Bacillus subtilis has been studied as a model organism for biofilm formation. In this study, I explored why the cysL deletion mutant was defective in biofilm formation. I demonstrated that the ΔcysL mutation activated the disulfide stress response regulator Spx, which inhibits biofilm formation by repressing biofilm matrix genes. Homologs of Spx are highly conserved among Gram-positive bacteria with low G+C contents. In some pathogens, Spx is also reported to inhibit biofilm formation by repressing biofilm matrix genes, even though these genes and their regulation are quite different from those of B. subtilis. Thus, the negative regulation of biofilm formation by Spx is likely to be well conserved across species and may be an appropriate target for control of biofilm formation.
KEYWORDS: Bacillus subtilis, Spx, biofilms, cysteine biosynthesis, disulfide stress
ABSTRACT
Bacillus subtilis forms biofilms in response to internal and external stimuli. I previously showed that the cysL deletion mutant was defective in biofilm formation, but the reason for this remains unidentified. CysL is a transcriptional activator of the cysJI operon, which encodes sulfite reductase, an enzyme involved in cysteine biosynthesis. Decreased production of sulfite reductase led to biofilm formation defects in the ΔcysL mutant. The ΔcysL mutation was suppressed by disrupting cysH operon genes, whose products function upstream of sulfite reductase in the cysteine biosynthesis pathway, indicating that defects in cysteine biosynthesis were not a direct cause for the defective biofilm formation observed in the ΔcysL mutant. The cysH gene encodes phosphoadenosine phosphosulfate reductase, which requires a reduced form of thioredoxin (TrxA) as an electron donor. High expression of trxA inhibited biofilm formation in the ΔcysL mutant but not in the wild-type strain. Northern blot analysis showed that trxA transcription was induced in the ΔcysL mutant in a disulfide stress-induced regulator Spx-dependent manner. On the basis of these results, I propose that the ΔcysL mutation causes phosphoadenosine phosphosulfate reductase to consume large amounts of reduced thioredoxin, inducing disulfide stress and activating Spx. The spx mutation restored biofilm formation to the ΔcysL mutant. The ΔcysL mutation reduced expression of the eps operon, which is required for exopolysaccharide production. Moreover, overexpression of the eps operon restored biofilm formation to the ΔcysL mutant. Taken together, these results suggest that the ΔcysL mutation activates Spx, which then inhibits biofilm formation through repression of the eps operon.
IMPORTANCE Bacillus subtilis has been studied as a model organism for biofilm formation. In this study, I explored why the cysL deletion mutant was defective in biofilm formation. I demonstrated that the ΔcysL mutation activated the disulfide stress response regulator Spx, which inhibits biofilm formation by repressing biofilm matrix genes. Homologs of Spx are highly conserved among Gram-positive bacteria with low G+C contents. In some pathogens, Spx is also reported to inhibit biofilm formation by repressing biofilm matrix genes, even though these genes and their regulation are quite different from those of B. subtilis. Thus, the negative regulation of biofilm formation by Spx is likely to be well conserved across species and may be an appropriate target for control of biofilm formation.
INTRODUCTION
Biofilms are structured, multicellular communities of bacteria in which bacterial cells adhere to each other on a surface via a biofilm matrix. The biofilm matrix consists of polymeric exopolysaccharides, proteins, and/or nucleic acids (1, 2) produced by the bacteria that make up the biofilm. Biofilms are resistant to environmental stresses, including antibiotics, bactericidal chemicals, metals, and host defense mechanisms (3, 4). Most bacteria in nature are capable of forming biofilms and surviving. Some biofilms cause serious problems in man-made environments, such as those associated with contamination and infectious diseases, and therefore reagents and treatments to effectively remove, prevent, or inhibit biofilms are in demand.
The Gram-positive bacterium Bacillus subtilis forms floating biofilms (pellicles) on the surface of liquid medium under static culture conditions or colony biofilms on solid medium (5). B. subtilis biofilms produce a biofilm matrix composed of exopolysaccharides, TasA amyloid fibers, and BslA hydrophobins produced by proteins of the epsABCDEFGHIJKLMNO operon, the tapA-sipW-tasA operon, and bslA, respectively (6–10). These genes are directly or indirectly repressed by the transcriptional repressors AbrB and SinR (11–14). One of the triggers for the initiation of biofilm formation is a self-produced and secreted antibiotic, surfactin, which is considered to stimulate autophosphorylation of histidine kinase KinC, leading to the activation of antagonistic regulatory mechanisms against AbrB and SinR repression (15, 16). Although regulatory mechanisms for B. subtilis biofilm formation have been extensively studied, unanswered questions still remain.
I previously tested the biofilm formation ability of 285 transcriptional regulation mutants and identified ninety regulators involved in biofilm formation (17). One of these is cysL, which encodes a transcriptional activator of the cysteine biosynthesis genes, cysJI (18). However, why inactivation of cysL causes biofilm formation defects remains unexplained. In this study, I investigated the phenotype of the ΔcysL mutant and identified a previously unknown mechanism for the regulation of B. subtilis biofilm formation.
RESULTS AND DISCUSSION
The ΔcysL mutant blocks biofilm formation.
I previously found that inactivation of cysL impaired the ability of B. subtilis to form pellicle biofilms under static culture conditions in 2× Schaeffer's sporulation medium supplemented with glucose and glycerol (2×SGG) medium, which is a nutrient broth (Difco)-based complex medium supplemented with sugars and metals (17). Since the previous study was conducted using B. subtilis strain ATCC 6051, I reexamined the effects of the ΔcysL mutation on biofilm formation using another B. subtilis strain, NCIB3610 (5), which has been widely used in studies of B. subtilis biofilm formation. As shown in Fig. 1A, the wild-type strain NCIB3610 formed thick pellicles with densely wrinkled morphology in 2×SGG medium, whereas the ΔcysL mutant of NCIB3610 formed thin pellicles lacking macroscopic structures, such that the brown color of the medium was seen through them. The addition of cysteine to the medium suppressed the biofilm formation-defective phenotype of the ΔcysL mutant (Fig. 1A). Since cysL encodes an LysR-type transcriptional regulator that activates the transcription of the cysJI operon encoding sulfite reductase (18, 19), decreased expression of cysJI was thought to cause the biofilm formation-defective phenotype of the ΔcysL mutant. To test this idea, the ΔcysI mutant was cultured in 2×SGG medium without agitation. The ΔcysI mutant formed very thin pellicles similar to those of the ΔcysL mutant (Fig. 1A). The addition of cysteine to the medium restored pellicle formation to the ΔcysI mutant (Fig. 1A). Thus, ΔcysL and ΔcysI mutants displayed the same phenotype, supporting my assumption.
FIG 1.
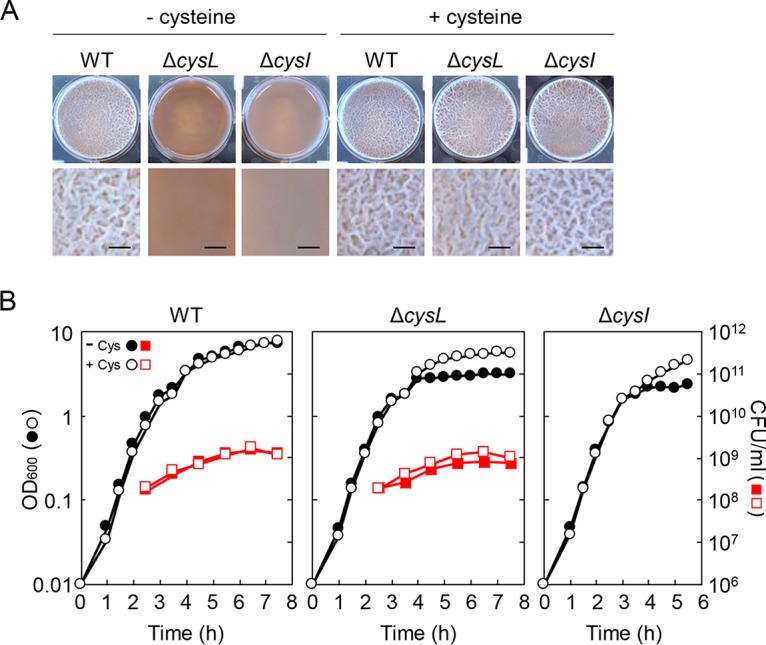
The ΔcysL mutant is defective in biofilm formation. (A) Biofilm formation ability of ΔcysL and ΔcysI mutants in the presence or absence of 100 μg/ml cysteine. B. subtilis strains were grown at 30°C for 48 h in 2×SGG medium with or without cysteine under static conditions. Top-down photographs of pellicle biofilms are shown. The well diameter is 34 mm. Extended images of each pellicle are also shown. Bars, 2 mm. (B) Growth profiles of the ΔcysL and ΔcysI mutants. B. subtilis strains were grown with vigorous shaking in 2×SGG medium or 2×SGG medium plus 100 μg/ml cysteine. CFU/ml were determined only for the wild-type strain and the ΔcysL mutant.
The ΔcysL mutant has cysteine auxotrophy and cannot grow in Spizizen minimal medium (SMM) due to decreased expression of cysJI (18) (Fig. 2D). To determine whether the ΔcysL mutation had severe effects on growth in 2×SGG complex medium, the wild-type strain and the ΔcysL mutant were grown in 2×SGG medium with vigorous shaking, and optical density at 600 nm (OD600) was measured over time. The ΔcysL mutant showed a comparable growth rate to the wild-type strain during the exponential growth phase, but reached a lower OD600 than that of the wild-type strain in the stationary phase (Fig. 1B). The addition of cysteine to the medium raised the OD600 of the ΔcysL mutant culture in the stationary phase to the wild-type strain level (Fig. 1B). The ΔcysI mutant also showed the same growth phenotype. These results indicate that the ΔcysL mutation does not severely affect growth during the exponential phase but does suppress growth in the stationary phase, likely caused by decreased cysteine in the medium.
FIG 2.
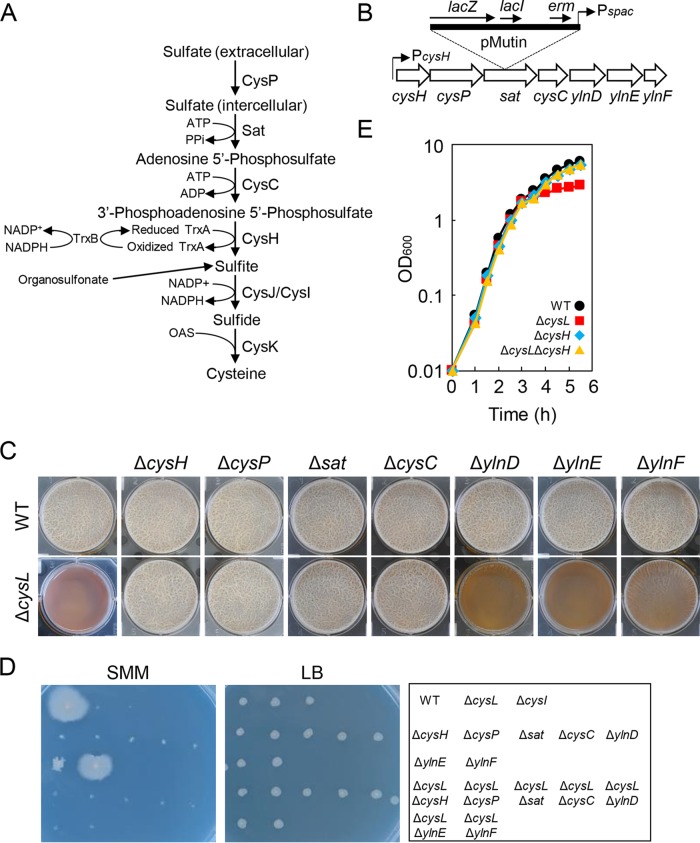
Null mutations of the cysH operon genes suppress the ΔcysL mutation. (A) Diagram of the cysteine biosynthesis pathway of B. subtilis. The proteins believed to function in the process are shown. (B) Gene organization of the cysH operon. A pMutin insertion into sat is shown above the gene map as an example. (C) Biofilm formation of cysH operon mutants. The wild-type and mutant strains were statically grown for 48 h in 2×SGG medium with 1 mM IPTG. (D) Viability of cysH operon mutants on Spizizen minimal medium (SMM) and LB. Strains were grown at 37°C for 48 h on indicated medium supplemented with 1 mM IPTG. Strain positions on the media are shown in the right panel. (E) Growth profiles of ΔcysH and ΔcysH ΔcysL mutants. Strains were grown at 37°C in 2×SGG medium with vigorous shaking.
The ΔcysL mutation drastically reduces transcription of sulfite reductase (CysJ/CysI) (18), which is required for the reduction of sulfite to sulfide in cysteine biosynthesis (19) (Fig. 2A). I tested whether disrupting genes involved in earlier steps in the cysteine biosynthesis pathway inhibited biofilm formation. Sulfite is synthesized from sulfate through four steps catalyzed by proteins of the cysH operon (Fig. 2B) (20). The first four genes of the cysH operon belong to the cysteine biosynthesis pathway; cysP encodes sulfate permease (21), while sat, cysC, and cysH encode sulfate adenylyl transferase, adenylyl sulfate kinase, and phosphoadenosine phosphosulfate reductase, respectively, all of which are responsible for synthesizing sulfite from incorporated sulfate in the cysteine biosynthesis pathway (20, 21). The remaining genes, ylnD, ylnE, and ylnF, are probably involved in the synthesis of siroheme, a cofactor of sulfite reductase (22, 23). The cysH operon genes were disrupted by the insertion of an integration vector, pMutin (24). The pMutin vector contains a LacI-repressive and isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible spac promoter, which alleviates the polar effect of the pMutin insertion on downstream genes by expressing them from the spac promoter in an IPTG-dependent fashion (Fig. 2B). The pMutin insertion mutants of the cysH operon genes were grown statically in 2×SGG medium supplemented with IPTG. Unlike the ΔcysL mutant, ΔcysH, ΔcysP, Δsat, ΔcysC, ΔylnD, ΔylnE, and ΔylnF mutants all formed pellicle biofilms comparable to those of the wild-type strain (Fig. 2C). Except for ΔylnE and ΔylnF, these mutants showed cysteine auxotrophy on SMM (Fig. 2D). These results indicate that cysteine biosynthesis deficiency does not always inhibit biofilm formation, and that siroheme synthesis by YlnD, YlnE, and YlnF is not required for sulfite reductase activity in 2×SGG medium.
These insertion mutations of the cysH operon genes were transferred to the ΔcysL mutant, and the resultant double mutants were tested for their ability to form biofilms. ΔcysH ΔcysL, ΔcysP ΔcysL, Δsat ΔcysL, and ΔcysC ΔcysL double mutants formed normal pellicles, while ΔylnD ΔcysL, ΔylnE ΔcysL, and ΔylnF ΔcysL double mutants did not (Fig. 2C). These double mutants showed cysteine auxotrophy on SMM (Fig. 2D). Growth of ΔcysH and ΔcysH ΔcysL mutants was tested in 2×SGG medium with vigorous shaking. ΔcysH and ΔcysH ΔcysL mutants showed growth comparable to that of the wild-type strain from the exponential phase to the stationary phase (Fig. 2E). Thus, disrupting genes involved in steps earlier than sulfite reductase in the cysteine biosynthesis pathway restored biofilm formation and stationary-phase growth to the ΔcysL mutant. These results indicate that the biofilm formation-defective phenotype of ΔcysL mutant is caused by the action of the cysH operon proteins, CysH, CysP, Sat, and CysC. As described above, the ΔcysL mutation was suppressed by the addition of cysteine. Since expression of cysteine biosynthetic genes, including that of cysH, is repressed in the presence of cysteine (20), this suppression is likely to be caused by decreased expression of the cysH operon.
Sulfite accumulation is not responsible for the biofilm formation-defective phenotype of the ΔcysL mutant.
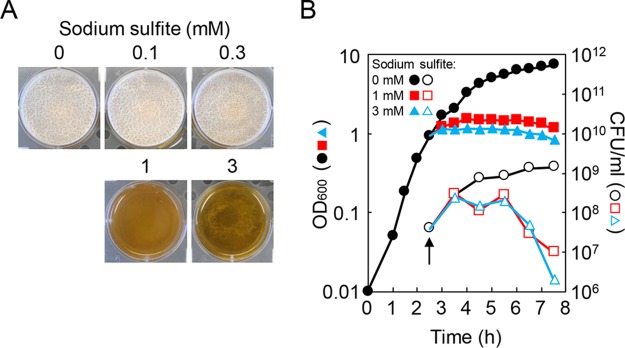
CysH, CysP, Sat, and CysC are required for the biosynthesis of sulfite, which is expected to accumulate in the ΔcysL mutant due to decreased expression of sulfite reductase (Fig. 2A). Sulfite is often used as a food preservative and an antioxidant and is toxic to bacteria. I therefore hypothesized that accumulated sulfite might inhibit biofilm formation in the ΔcysL mutant. To address this hypothesis, the effects of various concentrations of sulfite on biofilm formation were tested. The wild-type strain was statically grown to just before formation of pellicle biofilms, and then the cultures were treated with sulfite. Although the low concentrations of sulfite did not affect biofilm formation, sulfite completely inhibited biofilm formation at 1 mM and 3 mM (Fig. 3A). I examined the effects of those concentrations of sulfite on growth. The wild-type strain was grown to the end of the exponential phase with vigorously shaking. The culture was then divided into three parts, and each of the cultures was treated with 0, 1, or 3 mM sulfite (Fig. 3A). The addition of 1 mM or 3 mM sulfite immediately inhibited growth and caused a decrease in the number of viable cells (Fig. 3B). These results indicate that sulfite inhibits biofilm formation by killing cells. This contradicted the observation that the ΔcysL mutation had moderate effects on the number of viable cells (Fig. 1B). Thus, sulfite accumulation is unlikely to be the primary cause of the biofilm formation-defective phenotype of the ΔcysL mutant.
FIG 3.
Sulfite inhibits biofilm formation by killing cells. (A) Impacts of sulfite on biofilm formation. The wild-type strain was statically grown at 30°C for 18 h in 2×SGG medium, and then sulfite was added to the cultures. The cultures were further incubated at 30°C for 30 h. (B) Impact of sulfite on growth. The wild-type strain was grown to an OD600 of 1.0, and then the culture was divided into three parts. Each of the cultures was treated with 0, 1, or 3 mM sulfite. An arrow indicates the time point at which sulfite was added to the cultures.
Excessive activity of phosphoadenosine phosphosulfate reductase activates Spx in the ΔcysL mutant.
I hypothesized that the biofilm formation-defective phenotype of the ΔcysL mutant might be caused by the action of enzymes for sulfite synthesis rather than by their reaction products. I focused on phosphoadenosine phosphosulfate reductase (CysH), which catalyzes the most downstream reaction among the enzymes involved in sulfite synthesis (Fig. 2A). Phosphoadenosine phosphosulfate reductase, which catalyzes the reduction of 3′-phosphoadenosine 5-phosphosulfate to sulfite, requires a small protein, called thioredoxin, encoded by trxA (25). The reduced form of thioredoxin (thioredoxin disulfide) serves as an electron donor to phosphoadenosine phosphosulfate reductase. Thioredoxin reductase TrxB reduces oxidized thioredoxin using NADPH and maintains cellular levels of thioredoxin disulfide (25). Because the thioredoxin system contributes to a wide variety of physiological functions as a major thiol-disulfide oxidoreductase, trxA and trxB are essential for growth in B. subtilis (26). I examined the effects of modulating thioredoxin levels on biofilm formation. For this purpose, the trxA gene was placed under the control of the spac promoter, and the resultant conditional mutant Pspac-trxA was grown statically in 2×SGG medium supplemented with various concentrations of IPTG. The Pspac-trxA mutant was able to grow and form biofilms at 10 μM or greater IPTG (Fig. 4A). In contrast, the Pspac-trxA ΔcsyL mutant was able to grow at 10 μM IPTG or more but formed normal biofilms only at 60 to 90 μM IPTG (Fig. 4A). In particular, its pellicles completely lost macroscopic structures, as seen in the pellicles of the ΔcysL mutant, at high concentrations of IPTG (360, 720, and 1,000 μM) (Fig. 4A). Thus, unlike in the wild-type strain, low and high expression of trxA inhibits biofilm formation in the ΔcysL mutant.
FIG 4.
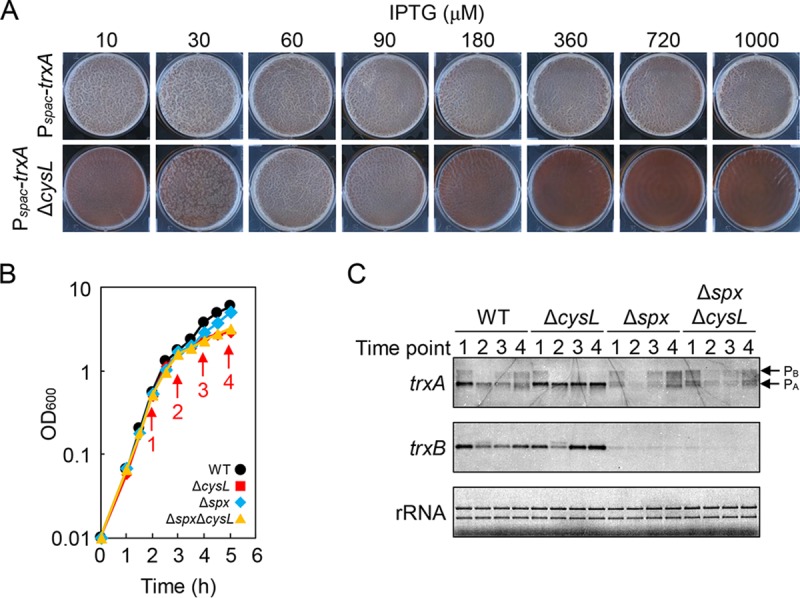
The ΔcysL mutation activates Spx. (A) Low and high levels of trxA inhibit biofilm formation in the ΔcysL mutant. Pspac-trxA and Pspac-trxA ΔcysL mutants were grown at 30°C for 48 h in 2×SGG with the indicated concentrations of IPTG. (B) Growth profiles of ΔcysL and Δspx mutants. Strains were grown in 2×SGG medium with vigorous shaking. Arrows indicate the time points at which samples were taken for RNA isolation. (C) Northern blot analysis of trxA and trxB. Transcripts were detected with gene-specific digoxigenin (DIG)-labeled RNA probes. Time point numbers correspond to the time points shown in panel B. trxA transcripts from PB and PA promoters are indicated by arrows. rRNA stained with methylene blue is shown as a loading control.
I next explored the possibility that transcription of trxA and trxB is altered in the ΔcysL mutant. The wild-type strain and the ΔcysL mutant were grown in 2×SGG medium with vigorous shaking. Cells were taken at 1 h intervals from the mid-exponential phase to the stationary phase, and total RNA was isolated (Fig. 4B). Transcription of trxA and trxB was analyzed by Northern blotting (Fig. 4C). trxA is transcribed from two promoters, PB and PA (26). The upstream promoter PB is induced by the general stress sigma factor σB, and the downstream promoter PA is recognized by the major sigma factor σA. Under my culture conditions, trxA was mainly transcribed from PA, and its transcription levels were almost the same between the wild-type strain and the ΔcysL mutant during the exponential phase (time point 1 in Fig. 4C). However, trxA transcription from PA decreased during the stationary phase in the wild-type strain, whereas it was observed consistently throughout the growth period in the ΔcysL mutant (time points 3 and 4 in Fig. 4C). Thus, trxA transcription levels were higher in the ΔcysL mutant than in the wild-type strain in the stationary phase. Likewise, trxB transcription levels were higher in the ΔcysL mutant than in the wild-type strain in the stationary phase (Fig. 4C). Transcription of trxA from PA and trxB is induced by the transcription factor Spx (27). The effect of the Δspx mutation on trxA and trxB transcription was then examined under the same culture conditions (Fig. 4B). The Δspx mutation drastically reduced transcription of trxA from PA and trxB in both the wild-type strain and the ΔcysL mutant throughout the growth period (Fig. 4C). In the Northern blot of trxA, some faint bands were observed around the position of trxA transcripts from PA in Δspx and Δspx ΔcysL mutants. However, those transcripts seemed to be degradative products of trxA transcripts from PB because (i) the size of those transcripts was slightly larger or smaller than that of trxA transcripts from PA, and (ii) the intensity of those transcripts was proportional to that of trxA transcripts from PB. These results indicate that Spx regulates transcription of trxA from PA and trxB in both the wild-type strain and in the ΔcysL mutant. Unlike in the wild-type strain, Spx activity does not decrease during the stationary phase and continues to induce transcription of trxA and trxB in the ΔcysL mutant.
Spx activates or represses transcription of a large number of genes in response to thiol-specific oxidative (disulfide) stress (27, 28). To determine whether Spx inhibited biofilm formation in the ΔcysL mutant, I tested the ability of the Δspx and Δspx ΔcysL mutants to form biofilms. The Δspx mutant formed biofilms, but its pellicles were slightly thinner than pellicles formed by the wild-type strain (Fig. 5A). The Δspx ΔcysL double mutant also formed slightly thin pellicles, as observed in the Δspx mutant. Thus, the Δspx mutation largely restored biofilm formation to the ΔcysL mutant. I tested the biofilm formation ability of the ΔyjbH mutant, which has high levels of Spx (47). The ΔyjbH mutant formed only very thin pellicles, and this phenotype was suppressed by the Δspx mutation (Fig. 5A). These results indicate that activated or increased Spx inhibits biofilm formation. Based on these results, I concluded that the ΔcysL mutation leads to activation of Spx, causing the biofilm formation-defective phenotype. I propose that excessive activity of phosphoadenosine phosphosulfate reductase (CysH) induces disulfide stress and then activates Spx in the ΔcysL mutant as in the following description. Cysteine biosynthesis genes, including the cysH operon, are expected to be induced in the ΔcysL mutant in 2×SGG medium, as cysteine levels in the medium decrease with growth (29). However, the induction of cysteine biosynthesis genes does not bring about cysteine synthesis in the ΔcysL mutant. Consequently, phosphoadenosine phosphosulfate reductase continues to work and consume thioredoxin disulfide. Since thioredoxin disulfide maintains intracellular thiol-disulfide redox homeostasis and reduces abnormal disulfide bonds, the consumption of significant amounts of thioredoxin disulfide or a shortage of thioredoxin disulfide could have great impacts on intracellular thiol-disulfide redox balance and induce disulfide stress, which activates Spx.
FIG 5.
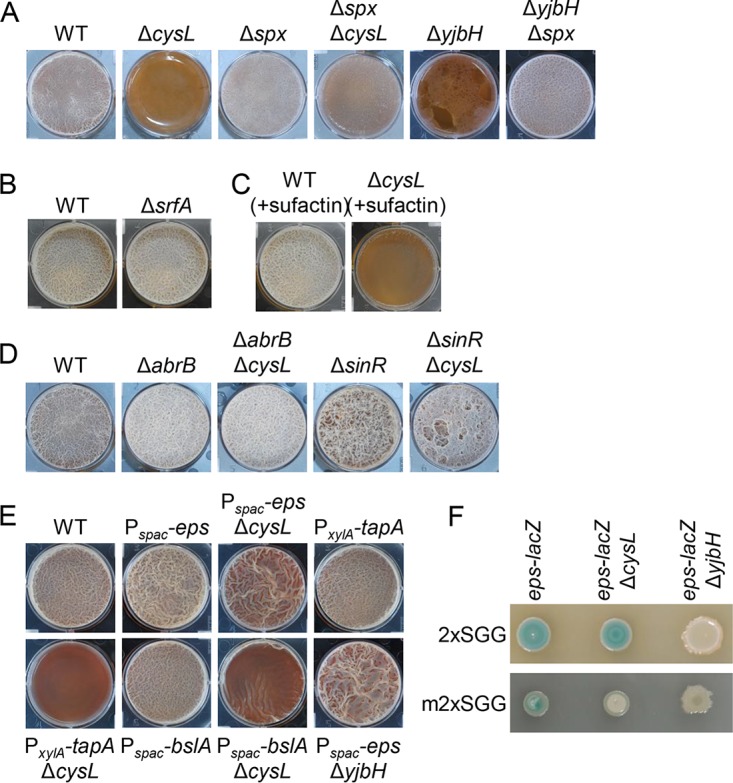
ΔcysL mutation inhibits expression of biofilm matrix genes. (A) Biofilm formation of Δspx and ΔyjbH mutants. (B) The ΔsrfA mutation has little or no effect on biofilm formation in in 2×SGG. (C) Surfactin does not restore biofilm formation to the ΔcysL mutant. The wild-type strain and ΔcysL mutants were grown in 2×SGG medium with 0.1 μg/ml surfactin. (D) ΔabrB and ΔsinR mutations restore biofilm formation to the ΔcysL mutant. (E) Effects of the artificial expression of biofilm matrix genes on biofilm formation of the ΔcysL mutant. Pspac-eps operon and Pspac-bslA strains were grown in 2×SGG medium with 1 mM IPTG. (F) Expression of the eps operon. epsH::pMutin strains harboring the eps-lacZ reporter were grown for 24 h on 2×SGG medium or modified 2×SGG (m2×SGG) medium with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). m2×SGG medium contains four times less nutrient broth than does 2×SGG medium. All photographs of biofilms were taken 48 h after inoculation.
Spx negatively regulates biofilm matrix genes.
I investigated which critical genes for biofilm formation were inhibited by the ΔcysL mutation or activated Spx. Spx is a global regulator, but little is known about its function in B. subtilis biofilm formation (27, 30). One of the known targets of Spx is the srfA operon; Spx inhibits its transcription (27). The srfA operon is required for the production of the lipopeptide antibiotic surfactin, which serves as a self-produced trigger for biofilm formation and induces biofilm matrix genes, the eps operon and the tapA operon (15). However, the deletion of the srfA operon had little or no effect on biofilm formation in 2×SGG medium (Fig. 5B), and the addition of surfactin to medium did not restore biofilm formation to the ΔcysL mutant (Fig. 5C), indicating that Spx may inhibit other important genes for biofilm formation.
Two transcriptional repressors, AbrB and SinR, negatively regulate biofilm formation in B. subtilis (11–14). Double mutants ΔabrB ΔcysL and ΔsinR ΔcysL formed thick pellicles in standing culture, as observed for ΔabrB and ΔsinR mutants (Fig. 5D), showing that both ΔabrB and ΔsinR mutations can suppress the ΔcysL mutation in biofilm formation. Since these repressors directly and indirectly repress biofilm matrix genes, the eps operon, the tapA operon, and bslA (11–14), I examined whether induction of one of these biofilm matrix genes restored biofilm formation to the ΔcysL mutant. For this purpose, the IPTG-inducible spac promoter was inserted upstream of the eps operon and bslA, and a constitutive xylA promoter from Bacillus megaterium (31) was inserted upstream of the tapA operon. These constructs, the Pspac-eps operon, the PxylA-tapA operon, and Pspac-bslA, were transferred to the ΔcysL mutant. Induction of the eps operon largely restored biofilm formation to the ΔcysL mutant, while induction of the tapA operon or bslA did not (Fig. 5E). Moreover, induction of the eps operon restored biofilm formation to the ΔyjbH mutant (Fig. 5E). To determine whether ΔcysL and ΔyjbH mutations reduced expression of the eps operon, expression of the eps operon was analyzed using the ΔepsH::pMutin mutant harboring the lacZ reporter within epsH. The ΔepsH::pMutin mutant formed colonies with clear blue pigmentation on 2×SGG solid medium supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), whereas the ΔepsH::pMutin ΔyjbH mutant formed white colonies (Fig. 5F). However, in contrast to the above results, the ΔepsH::pMutin ΔcysL mutant formed blue colonies similar to those of the ΔepsH::pMutin mutant. Since the ΔcysL mutant phenotype appeared after cysteine levels in medium decreased with growth, I suspected that the ΔcysL mutant might form blue colonies before this decrease. Accordingly, eps-lacZ activity was examined on a modified 2×SGG (m2×SGG) medium, which contained four times less nutrient broth as a nitrogen source than 2×SGG medium. On the modified 2×SGG medium, the ΔepsH::pMutin mutant formed pale blue colonies, whereas the ΔepsH::pMutin ΔcysH mutant and the ΔepsH::pMutin ΔyjbH mutant formed white colonies (Fig. 5F). These results indicate that the eps operon is the primary target of Spx in biofilm formation, although my results did not show whether Spx directly represses the eps operon.
The effects of disulfide stress on biofilm formation.
My finding that the disulfide stress response regulator Spx inhibited biofilm formation suggested the possibility that inducing biofilm formation is detrimental to cells during disulfide stress. If this were the case, biofilm-inducing conditions or overexpression of the eps operon should adversely affect resistance to disulfide stress. To examine this possibility, stationary-phase cultures of the wild-type strain and the Pspac-eps operon mutant in LB medium were 10-fold serially diluted, and the dilutions were spotted onto biofilm formation medium (2×SGG) or non-biofilm formation medium (LB) supplemented with IPTG and diamide, the latter of which is a thiol-specific oxidant used to induce disulfide stress (Fig. 6A). I did not observe a significant difference between the two media or between the two strains in diamide resistance. Thus, inducing biofilm formation appears not to have a negative effect on diamide resistance. At present, I cannot answer the question of why biofilm formation is inhibited by Spx in response to disulfide stress.
FIG 6.
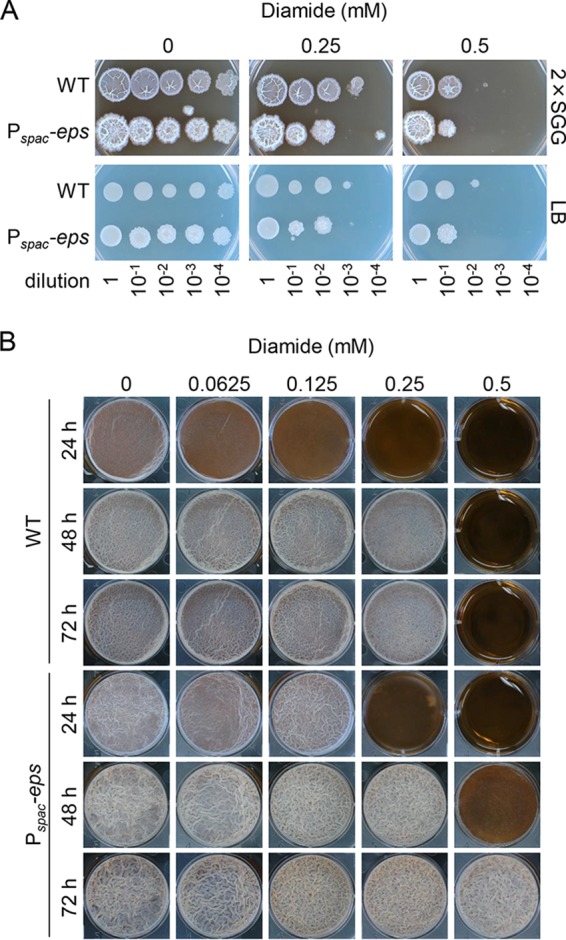
The effects of diamide on biofilm formation. (A) Diamide resistance. Stationary-phase cultures of the wild-type strain and the Pspac-eps operon mutant in LB were serially diluted 10-fold, and 2-μl aliquots of the indicated dilutions were spotted on 2×SGG or LB solid medium supplemented with 1 mM IPTG. (B) Biofilm formation in the presence of diamide. Stationary-phase cultures of the wild-type strain and the Pspac-eps operon mutant were diluted 10-fold, and 10 μl of the dilutions was added to each well containing 10 ml of 2×SGG medium with 1 mM IPTG and the indicated concentrations of diamide.
I further examined whether overexpression of the eps operon led to disulfide stress-resistant biofilm formation. The wild-type strain and the Pspac-eps operon mutant were statically grown in 2×SGG medium supplemented with IPTG and diamide (Fig. 6B). The addition of diamide to medium delayed or inhibited biofilm formation of the wild-type strain as its concentration increased. Specifically, a slight delay was observed even in the presence of a low concentration (0.0625 mM) of diamide, and its biofilm formation was completely inhibited for 72 h by 0.5 mM diamide. Low concentrations of diamide did not affect biofilm formation of the Pspac-eps operon mutant. The Pspac-eps operon mutant formed biofilms even in the presence of 0.5 mM diamide. These observations supported my findings that Spx inhibits biofilm formation through repressing the eps operon.
Concluding remarks.
In this study, I investigated why the ΔcysL mutant was defective in biofilm formation. I showed that the ΔcysL mutation resulted in the overworking of phosphoadenosine phosphosulfate reductase and thereby induced disulfide stress, leading to the activation of Spx. Spx was previously shown to negatively regulate cysteine biosynthesis genes in B. subtilis (32, 33). Moreover, since B. subtilis has a large cysteine pool (34), cysteine biosynthesis is expected to be highly active in B. subtilis. Taken together, my findings suggest that the negative regulation of cysteine biosynthesis by Spx plays a vital role in coordinating cysteine biosynthesis, a potential producer of disulfide stress, with the cellular redox state. I demonstrated that Spx negatively regulates biofilm formation by repressing the eps operon in B. subtilis. Spx is highly conserved among low-G+C-content Gram-positive bacteria or members of the Firmicutes (35). The negative regulation of biofilm formation by Spx has also been reported in several pathogens of Firmicutes, including Staphylococcus aureus (36), Staphylococcus epidermidis (37), and Streptococcus mutans (38). In these pathogens, Spx also inhibits transcription of biofilm matrix genes, despite the fact that these bacteria have different biofilm matrix genes with different regulatory systems from those of B. subtilis (36, 37). Bacterial pathogens have evolved to adapt to their host environment and robustly respond to reactive oxygen species (ROS) generated by host immune cells to kill invading pathogens (38). In many pathogens of Firmicutes, Spx has also evolved to play a critical role in virulence and survival within the host (39–44). However, my findings indicate that the negative regulation of biofilm formation by Spx is not specific to those pathogens but widespread among many Firmicutes species. Drugs that induce Spx may offer a promising new method for controlling biofilm formation in Firmicutes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. subtilis strain NCIB3610 and its derivatives used in this study are listed in Table 1. Construction of B. subtilis mutants is described in the supplemental material. Primers used for strain construction are listed in Table S1 in the supplemental material. B. subtilis strains were grown in LB (LB Lennox; BD Difco, Franklin Lakes, NJ), 2×SGG (17), or SMM (45). Cysteine was added to medium at a final concentration of 100 μg/ml where indicated. For the biofilm formation test, a fresh single colony of tested strains was inoculated into 10 ml of 2×SGG medium in a well of a 6-well plate and cultured at 30°C for 48 h without agitation. Escherichia coli strains HB101 and JM105 were used for construction and maintenance of plasmids. Sodium sulfite and surfactin were obtained from Fuji Wako Pure Chemicals (Osaka, Japan), and diamide was obtained from Sigma-Aldrich (St. Louis, MO).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant feature(s) | Reference or constructiona |
|---|---|---|
| NCIB3610 | Prototroph | 5 |
| NTF234 | ΔcysL::cat | WTF237 (17)→ NCIB3610 |
| N502 | ΔcysI::pMutin (erm) | YVGRd (48)→NCIB3610 |
| N503 | ΔcysH::pMutin (erm) | CYSHd (48)→NCIB3610 |
| N504 | ΔcysP::pMutin (erm) | CYSPd (48)→NCIB3610 |
| N505 | Δsat::pMutin (erm) | SATd (48)→NCIB3610 |
| N506 | ΔcysC::pMutin (erm) | CYSCd (48)→NCIB3610 |
| N507 | ΔylnD::pMutin (erm) | YLNDd (48)→NCIB3610 |
| N508 | ΔylnE::pMutin (erm) | YLNEd (48)→NCIB3610 |
| N509 | ΔylnF::pMutin (erm) | YLNFd (48)→NCIB3610 |
| N510 | ΔcysH::pMutin (erm) ΔcysL::cat | N503→NTF234 |
| N511 | ΔcysP::pMutin (erm) ΔcysL::cat | N504→NTF234 |
| N512 | Δsat::pMutin (erm) ΔcysL::cat | N505→NTF234 |
| N513 | ΔcysC::pMutin (erm) ΔcysL::cat | N506→NTF234 |
| N514 | ΔylnD::pMutin (erm) ΔcysL::cat | N507→NTF234 |
| N515 | ΔylnE::pMutin (erm) ΔcysL::cat | N508→NTF234 |
| N515 | ΔylnF::pMutin (erm) ΔcysL::cat | N509→NTF234 |
| N519 | trxA:pMutinNC (Pspac-trxA erm) | TRXAp (48)→NCIB361 |
| N520 | trxA:pMutinNC (Pspac-trxA erm) ΔcysL::cat | N519→NTF234 |
| N523 | Δspx::kan | This study |
| N524 | Δspx::kan ΔcysL::cat | N520→NTF234 |
| N744 | ΔyjbH::pMutinNC (erm) | This study |
| N751 | ΔyjbH::pMutinNC (erm) Δspx::kan | N744→N523 |
| N743 | ΔyjbH::kan | This study |
| N440 | ΔsrfA operon | This study |
| NTF2 | ΔabrB::kan | WTF2 (17)→NCIB3610 |
| N552 | ΔabrB::kan ΔcysL::cat | NTF2→NTF234 |
| NTF92 | ΔsinR::cat | WTF92 (17)→NCIB3610 |
| N553 | ΔsinR::kan ΔcysL::cat | NTF92→NTF234 |
| N901 | epsA::pMutinT3-hy (Pspac-hy-epsABCDEFGHIJKLMNO erm) | This study |
| N902 | epsA::pMutinT3-hy (Pspac-hy-epsABCDEFGHIJKLMNO erm) ΔcysL::cat | NTF234→N901 |
| N903 | epsA::pMutinT3-hy (Pspac-hy-epsABCDEFGHIJKLMNO erm) ΔyjbH::kan | N743→N901 |
| N963 | tapA::pCAxylAtapA (PxylA-tapA-sipW-tasA spc) | This study |
| N964 | tapA::pCAxylAtapA (PxylA-tapA-sipW-tasA spc) ΔcysL::cat | NTF234→N963 |
| N904 | bslA::pMutinT3-hy (Pspac-hy-bslA tet) | This study |
| N905 | bslA::pMutinT3-hy (Pspac-hy-bslA tet) ΔcysL::cat | NTF234→N904 |
| N978 | ΔepsH::pMutin (epsA-H-lacZ erm) | YVERd (48) →NCIB3610 |
| N979 | ΔepsH::pMutin (epsA-H-lacZ erm) ΔcysL::cat | NTF234→N978 |
| N980 | ΔepsH::pMutin (epsA-H-lacZ erm) ΔyjbH::kan | N743→N978 |
Arrows indicate B. subtilis transformation, donor strain → recipient strain.
Northern blot analysis.
RNA samples were prepared from cells grown at 37°C in 2×SGG medium with vigorous shaking, as described previously (46). Northern blot analysis was carried out as described previously (46). Primers trxA-N-F, trxA-N-T7R, trxB-N-F, and trxB-N-T7R were used for RNA probe synthesis. Primer sequences are shown in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
I acknowledge Hisaji Maki and members of the Maki lab for their support.
This work was supported in part by JSPS KAKENHI (grant 17K07721).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00712-18.
REFERENCES
- 1.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 4.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 7.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi K, Iwano M. 2012. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 10.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol 52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 13.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 14.Verhamme DT, Murray EJ, Stanley-Wall NR. 2009. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J Bacteriol 191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mielich-Süss B, Lopez D. 2015. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. doi: 10.1111/1462-2920.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi K. 2007. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol 189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillouard I, Auger S, Hullo MF, Chetouani F, Danchin A, Martin-Verstraete I. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J Bacteriol 184:4681–4689. doi: 10.1128/JB.184.17.4681-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Ploeg JR, Barone M, Leisinger T. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol Lett 201:29–35. doi: 10.1111/j.1574-6968.2001.tb10728.x. [DOI] [PubMed] [Google Scholar]
- 20.Mansilla MC, Albanesi D, de Mendoza D. 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. J Bacteriol 182:5885–5892. doi: 10.1128/JB.182.20.5885-5892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansilla MC, de Mendoza D. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiol 146:815–821. doi: 10.1099/00221287-146-4-815. [DOI] [PubMed] [Google Scholar]
- 22.Johansson P, Hederstedt L. 1999. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiol 145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- 23.Raux E, Leech HK, Beck R, Schubert HL, Santander PJ, Roessner CA, Scott AI, Martens JH, Jahn D, Thermes C, Rambach A, Warren MJ. 2003. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem J 370:505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiol 144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Holmgren A. 2014. The thioredoxin antioxidant system. Free Radic Biol Med 66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 26.Scharf C, Riethdorf S, Ernst H, Engelmann S, Völker U, Hecker M. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J Bacteriol 180:1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano S, Küster-Schöck E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A 100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano S, Erwin KN, Ralle M, Zuber P. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol 55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I. 2008. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem 283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 30.Rochat T, Nicolas P, Delumeau O, Rabatinová A, Korelusová J, Leduc A, Bessières P, Dervyn E, Krásny L, Noirot P. 2012. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res 40:9571–9583. doi: 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhavsar AP, Zhao X, Brown ED. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol 67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erwin KN, Nakano S, Zuber P. 2005. Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx. J Bacteriol 187:4042–4049. doi: 10.1128/JB.187.12.4042-4049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi SY, Reyes D, Leelakriangsak M, Zuber P. 2006. The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis. J Bacteriol 188:5741–57451. doi: 10.1128/JB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol 178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol 186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol 188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Fan J, Niu C, Wang C, Villaruz AE, Otto M, Gao Q. 2010. Role of spx in biofilm formation of Staphylococcus epidermidis. FEMS Immunol Med Microbiol 59:152–160. doi: 10.1111/j.1574-695X.2010.00673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reniere ML. 2018. Reduce, induce, thrive: bacterial redox sensing during pathogenesis. J Bacteriol 200:e00128-18. doi: 10.1128/JB.00128-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kajfasz JK, Martinez AR, Rivera-Ramos I, Abranches J, Koo H, Quivey RG Jr, Lemos JA. 2009. Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J Bacteriol 191:2060–2068. doi: 10.1128/JB.01609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajfasz JK, Mendoza JE, Gaca AO, Miller JH, Koselny KA, Giambiagi-Demarval M, Wellington M, Abranches J, Lemos JA. 2012. The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect Immun 80:2265–2275. doi: 10.1128/IAI.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng C, Xu J, Li J, Hu L, Xia J, Fan J, Guo W, Chen H, Bei W. 2014. Two Spx regulators modulate stress tolerance and virulence in Streptococcus suis serotype 2. PLoS One 9:e108197. doi: 10.1371/journal.pone.0108197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Ge X, Wang X, Patel JR, Xu P. 2012. SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One 7:e40034. doi: 10.1371/journal.pone.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteley AT, Ruhland BR, Edrozo MB, Reniere ML. 2017. A redox-responsive transcription factor is critical for pathogenesis and aerobic growth of Listeria monocytogenes. Infect Immun 85:e00978-16. doi: 10.1128/IAI.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci U S A 44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 47.Larsson JT, Rogstam A, von Wachenfeldt C. 2007. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol Microbiol 66:669–684. doi: 10.1111/j.1365-2958.2007.05949.x. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Débarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauël C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Séror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. 2003. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.