10:288. 10.1186/s13071-017-2229-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra R., Thiel S., Reid K. B., Sim R. B. (1990). Human leukocyte C1q receptor binds other soluble proteins with collagen domains. J. Exp. Med. 172 955–959. 10.1084/jem.172.3.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikovsky M., Levi-Schaffer F., Arnon R., Fishelson Z. (1986). Schistosoma mansoni: killing of transformed schistosomula by the alternative pathway of human complement. Exp. Parasitol. 61 86–94. 10.1016/0014-4894(86)90138-4 [DOI] [PubMed] [Google Scholar]
- Martin M., Blom A. M. (2016). Complement in removal of the dead - balancing inflammation. Immunol. Rev. 274 218–232. 10.1111/imr.12462 [DOI] [PubMed] [Google Scholar]
- McGrath F. D., Brouwer M. C., Arlaud G. J., Daha M. R., Hack C. E., Roos A. (2006). Evidence that complement protein C1q interacts with C-reactive protein through its globular head region. J. Immunol. 176 2950–2957. 10.4049/jimmunol.176.5.2950 [DOI] [PubMed] [Google Scholar]
- Mejia P., Diez-Silva M., Kamena F., Lu F., Fernandes S. M., Seeberger P. H., et al. (2016). Human C1-inhibitor suppresses malaria parasite invasion and cytoadhesion via binding to parasite glycosylphosphatidylinositol and host cell receptors. J. Infect. Dis. 213 80–89. 10.1093/infdis/jiv439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Sousa A. F., Vale V. F., Queiroz D. C., Pereira-Filho A. A., da Silva N. C. S., Koerich L. B., et al. (2018). Inhibition of the complement system by saliva of Anopheles (Nyssorhynchus) aquasalis. Insect Biochem. Mol. Biol. 92 12–20. 10.1016/j.ibmb.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle N. S., Church S. E., Fremeaux-Bacchi V., Roumenina L. T. (2015a). Complement system part I - molecular mechanisms of activation and regulation. Front. Immunol. 6:262. 10.3389/fimmu.2015.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle N. S., Noe R., Halbwachs-Mecarelli L., Fremeaux-Bacchi V., Roumenina L. T. (2015b). Complement system part II: role in immunity. Front. Immunol. 6:257. 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais S. B., Figueiredo B. C., Assis N. R. G., Homan J., Mambelli F. S., Bicalho R. M., et al. (2018). Schistosoma mansoni SmKI-1 or Its C-terminal fragment induces partial protection against S. mansoni infection in mice. Front. Immunol. 9:1762. 10.3389/fimmu.2018.01762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motran C. C., Ambrosio L. F., Volpini X., Celias D. P., Cervi L. (2017). Dendritic cells and parasites: from recognition and activation to immune response instruction. Semin. Immunopathol. 39 199–213. 10.1007/s00281-016-0588-7 [DOI] [PubMed] [Google Scholar]
- Naresha S., Suryawanshi A., Agarwal M., Singh B. P., Joshi P. (2009). Mapping the complement C1q binding site in Haemonchus contortus calreticulin. Mol. Biochem. Parasitol. 166 42–46. 10.1016/j.molbiopara.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Ninomiya H., Stewart B. H., Rollins S. A., Zhao J., Bothwell A. L., Sims P. J. (1992). Contribution of the N-linked carbohydrate of erythrocyte antigen CD59 to its complement-inhibitory activity. J. Biol. Chem. 267 8404–8410. [PubMed] [Google Scholar]
- Noris M., Remuzzi G. (2013). Overview of complement activation and regulation. Semin. Nephrol. 33 479–492. 10.1016/j.semnephrol.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K. A. (1998). Stable transfection of Trypanosoma cruzi epimastigotes with the trypomastigote-specific complement regulatory protein cDNA confers complement resistance. Infect. Immun. 66 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K. A., Bradt B., Cooper N. R., So M. (1991). Characterization of a Trypanosoma cruzi C3 binding protein with functional and genetic similarities to the human complement regulatory protein, decay-accelerating factor. J. Immunol. 147 2240–2247. [PubMed] [Google Scholar]
- Norris K. A., Harth G., So M. (1989). Purification of a Trypanosoma cruzi membrane glycoprotein which elicits lytic antibodies. Infect. Immun. 57 2372–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K. A., Schrimpf J. E. (1994). Biochemical analysis of the membrane and soluble forms of the complement regulatory protein of Trypanosoma cruzi. Infect. Immun. 62 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K. A., Schrimpf J. E., Szabo M. J. (1997). Identification of the gene family encoding the 160-kilodalton Trypanosoma cruzi complement regulatory protein. Infect. Immun. 65 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., et al. (2001). C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194 781–795. 10.1084/jem.194.6.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogun S. A., Dumon-Seignovert L., Marchand J. B., Holder A. A., Hill F. (2008). The oligomerization domain of C4-binding protein (C4bp) acts as an adjuvant, and the fusion protein comprised of the 19-kilodalton merozoite surface protein 1 fused with the murine C4bp domain protects mice against malaria. Infect. Immun. 76 3817–3823. 10.1128/iai.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladiran A., Belosevic M. (2010). Trypanosoma carassii calreticulin binds host complement component C1q and inhibits classical complement pathway-mediated lysis. Dev. Comp. Immunol. 34 396–405. 10.1016/j.dci.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Parizade M., Arnon R., Lachmann P. J., Fishelson Z. (1994). Functional and antigenic similarities between a 94-kD protein of Schistosoma mansoni (SCIP-1) and human CD59. J. Exp. Med. 179 1625–1636. 10.1084/jem.179.5.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. L., Sodetz J. M. (2002). Role of the human C8 subunits in complement-mediated bacterial killing: evidence that C8 gamma is not essential. Mol. Immunol. 39 453–458. 10.1016/S0161-5890(02)00121-9 [DOI] [PubMed] [Google Scholar]
- Pearce E. J., James S. L., Hieny S., Lanar D. E., Sher A. (1988). Induction of protective immunity against Schistosoma mansoni by vaccination with schistosome paramyosin (Sm97), a nonsurface parasite antigen. Proc. Natl. Acad. Sci. U.S.A. 85 5678–5682. 10.1073/pnas.85.15.5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pednekar L., Pandit H., Paudyal B., Kaur A., Al-Mozaini M. A., Kouser L., et al. (2016). Complement protein C1q interacts with DC-SIGN via its globular domain and thus may interfere with HIV-1 transmission. Front. Immunol. 7:600. 10.3389/fimmu.2016.00600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Li K., Anderson K., Farrar C. A., Lu B., Smith R. A., et al. (2008). Local production and activation of complement up-regulates the allostimulatory function of dendritic cells through C3a-C3aR interaction. Blood 111 2452–2461. 10.1182/blood-2007-06-095018 [DOI] [PubMed] [Google Scholar]
- Peng Q., Li K., Wang N., Li Q., Asgari E., Lu B., et al. (2009). Dendritic cell function in allostimulation is modulated by C5aR signaling. J. Immunol. 183 6058–6068. 10.4049/jimmunol.0804186 [DOI] [PubMed] [Google Scholar]
- Poulin R. (2006). Evolutionary Ecology of Parasites, 2nd Edn. Princeton, NJ: Princeton University Press. [Google Scholar]
- Prellner K. (1981). C1q binding and complement activation by capsular and cell wall components of S. pneumoniae type XIX. Acta Pathol. Microbiol. Scand. C 89 359–364. [DOI] [PubMed] [Google Scholar]
- Pritchard D. I., Brown A., Kasper G., McElroy P., Loukas A., Hewitt C., et al. (1999). A hookworm allergen which strongly resembles calreticulin. Parasite Immunol. 21 439–450. 10.1046/j.1365-3024.1999.00238.x [DOI] [PubMed] [Google Scholar]
- Ramalho-Pinto F. J., Carvalho E. M., Horta M. F. (1992). Mechanisms of evasion of Schistosoma mansoni schistosomula to the lethal activity of complement. Mem. Inst. Oswaldo Cruz. 87(Suppl. 4), 111–116. 10.1590/S0074-02761992000800016 [DOI] [PubMed] [Google Scholar]
- Ramirez-Toloza G., Ferreira A. (2017). Trypanosoma cruzi evades the complement system as an efficient strategy to survive in the mammalian host: the specific roles of host/parasite molecules and Trypanosoma cruzi calreticulin. Front. Microbiol. 8:1667. 10.3389/fmicb.2017.01667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K. R., Kemp W. M. (1987). Schistosoma mansoni: interactions of adult parasites with the complement system. Parasite Immunol. 9 235–248. 10.1111/j.1365-3024.1987.tb00503.x [DOI] [PubMed] [Google Scholar]
- Rawal N., Pangburn M. K. (2001). Structure/function of C5 convertases of complement. Int. Immunopharmacol. 1 415–422. 10.1016/S1567-5769(00)00039-4 [DOI] [PubMed] [Google Scholar]
- Reynolds S. L., Pike R. N., Mika A., Blom A. M., Hofmann A., Wijeyewickrema L. C., et al. (2014). Scabies mite inactive serine proteases are potent inhibitors of the human complement lectin pathway. PLoS Negl. Trop. Dis. 8:e2872. 10.1371/journal.pntd.0002872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C. H., Lynch N. J., Stover C. M., Ali Y. M., Valck C., Noya-Leal F., et al. (2015). Deficiency in mannose-binding lectin-associated serine protease-2 does not increase susceptibility to Trypanosoma cruzi infection. Am. J. Trop. Med. Hyg. 92 320–324. 10.4269/ajtmh.14-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich S. M., Leendertz F. H., Xu G., LeBreton M., Djoko C. F., Aminake M. N., et al. (2009). The origin of malignant malaria. Proc. Natl. Acad. Sci. U.S.A. 106 14902–14907. 10.1073/pnas.0907740106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010). Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11 785–797. 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D., Reis E. S., Lambris J. D. (2016). Complement in disease: a defence system turning offensive. Nat. Rev. Nephrol. 12 383–401. 10.1038/nrneph.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S., Murugavel S., Devi I. K., Vedamurthy G. V., Gupta S. C., Singh B. P., et al. (2013). Glyceraldehyde-3-phosphate dehydrogenase of the parasitic nematode Haemonchus contortus binds to complement C3 and inhibits its activity. Parasite Immunol. 35 457–467. 10.1111/pim.12058 [DOI] [PubMed] [Google Scholar]
- Schmidt C. Q., Lambris J. D., Ricklin D. (2016). Protection of host cells by complement regulators. Immunol. Rev. 274 152–171. 10.1111/imr.12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte L. L. S., Pascoal-Xavier M. A., Nahum L. A. (2018). Helminths and cancers from the evolutionary perspective. Front. Med. 5:90. 10.3389/fmed.2018.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraufstatter I. U., Trieu K., Sikora L., Sriramarao P., DiScipio R. (2002). Complement c3a and c5a induce different signal transduction cascades in endothelial cells. J. Immunol. 169 2102–2110. 10.4049/jimmunol.169.4.2102 [DOI] [PubMed] [Google Scholar]
- Shi Y., Toet H., Rathinasamy V., Young N. D., Gasser R. B., Beddoe T., et al. (2014). First insight into CD59-like molecules of adult Fasciola hepatica. Exp. Parasitol. 144 57–64. 10.1016/j.exppara.2014.06.012 [DOI] [PubMed] [Google Scholar]
- Silva E. E., Clarke M. W., Podesta R. B. (1993). Characterization of a C3 receptor on the envelope of Schistosoma mansoni. J. Immunol. 151 7057–7066. [PubMed] [Google Scholar]
- Silva-Barrios S., Stager S. (2017). Protozoan parasites and type I IFNs. Front. Immunol. 8:14. 10.3389/fimmu.2017.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M., Santiago-Schwarz F., Al-Abed Y., Diamond B. (2012). C1q limits dendritic cell differentiation and activation by engaging LAIR-1. Proc. Natl. Acad. Sci. U.S.A. 109 E3160–E3167. 10.1073/pnas.1212753109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. Y., Zhang Y., Yang D., Ren H. N., Sun G. G., Jiang P., et al. (2018). The immune protection induced by a serine protease inhibitor from the foodborne parasite Trichinella spiralis. Front. Microbiol. 9:1544. 10.3389/fmicb.2018.01544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosoniuk E., Vallejos G., Kenawy H., Gaboriaud C., Thielens N., Fujita T., et al. (2014). Trypanosoma cruzi calreticulin inhibits the complement lectin pathway activation by direct interaction with L-Ficolin. Mol. Immunol. 60 80–85. 10.1016/j.molimm.2014.03.014 [DOI] [PubMed] [Google Scholar]
- Sosoniuk-Roche E., Vallejos G., Aguilar-Guzman L., Pizarro-Bauerle J., Weinberger K., Rosas C., et al. (2017). Exogenous Calreticulin, incorporated onto non-infective Trypanosoma cruzi epimastigotes, promotes their internalization into mammal host cells. Immunobiology 222 529–535. 10.1016/j.imbio.2016.10.020 [DOI] [PubMed] [Google Scholar]
- Strainic M. G., Liu J., Huang D., An F., Lalli P. N., Muqim N., et al. (2008). Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity 28 425–435. 10.1016/j.immuni.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Zhao X., Wang Z., Yang J., Zhao L., Zhan B., et al. (2015). Trichinella spiralis paramyosin binds human complement C1q and inhibits classical complement activation. PLoS Negl. Trop. Dis. 9:e0004310. 10.1371/journal.pntd.0004310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambourgi D. V., Kipnis T. L., da Silva W. D., Joiner K. A., Sher A., Heath S., et al. (1993). A partial cDNA clone of trypomastigote decay-accelerating factor (T-DAF), a developmentally regulated complement inhibitor of Trypanosoma cruzi, has genetic and functional similarities to the human complement inhibitor DAF. Infect. Immun. 61 3656–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen J. M., Isenman D. E. (2011). A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science 332 608–611. 10.1126/science.1201954 [DOI] [PubMed] [Google Scholar]
- Vedamurthy G. V., Sahoo S., Devi I. K., Murugavel S., Joshi P. (2015). The N-terminal segment of glyceraldehyde-3-phosphate dehydrogenase of Haemonchus contortus interacts with complements C1q and C3. Parasite Immunol. 37 568–578. 10.1111/pim.12273 [DOI] [PubMed] [Google Scholar]
- Venneker G. T., Asghar S. S. (1992). CD59: a molecule involved in antigen presentation as well as downregulation of membrane attack complex. Exp. Clin. Immunogenet. 9 33–47. [PubMed] [Google Scholar]
- Verma S., Mandal A., Ansari M. Y., Kumar A., Abhishek K., Ghosh A. K., et al. (2018). Leishmania donovani inhibitor of serine peptidases 2 mediated inhibition of lectin pathway and upregulation of C5aR signaling promote parasite survival inside host. Front. Immunol. 9:63. 10.3389/fimmu.2018.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang C., Zhao M. H., Chen M. (2015). Neutrophil extracellular traps can activate alternative complement pathways. Clin. Exp. Immunol. 181 518–527. 10.1111/cei.12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun X., Huang J., Zhan B., Zhu X. (2017). Heterologous prime-boost vaccination enhances TsPmy’s protective immunity against Trichinella spiralis infection in a murine model. Front. Microbiol. 8:1394. 10.3389/fmicb.2017.01394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang X., Bi K., Sun X., Yang J., Gu Y., et al. (2016). Oral vaccination with attenuated Salmonella typhimurium-delivered TsPmy DNA vaccine elicits protective immunity against Trichinella spiralis in BALB/c Mice. PLoS Negl. Trop. Dis. 10:e0004952. 10.1371/journal.pntd.0004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Gu Y., Yang J., Yang Y., Wang S., Cui S., et al. (2011). Identification and characterization of protective epitope of Trichinella spiralis paramyosin. Vaccine 29 3162–3168. 10.1016/j.vaccine.2011.02.072 [DOI] [PubMed] [Google Scholar]
- Wiesner J., Jomaa H., Wilhelm M., Tony H. P., Kremsner P. G., Horrocks P., et al. (1997). Host cell factor CD59 restricts complement lysis of Plasmodium falciparum-infected erythrocytes. Eur. J. Immunol. 27 2708–2713. 10.1002/eji.1830271034 [DOI] [PubMed] [Google Scholar]
- Wilson R. A., Coulson P. S. (2009). Immune effector mechanisms against schistosomiasis: looking for a chink in the parasite’s armour. Trends Parasitol. 25 423–431. 10.1016/j.pt.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Gupta S., Saxena J. K. (2017). Monitoring thermal and chemical unfolding of Brugia malayi calreticulin using fluorescence and Circular Dichroism spectroscopy. Int. J. Biol. Macromol. 102 986–995. 10.1016/j.ijbiomac.2017.04.053 [DOI] [PubMed] [Google Scholar]
- Yadav S., Gupta S., Selvaraj C., Doharey P. K., Verma A., Singh S. K., et al. (2014). In silico and in vitro studies on the protein-protein interactions between Brugia malayi immunomodulatory protein calreticulin and human C1q. PLoS One 9:e106413. 10.1371/journal.pone.0106413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Gu Y., Yang Y., Wei J., Wang S., Cui S., et al. (2010). Trichinella spiralis: immune response and protective immunity elicited by recombinant paramyosin formulated with different adjuvants. Exp. Parasitol. 124 403–408. 10.1016/j.exppara.2009.12.010 [DOI] [PubMed] [Google Scholar]
- Yao C. (2010). Major surface protease of trypanosomatids: one size fits all? Infect. Immun. 78 22–31. 10.1128/iai.00776-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Donelson J. E., Wilson M. E. (2003). The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132 1–16. 10.1016/S0166-6851(03)00211-1 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Kato H., Ikeda Y., Nangaku M. (2018). Pathogenesis of atypical hemolytic uremic syndrome. J. Atheroscler. Thromb. 26 99–110. 10.5551/jat.RV17026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. R., Jr., Ambrus J. L., Jr., Malbran A., Fauci A. S., Tenner A. J. (1991). Complement subcomponent C1q stimulates Ig production by human B lymphocytes. J. Immunol. 146 3356–3364. [PubMed] [Google Scholar]
- Zhang Z., Yang J., Wei J., Yang Y., Chen X., Zhao X., et al. (2011). Trichinella spiralis paramyosin binds to C8 and C9 and protects the tissue-dwelling nematode from being attacked by host complement. PLoS Negl. Trop. Dis. 5:e1225. 10.1371/journal.pntd.0001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Shao S., Chen Y., Sun X., Sun R., Huang J., et al. (2017). Trichinella spiralis calreticulin binds human complement C1q as an immune evasion strategy. Front. Immunol. 8:636. 10.3389/fimmu.2017.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Hao Y., Yang J., Gu Y., Zhu X. (2014). Mapping of the complement C9 binding domain on Trichinella spiralis paramyosin. Parasit. Vectors 7:80. 10.1186/1756-3305-7-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel P. F., Skerka C. (2009). Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 9 729–740. 10.1038/nri2620 [DOI] [PubMed] [Google Scholar]