Of five functional ethylene biosynthesis genes in the rice genome, OsACS1 plays a more important role in regulating Pi deficiency-responsive root architecture modulation, Pi uptake, and Pi starvation responses
Keywords: Adaptive response, CRISPR/Cas, ethylene biosynthesis, OsACS, phosphate deficiency, rice
Abstract
Phosphate (Pi) deficiency severely influences the growth and reproduction of plants. To cope with Pi deficiency, plants initiate morphological and biochemical adaptive responses upon sensing low Pi in the soil, and the plant hormone ethylene plays a crucial role during this process. However, how regulation of ethylene biosynthesis influences the Pi-induced adaptive responses remains unclear. Here, we determine the roles of rice 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS), the rate-limiting enzymes in ethylene biosynthesis, in response to Pi deficiency. Through analysis of tissue-specific expression of OsACS in response to Pi deficiency and OsACS mutants generated by CRISPR/Cas9 [clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9] genome editing, we found that two members of the OsACS family, i.e. OsACS1 and OsACS2, are involved but differed in their importance in controlling the remodeling of root system architecture, transcriptional regulation of Pi starvation-induced genes, and cellular phosphorus homeostasis. Interestingly, in contrast to the known inhibitory role of ethylene on root elongation, both OsACS mutants, especially OsACS1, almost fail to promote lateral root growth in response to Pi deficiency, demonstrating a stimulatory role for ethylene in lateral root development under Pi-deficient conditions. Together, this study provides new insights into the roles of ethylene in Pi deficiency response in rice seedlings and the isoform-specific function of OsACS genes in this process.
Introduction
Rice is an important staple crop that feeds more than half of the world’s population. However, the yield and sustainability of rice suffer significantly from low phosphorus stress due to a high phosphorus fixation rate and naturally low phosphorus content in its natural habitats (Raghothama, 1999; Gamuyao et al., 2012). Phosphorus is an essential macro-element for a plant throughout the whole life cycle (Lopez-Arredondo et al., 2014). It serves as an important structural element of nucleic acids, energy currency ATP, and phospholipids (Raghothama, 1999; Lin et al., 2009; Zhang et al., 2014). Whereas a wide range of phosphorus forms exist in the soil, only inorganic phosphate anions (Pi) are exclusively assimilated by plants (Holford, 1997). However, the low mobility and solubility of Pi become a serious constraint, which affects the overall growth and reproduction of the plants. To cope with the low availability of Pi in the soil, higher plants, including rice, undergo significant morphological and biochemical changes through local and systemic low Pi-sensing mechanisms (Neumann, 2015). The most common morphological changes in response to Pi deficiency include the alteration of root system architecture, such as inhibited primary root growth, increase in lateral and secondary root growth (Williamson et al., 2001; Perez-Torres et al., 2008), and the enhancement of fine root structures (e.g. root hairs) through a local adaptive response (Forde and Lorenzo, 2001; Lynch and Brown, 2001; Williamson et al., 2001; Linkohr et al., 2002; Lopez-Bucio et al., 2002; Nacry et al., 2005). Pi-deficient plants also induce the systemic transcriptional response of Pi starvation-induced genes that are involved in ethylene biosynthesis, Pi transportation, and Pi homeostasis in the shoots (Song and Liu, 2015). Accumulating studies indicate that plant hormones play a pivotal role in these adaptive responses by integrating low Pi signals into plants (Rouached et al., 2010; Nagarajan and Smith, 2012; Kazan, 2013). Among plant hormones, the gaseous plant hormone ethylene has been considered as an important modulator for both local and systemic Pi starvation-induced responses (Neumann, 2015; Song and Liu, 2015), yet the underlying molecular mechanism remains unclear.
Ethylene is derived from the amino acid methionine via two intermediates, S-adenosyl-l-methionine (SAM) and 1-aminocyclopropane-1-carboxylic acid (ACC) (Yang and Hoffman, 1984). SAM synthetase converts methionine to SAM, which is subsequently converted to ACC by ACC synthase (ACS). ACC is then oxidized to ethylene by ACC oxidase (ACO). The conversion of SAM to ACC by ACS is generally considered as the rate-limiting step of the pathway. Numerous studies have demonstrated that ACS is the primary target for controlling ethylene production in response to developmental changes or various biotic and abiotic stresses (Argueso et al., 2007). In higher plants, ACS is encoded by a multigene family, and Arabidopsis contains 12 ACS genes in the genome, only eight of which encode functional ACS proteins (Yamagami et al., 2003). The eight functional ACS proteins are further classified into type-1, type-2, and type-3 ACS based on the phosphorylation sites in the C-terminal extension (Chae and Kieber, 2005). The type-1 ACS protein has a single calcium-dependent protein kinase (CDPK) phosphorylation site and three mitogen-activated protein kinase (MAPK) phosphorylation sites in the C-terminal region. The type-2 ACS protein has a single CDPK phosphorylation site and a unique regulatory motif called a Target of ETO1 (TOE) in the C-terminus. TOE is the binding site for ETHYLENE OVERPRODUCER1 (ETO1) and its two paralogs ETO1-Like1 (EOL1) and EOL2 E3 ligases (Wang et al., 2004). ETO1/EOL1/EOL2 are BTB/TRP-containing E3 ligases that control the degradation of type-2 ACS proteins via the 26S proteasome (Wang et al., 2004; Yoshida et al., 2006). In contrast to both type-1 and type-2 ACS, type-3 ACS protein does not contain known regulatory sites or motifs, including phosphorylation sites (Chae and Kieber, 2005). The completion of the rice genome sequence has revealed that rice (Oryza sativa L.) also has a multigene family of ACS genes containing at least five putative members (OsACS1–OsACS5) (Zarembinski and Theologis, 1994). The sequence homology analysis with Arabidopsis ACS proteins showed that rice ACS proteins can also be classified into three types of ACS (OsACS2 for type-1; OsACS1 for type-2; and OsACS3 to 5 for type-3) in a similar manner to those in Arabidopsis (Yoshida et al., 2006; Lee and Yoon, 2018).
A number of studies have demonstrated that Pi deficiency induces the modulation of ethylene biosynthesis or alters the sensitivity of plants to ethylene in many plant species, including rice (He et al., 1992; Song et al., 2016; Zhu et al., 2016; Shukla et al., 2017). Upon sensing low Pi in the soil, plant species undergo alterations for ethylene biosynthesis, though there is discrepancy as to whether the Pi deficiency stimulates or inhibits the biosynthesis of ethylene (Drew et al., 1989; Borch et al., 1999; Kim et al., 2008; Roldan et al., 2016). Nevertheless, the level of ethylene in plants is mostly regulated via altering the expression of ACS or ACO (Tsuchisaka and Theologis, 2004; Graham et al., 2006; Hernandez et al., 2007; Thibaud et al., 2010; Roldan et al., 2013). Multiple studies have demonstrated that both local and systemic sensing of low Pi stress result in modulation of the transcript levels of ACS and ACO, thus affecting ethylene biosynthesis. For example, a transcript analysis of Pi-starved Arabidopsis roots has demonstrated that low Pi stress increases the transcripts of ethylene biosynthetic genes (Misson et al., 2005; Thibaud et al., 2010; Chacon-Lopez et al., 2011). Furthermore, a split-root transcriptome analysis of Arabidopsis under low Pi stress has shown that the expression of a subset of ACO genes increased via the local sensing of Pi stress (Thibaud et al., 2010). Similarly, the systemic response to low Pi stress increases the abundance of ACS transcripts in Arabidopsis leaves, resulting in enhanced ethylene biosynthesis in this tissue (Misson et al., 2005). A study has also shown that the increased transcript levels of ACS genes were readily reduced to the levels before Pi starvation upon provision of Pi (Morcuende et al., 2007), suggesting the tight correlation between ethylene biosynthesis and Pi availability. While the Pi-induced transcriptional regulation of ACS genes is recognized, the role of different ACS isoforms in Pi deficiency remains to be determined.
In addition to the regulation of ethylene biosynthesis at the transcriptional level, the post-translational modification of ACS is an important alternative to modulate the levels of ethylene rapidly in response to various stresses (Chae and Kieber, 2005). A recent work from the analysis of the hypersensitive to Pi starvation3 (hsp3) mutant (Wang et al., 2012) has suggested that regulation of the protein stability of ACS plays a role in the adaptation of a plant in Pi stress. The molecular cloning of hsp3 has revealed that the Pi-hypersensitive phenotype of hsp3 results from a mutation in ETO1 E3 ligases. As ETO1 E3 ligase specifically targets type-2 ACS proteins for their degradation (Wang et al., 2004), this study implies that the post-translational control of type-2 ACS plays a role in the Pi-mediated stress response. Studies from the various combination of Arabidopsis acs knockout mutants and the biochemical analysis of ACS proteins have shown that different ACS isoforms share an overlapping function, yet each ACS isoform possesses a unique role in a specific developmental or stress response (Yamagami et al., 2003; Tsuchisaka et al., 2009). For example, type-1 ACS isoforms have a role in responding to pathogen invasion through the stress-activated MAPK cascade (Liu and Zhang, 2004). Type-2 ACS has a primary role in controlling ethylene biosynthesis in the dark through the crosstalk with other plant hormones (Lee et al., 2017). In contrast, type-3 ACS appears not to be involved in stress responses by itself, but rather it works co-operatively with other types of ACS through formation of a heterodimer (Lee et al., 2017). Similar to Arabidopsis, rice possesses three different types of ACS isoforms (Yoshida et al., 2006; Lee and Yoon, 2018). Interestingly, the transcript analysis of OsACS has demonstrated that only a subset of OsACS genes is differentially regulated under Pi-deficient conditions (Zhu et al., 2016), indicating an isoform-specific role for OsACS genes in low Pi stress conditions. The existence of the ETO1-Like gene, OsETOL1, in the rice genomealso supports the possible type-specific role of OsACS in stress responses (Du et al., 2014). Thus, it is of great interest to investigate the role of each isoform of OsACS in regulating Pi stress-induced adaptive responses.
In this study, we specifically investigate the role of OsACS1 and OsACS2 in Pi deficiency-induced adaptive responses in rice seedlings. Our results demonstrate that OsACS1 and OsACS2 are involved in several Pi deficiency-induced adaptive responses with a significant role in altering root system architecture. The results also show that OsACS1 and OsACS2 have an overlapping function in Pi starvation-induced responses, yet OsACS1 plays a larger role in controlling Pi deficiency-induced adaptive responses, indicating the isoform-specific role of OsACS in this process.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa L. cv. Nipponbare) seeds were dehusked and sterilized with 30% sodium hypochlorite for 10 min, followed by rinsing 5–6 times with deionized water. The sterilized seeds were subsequently placed onto solid Murashige and Skoog (MS) medium in a sterilized magenta box. The seedlings were grown in a growth chamber with a 16 h/8 h light/dark cycle at 28 °C. After 2 weeks, the seedlings were transferred to soil and grown in a greenhouse with a similar light/dark cycle and temperature conditions.
Construction of recombinant DNA plasmid for clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated OsACS mutagenesis
A 20 nt guide RNA (gRNA) sequence for OsACS1 and OsACS2 was designed using CRISPR-PLANT (https://www.genome.arizona.edu/crispr/). Using an overlapping PCR, a fragment containing a rice U3 promoter and the gRNA for OsACS1 or OsACS2 was generated and subcloned into a plant binary vector, pARS3-MUb-Cas9, at the PmeI site using an infusion method. The final constructs were sequenced using the Sanger sequencing method.
Rice transformation and selection
The rice transformation was performed by the Agrobacterium tumefaciens-mediated co-cultivation method, as described previously (Hiei et al., 1994). T0 generation lines of Cr-OsACS1 and Cr-OsACS2 were selected on media with hygromycin, and the hygromycin-resistant T0 transgenic plants were analyzed to examine CRISPR/Cas9-induced mutations in the OsACS loci and the insertion of T-DNA containing the gRNA and the Cas9 transgene using T7 endonuclease I (T7E1) analysis, PCR, and Sanger sequencing. The selected T0 lines with the Cas9 transgene and mutations at the OsACS loci were self-pollinated to collect T1 seeds in a greenhouse with a 16 h/8 h light/dark cycle at 20 °C. The T1 plants were further analyzed to determine the zygosity of the mutation and the insertion of the Cas9 transgene using the same method as described above.
The identification of OsACS mutants by T7E1 and sequencing
T7E1 assay was conducted as previously described. PCR was performed by using gene-specific primers for each OsACS gene (Shan et al., 2013; Wang et al., 2014). To make heteroduplexes, the PCR product was reacted as follows: 95 °C for 10 min, 95–85 °C (ramp rate of –2 °C s–1), and 85–25 °C (ramp rate of –0.3 °C s–1). One unit of T7E1 was added to the PCR heteroduplexes and the reaction was incubated at 37 °C for 1 h. The T7 endonuclease-treated PCRs were separated by 2% agarose gel and visualized by ethidium bromide staining. Sequencing analyses were performed at the Purdue Genomics Core Facility by using ABI3730xl (Applied Biosystems™). Primers used are listed in Supplementary Table S1 at JXB online).
Measurements of ethylene production
Ethylene measurements were performed as previously described (Hansen et al., 2009) with a small modification. Surface-sterilized seeds were germinated in 22 ml GC vials containing 3 ml of MS with 1% sucrose and 1% bactoagar. After 2 d of dark treatment, the vials were capped and incubated at 28 °C for 3 d in the dark for ethylene measurement. For ethylene measurement of light-grown rice seedlings grown on either Pi-sufficient or -deficient media, capped vials were placed in a growth chamber with a 16 h/8 h light/dark cycle at 28 °C for 7 d. The accumulated ethylene was measured by GC using a Shimadzu GC2010 Plus capillary GC system with an HS-20 headspace autosampler. All treatments were measured from three vials each.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using an RNeasy Plus kit (Qiagen). cDNA was prepared from the total RNA using SuperScript II reverse transcriptase (Invitrogen) as described by the manufacturer. Quantitative real-time PCR (qRT-PCR) was performed using PowerUP™ SYBR® green Master Mix (Applied Biosystems) and the CFX Connect™ Real-Time System (Bio-Rad). The relative expression of OsACS genes was normalized to the reference gene OsActin1 and was calculated using the 2ΔΔCT method. Primers used are listed in Supplementary Table S1.
Hydroponic experiment
The modified hydroponic system was assembled as previously described (Negi et al., 2016). Pi-sufficient (0.3 mM NaH2PO4) and Pi-deficient (0.015 mM NaH2PO4) media were prepared as previously described (Jain et al., 2009) and buffered to pH 5.7 using 0.5 mM MES. Germinated seeds were placed onto sieves with Pi-sufficient or Pi-deficient media, and subsequently grown in a growth chamber with a 16 h/8 h light/dark cycle at 28 °C for 7 d.
Soluble Pi content
Measurement of soluble Pi content was performed as previously described (Negi et al., 2016). Rice plants were rinsed 5–6 times with deionized water and placed on 3MM paper for drying. Dried plants were ground in liquid nitrogen and further homogenized with 250 μl of 1% acetic acid, and vortexed for 1 min. After centrifugation at 10 000 rpm for 5 min, the supernatant was collected for assaying Pi content by using phosphomolybdate colorimetric assay as described (Ames, 1966).
Results
ACC promotes the alteration of rice root architecture in response to phosphorus
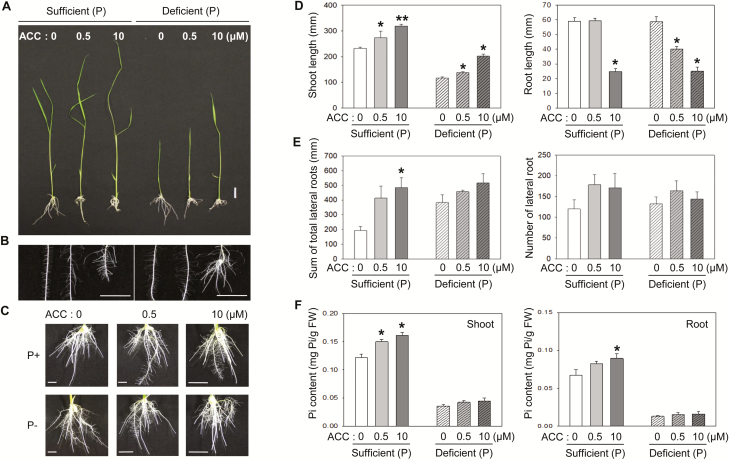
Pi deficiency changes ethylene biosynthesis, resulting in the modulation of rice root architecture for better uptake of Pi in the soil (Peret et al., 2014). To determine a dose effect of ethylene in relation to its function in controlling root architecture, wild-type (WT) rice seedlings were grown in hydroponic solutions supplemented with different concentrations of ACC, the direct precursor of ethylene (Adams and Yang, 1979), in either Pi-sufficient or -deficient conditions for 7 d. Longer lateral roots were observed in seedlings grown under Pi-deficient conditions than in those under Pi-sufficient conditions (Fig. 1B, C, E). Addition of ACC further accelerated the elongation of lateral roots regardless of the presence or absence of Pi, although the effect was dramatically enhanced in the Pi-sufficient plants (Fig. 1B, C); while the total lateral root lengths of both Pi-sufficient and -deficient seedlings were similar at the same concentration of ACC used in the growth medium, greater changes were found in the Pi-sufficient roots than in the Pi-deficient roots. Unlike the root length, ACC did not affect the total lateral root number from both conditions. Intriguingly, the primary root lengths of rice seedlings in either Pi-sufficient or -deficient conditions without ACC were comparable. However, addition of ACC led to an inhibited growth of primary root, and the primary root length was reduced in inverse relation to the increasing concentrations of ACC introduced in both the Pi-sufficient and -deficient media (Fig. 1A, C, D). Similar to the roots, we did not observe a prominent effect of ACC on the shoot growth of seedlings grown in Pi-deficient growth media compared with that of seedlings grown under Pi-sufficient conditions; Pi deficiency resulted in an inhibited shoot growth in seedlings grown in Pi-deficient media, yet seedlings in both growth conditions showed similar levels of enhancement in the shoot growth proportional to the increasing concentrations of ACC used in the growth media (Fig. 1A, D). Next, we examined the cellular Pi contents because ACC altered the root morphology of WT seedlings in both growth conditions (Fig. 1F). The Pi contents in both roots and shoots under the Pi-sufficient condition increased proportionally to the concentrations of ACC, indicating the positive role of ACC in Pi uptake. However, no obvious change in the Pi contents was found in the seedlings under Pi-deficient conditions. Taken together, these results indicate that ethylene plays a role in the alteration of shoot and root architecture, particularly lateral root length, and there is a positive correlation between the quantity of ethylene and the effect it exerts on the alteration of root morphology. The lack of response of the primary root to Pi deficiency may reflect the characteristic of a short-term induction period and/or the low ethylene content in the rice seedlings as the reduction in the primary root length of seedlings was correlated to the increasing dose of ACC used. A different response of primary root growth to Pi deficiency among different rice varieties could also be a reason for the lack of the primary root response in our study (Negi et al., 2016).
Fig. 1.
ACC accentuates the remodeling of root architecture in response to phosphorus. (A) Representative images of 7-day-old light-grown seedlings grown in a Pi-sufficient (0.3 mM) or -deficient (0.015 mM) condition in the presence of different concentrations of ACC. Scale bar=20 mm. (B, C) Representative images of the enlarged roots (B) or full root images (C) from seedlings in (A). Scale bars=10 mm. (D, E) Statistical analysis of the length of shoot and root (D) and total lateral root length and lateral root number (E). (F) Cellular Pi content of the shoot and root of rice seedlings grown in Pi+ or Pi– conditions with different ACC concentrations. Error bars indicate the SD; n=5, *P<0.05, **P<0.005, Student’s t-test.
OsACS1 and OsACS2 are predominantly expressed in rice seedlings, and Pi deficiency leads to an increase in the transcript levels of both genes
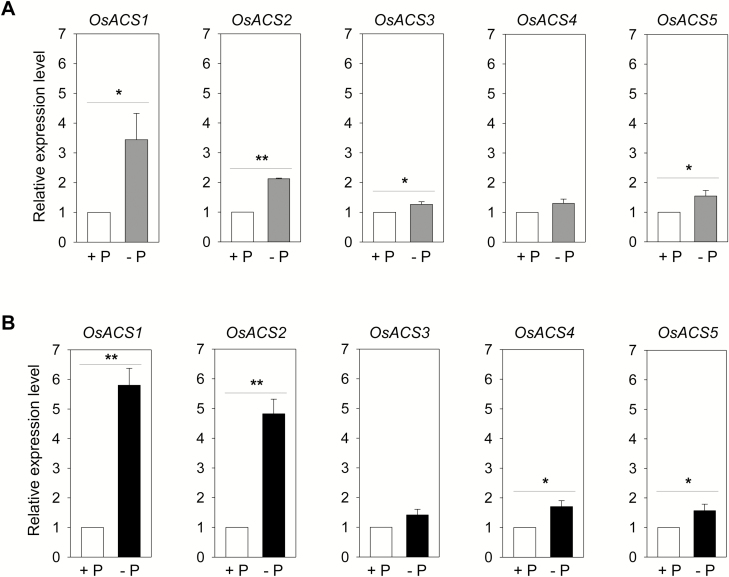
The additive effect of ACC on the morphological changes in rice seedlings in response to phosphorus (Fig. 1) suggests that ACS is likely to play a role in this process because ACS controls the rate of ethylene biosynthesis through controlling the formation of ACC. The transcriptional regulation of ACS is the primary mechanism to control ethylene biosynthesis under many stress conditions in several plant species (Argueso et al., 2007). To investigate whether Pi deficiency affects the expression of OsACS genes, the transcript levels of five OsACS genes from 7-day-old WT rice seedlings grown under Pi-sufficient or -deficient conditions were examined. The majority of OsACS genes, particularly OsACS1 and OsACS2, were predominantly expressed in the shoots, except OsACS3 under Pi-sufficient growth conditions (Supplementary Fig. S1). Similar transcript levels of OsACS1, 2, 3, and 4 were found in the root, but almost no expression of OsACS5 was detected in this tissue (Supplementary Fig. S1). Pi deficiency significantly increased the transcript levels of OsACS1 and 2, but only led to a marginal or no increase in the transcript levels of the rest of the OsACS genes in both root and shoot tissues (Fig. 2). Moreover, the induction of the OsACS1 and 2 transcripts in the root was ~1.7–2.4 times higher than that in the shoot under Pi deficiency. Taken together, these results suggest that the regulation of ethylene biosynthesis via OsACS genes, particularly OsACS1 and 2 in the root, is probably one of the strategies that rice plants use to cope with Pi-deficient stress conditions.
Fig. 2.
Transcript analysis of OsACS genes in light-grown rice seedlings in Pi-sufficient or -deficient conditions. (A, B) Relative gene expression of different OsACS genes in the shoots (A) or roots (B) in 7-day-old WT rice seedlings grown in the light. ‘+’ or ‘–’ indicate Pi-sufficient or Pi-deficient conditions, respectively. Error bars indicate the SD of three biological replicates. *P<0.05, **P<0.005, Student’s t-test.
Generation of OsACS1 and OsACS2 mutants using CRISPR/Cas9-based genome editing
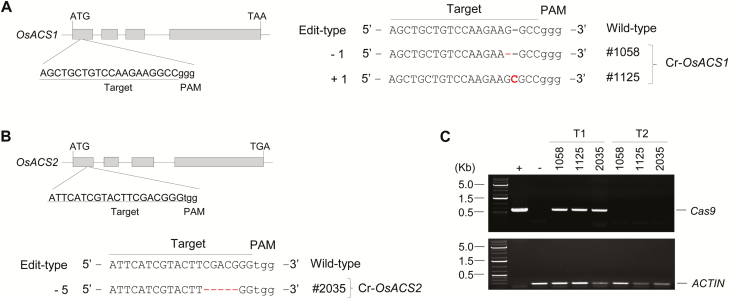
To investigate further the role of ethylene in rice under Pi deficiency, we generated knockout lines of OsACS1 or OsACS2 using the CRISPR/Cas-based genome editing method (Supplementary Fig. S3). Two independent homozygous Cas9 transgene-free Cr-OsACS1 mutants (#1058 and #1125) and one homozygous Cr-OsACS2 mutant (#2035) were selected for further studies (Fig. 3). The two selected independent Cr-OsACS1 mutants have either a 1 bp deletion or a 1 bp insertion at 3 bp upstream of the protospacer adjacent motif (PAM) sequence, which results in a premature stop codon in the first exon of the OsACS1 (Supplementary Fig. S4). The selected Cr-OsACS2 has a 5 bp deletion at 2 bp upstream of the PAM sequence (Supplementary Fig. S4), resulting in a premature stop codon in the first exon of OsACS2. The off-target effects of the editing of OsACS1 and OsACS2 were examined by identifying potential off-target sequences using CRISPR-P v.20 (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR). The top three highest potential off-target candidates of each OsACS gRNA were selected and examined using PCR amplification and Sanger sequencing. No mutations were found in the potential off-target loci in the Cr-OsACS1 and Cr-OsACS2 mutants (Supplementary Fig. S5).
Fig. 3.
CRISPR/Cas9-based mutagenesis on OsACS1 or OsACS2. (A, B) Schematic gene structure of OsACS1 (A) or OsACS2 (B) and detection of targeted mutation(s) (deletion or insertion) in the OsACS1 (A) or OsACS2 (B) locus. The target site nucleotides are shown in upper case letters and the protospacer adjacent motif (PAM) is indicated as lower case letters. ‘–’ and ‘+’ indicate either the deletion or insertion of nucleotide(s), respectively. (C) PCR-based identification of Cas-free OsACS1 (Cr-OsACS1) and OsACS2 (Cr-OsACS2) mutant plants. The gel image shows the presence or absence of Cas in the genomic DNA of T1 and T2 lines of Cr-OsACS1 and Cr-OsACS2. ‘+’ indicates a positive plasmid control used in this study containing cloned Cas9. ‘–’ indicates a negative control genomic DNA isolated from WT rice.
A mutation in OsACS1 or OsACS2 results in reduced ethylene biosynthesis and morphological changes in the etiolated rice seedlings
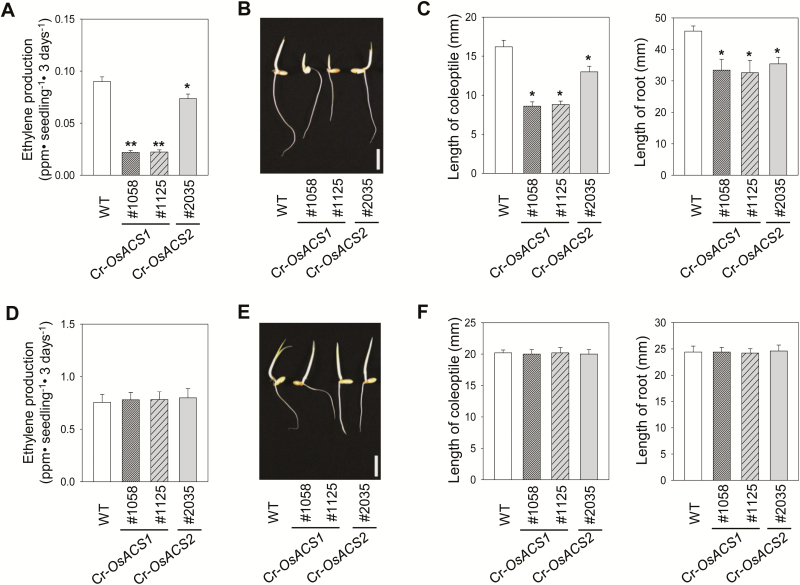
A successful disruption of ACS may cause a reduction of ethylene production in a plant as a result of a decrease in the concentration of ACC. We found that both Cr-OsACS1 and Cr-OsACS2 mutant lines produced less ethylene than WT seedlings in the dark (Fig. 4A). Interestingly, the levels of ethylene produced by two Cr-OsACS1 mutants were significantly lower than the levels of ethylene produced in Cr-OsACS2 (Fig. 4A), indicating a more important role for OsACS1 in ethylene production than for OsACS2 in the dark. The decreased ethylene biosynthesis in both Cr-OsACS1 and Cr-OsACS2 mutants was rescued by exogenously added ACC (Fig. 4D). This further confirms that the reduced levels of ethylene biosynthesis in both Cr-OsACS mutants were due to the inability of the mutants to produce a sufficient amount of ACC owing to the disruption of OsACS1 or OsACS2.
Fig. 4.
Morphological analysis of Cr-OsACS mutants and ethylene biosynthesis in the mutants. (A) Ethylene production of 3-day-old dark-grown WT and Cr-OsACS mutant seedlings. (B) A representative image of dark-grown Cr-OsACS1 (#1058 and #1125) and Cr-OsACS2 (#2035) mutant seedlings. Scale bar=10 mm. (C) Measurement of the coleoptiles and roots length of Cr-OsACS1 and Cr-OsACS2 mutant seedlings. Error bars indicate the SD; n=5; *P<0.05, Student’s t-test. (D) Ethylene production in dark-grown Cr-OsACS mutant seedlings grown in media supplemented with 10 µM ACC. (E) A representative image of dark-grown Cr-OsACS1 and Cr-OsACS2 mutant seedlings that were grown in a hydroponic solution containing 10 µM ACC. Scale bar indicates 10 mm. (F) Measurement of the coleoptiles and roots length of the mutant seedlings in the presence of 10 µM ACC. Error bars indicate the SD; n=5; *P<0.05, **P<0.005, Student’s t-test.
The reduced ethylene biosynthesis in the Cr-OsACS1 and Cr-OsACS2 mutants appeared to affect the overall morphology of the mutant seedlings in the dark (Fig. 4B, C). Ethylene stimulates a coleoptile growth of rice seedlings in the dark. Therefore, examining the coleoptile growth is a good proxy for inferring defects associated with ethylene function in the mutants (Lee and Yoon, 2018). Both Cr-OsACS1 and Cr-OsACS2 mutants produced shorter coleoptiles than the WT in hydroponic solutions, although the phenotype is more rigorous in Cr-OsACS1 plants. Additionally, Cr-OsACS1 and Cr-OsACS2 produced shorter roots compared with the WT, indicating the role of OsACS1 and 2 in controlling root elongation. Similar to the ACC-mediated rescue of ethylene production in the Cr-OsACS mutants (Fig. 4D), ACC treatment rescued the growth inhibition on the coleoptiles and roots of the mutant seedlings (Fig. 4E, F). These phenotypes and the reduced ethylene production in the dark were successfully transmitted to the T3 generation in both Cr-OsACS mutants with the same mutation in the OsACS loci, corroborating the inheritability of the mutation by the next generation (Supplementary Figs S6, S7).
In addition to regulation of ethylene biosynthesis at the transcriptional level for ACS genes, our studies and those of others have demonstrated that controlling ACS protein stability via the interaction with other plant hormones is another strategy for regulating tissue ethylene content (Chae and Kieber, 2005; Argueso et al., 2007; Yoon, 2015). For example, we have recently shown that cytokinin and brassinosteroid (BR) increase ethylene biosynthesis in etiolated rice seedlings without affecting transcript levels of all five OsACS genes (Lee and Yoon, 2018). As another proxy to confirm the disruption of OsACS function, ethylene biosynthesis in etiolated Cr-OsACS mutant seedlings in response to cytokinin and BR was measured (Supplementary Fig. S8). We found that unlike WT rice seedlings, Cr-OsACS1 mutants, in which OsACS2 is still active, almost completely failed to increase ethylene production in response to the treatment of cytokinin or BR. Different from the Cr-OsACS1 mutant, cytokinin and BR slightly increased ethylene biosynthesis in the Cr-OsACS2 mutant in which the OsACS1 is still active, though the degree of increment was significantly smaller than that in WT seedlings. The lower reduction of ethylene biosynthesis in Cr-OsACS2 in which OsACS1 protein (type-2 ACS) is presumably still functional aligns well with our previous studies demonstrating that type-2 ACS plays a predominant role in responding to cytokinin and BR for increasing ethylene biosynthesis (Lee et al., 2017). These results associated with protein stability further support that CRISPR/Cas9-mediated mutagenesis successfully disrupted OsACS1 and OsACS2 genes, which inhibits cytokinin- or BR-induced ethylene biosynthesis due to the absence of functional OsACS1 or OsACS2 protein. We also confirmed that the T3 generation of both Cr-OsACS mutants showed reduced induction of ethylene biosynthesis in response to cytokinin (Supplementary Fig. S9). Together, these results demonstrate that the Cr-OsACS mutants generated by the CRISPR/Cas9-mediated targeted mutagenesis have a compromised ACS function and reduced ethylene content in rice, which led to morphological changes in the mutant seedlings.
Lateral root elongation is decreased in Cr-OsACS1 and Cr-OsACS2 mutants under Pi-deficient conditions
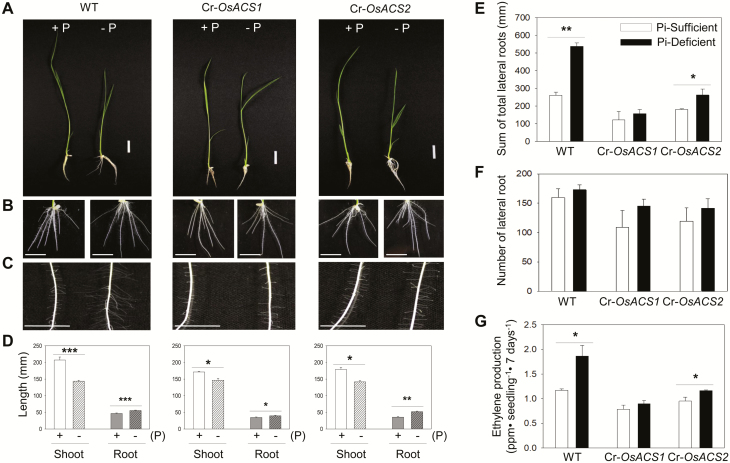
Pi deficiency affects the overall architecture of a plant as part of the adaptive responses to cope with Pi limitation. Upon growth in Pi-deficient conditions, WT seedlings exhibited typical Pi deficiency-induced phenotypes which include inhibited shoot growth and an increase in secondary root and lateral root growth (Fig. 5A, E). To examine the roles of OsACS1 and OsACS2 in Pi deficiency-mediated morphological changes in rice, both Cr-OsACS mutant seedlings were grown under Pi-sufficient or -deficient conditions and their morphologies were examined. Since the two independent Cr-OsACS1 mutant lines (#1058 and #1125) showed almost identical phenotypes and a similar reduction in ethylene production (Fig. 4), further studies were performed using only the #1058 mutant line for Cr-OsACS1. Under Pi-sufficient conditions, both Cr-OsACS mutants produced significantly shorter shoots (P<0.05) than WT plants (Fig. 5A, D; Supplementary Fig. S10). In addition, the growth of the primary root in both Cr-OsACS mutants was also inhibited (P<0.005) (Fig. 5A, D; Supplementary Fig. S10). Although both Cr-OsACS mutants displayed reduced shoot and root lengths compared with the WT in Pi-sufficient conditions, they still responded to Pi deficiency like the WT (Fig. 5D), resulting in shorter shoots and roots. It is worth noting that the degree of reduction in the shoot length was smaller in the Cr-OSACS mutants than in the WT. Under Pi-deficient conditions, both Cr-OsACS mutants displayed a significantly inhibited lateral root elongation in response to Pi deficiency (Fig. 5C, E). The Cr-OsACS2 mutant showed a reduced sensitivity to Pi deficiency for lateral root elongation, but a slight increment of Pi-induced lateral root elongation was still observed (P<0.05) (Fig. 5E). Unlike the Cr-OsACS2 mutant, the Cr-OsACS1 mutant seedlings were almost unresponsive to Pi deficiency with regard to lateral root elongation (Fig. 5C, E). Although the induction of the lateral root elongation in Cr-OsACS1 mutant seedlings was not statistically significant, the tendency for a slightly increased lateral root length in response to Pi deficiency indicated that OsACS2 may also play a role in this process. In contrast to lateral root elongation, we did not observe differences in lateral root numbers between WT and both Cs-OsACS seedlings. These results suggest that the Pi deficiency-induced lateral root elongation is mainly a result of the overlapping function between OsACS1 and OsACS2, although OsACS1 plays a more significant role in this process.
Fig. 5.
Altered morphologies and ethylene biosynthesis of light-grown Cr-OsACS mutants in Pi-sufficient or -deficient growth conditions. (A, B) Representative images of whole seedling of WT and Cr-OsACS mutants grown in either Pi-sufficient (+P) or -deficient (–P) conditions (A) and the enlarged root images of seedlings (B) in (A). Scale bars=20 mm. (C) Representative images of the enlarged lateral roots of WT and Cr-OsACS mutant seedlings in (A). Scale bars=10 mm. (D) The length of shoots and roots of WT and Cr-OsACS mutant seedlings grown in P-sufficient or -deficient conditions. (E, F) Measurement of the total lateral root length (E) and lateral root number (F) of WT and Cr-OsACS mutants grown in Pi-sufficient or -deficient conditions. (G) Pi deficiency-induced ethylene biosynthesis in the Cr-OsACS mutants. Seedlings were grown in either Pi-sufficient or -deficient media for 7 d and the accumulated ethylene was measured. Error bars indicate the SD; n=3, *P<0.05, **P<0.01, ***P<0.005, Student’s t-test.
Light-grown Cr-OsACS1 and Cr-OsACS2 mutants have compromised ethylene biosynthesis in response to Pi deficiency
Pi deficiency results in an increase in ethylene production in several plant species, including rice, bean (Phaseolus vulgaris), white lupin (Lupinus albus), and Medicago falcate (Borch et al., 1999; Gilbert et al., 2000; Lin et al., 2009). To examine the role of OsACS1 and OsACS2 in Pi deficiency-induced ethylene biosynthesis, the levels of ethylene in Cr-OsACS1 and Cr-OsACS2 mutants grown in Pi-sufficient or -deficient growth media with a 16 h/8 h light/dark cycle were measured. Consistent with previous studies, WT rice seedlings showed an enhanced ethylene biosynthesis under Pi-deficient conditions (P<0.05). However, the degrees of ethylene biosynthesis induced by Pi deficiency in both Cr-OsACS1 and Cr-OsACS2 mutants were considerably reduced, although the reduction was more dramatic in Cr-OsACS1 (Fig. 5G). We found that the reduced ethylene biosynthesis in Cr-OsACS mutants is somewhat correlated to the reduced lateral root length in the mutant seedlings, indicating the positive relationship between lateral root elongation and ethylene biosynthesis in Pi deficiency. This result indicates that both OsACS1 and OsACS2 contribute to an enhanced ethylene production in rice seedlings in response to Pi deficiency.
The expression of auxin-related genes is reduced in the roots of Cr-OsACS mutants
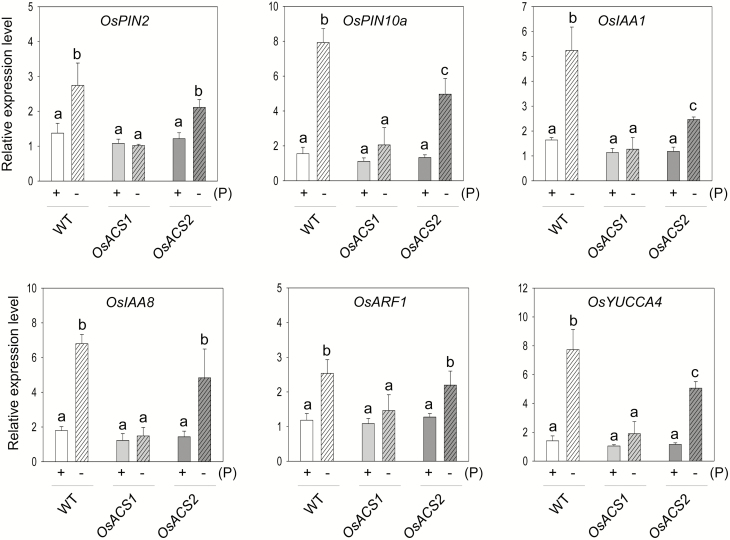
The reduced total length of lateral roots in both Cr-OsACS mutants (Fig. 5) suggests that auxin might be involved in this process under Pi deficiency. To examine this, we determined the expression levels of a subset of auxin-related genes from auxin biosynthesis (OsYUCCA4), transport (OsPIN2 and OsPIN10a), and signaling (OsIAA1, OsIAA8, and OsARF1), all of which have been shown to have increased expression in the roots of WT seedlings under Pi deficiency. As expected, Pi deficiency significantly induced the expression of all six genes in the WT roots (Fig. 6). The Cr-OsACS2 mutant also showed increased expression of these genes to some extent, albeit at a minor level. In contrast, Cr-OsACS1 almost failed to respond to low Pi stress to induce the expression of these genes. Taken together, these results indicate that auxin plays a role in lateral root elongation of rice seedlings via OsACS1 and 2.
Fig. 6.
Pi deficiency-induced expression of auxin-related genes is altered in Cr-OsACS mutants. Seven-day-old light-grown seedlings were grown in either Pi-sufficient (+) or -deficient (–) conditions. OsActin1 was used as an internal control. Error bars indicate the SD from three biological replicates. Different letters indicate a significant difference at P<0.05 (ANOVA, Tukey’s HSD post-hoc test).
Pi homeostasis is perturbed in the Cr-OsACS mutants
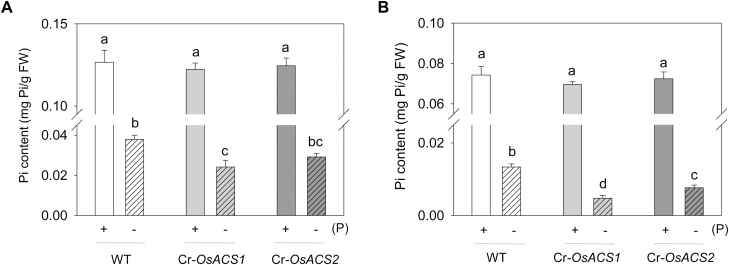
Under Pi-deficient conditions, rice alters root system architecture to ensure the efficient uptake of available soluble Pi from the soil. To examine the relationship between Pi deficiency-induced morphological changes and Pi uptake and distribution in the Cr-OsACS mutants, the soluble cellular Pi content in both Cr-OsACS mutants grown under Pi-sufficient or -deficient conditions was measured. Compared with the Pi-sufficient roots and shoots, the Pi content in the Pi-deficient shoots and roots of WT seedlings were significantly reduced (P<0.01) (Fig. 7). Under the Pi-sufficient conditions, the soluble Pi content in the shoots and roots of Cr-OsACS mutants was comparable with that in the WT. However, the shoot Pi content in the Cr-OsACS1 mutant was significantly lower than that in WT seedlings after Pi deprivation for 7 d (Fig. 7A); a similar trend was observed in Cr-OsACS2 although not statistically significant. Pi deprivation results in a significant reduction of Pi content in the roots of Cr-OsACS1 and Cr-OsACS2 mutants compared with that of WT seedlings, and the effect is more severe in Cr-OsACS1. Overall, these results demonstrate that OsACS genes, particularly OsACS1, are involved in regulating root architecture response and the associated Pi uptake and translocation.
Fig. 7.
Cr-OsACS mutants have lower cellular Pi content than the wild type in Pi-deficient conditions. (A, B) Soluble cellular Pi contents in the shoots (A) or roots (B) of 7-day-old light-grown WT and Cr-OsACS mutants grown in Pi-sufficient (+P) or -deficient (–P) growth media. Error bars indicate the SD; n=3. Different letters indicate a significant difference at P<0.01 (ANOVA, Tukey’s HSD post-hoc test).
Expression of Pi deficiency-induced genes is down-regulated in the Cr-OsACS1 and Cr-OsACS2 mutants
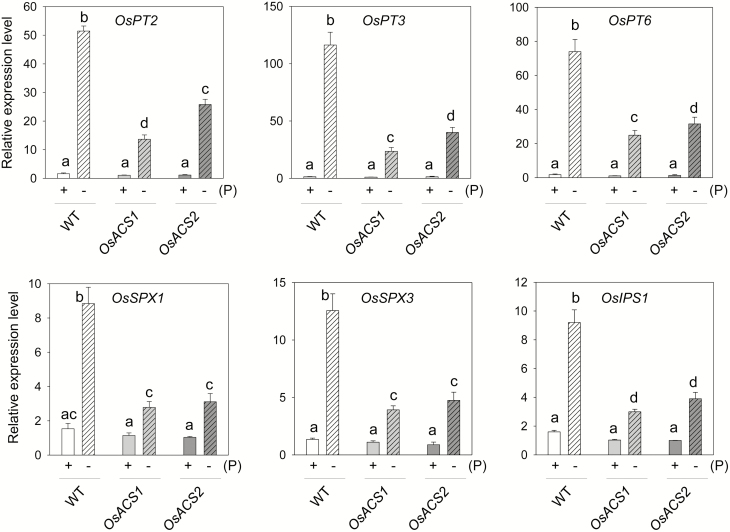
The disturbed cellular Pi content in the Cr-OsACS mutants indicates that the transcriptional regulation of genes involved in the maintenance of Pi homeostasis may also change. To test this hypothesis, we examined the expression levels of a few representative Pi metabolism-related genes, namely those encoding three Pi transporter (OsPT), two SPX domain-containing protein (OsSPX), and one induced by phosphate starvation proteins (OsIPS). To this end, Cr-OsACS mutant seedlings were grown in Pi-sufficient or Pi-deficient conditions for 7 d. qRT-PCR results showed that both Cr-OsACS1 and Cr-OsACS2 expressed transcript levels of the six genes examined similar to those in WT seedlings under Pi-sufficient conditions. Under Pi-deficient conditions, the expression levels of the six genes in the WT seedlings substantially increased, which is consistent with previous studies (Zhou et al., 2008; Ai et al., 2009; Wang et al., 2009; Wu et al., 2013; Negi et al., 2016) (Fig. 8). However, in contrast to the WT seedlings, both Cr-OsACS mutants, particularly Cr-OsACS1, showed a reduced sensitivity to Pi deficiency with respect to an increase in the transcript levels of most genes examined. For example, OsPT2, OsPT3, and OsPT6 were induced by ~50- to 100-fold in the WT by Pi deficiency, but in Cr-OsACS1 and Cr-OsACS2, the induction only ranged ~15- to 30-fold. Similar to the OsPT genes, Pi deficiency highly induced the expression of OsSPX1, OsSPX3, and OsIPS1 genes in the WT, but not in CR-OsACS1 and Cr-OsACS2 mutants. Together, these results suggest that OsACS1 and 2 are required for Pi acquisition to maintain Pi homeostasis via the induction of known Pi starvation-induced genes. Moreover, the greater decline in the Pi deficiency-induced expression of the OsPT genes in Cr-OsACS1 suggests that OsACS1 plays a more important role than OsACS2 in the uptake and translocation of Pi under Pi-deficient conditions.
Fig. 8.
Cr-OsACS mutants exhibited a reduced expression of Pi deficiency-induced genes under low Pi stress. Seven-day-old light-grown seedlings were grown in either Pi-sufficient (+) or -deficient (–) media. OsActin1 was used as an internal control. Error bars indicate the SD from three biological replicates. Different letters indicate a significant difference at P<0.05 (ANOVA, Tukey’s HSD post-hoc test).
Discussion
Lateral root elongation is one of the key changes that are induced upon the sensing of Pi deficiency (Sato and Miura, 2011). This branching of roots enables a plant to take up more Pi by increasing root surface areas and maximizing foraging of topsoil where Pi accumulates. The severe reduction on the lateral root elongation in both Cr-OsACS mutants in response to Pi deficiency indicates the positive effect of ethylene on root elongation. The stimulatory role of ethylene in lateral root elongation is intriguing as multiple studies have shown that ethylene generally inhibits root elongation during root development (Le et al., 2001; De Cnodder et al., 2005; Ruzicka et al., 2007). However, recent works have demonstrated a biphasic ethylene response in which ethylene has either a stimulatory or an inhibitory role in regulating root elongation depending on ethylene concentration and plant species (Pierik et al., 2006; Pierik and Sasidharan, 2007). In sugar beet, a low concentration of ACC and the associated ethylene production promoted root elongation, while root growth was inhibited at high concentrations of ACC (Abts et al., 2014). The biphasic mode of hormone response pertaining to plant growth regulation is not uncommon. BR or abscisic acid is known to either stimulate or inhibit root meristem size or root elongation, respectively, depending on its concentration (Prokic et al., 2006; Gonzalez-Garcia et al., 2011; Li et al., 2017). Compared with the known inhibitory role of ethylene in lateral root initiation, the stimulatory role of ethylene in lateral root elongation has not been well described. One possibility for the stimulatory role of ethylene in lateral root elongation in response to Pi deficiency in rice seedlings is through a crosstalk with other plant hormones such as cytokinin. Cytokinin increases ethylene biosynthesis by stabilizing ACS proteins in Arabidopsis, and our recent studies showed that cytokinin probably plays a similar role in controlling ethylene levels in rice seedlings (Hansen et al., 2009; Lee et al., 2017; Lee and Yoon, 2018). A recent study has demonstrated that cytokinin stimulates lateral root elongation in rice (Rani Debi et al., 2005). Together with the reduced auxin function in the mutants, this may explain the reduced lateral root elongation in both Cr-OsACS mutants in Pi-deficient growth condition as both mutants showed defects in enhancing ethylene levels in response to cytokinin treatment (Supplementary Figs S8, S9). Further studies on the crosstalk between ethylene and other phytohormones, including cytokinin, in the phosphate signaling pathway would shed light on Pi-induced lateral root development in rice.
The reduced expression levels of the selected PSI genes in Cr-OsACS mutants confirmed that OsACS is involved in the process. It also showed an OsACS isoform-specific role in the process because a greater reduction of OsPT genes was found in the Cr-OsACS1 mutant. The reduction in the expression of OsSPX and OsIPS further suggests the potential role of ethylene in Pi sensing that is operated by a Pi concentration-dependent interaction between OsSPX1/2 and Phosphate Starvation Response Regulator 1 (OsPHR1) to control the expression of various PSI genes (Wang et al., 2014). It would be of great interest to examine whether OsPHR1 is a direct target of the ethylene signaling pathway.
An intriguing observation from our studies is that OsACS1 appears to play a larger role than OsACS2, while there are clear overlapping functions between two types of OsACS in controlling several Pi-induced adaptive responses. The disruption of OsACS1 has more prominent effects on Pi deficiency-induced responses, such as inhibited lateral root elongation, reduced expression levels of Pi transport genes, and perturbed cellular Pi homeostasis. This might be related to the tissue- or cell type-specific gene expression characteristic of different isoforms of OsACS genes. We examined the gene expression of all five OsACS genes (OsACS1–OsACS5) in 7-day-old light-grown rice seedlings grown in either Pi-sufficient or -deficient growth conditions (Fig. 2; Supplementary Fig. S1). Among the OsACS genes examined, all the OsACS genes, except OsACS3, have higher expression in the shoot than in the root. A similar expression pattern was also found in 2-week-old rice seedlings, although higher expression of OsACS genes was generally found in the leaf blade (Supplemntary Fig. S2). There are no data available for OsACS3 and 4 in RiceXPro, but the available microarray data for OsACS1, 2, and 5 show that the expression of OsACS1 and 2 is higher in roots than in shoots, while the expression of OsACS5 remains high in the shoot. This suggests that the role of OsACS1 and 2 is shifted from shoots to roots in mature rice (Sato and Miura, 2011). Despite the similar expression patterns among the OsACS genes in our study (Supplementary Figs S1, S2), only OsACS1 and 2 were significantly induced under Pi deficiency and the changes in gene expression was more dramatic in the root than the shoot (Fig. 2). This is consistent with the general understanding that root architecture alteration in response to Pi deficiency is mainly regulated in roots rather than the systemic signaling from the shoot (Zhang et al., 2014). The isoform-specific role of OsACS has been found in other stress conditions. For example, OsACS1 and OsACS5 are induced under submergence stress (Zarembinski and Theologis, 1997; Van Der Straeten et al., 2001) and the transcript levels of OsACS1 and OsACS2 are up-regulated when rice plants are infected by Magnaporthe oryzae (Iwai et al., 2006). The differential regulation of OsACS1 and OsACS2 in response to Pi deficiency can also be derived from regulation of different ACS transcription regulators. It has been shown that the disruption of MYB-type transcription factors PHR1 and its closest paralog PHL1, specifically down-regulated ACS6 and ACS7 in response to Pi deficiency (Bustos et al., 2010).
Another possibility that could account for the larger role of OsACS1 in Pi starvation-induced adaptive responses is the post-translational control of OsACS proteins such as protein stability. Previous studies and our recent studies on the protein stability regulation of Arabidopsis ACS proteins have demonstrated that input signals including plant hormones differentially regulate the stability of ACS in a type-specific manner (Lee et al., 2017). For example, auxin rapidly enhances the protein stability of type-2 ACS, resulting in an increase in the steady-state levels of the protein by ~7-fold within 1 h of auxin treatment (Lee et al., 2017). Auxin also extends the steady-state levels of type-1 ACS as well, yet it does not have any effect on type-3 ACS protein. The regulation of protein stability is one of the fastest ways for a plant to adapt to rapid developmental shifts or to respond to environmental stresses as it does not require gene expression. Pi deficiency is a severe environmental stress that requires the rapid adaptation of the plant to the surroundings, and thus it may require rapid mechanisms that initially enable the plant to sense the deficiency of Pi. A recent study on the Arabidopsis hps3 mutant, which has a single amino acid substitution in ETO1 E3 ligase (Wang et al., 2012), also supports the involvement of the post-translational control of ACS proteins in Pi deficiency-induced adaptive responses. This study underpinned the possible type-specific role of ACS proteins in Pi stress response because ETO1 E3 ligase specifically targets type-2 ACS proteins for degradation via the 26S proteasome. Further studies on the protein stability of OsACS proteins in relation to Pi deficiency will throw light on the roles of post-translational control of the OsACS proteins during Pi stress.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Transcript analysis of OsACS genes in light-grown seedlings under Pi-sufficient growth condition.
Fig. S2. Gene expression analysis of OsACS in 2-week-old rice seedlings.
Fig. S3. Schematic diagrams of the pARS-MUbCAS9-OsACS constructs used in this study.
Fig. S4. Sequencing results of the Cr-OsACS mutants.
Fig. S5. Sequencing results of the potential off-target sites.
Fig. S6. Morphological analysis of Cr-OsACS T3 mutants and ethylene biosynthesis in the mutants.
Fig. S7. Sequencing results of the T3 generation of Cr-OsACS mutants.
Fig. S8. Cr-OsACS mutants have impaired hormone-induced ethylene biosynthesis.
Fig. S9. T3 generation of Cr-OsACS mutants have impaired cytokinin-induced ethylene biosynthesis.
Fig. S10. Altered morphology of Cr-OsACS mutants in Pi-sufficient condition.
Table S1. List of the primers used in this study.
Acknowledgements
We thank Dr Zachary L. Nimchuk’s laboratory for providing the pARS3-MUb-Cas9 vector. This work was supported by Purdue University start-up funds to GMY, and the National Science Foundation (IOS-1502141) to CZ.
Conflict of interest
The authors declare no conflict of interest.
References
- Abts W, Van de Poel B, Vandenbussche B, De Proft MP. 2014. Ethylene is differentially regulated during sugar beet germination and affects early root growth in a dose-dependent manner. Planta 240, 679–686. [DOI] [PubMed] [Google Scholar]
- Adams DO, Yang SF. 1979. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proceedings of the National Academy of Sciences, USA 76, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai P, Sun S, Zhao J, et al. 2009. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. The Plant Journal 57, 798–809. [DOI] [PubMed] [Google Scholar]
- Ames BN. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods in Enzymology 8, 115–118. [Google Scholar]
- Argueso CT, Hansen M, Kieber JJ. 2007. Regulation of ethylene biosynthesis. Journal of Plant Growth Regulation 262, 92–105. [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. 1999. A regulator of root architectural responses to soil phosphorus availability. Plant, Cell & Environment 22, 425–431. [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-López A, Ibarra-Laclette E, Sánchez-Calderón L, Gutiérrez-Alanis D, Herrera-Estrella L. 2011. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signaling & Behavior 6, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ. 2005. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends in Plant Science 10, 291–296. [DOI] [PubMed] [Google Scholar]
- De Cnodder T, Vissenberg K, Van Der Straeten D, Verbelen JP. 2005. Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid: a matter of apoplastic reactions. New Phytologist 168, 541–550. [DOI] [PubMed] [Google Scholar]
- Drew MC, He CJ, Morgan PW. 1989. Decreased ethylene biosynthesis, and induction of aerenchyma, by nitrogen- or phosphate-starvation in adventitious roots of Zea mays L. Plant Physiology 91, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wu N, Cui F, You L, Li X, Xiong L. 2014. A homolog of ETHYLENE OVERPRODUCER, OsETOL1, differentially modulates drought and submergence tolerance in rice. The Plant Journal 78, 834–849. [DOI] [PubMed] [Google Scholar]
- Forde B, Lorenzo H. 2001. The nutritional control of root development. Plant and Soil 232, 51–68. [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. 2012. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539. [DOI] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. 2000. Proteoid root development of phosphorus deficient Lupin is mimicked by auxin and phosphonate. Annals of Botany 85, 921–928. [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. 2011. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. [DOI] [PubMed] [Google Scholar]
- Graham MA, Ramirez M, Valdes-Lopez O, Lara M, Tesfaye M, Vance C, Hernandez G. 2006. Identification of candidate phosphorus stress induced genes in Phaseolus vulgaris through clustering analysis across several plant species. Functional Plant Biology 33, 789–797. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chae HS, Kieber JJ. 2009. Regulation of ACS protein stability by cytokinin and brassinosteroid. The Plant Journal 57, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. 1992. Enhanced sensitivity to ethylene in nitrogen- or phosphate-starved roots of Zea mays L. during aerenchyma formation. Plant Physiology 98, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, et al. 2007. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiology 144, 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Holford ICR. 1997. Soil phosphorus: its measurement, and its uptake by plants. Australian Journal of Soil Research 35, 227–240. [Google Scholar]
- Iwai T, Miyasaka A, Seo S, Ohashi Y. 2006. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiology 142, 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Smith AP, Nagarajan VK, Lahner B, Meagher RB, Raghothama KG. 2009. Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiology 150, 1033–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. 2013. Auxin and the integration of environmental signals into plant root development. Annals of Botany 112, 1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lynch JP, Brown KM. 2008. Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant, Cell & Environment 31, 1744–1755. [DOI] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van Der Straeten D, Verbelen JP. 2001. In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiology 125, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Chen YC, Kieber JJ, Yoon GM. 2017. Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. The Plant Journal 91, 491–504. [DOI] [PubMed] [Google Scholar]
- Lee HY, Yoon GM. 2018. Regulation of ethylene biosynthesis by phytohormones in etiolated rice (Oryza sativa L.) seedlings. Molecules and Cells 41, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen L, Forde BG, Davies WJ. 2017. The biphasic root growth response to abscisic acid in Arabidopsis involves interaction with ethylene and auxin signalling pathways. Frontiers in Plant Science 8, 1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lin SI, Chiou TJ. 2009. Molecular regulators of phosphate homeostasis in plants. Journal of Experimental Botany 60, 1427–1438. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. The Plant Journal 29, 751–760. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S. 2004. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. The Plant Cell 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. 2014. Phosphate nutrition: improving low-phosphate tolerance in crops. Annual Review of Plant Biology 65, 95–123. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. 2002. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiology 129, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2001. Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant and Soil 237, 225–237. [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. 2005. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA 102, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, et al. 2007. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell & Environment 30, 85–112. [DOI] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P. 2005. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiology 138, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan VK, Smith AP. 2012. Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant & Cell Physiology 53, 277–286. [DOI] [PubMed] [Google Scholar]
- Negi M, Sanagala R, Rai V, Jain A. 2016. Deciphering phosphate deficiency-mediated temporal effects on different root traits in rice grown in a modified hydroponic system. Frontiers in Plant Science 7, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G. 2015. The role of ethylene in plant adaptations for phosphate acquisition in soils—a review. Frontiers in Plant Science 6, 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L. 2014. Root architecture responses: in search of phosphate. Plant Physiology 166, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell 20, 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Sasidharan R. 2007. Growth control by ethylene: adjusting phenotypes to the environment. Journal of Plant Growth Regulation 26, 188–200. [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA. 2006. The Janus face of ethylene: growth inhibition and stimulation. Trends in Plant Science 11, 176–183. [DOI] [PubMed] [Google Scholar]
- Prokic L, Jovanovic Z, McAinsh MR, Vucinic Z, Stikic R. 2006. Species-dependent changes in stomatal sensitivity to abscisic acid mediated by external pH. Journal of Experimental Botany 57, 675–683. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50, 665–693. [DOI] [PubMed] [Google Scholar]
- Rani Debi B, Taketa S, Ichii M. 2005. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). Journal of Plant Physiology 162, 507–515. [DOI] [PubMed] [Google Scholar]
- Roldan M, Dinh P, Leung S, McManus MT. 2013. Ethylene and the responses of plants to phosphate deficiency. AoB Plants 5, plt013. [Google Scholar]
- Roldan M, Islam A, Dinh PTY, Leung S, McManus MT. 2016. Phosphate availability regulates ethylene biosynthesis gene expression and protein accumulation in white clover (Trifolium repens L.) roots. Bioscience Reports 36, e00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y. 2010. Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Molecular Plant 3, 288–299. [DOI] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. The Plant Cell 19, 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Miura K. 2011. Root architecture remodeling induced by phosphate starvation. Plant Signaling & Behavior 6, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, et al. 2013. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnology 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Shukla D, Rinehart CA, Sahi SV. 2017. Comprehensive study of excess phosphate response reveals ethylene mediated signaling that negatively regulates plant growth and development. Scientific Reports 7, 3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Liu D. 2015. Ethylene and plant responses to phosphate deficiency. Frontiers in Plant Science 6, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, Che X, Jiao Y, Liu D. 2016. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genetics 12, e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. 2010. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. The Plant Journal 64, 775–789. [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. 2004. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiology 136, 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A. 2009. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Straeten D, Zhou Z, Prinsen E, Van Onckelen HA, Van Montagu MC. 2001. A comparative molecular–physiological study of submergence response in lowland and deepwater rice. Plant Physiology 125, 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. 2004. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950. [DOI] [PubMed] [Google Scholar]
- Wang L, Dong J, Gao Z, Liu D. 2012. The Arabidopsis gene hypersensitive to phosphate starvation 3 encodes ethylene overproduction 1. Plant & Cell Physiology 53, 1093–1105. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. 2009. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. Journal of Integrative Plant Biology 51, 663–674. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, et al. 2014. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 14953–14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. 2001. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology 126, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X. 2013. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Current Opinion in Plant Biology 16, 205–212. [DOI] [PubMed] [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. 2003. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. Journal of Biological Chemistry 278, 49102–49112. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 35, 155–189. [Google Scholar]
- Yoon GM. 2015. New insights into the protein turnover regulation in ethylene biosynthesis. Molecules and Cells 38, 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang KL, Chang CM, Mori K, Uchida E, Ecker JR. 2006. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Molecular Biology 62, 427–437. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. 1994. Ethylene biosynthesis and action: a case of conservation. Plant Molecular Biology 26, 1579–1597. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. 1997. Expression characteristics of OS-ACS1 and OS-ACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Molecular Biology 33, 71–77. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. 2014. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. Journal of Integrative Plant Biology 56, 192–220. [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Zhu CQ, Zhao XS, Zheng SJ, Shen RF. 2016. Ethylene is involved in root phosphorus remobilization in rice (Oryza sativa) by regulating cell-wall pectin and enhancing phosphate translocation to shoots. Annals of Botany 118, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.