Abstract
Background
Genetic mechanisms are associated with male infertility, but the association with non-obstructive azoospermia (NOA) remains unclear. Mutations in the chloride channel accessory 4 (CLCA4) gene have been shown to have a role in male infertility. The aim of this study was to investigate the associations between single nucleotide polymorphisms (SNPs) of the CLCA4 gene and NOA in a Chinese Han population of Northeast China using combined targeted gene capture next-generation sequencing and bioinformatics analysis.
Material/Methods
The study group included 100 men with NOA, and there were 100 normal controls. Targeted gene capture next-generation sequencing was performed combined with bioinformatics analysis. Ten CLCA4 SNPs were screened in the cases of NOA and control subjects. The associations between SNPs and NOA were analyzed.
Results
Six SNPs, c.390C>T (rs190628533), c.1474A>G (rs2231599), c.2105C>G (rs757773924), c.2371A>T) (rs759981524), c.956G>A (rs763334876), and c.895T>C (rs79822589) were identified in the study group of cases in NOA but not in control subjects. All CLCA4 SNPs were in Hardy-Weinberg equilibrium. The allele and genotype frequencies of the six SNPs were not significantly different between the study group and the controls. Haplotype analysis showed the existence of two haplotypes, CTAGACTACG and CTCGACTACG, which showed statistical significance of 0.074, and 0.088 between cases of NOA and the controls, respectively.
Conclusions
There were no significant associations between CLCA4 SNPs and NOA in men in a Chinese Han population of Northeast China.
MeSH Keywords: Azoospermia; Infertility, Male; Polymorphism, Single Nucleotide
Background
Genetic mechanisms are associated with male infertility, but the association with non-obstructive azoospermia (NOA) remains unclear [1]. Azoospermia is the most severe form of male infertility, in which genetic abnormalities have been identified in between 15–20% of cases [1]. Compared with obstructive azoospermia, patients with non-obstructive azoospermia (NOA) are less likely to be fertile. The genetic mechanism of NOA remains unclear, and has recently become a focus for research in reproductive medicine [2]. Currently, the genetic causes of azoospermia are known to include chromosomal abnormalities, azoospermia factor (AZF) microdeletion of the Y chromosome, copy number variations (CNVs), and monogenic mitochondrial and epigenetic abnormalities [3]. Recently, associations between gene polymorphisms and male infertility have been identified [4–7]. The relationships between additional genes and abnormal spermatogenesis require further investigation.
The chloride channel accessory 4 (CLCA4) gene (MIM: 616857) maps to a cluster of CLCA genes on chromosome 1p22.3 [8]. CLCA4 belongs to the calcium-dependent chloride channel family and modulates the basic defect in cystic fibrosis [9]. Kolbe et al. [10] also reported that the basic defect of cystic fibrosis was associated with markers that included the CLCA4 gene promotor. Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene are known to cause cystic fibrosis and are also associated with male infertility [11]. Using DNA arrays to identify novel CNVs, Frühmesser et al. [12] found that CLCA4 plays an important role in human spermatogenesis, and may act by mediating CFTR gene expression. Therefore, disrupted CLCA4 function may play a role in male infertility. However, these results require further confirmation.
Therefore, the aim of this study was to investigate the associations between single nucleotide polymorphisms (SNPs) of the CLCA4 gene and NOA in a Chinese Han population of men in Northeast China using combined targeted gene capture next-generation sequencing and bioinformatics analysis.
Material and Methods
Subjects
A case-control study included 100 subjects from the Center for Reproductive Medicine of the First Hospital, Jilin University, China, enrolled between January 2015 and December 2016. Patients were diagnosed with NOA by semen analysis and testicular fine needle aspiration cytology. One hundred control subjects were randomly selected from sperm donors at the Human Sperm Bank of Jilin Province between January 2015 and December 2016. All patients and controls were of Han ethnicity and were from Northeast China. Karyotype analysis and Y chromosome AZF microdeletion detection were performed for all cases, and those with chromosomal abnormalities or AZF microdeletions were excluded. Written informed consent was provided by each participant. The present study was approved by the Ethics Committee of the First Hospital of Jilin University (no. 2016-419, dated 10th Dec. 2016). All participants signed informed consent.
DNA extraction
Peripheral venous blood was collected from each participant into anticoagulant tubes containing ethylenediaminetetraacetic acid using standard protocols. Total DNA was extracted from blood using a blood genomic DNA extraction kit (TIANGEN Biotech Co., Ltd, Beijing) according to the manufacturer’s instructions and then kept at −20°C until use.
Targeted gene capture sequencing
Targeted gene capture sequencing was performed by MyGenostics (Beijing, China). Genomic DNA samples were fragmented and prepared for standard Illumina library construction. Biotinylated capture probes were designed for the exons of CLCA4 gene and then sequenced using Illumina HiSeq2000 Next-Generation Sequencing platform and bioinformatical analyses (MyGenostics, Beijing, China). Data analysis was performed according to MyGenostics protocols.
Statistical analysis
Clinical data were assessed using the t-test with SPSS software version 17.0 (IBM Corporation, Armonk, NY, USA). Assessment of the Hardy-Weinberg equilibrium (HWE) was determined using SHE software (http://analysis.bio-x.cn) for each single nucleotide polymorphism (SNP) of the CLCA4 gene in the cases and controls separately. The allele frequencies of CLCA4 polymorphisms were compared using the chi-squared (χ2) test and Fisher’s exact test (two-sided). SNP genotype frequencies and dominant/recessive model analysis of cases and controls were calculated using logistic regression analysis. Haplotype analysis was conducted using SHE software (http://analysis.bio-x.cn/SHEsisMain.htm). Linkage disequilibrium analysis was performed using Haploview bioinformatics software (http://www.broadinstitute.org/haploview/). P-values <0.05 were considered as statistically significant.
Results
Subject characteristics
The clinical characteristics of the subjects are shown in Table 1. Clinical information includes age, body mass index (BMI), semen volume, pH value, and sperm concentration. The study group was statistically older than the control group because for the control group we enrolled fertile men who were donating to the human sperm bank (P<0.001). BMI and semen volume were also significantly different between the two groups (P<0.001).
Table 1.
Clinical characteristics of the study population.
| Variables | Cases group (100) | Control group (100) | P value |
|---|---|---|---|
| Age(years) | 29.14±4.40 | 25.10±5.68 | <0.001* |
| BMI(kg/m2) | 25.51±4.17 | 22.79±3.70 | <0.001* |
| Semen volume | 2.84±1.37 | 3.66±1.26 | <0.001* |
| Semen pH | 7.48±0.32 | 7.50±0.03 | 0.537 |
| Sperm concentration (106/mL) | 0±0 | 62.95±6.49 | NA |
NA – not available;
P<0.05 has statistical significance.
Genotype and frequency distribution
The genotype distribution of the CLCA4 single nucleotide polymorphisms (SNPs) are shown in Table 2. In the study group, c.390C>T was present in one case, c.907A>C was present in 20 cases, c.1474A>G was present in two cases, c.2398C>A was present in one case, c.2105C>G was present in one case, c. 2371A>T was present in two cases, c.956G>A was present in one case, and c.895T>C was present in one case. In the control group, c.2151T>A was present in two cases, c.907A>C was present in 32 cases, c.2428G>C was present in two cases, and c.2398C>A was present in three cases. There was no significant difference in genotype distribution between the two groups (P>.05). However, six sites, c.390C>T (rs190628533), c.1474A>G (rs2231599), c.2105C>G (rs757773924), c.2371A>T (rs759981524), c.956G>A (rs763334876), and c.895T>C (rs79822589) appeared only in the study group, and their role in azoospermia requires further study.
Table 2.
Distribution of CLCA4 SNPs.
| Number | SNV | Site | dbSNP code | n | OR (95%) | P value | |
|---|---|---|---|---|---|---|---|
| Case group | Control group | ||||||
| 1 | c.2151T>A | 1p22.3-87045065 | rs185369520 | 0 | 2 | – | 0.999 |
| 2 | c.390C>T | 1p22.3-87025983 | rs190628533 | 1 | 0 | – | 1 |
| 3 | c.907A>C | 1p22.3-87031656 | rs2231592 | 20 | 32 | 0.583 (0.321–1.060) | 0.077 |
| 4 | c.1474A>G | 1p22.3-87040229 | rs2231599 | 2 | 0 | – | 0.999 |
| 5 | c.2428G>C | 1p22.3-87045696 | rs2231604 | 0 | 2 | – | 0.999 |
| 6 | c.2398C>A | 1p22.3-87045666 | rs539177280 | 1 | 3 | 0.330 (0.034–3.200) | 0.339 |
| 7 | c.2105C>G | 1p22.3-87043738 | rs757773924 | 1 | 0 | – | 1 |
| 8 | c.2371A>T | 1p22.3-87045639 | rs759981524 | 2 | 0 | – | 0.999 |
| 9 | c.956G>A | 1p22.3-87033108 | rs763334876 | 1 | 0 | – | 1 |
| 10 | c.895T>C | 1p22.3-87031644 | rs79822589 | 1 | 0 | – | 1 |
Correlation analysis between SNPs and non-obstructive azoospermia (NOA)
This study classified the minimum genotype frequencies of the ten polymorphic loci and a Hardy-Weinberg equilibrium test was performed on the genotype distribution of each polymorphic locus. The results are shown in Table 3. There was no statistical difference between the two groups at any site (P>0.05). These results indicated that the CLCA4 SNPs were not associated with the occurrence of NOA.
Table 3.
Correlation analysis between CLCA4 and NOA.
| SNV | dbSNP code | Case group (n=100) | Control group (n=100) | P value | PHWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| het | hom | MAF% | het | hom | MAF% | Case group | Control group | |||
| c.2151T>A | rs185369520 | 0 | 0 | 0.00 | 2 | 0 | 1.00 | 0.156 | 1 | 0.919 |
|
| ||||||||||
| c.390C>T | rs190628533 | 1 | 0 | 0.50 | 0 | 0 | 0.00 | 0.316 | 0.959 | 1 |
|
| ||||||||||
| c.907A>C | rs2231592 | 18 | 1 | 10.00 | 28 | 2 | 16.00 | 0.074 | 1 | 0.677 |
|
| ||||||||||
| c.1474A>G | rs2231599 | 2 | 0 | 1.00 | 0 | 0 | 0.00 | 0.156 | 0.919 | 1 |
|
| ||||||||||
| c.2428G>C | rs2231604 | 0 | 0 | 0.00 | 2 | 0 | 1.00 | 0.156 | 1 | 0.919 |
|
| ||||||||||
| c.2398C>A | rs539177280 | 1 | 0 | 0.50 | 3 | 0 | 1.50 | 0.315 | 0.959 | 0.878 |
|
| ||||||||||
| c.2105C>G | rs757773924 | 1 | 0 | 0.50 | 0 | 0 | 0.00 | 0.316 | 0.959 | 1 |
|
| ||||||||||
| c.2371A>T | rs759981524 | 2 | 0 | 1.00 | 0 | 0 | 0.00 | 0.156 | 0.919 | 1 |
|
| ||||||||||
| c.956G>A | rs763334876 | 1 | 0 | 0.50 | 0 | 0 | 0.00 | 0.316 | 0.959 | 1 |
|
| ||||||||||
| c.895T>C | rs79822589 | 1 | 0 | 0.50 | 0 | 0 | 0.00 | 0.316 | 0.959 | 1 |
Genotype and the correlation with the dominant/recessive model and NOA
In the genotype analysis of the ten SNPs, the genotypes for each group were classified into dominant and recessive genotypes, and binary logistic regression analysis methods were applied. Neither the dominant nor the recessive model were associated with the occurrence of NOA. The results are shown in Table 4, and there was no statistical difference between the two groups (P>0.05). These results indicate that there is no significant correlation between the dominant or recessive models of the genotypes of the ten SNPs and the occurrence of NOA.
Table 4.
Genotypes and dominant/recessive model analysis of NOA-associated SNPs.
| SNV | dbSNP code | Model | Genotypes | n | OR (95%) | P value | |
|---|---|---|---|---|---|---|---|
| Case group | Control group | ||||||
| c.2151T>A | rs185369520 | Dominant | AT/AA+TT | 0/100 | 2/98 | – | 0.999 |
| Recessive | AA/AT+TT | – | – | NA | NA | ||
| c.390C>T | rs190628533 | Dominant | TC/TT+CC | 1/99 | 0/100 | – | 1 |
| Recessive | TT/TC+CC | – | – | NA | NA | ||
| c.907A>C | rs2231592 | Dominant | CA/CC+AA | 19/81 | 30/70 | 0.547 (0.284–1.056) | 0.072 |
| Recessive | CC/CA+AA | 1/99 | 2/98 | 0.495 (0.044–5.548) | 0.568 | ||
| c.1474A>G | rs2231599 | Dominant | GA/GG+AA | 2/98 | 0/100 | – | 0.999 |
| Recessive | GG/GA+AA | – | – | NA | NA | ||
| c.2428G>C | rs2231604 | Dominant | CG/CC+GG | 0/100 | 2/98 | – | 0.999 |
| Recessive | CC/CG+GG | – | – | NA | NA | ||
| c.2398C>A | rs539177280 | Dominant | AC/AA+CC | 1/99 | 3/97 | 0.327 (0.033–3.194) | 0.336 |
| Recessive | AA/AC+CC | – | – | NA | NA | ||
| c.2105C>G | rs757773924 | Dominant | GC/GG+CC | 1/99 | 0/100 | – | 1 |
| Recessive | GG/GC+CC | – | – | NA | NA | ||
| c.2371A>T | rs759981524 | Dominant | TA/TT+AA | 2/98 | 0/100 | – | 0.999 |
| Recessive | TT/TA+AA | – | – | NA | NA | ||
| c.956G>A | rs763334876 | Dominant | AG/AA+GG | 1/99 | 0/100 | – | 1 |
| Recessive | AA/GA+GG | – | – | NA | NA | ||
| c.895T>C | rs79822589 | Dominant | CT/CC+TT | 1/99 | 0/100 | – | 1 |
| Recessive | CC/CT+TT | – | – | NA | NA | ||
Binary logistic regression analysis; P<0.05 has statistical significance.
Candidate SNP haplotype analysis
The ten candidate SNPs were distributed on chromosome 1. Haplotype analysis was performed using Haploview bioinformatics software and the results are shown in Table 5. The CLCA4 SNPs, rs185369520, rs190628533, rs2231592, rs2231599, rs2231604, rs539177280, rs757773924, rs759981524, rs763334876 and rs79822589 form a haploid block; haplotypes were CTAGACTACG and CTCGACTACG. Haplotype frequency results are shown in Figure 1. There was no significant difference between the two groups for the two CLCA4 haplotypes (P>0.005). This finding showed that haplotypes, CTAGACTACG and CTCGACTACG were not associated with NOA.
Table 5.
Haplotype analysis of NOA-associated SNPs.
| Haplotype | Frequency (%) | P value | |
|---|---|---|---|
| Case group | Control group | ||
| BLOCK 1 | |||
| CTAGACTACG | 0.900 | 0.840 | 0.074 |
| CTCGACTACG | 0.090 | 0.145 | 0.088 |
P<0.05 has statistical significance.
Figure 1.
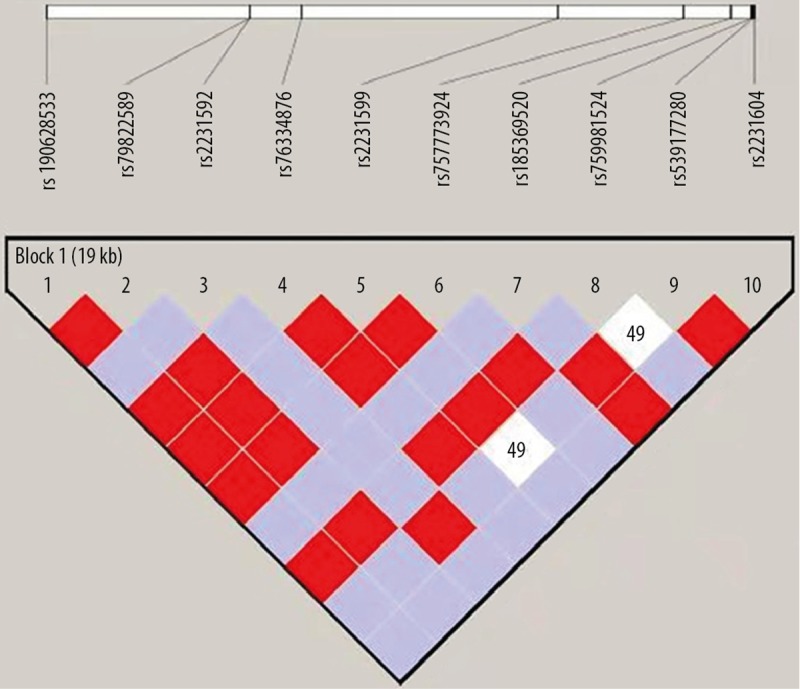
Haplotype analysis of non-obstructive azoospermia (NOA)-associated single nucleotide polymorphisms (SNPs) on chromosome 1.
Discussion
Male infertility is a complex multifactorial condition, in which azoospermia is the most severe cause. Genetic factors are major contributors to non-obstructive azoospermia (NOA) [13]. Some studies have suggested that up to 2,300 genes are involved in spermatogenesis, and change in any one of these genes may lead to male infertility. These genes can provide targets for diagnostic tests of male factor infertility [14]. Next-generation sequencing technology has been applied to many genomic features of physiology and disease, including male infertility [15]. Targeted gene sequencing in cohorts of infertile men has identified several gene polymorphisms that are associated with male infertility [15]. This study explored the relationship between CLCA4 polymorphisms and NOA in a Northeastern Han Chinese population.
In this study, ten CLCA4 SNPs were examined in 100 infertile men and 100 fertile controls, and c.907A>C (rs2231592) and c.2398C>A (rs539177280) were detected in both the case and control groups. These two SNPs showed no significant association between infertile men and controls in the study cohort. Also, c.2151T>A (rs185369520) and c.2428G>C (rs2231604) were only detected in the control group. The other six SNPs were only detected in the study group. Further statistical analysis indicated that the SNPs are not associated with NOA occurrence. There was no significant correlation between dominant or recessive models of the SNPs and the occurrence of NOA. Using Haploview bioinformatics software to analyze the SNPs, haplotype was not associated with NOA. Together these results indicated that the candidates SNPs (under dominant and recessive models and haplotypes) are not related to the occurrence of NOA. This result is inconsistent with previous findings [12]. This discrepancy might be related to sample size, detection or analysis methods used, or, more likely, to the ethnicity of the study populations. Further study is required to understand the full significance of CLCA4 in infertile men.
The major limitation of this study was the relatively small number of men included in the study with NOA. Also, this study only included a Han ethnic population from Northeast China. However, linkage disequilibrium structures associated with the tested markers are known to be different between ethnic groups [16].
Conclusions
This study showed no significant differences in genotype and allele frequencies or haplotype for CLCA4 gene single nucleotide polymorphisms (SNPs) between the men with non-obstructive azoospermia (NOA) and healthy controls in a Chinese Han population from Northeast China. These findings indicate that the CLCA4 gene is not involved in genetic susceptibility to infertility in Chinese men.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the Science and Technology Funds of Education Department of Jilin Province, Peoples’ Republic of China (JJKH20170846KJ)
References
- 1.Flannigan R, Schlegel PN. Genetic diagnostics of male infertility in clinical practice. Best Pract Res Clin Obstet Gynaecol. 2017;44:26–37. doi: 10.1016/j.bpobgyn.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Zhang H, Jiang Y, et al. Association study between MTRR, TAF4B, PIWIL1 variants and non-obstructive azoospermia in Northeast Chinese Han population. Clin Lab. 2018;64:1731–38. doi: 10.7754/Clin.Lab.2018.180525. [DOI] [PubMed] [Google Scholar]
- 3.Halder A, Kumar P, Jain M, et al. Genomics: Tool to predict and prevent male infertility. Front Biosci (Schol Ed) 2017;9:448–508. doi: 10.2741/s496. [DOI] [PubMed] [Google Scholar]
- 4.Cortés-Rodriguez M, Royo JL, Reyes-Palomares A, et al. Sperm count and motility are quantitatively affected by functional polymorphisms of HTR2A, MAOA and SLC18A. Reprod Biomed Online. 2018;36:560–67. doi: 10.1016/j.rbmo.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Wei Y, Zhang Z, Chen G, et al. Lack of association between aryl hydrocarbon receptor gene Arg554Lys polymorphism and male infertility risk: A systematic review and meta-analysis. Eur J Obstet Gynecol Reprod Biol. 2018;223:1–7. doi: 10.1016/j.ejogrb.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Aydos OSE, Hekmatshoar Y, Altınok B, et al. Genetic Polymorphisms in PRM1, PRM2, and YBX2 genes are associated with male factor infertility. Genet Test Mol Biomarkers. 2018;22:55–61. doi: 10.1089/gtmb.2017.0040. [DOI] [PubMed] [Google Scholar]
- 7.Sato Y, Hasegawa C, Tajima A, et al. Association of TUSC1 and DPF3 gene polymorphisms with male infertility. J Assist Reprod Genet. 2018;35:257–63. doi: 10.1007/s10815-017-1052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauli BU, Abdel-Ghany M, Cheng HC, et al. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol. 2000;27:901–5. doi: 10.1046/j.1440-1681.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- 9.Ritzka M, Stanke F, Jansen S, et al. The CLCA gene locus as a modulator of the gastrointestinal basic defect in cystic fibrosis. Hum Genet. 2004;115:483–91. doi: 10.1007/s00439-004-1190-y. [DOI] [PubMed] [Google Scholar]
- 10.Kolbe EW, Tamm S, Hedtfeld S, et al. CLCA4 variants determine the manifestation of the cystic fibrosis basic defect in the intestine. Eur J Hum Genet. 2013;21:691–94. doi: 10.1038/ejhg.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Ruan YC, Xu WM, et al. Regulation of male fertility by CFTR and implications in male infertility. Hum Reprod Update. 2012;18:703–13. doi: 10.1093/humupd/dms027. [DOI] [PubMed] [Google Scholar]
- 12.Frühmesser A, Vogt PH, Zimmer J, et al. Single nucleotide polymorphism array analysis in men with idiopathic azoospermia or oligoasthenozoospermia syndrome. Fertil Steril. 2013;100:81–87. doi: 10.1016/j.fertnstert.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: Current applications and future directions. Andrology. 2014;2:339–50. doi: 10.1111/j.2047-2927.2014.00200.x. [DOI] [PubMed] [Google Scholar]
- 14.Gershoni M, Hauser R, Yogev L, et al. A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet Med. 2017;19:998–1006. doi: 10.1038/gim.2016.225. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S, Miyado M, Saito K, et al. Next-generation sequencing for patients with non-obstructive azoospermia: implications for significant roles of monogenic/oligogenic mutations. Andrology. 2017;5:824–31. doi: 10.1111/andr.12378. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y, Tajima A, Tsunematsu K, et al. Lack of replication of four candidate SNPs implicated in human male fertility traits: a large-scale population-based study. Hum Reprod. 2015;30:1505–9. doi: 10.1093/humrep/dev081. [DOI] [PubMed] [Google Scholar]


