The global regulator H-NS represses the expression of acquired genes and thus avoids possible detrimental effects on bacterial fitness. Regulatory mechanisms are adapted to induce expression of the acquired genes in particular niches to obtain a benefit from the information encoded in the foreign DNA, as for pathogenesis. Here, we show two mechanisms that were integrated for the expression of virulence genes in Salmonella Typhimurium. One involves the additive action of the regulators SlyA and HilD, whereas the other involves SlyA, but not HilD, to counteract H-NS-mediated repression on the ssrAB operon, thus favoring its activation by the OmpR regulator. To our knowledge, this is the first report involving the coordinated action of two regulators to counteract H-NS-mediated repression.
KEYWORDS: H-NS, HilD, OmpR, SPI, Salmonella, SlyA, transcriptional regulation, ssrAB
ABSTRACT
H-NS-mediated repression of acquired genes and the subsequent adaptation of regulatory mechanisms that counteract this repression have played a central role in the Salmonella pathogenicity evolution. The Salmonella pathogenicity island 2 (SPI-2) is an acquired chromosomal region containing genes necessary for Salmonella enterica to colonize and replicate in different niches of hosts. The ssrAB operon, located in SPI-2, encodes the two-component system SsrA-SsrB, which positively controls the expression of the SPI-2 genes but also other many genes located outside SPI-2. Several regulators have been involved in the expression of ssrAB, such as the ancestral regulators SlyA and OmpR, and the acquired regulator HilD. In this study, we show how SlyA, HilD, and OmpR coordinate to induce the expression of ssrAB under different growth conditions. We found that when Salmonella enterica serovar Typhimurium is grown in nutrient-rich lysogeny broth (LB), SlyA and HilD additively counteract H-NS-mediated repression on ssrAB, whereas in N-minimal medium (N-MM), SlyA antagonizes H-NS-mediated repression on ssrAB independently of HilD. Interestingly, our results indicate that OmpR is required for the expression of ssrAB independently of the growth conditions, even in the absence of repression by H-NS. Therefore, our data support two mechanisms adapted for the expression of ssrAB under different growth conditions. One involves the additive action of SlyA and HilD, whereas the other involves SlyA, but not HilD, to counteract H-NS-mediated repression on ssrAB, thus favoring in both cases the activation of ssrAB by OmpR.
IMPORTANCE The global regulator H-NS represses the expression of acquired genes and thus avoids possible detrimental effects on bacterial fitness. Regulatory mechanisms are adapted to induce expression of the acquired genes in particular niches to obtain a benefit from the information encoded in the foreign DNA, as for pathogenesis. Here, we show two mechanisms that were integrated for the expression of virulence genes in Salmonella Typhimurium. One involves the additive action of the regulators SlyA and HilD, whereas the other involves SlyA, but not HilD, to counteract H-NS-mediated repression on the ssrAB operon, thus favoring its activation by the OmpR regulator. To our knowledge, this is the first report involving the coordinated action of two regulators to counteract H-NS-mediated repression.
INTRODUCTION
Salmonella enterica serovar Typhimurium, a facultative intracellular pathogen, generally causes mild self-limiting gastroenteritis in humans and several animals, but it can also produce severe systemic infections in different hosts, including humans (1, 2). Thus, S. Typhimurium has been extensively used as a model for studying the molecular mechanisms governing S. enterica virulence (1, 3, 4).
Around one-quarter of the S. Typhimurium genome was shaped by the gain of DNA through several horizontal gene transfer (HGT) events that occurred at different evolutionary times (5). The Salmonella pathogenicity island 2 (SPI-2) is an acquired chromosomal region containing 44 genes which encode a type III secretion system (T3SS-2), their chaperones and effector proteins, and the two-component system SsrA-SsrB (4). The SPI-2 genes are required for survival and replication of S. Typhimurium inside host cells, such as macrophages, which leads to the systemic disease (1, 4). Additionally, the SPI-2 genes contribute to the induction of the intestinal inflammatory response (6–8). Consistently, the SPI-2 genes are expressed when S. Typhimurium is within host cells and in the intestinal lumen (9–15). In vitro, the expression of SPI-2 is induced when S. Typhimurium is grown in minimal medium containing low concentrations of phosphate, calcium, and magnesium (11, 16, 17), as well as during the late-stationary phase of growth in nutrient-rich medium (18, 19), conditions that somehow resemble the intracellular and the intestinal lumen environments, respectively.
The ssrA and ssrB genes, located in SPI-2, code for the two-component system SsrA-SsrB, with SsrA being the sensor kinase and SsrB the response regulator, which directly induces the expression of the SPI-2 genes and many other virulence genes located outside SPI-2 (4, 20–22). The ssrA and ssrB genes are transcribed as an operon (18); also, transcription of ssrB independent of ssrA has been reported (17, 23), which seems to be dependent on the growth conditions tested. To simplify, these genes are referred to here as the ssrAB operon. Multiple regulators have been involved in the expression of ssrAB, including SlyA, HilD, OmpR, PhoP, and SsrB, which act positively and directly on this operon (4, 23–29). SlyA, OmpR, and PhoP are transcriptional regulators that control the expression of a large number of genes, encoding distinct cellular functions in different bacteria; SlyA is a member of the MarR family of transcriptional factors, whereas OmpR and PhoP are the response regulators of the two-component systems EnvZ-OmpR and PhoQ-PhoP, respectively (24, 27, 30–40). HilD is an AraC-like transcriptional regulator, present only in Salmonella spp. and encoded in the Salmonella pathogenicity island 1 (SPI-1), which controls the expression of the SPI-1 genes and many other virulence genes (4, 18, 19, 41–51). On the other hand, the expression of ssrAB is also controlled by negative regulators, such as the nucleoid-associated protein H-NS (18, 19, 52–54), which acts as a global transcriptional factor in many bacteria (55, 56). H-NS is considered a genome sentinel that has played an important role during the evolution of Salmonella pathogenicity by preventing uncontrolled expression of acquired DNA that could be deleterious to bacterial fitness (52, 53, 56). Different regulatory proteins have been adapted to antagonize H-NS-mediated repression in specific promoters, which allows the expression of acquired genes only under those conditions where the encoded information is beneficial for bacteria, as in particular niches during infection of a host (57, 58). For instance, HilD induces the expression of ssrAB by directly displacing H-NS-mediated repression on the promoter upstream of ssrA under in vitro growth conditions that resemble the intestinal environment (18, 19).
In this work, we determine how the ancestral regulators SlyA and OmpR and the acquired regulator HilD induce the expression of the S. Typhimurium ssrAB operon under different growth conditions. During growth in nutrient-rich lysogeny broth (LB), SlyA and HilD additively counteract H-NS-mediated repression on ssrAB, whereas during growth in N-minimal medium (N-MM), SlyA antagonizes the H-NS-mediated repression on ssrAB independently of HilD. In both cases, the expression of ssrAB also requires the action of OmpR, even in the absence of the repression by H-NS.
RESULTS
SlyA is required for the expression of ssrAB under both rich and minimal growth conditions.
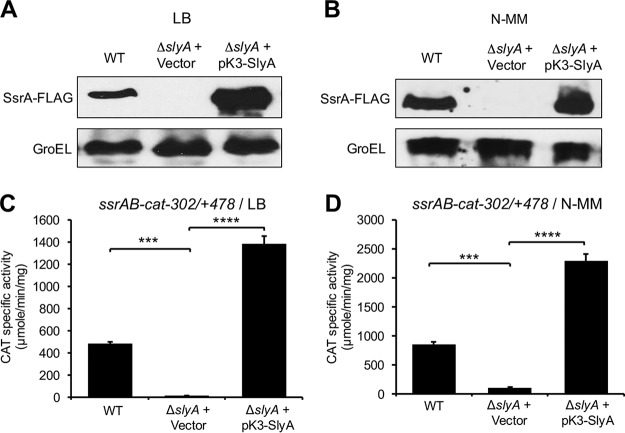
The ssrAB operon, and thus the SPI-2 genes, are expressed when S. Typhimurium is grown in nutrient-rich media, such as LB, as well as in minimal media like N-MM (18, 19), which mimic different niches that S. Typhimurium finds in its hosts. Several studies have shown that SlyA is required for the expression of ssrAB in minimal medium (42, 59). Additionally, some reports indicate that overexpression of SlyA induces the expression of ssrAB in LB (25, 28). To confirm whether physiological levels of SlyA regulate the expression of ssrAB in LB, we examined the chromosomal expression of the C-terminal 3×FLAG-tagged SsrA protein (SsrA-FLAG) in the wild-type (WT) S. Typhimurium strain SL1344 and its isogenic ΔslyA mutant, grown in LB at 37°C. As a control, the expression of SsrA-FLAG was also analyzed in these same strains grown in N-MM. In both LB and N-MM, SsrA-FLAG was detected in the WT strain but not in the ΔslyA mutant; as expected, the pK3-SlyA plasmid expressing SlyA, but not the control vector pMPM-K3, restored the expression of SsrA-FLAG in the ΔslyA mutant at levels even higher than those reached in the WT strain (Fig. 1A and B). To confirm these results, similar assays were performed by analyzing the expression of the ssrAB-cat−302/+478 transcriptional fusion (ssrAB sequence from −302 to +478 and the cat reporter gene) that we have successfully used before (19), which carries the promoter upstream of ssrA. In agreement with our results obtained by assessing SsrA-FLAG, the activity of the ssrAB-cat−302/+478 fusion was drastically reduced in the ΔslyA mutant with respect to the WT strain, and it was induced in the presence of the pK3-SlyA plasmid (Fig. 1C and D). Taken together, these results show that SlyA is required for the expression of ssrAB under different growth conditions.
FIG 1.
SlyA is required for expression of the ssrAB operon in LB and N-MM. (A and B) Expression of SsrA-FLAG in WT S. Typhimurium and its ΔslyA derivative mutant containing the pMPM-K3 vector or the pK3-SlyA plasmid that expresses SlyA from a constitutive promoter was analyzed by Western blotting using monoclonal anti-FLAG antibodies. Whole-cell lysates were prepared from samples of bacterial cultures grown for 9 h in LB (A) or for 16 h in N-MM at pH 7.4 (B) at 37°C. As a control, the expression of GroEL was also determined using polyclonal anti-GroEL antibodies. (C and D) Expression of the ssrAB-cat−302/+478 transcriptional fusion carried by the pssrAB-cat-302/+478 plasmid was tested in WT S. Typhimurium and its isogenic ΔslyA mutant containing the pMPM-K3 vector or the pK3-SlyA plasmid. CAT-specific activity was determined from samples of bacterial cultures grown for 9 h in LB (C) or for 16 h in N-MM at pH 7.4 (D) at 37°C. Data represent the mean with standard deviation from the results from three independent experiments performed in duplicate. Statistically different values are indicated (***, P < 0.001; ****, P < 0.0001).
SlyA counteracts repression exerted by H-NS on ssrAB during growth in LB.
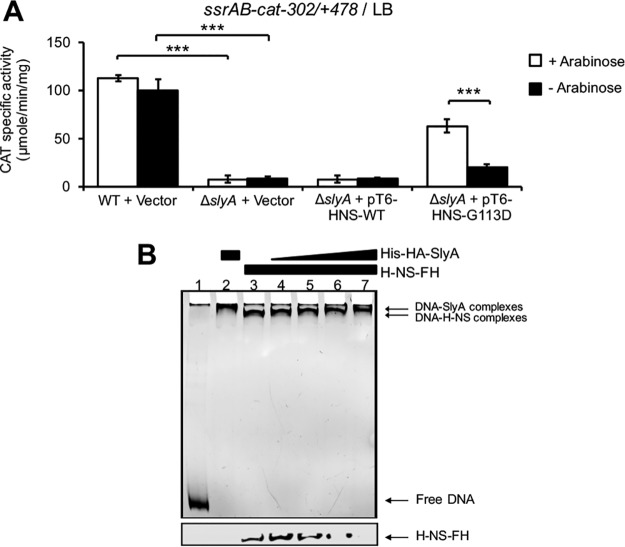
Previous studies indicate that SlyA induces gene expression mainly by counteracting the repression exerted by H-NS on target promoters (33, 39). Furthermore, we have shown that H-NS directly represses the expression of ssrAB when S. Typhimurium is grown in LB (18, 19). Therefore, in order to determine if SlyA counteracts H-NS-mediated repression on ssrAB, during growth in LB, we tested whether SlyA is required for the expression of ssrAB in the absence of H-NS activity. The S. Typhimurium Δhns mutant shows severe growth defects (53); however, activity of WT H-NS can be inactivated by overexpressing the H-NSG113D mutant, which does not have DNA binding activity but still forms heterodimers with WT H-NS monomers (60) and thus acts as a dominant negative mutant (61). In this way, the expression of the ssrAB-cat−302/+478 fusion was assessed in the WT S. Typhimurium strain and its derivative ΔslyA mutant containing the empty vector pMPM-T6Ω, or the pT6-HNS-WT or pT6-HNS-G113D plasmids, which express WT H-NS and the H-NSG113D mutant, respectively, from an arabinose-inducible promoter (44, 61). The strains were grown in LB at 37°C in the presence or absence of 0.1% arabinose to induce or not induce the expression of the H-NS proteins. As shown in Fig. 2A, overexpression of H-NSG113D, but not WT H-NS, induced the activity of the ssrAB-cat−302/+478 fusion in the ΔslyA mutant strain, indicating that the overproduction of the dominant negative H-NS mutant partially suppresses the need of SlyA for the expression of ssrAB. In similar experiments, we previously showed that overproduction of the H-NSQ92am dominant negative mutant also suppresses the need of HilD for the expression of ssrAB (19).
FIG 2.
SlyA directly displaces H-NS-mediated repression on ssrAB during growth in LB. (A) Expression of the ssrAB-cat−302/+478 transcriptional fusion carried by the pssrAB-cat-302/+478 plasmid was tested in WT S. Typhimurium and its isogenic ΔslyA mutant containing or not containing the pMPM-T6Ω vector or the pT6-HNS-WT or pT6-HNS-G113D plasmid, which expresses WT H-NS or the dominant negative H-NSG113D mutant, respectively, from an arabinose-inducible promoter. CAT-specific activity was determined from samples of bacterial cultures grown for 9 h in LB at 37°C. l-Arabinose (0.1%) was added (+) or not added (−) to the medium for inducing the expression of WT H-NS and H-NSG113D from pT6-HNS-WT and pT6-HNS-G113D, respectively. Data represent the mean with standard deviation of the results from three independent experiments performed in duplicate. Statistically different values are indicated (***, P < 0.001). (B) Competitive nonradioactive EMSAs between H-NS and SlyA on the −302/+478 region of ssrAB. Purified H-NS−FH protein was added at 0.5 μM (lanes 3 to 7), and purified His-HA-SlyA protein was added at 1, 2, 2.5, and 3 μM (lanes 4 to 7, respectively). No proteins were added in lane 1, and His-HA-SlyA was added at 3 μM in lane 2. The DNA-protein complexes were resolved in a nondenaturing 6% polyacrylamide gel. Top, protein-DNA complexes stained with ethidium bromide; bottom, immunoblot detection of H-NS−FH from the DNA-protein complexes. Similar results were obtained from three different experiments.
To investigate if SlyA counteracts directly the repression of H-NS on ssrAB, we performed competitive electrophoretic mobility shift assays (EMSAs) to examine the effect of SlyA on H-NS bound to the region −302/+478 of ssrAB. A DNA fragment spanning this region was first incubated with a constant concentration of purified H-NS-FLAG-His (H-NS−FH) protein, and then increasing amounts of purified His-HA-SlyA protein were added. The DNA-H-NS and DNA-SlyA complexes were detected by staining the DNA fragments. Furthermore, the presence of H-NS−FH or His-HA-SlyA on these complexes was monitored by immunoblotting with anti-FLAG and anti-HA antibodies, respectively. As shown in Fig. 2B, increasing amounts of His-HA-SlyA shifted the DNA-H-NS−FH complex to a slower-migrating complex, similar to that formed by only His-HA-SlyA; additionally, the immunoblots indicated that increasing amounts of His-HA-SlyA correlate with decreasing amounts of H-NS−FH bound to the DNA fragments. We were unable to detect His-HA-SlyA bound to the DNA fragments, probably due to a low sensitivity of the anti-HA antibodies used (data not shown).
In all, these results show that SlyA counteracts H-NS-mediated repression on ssrAB by directly displacing H-NS from the region −302/+478.
SlyA and HilD additively antagonize repression of H-NS on ssrAB during growth in LB.
In previous studies, we demonstrated that HilD also induces the expression of ssrAB during the growth of S. Typhimurium in LB by antagonizing H-NS-mediated repression (18, 19). To confirm that both SlyA and HilD act as antirepressors of H-NS to induce the expression of ssrAB, we analyzed if the inactivation of H-NS leads to the expression of ssrAB independently of both SlyA and HilD. For this, we determined the expression of the ssrAB-cat−302/+478 fusion in the WT S. Typhimurium strain and its derivative ΔhilD ΔslyA double mutant containing the empty vector pMPM-T6Ω or the plasmids pT6-HNS-WT or pT6-HNS-G113D in cultures grown in LB at 37°C. As could be expected, the activity of the ssrAB-cat−302/+478 fusion was drastically reduced in the ΔhilD ΔslyA mutant; in addition, overexpression of the H-NSG113D dominant negative mutant, but not WT H-NS, restored the activity of the ssrAB-cat−302/+478 fusion in this strain (Fig. 3). These results demonstrate that when the activity of H-NS is inactivated, the expression of ssrAB becomes independent of both SlyA and HilD.
FIG 3.
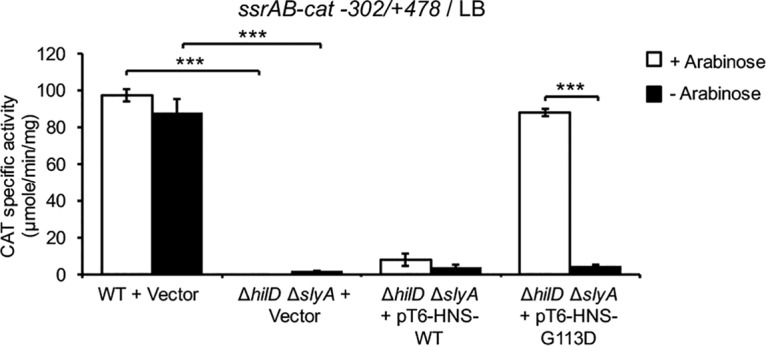
HilD and SlyA are not required for expression of ssrAB during growth in LB when H-NS is inactivated. Expression of the ssrAB-cat−302/+478 transcriptional fusion carried by the pssrAB-cat-302/+478 plasmid was determined in the WT S. Typhimurium strain and its derivative ΔhilD ΔslyA mutant containing the pMPM-T6Ω vector, as well as in the ΔhilD ΔslyA mutant containing the pT6-HNS-WT or pT6-HNS-G113D plasmid, which expresses WT H-NS or the H-NSG113D dominant negative mutant, respectively, from an arabinose-inducible promoter. CAT-specific activity was determined from samples of bacterial cultures grown for 9 h in LB at 37°C. Expression of WT H-NS or H-NSG113D from plasmids pT6-HNS-WT and pT6-HNS-G113D, respectively, was induced by adding 0.1% l-arabinose to the medium (+ arabinose). Data represent the mean with standard deviation of the results from three independent experiments done in duplicate. Statistically different values are indicated (***, P < 0.001).
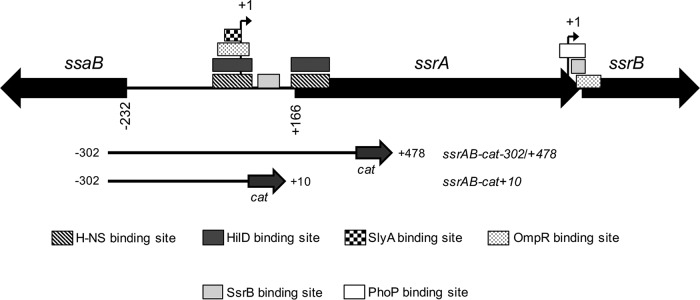
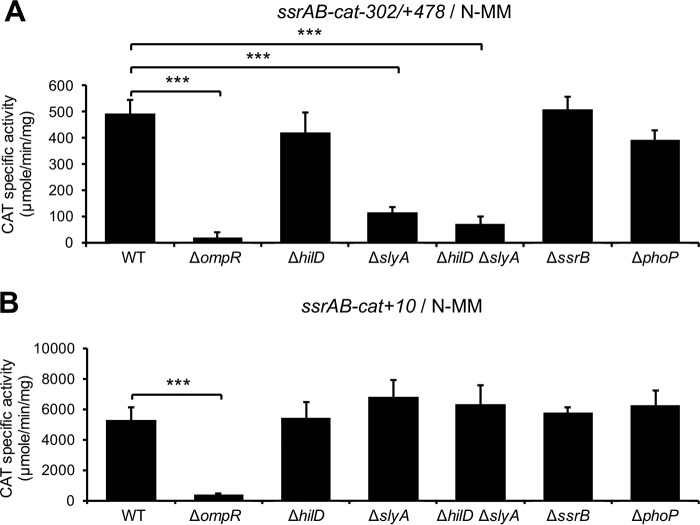
We next analyzed the expression of ssrAB in the absence of each SlyA or HilD, or both, when the repression of ssrAB by H-NS is blocked or not blocked. For this, the activities of the ssrAB-cat−302/+478 fusion and its derivative ssrAB-cat−302/+10 fusion were determined in the WT S. Typhimurium strain and its isogenic ΔhilD, ΔslyA, and ΔhilD ΔslyA mutants grown in LB. The ssrAB-cat−302/+10 fusion lacks H-NS-binding sites required for the repression by H-NS; repression of ssrAB by H-NS involves the H-NS-binding sites located on the promoter as well as those located on the translational start codon of ssrA (Fig. 4) (19). Since OmpR is required for the expression of ssrAB in LB, even in the absence of the H-NS activity (18), a ΔompR mutant was assessed as control in these assays.
FIG 4.
Schematic representation of ssrAB and the ssrAB-cat transcriptional fusions analyzed. The transcriptional start sites (tss) (+1) upstream ssrA and ssrB are indicated by a bent arrow. HilD-, OmpR-, SlyA-, SsrB-, PhoP- and H-NS-binding sites on ssrAB, which have been previously reported (19, 23–26, 28), are indicated. The DNA fragments carried by the ssrAB-cat−302/+478 and ssrAB-cat+10 transcriptional fusions are shown. All positions indicated are relative to the tss upsteam of ssrA.
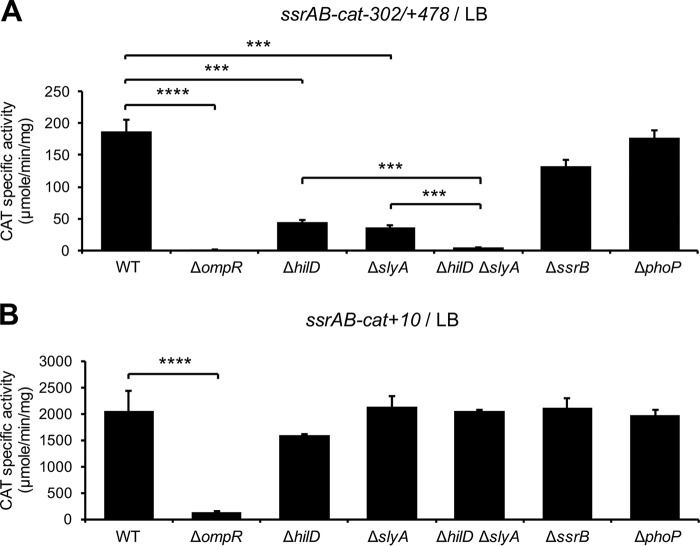
The activity of the ssrAB-cat−302/+478 fusion (repressed by H-NS) was reduced around 4-fold in the ΔhilD and ΔslyA mutants, with respect to the WT strain; interestingly, the ΔhilD ΔslyA double mutant showed a higher reduction (40-fold) in this activity (Fig. 5A) compared with the ΔhilD or ΔslyA single mutants, supporting the idea that SlyA and HilD have an additive effect on ssrAB. On the other hand, the activities of the ssrAB-cat−302/+10 fusion (not repressed by H-NS) were similar in the WT strain and its isogenic ΔhilD, ΔslyA, and ΔhilD ΔslyA mutants (Fig. 5B), whereas the activities of the two fusions analyzed were severely reduced in the ΔompR mutant (Fig. 5). These results confirm that SlyA and HilD act on ssrAB by counteracting H-NS-mediated repression and further support the idea that OmpR acts on ssrAB as a classical activator.
FIG 5.
HilD and SlyA act additively as anti-H-NS factors, whereas OmpR acts independently of H-NS, to induce expression of ssrAB during growth in LB. Expression of the ssrAB-cat−302/+478 (A) and ssrAB-cat+10 (B) transcriptional fusions carried by the pssrAB-cat-302/+478 and pssrAB-cat+10 plasmids, respectively, was tested in WT S. Typhimurium strain and its isogenic ΔompR, ΔhilD, ΔslyA, ΔhilD ΔslyA, ΔssrB, and ΔphoP mutants. CAT-specific activity was determined from samples of bacterial cultures grown for 9 h in LB at 37°C. Data represent the mean with standard deviation of the results from three independent experiments performed in duplicate. Statistically different values are indicated (***, P < 0.001; ****, P < 0.0001).
Collectively, these results indicate that SlyA and HilD act additively to counteract H-NS-mediated repression on ssrAB during the growth of S. Typhimurium in LB, which would favor the activation of ssrAB by OmpR.
SlyA, but not HilD or OmpR, counteracts H-NS-mediated repression on ssrAB during growth in N-MM.
In order to investigate if SlyA also antagonizes repression of ssrAB by H-NS in N-MM, a growth condition where HilD is not involved in the expression of ssrAB (18, 19), the activities of the ssrAB-cat−302/+478 and ssrAB-cat−302/+10 fusions were tested in the WT S. Typhimurium strain and the ΔslyA mutant, grown in N-MM at pH 5.8 and 37°C, growth conditions that somehow resemble the intracellular environment where S. enterica survives (11). As controls, the ΔhilD and ΔhilD ΔslyA mutants were also assessed in these assays. The activity of the ssrAB-cat−302/+478 fusion (repressed by H-NS), but not that of the ssrAB-cat−302/+10 fusion (not repressed by H-NS), was similarly decreased in the ΔslyA and ΔhilD ΔslyA mutants compared with the WT strain (Fig. 6). As expected, the activities of these fusions were not affected in the ΔhilD mutant (Fig. 6). Nearly the same results were obtained using N-MM at pH 7.4 (data not shown).
FIG 6.
SlyA, but not HilD, acts as an anti-H-NS factor, whereas OmpR acts independently of H-NS, to induce expression of ssrAB during growth in N-MM. Expression of the ssrAB-cat−302/+478 (A) and ssrAB-cat+10 (B) transcriptional fusions carried by the pssrAB-cat-302/+478 and pssrAB-cat+10 plasmids, respectively, was determined in the WT S. Typhimurium strain and its isogenic ΔompR, ΔhilD, ΔslyA, ΔhilD ΔslyA, ΔssrB, and ΔphoP mutants. The CAT-specific activity was determined from samples of bacterial cultures grown for 6 h in N-MM at pH 5.8 and 37°C. Data represent the mean with standard deviation of the results from three independent experiments performed in duplicate. Statistically different values are indicated (***, P < 0.001).
These results show that SlyA, but not HilD, counteracts the repression of ssrAB by H-NS during growth in N-MM.
On the other hand, OmpR is also required for the expression of ssrAB in N-MM (18); however, it is unknown whether, under these growth conditions, OmpR is still needed for the expression of ssrAB in the absence of repression by H-NS. Therefore, to determine this, we tested the expression of the ssrAB-cat−302/+478 and ssrAB-cat−302/+10 fusions in the ΔompR mutant grown in N-MM at pH 5.8. As shown in Fig. 6, the activities of both fusions were drastically reduced in the ΔompR mutant with respect to the WT strain. Similar results were obtained using N-MM at pH 7.4 (data not shown).
These results indicate that OmpR is required for the expression of ssrAB during growth in N-MM, independently of the repression by H-NS.
PhoP and SsrB are not involved in the transcription of ssrAB coordinated by SlyA, HilD, and OmpR.
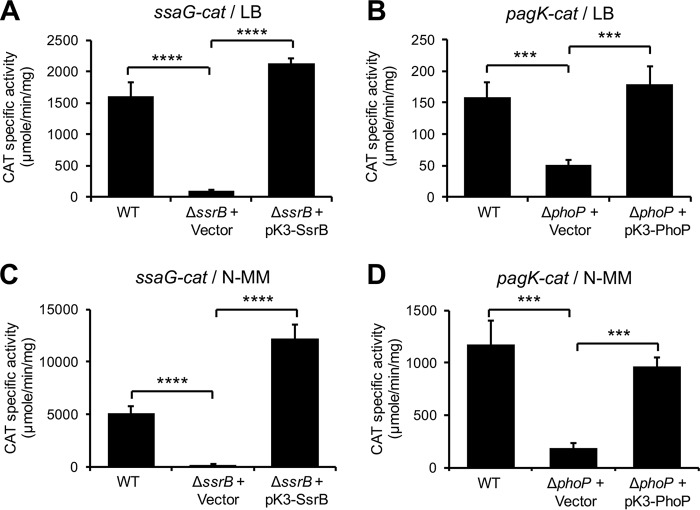
PhoP and SsrB seem to have no major effect on the promoter located upstream of ssrA but are required for the transcription of an additional promoter on ssrAB, located upstream of ssrB (23, 26), which is not contained in the ssrAB transcriptional fusions used in our study (Fig. 4). To discard a possible role of these regulators on the transcription of ssrAB mediated by SlyA, HilD, and OmpR, under the conditions tested in this work, where the ssrA and ssrB genes are transcribed as an operon (18), the activities of the ssrAB-cat−302/+478 and ssrAB-cat−302/+10 fusions were now analyzed in the ΔssrB and ΔphoP mutants grown in LB or N-MM at pH 5.8. As shown in Fig. 5 and 6, the expression of these fusions was not affected in the ΔssrB and ΔphoP mutants with respect to the WT strain. As controls for these assays, the expression of the ssaG-cat and pagK-cat transcriptional fusions, which is dependent on SsrB (62) and PhoP (44), respectively, was also analyzed in the respective ΔssrB or ΔphoP mutant. As expected, the expression of ssaG-cat and pagK-cat decreased specifically by the absence of SsrB or PhoP, respectively, in both LB and N-MM (Fig. 7).
FIG 7.
Positive controls for cat reporter assays in the ΔssrB and ΔphoP mutants. (A and C) Expression of the ssaG-cat transcriptional fusion carried by the pssaG-cat plasmid was determined in the WT S. Typhimurium strain and its isogenic ΔssrB mutant containing the pMPM-K3 vector or the pK3-SsrB plasmid that expresses SsrB from a constitutive promoter. (B and D) Expression of the pagK-cat transcriptional fusion carried by the ppagK-cat plasmid was determined in the WT S. Typhimurium strain and its isogenic ΔphoP mutant containing the pMPM-K3 vector or the pK3-PhoP plasmid that expresses PhoP from a constitutive promoter. The CAT-specific activity was determined from samples of bacterial cultures grown for 9 h in LB (A and B) or 6 h in N-MM (C and D) at pH 5.8 and 37°C. Data represent the mean with standard deviation of the results from three independent experiments performed in duplicate. Statistically different values are indicated (***, P < 0.001; ****, P < 0.0001).
Together, these results indicate that PhoP and SsrB are not involved in the coordinated regulation of ssrAB revealed in this study, mediated by SlyA, HilD, and OmpR.
DISCUSSION
Evolution of Salmonella pathogenicity has involved the adaptation of regulatory mechanisms for a tight control of the expression of virulence genes acquired through horizontal transfer. Different studies have shown that the nucleoid-associated protein H-NS plays a major role in these mechanisms by acting as a global transcriptional repressor (4, 52, 53, 55, 57, 63). For instance, H-NS represses the expression of SPI-1 and SPI-2, two chromosomal regions acquired in different evolutionary times which contain genes with essential roles for the Salmonella pathogenicity (1, 52, 53, 64, 65). Therefore, the expression of most acquired genes requires transcriptional regulators that antagonize the repression by H-NS.
SlyA, HilD, and OmpR have been involved in the expression of the ssrAB regulatory operon located in SPI-2, HilD by antagonizing H-NS-mediated repression (18, 19) and SlyA and OmpR by until-now unknown mechanisms, although it has been reported that the control of ssrAB expression by OmpR requires relaxation of DNA supercoiling (66). In this study, we show how these three regulators cooperate to induce the expression of ssrAB in LB and N-MM, two in vitro growth conditions that somehow mimic different niches that S. enterica colonizes in hosts, the intestinal and intracellular environments, respectively.
Previous studies have demonstrated that SlyA induces the expression of ssrAB and several other genes during growth in minimal medium (27, 42, 59). We found that SlyA induces the expression of ssrAB during growth in LB, which is consistent with previous reports indicating that the overexpression of SlyA induces the expression of ssrAB in LB (25, 28), as well as with results from a previous transcriptomic analysis supporting the idea that SlyA positively regulates ssrAB and several other genes during growth in LB (36). These data strongly suggest that SlyA is present and active under different growth conditions. Accordingly, the slyA gene is expressed during growth in rich and minimal media (17, 36, 67, 68). Furthermore, SlyA is required for the expression of the grhD1 virulence gene in both LB and N-MM (44). Our results show that SlyA induces the expression of ssrAB during growth in LB by counteracting H-NS-mediated repression. Previously, we reported that HilD also induces the expression of ssrAB in LB by counteracting H-NS-mediated repression (19). In vitro, both SlyA and HilD can independently displace H-NS from the promoter of ssrAB. However, our results indicate that the additive action of SlyA and HilD is required to antagonize the repression of ssrAB by H-NS during growth in LB. To our knowledge, this is the first report indicating a concerted action of SlyA and HilD to induce gene expression, and it is the first showing an additive action of two different regulators to antagonize H-NS-mediated repression.
Our results indicate that SlyA also induces the expression of ssrAB during growth in N-MM by counteracting H-NS-mediated repression. Interestingly, HilD is not required for the expression of ssrAB in N-MM (18, 19). It is tempting to speculate that another transcriptional factor replaces the action of HilD during growth in N-MM. Alternatively, a different H-NS repressor complex on ssrAB could be formed during growth in N-MM, probably caused by local changes in DNA structure, which could be displaced by only SlyA.
Our data show that even when the repression by H-NS is displaced, the expression of ssrAB requires OmpR. Several studies support the idea that OmpR mainly acts as a classical transcriptional activator on ssrAB and many other genes (18, 23–25, 66); classical activators favor RNA polymerase binding on promoters (69). Therefore, the expression of ssrAB mediated by SyA, HilD, and OmpR would involve two steps, as follows: the relief of H-NS-mediated repression by SlyA and HilD or only SlyA and the recruitment of the RNA polymerase by OmpR (Fig. 8). Similarly, expression of the ugtL and pagC genes of S. enterica first requires the action of SlyA to counteract H-NS-mediated repression and then the action of the PhoP classical activator that recruits the RNA polymerase on the promoters of these genes (38). The results from our study illustrate the integration of ancestral (H-NS, SlyA, and OmpR) and previously acquired (HilD) regulators into mechanisms that control the expression of newly acquired virulence genes, such as those from SPI-2.
FIG 8.
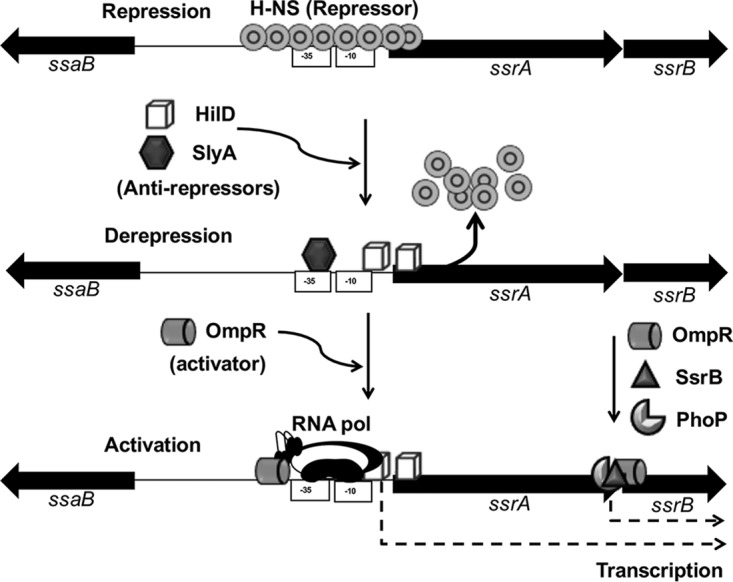
Model for the regulation of ssrAB by SlyA, HilD, OmpR, and H-NS. H-NS binds a region spanning the promoter upstream of ssrA and thus constitutively represses the expression of ssrAB, probably by blocking the access of both OmpR and the RNA polymerase. Under inducing conditions, SlyA and HilD (as during growth in LB) or only SlyA (as during growth in N-MM) displace the H-NS complex bound to the promoter upstream of ssrA. This allows binding of OmpR that recruits the RNA polymerase on this promoter, which finally induces the transcription of the ssrAB operon. The previously reported transcription of ssrB, from an additional promoter upstream of this gene, which involves the OmpR, PhoP, and SsrB regulators (23, 25, 26), is also indicated.
Important to note, under growth conditions other than those assessed in this study, there is transcription of ssrB independent of ssrA, which is directly mediated by OmpR, PhoP, and SsrB (23, 25, 26) (Fig. 4 and 8). Even more, in the absence of SsrA, unphosphorylated SsrB induces the expression of genes required for biofilm formation (70). We found that PhoP and SsrB do not play an evident role in transcription from the promoter upstream of ssrA, which is consistent with findings from previous reports (23, 26); however, PhoP controls the expression of ssrAB at the posttranscriptional level (26). Thus, our results, together with those from previous studies, show the high complexity of the mechanisms governing the expression of ssrAB, which favor the expression of the SPI-2 virulence genes in different in vivo niches.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this work are listed in Table 1. Bacterial cultures for chloramphenicol acetyltransferase (CAT) and Western blot assays were grown in LB or N-MM containing low Mg2+ (10 μM) at pH 5.8 or pH 7.4, as described previously (18, 19). Culture samples were taken after 9 h of growth in LB and 6 or 16 h of growth in N-MM at pH 5.8 or pH 7.4, respectively. When appropriate, antibiotics were used at the following final concentrations: ampicillin, 200 μg/ml; streptomycin, 100 μg/ml; tetracycline, 12 μg/ml; and kanamycin, 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. Typhimurium | ||
| SL1344 | Wild type; xyl hisG rpsL Smr | 74 |
| JPTM3 | ΔompR::kan | 18 |
| JPTM5 | ΔhilD::kan | 18 |
| JPTM8 | ssrA::3×FLAG-kan | 18 |
| JPTM25 | ΔhilD | 75 |
| JPTM28 | ΔompR | 75 |
| DTM99 | ΔssrB | 62 |
| DTM104 | ΔphoP | 44 |
| DTM115 | ΔslyA::kan | This study |
| DTM116 | ΔslyA | This study |
| DTM117 | ΔhilD ΔslyA::kan | This study |
| DTM118 | ΔhilD ΔslyA | This study |
| DTM119 | ΔslyA ssrA::3×FLAG-kan | This study |
| DTM120 | ΔslyA ssrA::3×FLAG | This study |
| E. coli | ||
| M15 | Strain for expression of recombinant proteins | Qiagen |
| DH10β | Laboratory strain | Invitrogen |
| Plasmids | ||
| pKK232-8 | pBR322 derivative containing a promoterless chloramphenicol acetyltransferase (cat) gene, Apr | 76 |
| pssrAB-cat-302 + 478 | pKK232-8 derivative containing a ssrAB-cat transcriptional fusion from nucleotides −302 to +478 | 19 |
| pssrAB-cat-302 + 10 | pKK232-8 derivative containing a ssrAB-cat transcriptional fusion from nucleotides −302 to +10 | 19 |
| pssaG-cat | pKK232-8 derivative containing a ssaG-cat transcriptional fusion from nucleotides −303 to +361 | 18 |
| ppagK-cat | pKK232-8 derivative containing a pagK-cat transcriptional fusion from nucleotides −880 to +251 | 44 |
| pMPM-K3 | p15A derivative low-copy-number cloning vector, lac promoter, Kanr | 71 |
| pK3-SlyA | pMPM-K3 derivative expressing SlyA from the lac promoter | This study |
| pK3-SsrB | pMPM-K3 derivative expressing SsrB from the lac promoter | 62 |
| pK3-PhoP | pMPM-K3 derivative expressing PhoP from the lac promoter | 44 |
| pMPM- T6Ω | p15A derivative low-copy-number cloning vector, arabinose-inducible promoter, Tcr | 71 |
| pT6-HNS-WT | pMPM-T6Ω derivative expressing promoter WT H-NS from the arabinose-inducible | 18 |
| pT6-HNS-G113D | pMPM-T6Ω derivative expressing H-NSG113D from the arabinose-inducible promoter | 44 |
| pQE30 | Vector for expression of recombinant proteins, lac promoter, Apr | Qiagen |
| pQE30-His-HA-SlyA | pQE30 derivative expressing His-HA-SlyA from the lac promoter, Apr | This study |
| pBAD-H-NS-FH | pBADMycHisC derivative expressing H-NS−FH from an ara promoter, Apr | 44 |
| pKD46 | pINT-ts derivative expressing red recombinase under the control of an arabinose-inducible promoter, Apr | 72 |
| pKD4 | pANTsγ derivative template plasmid containing the kanamycin cassette for λRed recombination, Apr | 72 |
| pCP20 | Plasmid expressing FLP recombinase from a temperature-inducible promoter, Apr | 72 |
The coordinates for the cat fusions are indicated with respect to the transcriptional start site of ssrA, ssaG, or pagK. Smr, streptomycin resistance; Apr, ampicillin resistance; Kanr, kanamycin resistance; Tcr, tetracycline resistance.
Construction of plasmids.
The plasmids and primers used in this study are listed in Tables 1 and 2, respectively. The pK3-SlyA plasmid was constructed by amplification of slyA from chromosomal DNA of S. Typhimurium SL1344, using the primers SlyA-RV11 and SlyA-FW22. This PCR product was digested with BamHI and HindIII restriction enzymes, purified, and then cloned into the same restriction sites of the pMPM-K3 vector (71). In Salmonella spp., the pK3-SlyA plasmid constitutively expresses SlyA under the control of a lac promoter (Plac), since both Salmonella spp. and the pMPM-K3 vector lack the gene encoding LacI, the repressor of Plac. The pQE30-His-HA-SlyA plasmid was generated by amplifying slyA from chromosomal DNA of S. Typhimurium, with primers SlyA/HA/His-F and SlyA/HA/His-R. This PCR product was digested with the BamHI and HindIII restriction enzymes, purified, and cloned into the same restriction sites of the vector pQE30. The pQE30-His-HA-SlyA plasmid expresses SlyA fused to the hemagglutinin (HA) epitope and 6×His (His-HA-SlyA) from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter.
TABLE 2.
Primers used in this work
| Primer by use | Sequence (5′–3′)a | Target gene | REb |
|---|---|---|---|
| Gene cloning | |||
| SlyA-RV11 | ACGGGATCCTCGGCAGGTCAGCGTGTCG | slyA | BamHI |
| SlyA-FW22 | TAAAAGCTTAGCAAGCTAATTATAAGGAG | slyA | HindIII |
| SlyA-HA-His-F | GATGGATCCTCTATCCGTATGATGTTCCTG ATTATGCTAGCCAAATTCGAATCGCCACTA GGTTC | slyA | BamHI |
| SlyA-HA-His-R | CTAAAGCTTTGTCGTGCTCGCCAGCAACG | slyA | HindIII |
| EMSAs | |||
| SsaBF (fw) | GGCTAAGATCTTCGGCCCTGATATCCTG | ssrAB | |
| SsrBRS6E (rv) | TTGGTCGACCGACAGATAGATGCCGG | ssrAB | |
| Gene deletions | |||
| slyA-H1P1 | GCTAATTATAAGGAGATGAAATTGGAATC GCCACTAGGTTGTAGGCTGGAGCTGCTT CG | slyA | |
| slyA-H2P2 | GTATGCCCCTGCACCTCAATCGTGAGAG TGCAATTCCATCATATGAATATCCTCCTT AG | slyA |
Underlined letters indicate the respective restriction enzyme site in the primer. The sequences corresponding to the template plasmid pKD4 (Table 1) are in italics.
RE, restriction enzyme for which a site was generated in the primer.
Construction of deletion mutant strains and strains expressing FLAG-tagged proteins.
The bacterial strains used in this work are listed in Table 1. Nonpolar deletion of the slyA gene in the S. Typhimurium SL1344 strain was performed with the λ Red recombinase system, as reported previously (72), using the respective primers described in Table 2, thus generating the strain DTM115. P22 transduction was used to transfer the ΔslyA::km allele from strain DTM115 into strain JPTM25, generating strain DTM117, as well as to transfer the ssrA::3×FLAG-kan allele from strain JPTM8 into strain DTM116, generating strain DTM119. The kanamycin resistance cassette was excised from strains DTM115, DTM117, and DTM119 by using the pCP20 plasmid expressing the FLP recombinase, as described previously (72), generating strains DTM116, DTM118, and DTM120, respectively. All mutant strains were verified by PCR amplification and sequencing.
Chloramphenicol acetyltransferase assays.
The chloramphenicol acetyltransferase (CAT) assay and protein determinations to calculate CAT specific activities were performed as described previously (73).
Statistical analysis.
Data from CAT assays were analyzed with Prism 5.0 software version 5.04 (GraphPad, Inc., San Diego, CA) using one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test. A P value of <0.05 was considered significant.
Expression and purification of His-HA-SlyA.
Escherichia coli M15(pREP4) containing pQE30-His-HA-SlyA was grown in 200 ml of LB at 37°C in a shaken water bath. At an optical density at 600 nm of 0.6, the expression of His-HA-SlyA was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were allowed to grow for an additional 4 h at 30°C. Bacterial cells were then collected by centrifugation at 4°C. The pellet was washed once with ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8]) and then resuspended in 10 ml of the same buffer. The bacterial suspension was sonicated for 8 min, combining 9.9-s pulses with 9.9-s resting cycles, in a Soniprep 150 sonicator (Sonics and Materials, Inc.). Bacterial debris was separated by centrifugation at 4°C, and the soluble extract was loaded into a Ni2+-nitrilotriacetic acid (Ni2+-NTA)–agarose affinity column equilibrated with lysis buffer; the column was then washed with 20 volumes of washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8]). His-HA-SlyA was eluted with washing buffer containing 250 mM imidazole. Fractions were analyzed by SDS-PAGE, and those containing the purified protein were loaded into a Slide-A-Lyzer G2 dialysis cassette (Thermo) and dialyzed at 4°C in a buffer containing 20 mM Tris (pH 8), 150 mM NaCl, 0.1 mM EDTA, 5 mM dithiothreitol (DTT), and 20% (vol/vol) glycerol. Protein concentration was determined by the Bradford procedure. Aliquots of the purified protein were stored at −70°C.
Expression and purification of H-NS−FH.
The His-tagged fusion protein H-NS−FH was expressed in E. coli BL21(DE3) containing the pBAD-H-NS-FH plasmid and purified by using a Ni2+-NTA–agarose affinity column, as described previously (18).
Competitive electrophoretic mobility shift assays.
The DNA fragment containing the regulatory region of ssrAB was amplified by PCR using the SsaBF/SsrBRS6E primer pair and chromosomal DNA of S. Typhimurium SL1344 as the template. PCR products were purified using the DNA Clean & Concentrator kit (Zymo Research). Binding reactions were performed by mixing ≈100 ng of the PCR product first with 0.5 μM H-NS–FH for 15 min and then incubated with increasing concentrations of His-HA-SlyA for an additional 40 min in binding buffer containing 10 mM Tris (pH 9), 50 mM KCl, and 0.1% of Triton X-100 in a total volume of 20 μl. Protein-DNA binding reactions were electrophoretically separated in 6% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA buffer at 4°C. The DNA fragments were stained with ethidium bromide and visualized with an Alpha-Imager UV transilluminator (Alpha Innotech Corp.).
Western blotting.
H-NS–FH–DNA complexes from EMSAs were transferred to 0.45-μm-pore-size nitrocellulose membranes (Bio-Rad) using a semidry transfer apparatus (Bio-Rad). Membranes were blocked with 5% nonfat milk and incubated with anti-FLAG M2 (Sigma) monoclonal antibodies at a dilution of 1:3,000. Horseradish peroxidase-conjugated anti-mouse antibody (Pierce), at a dilution of 1:10,000, was used as the secondary antibody. Bands on the blotted membranes were developed by incubation with the Western Lightning chemiluminescence reagent plus (PerkinElmer) and exposed to Kodak X-Omat films.
SsrA-FLAG and GroEL were detected from whole-cell extracts as described above, using anti-FLAG M2 monoclonal antibody (Sigma) or anti-GroEL polyclonal antibody (StressGen) at 1:2,000 and 1:100,000 dilutions, respectively. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody (Pierce), at a dilution of 1:10,000, was used as a secondary antibody.
ACKNOWLEDGMENTS
This work was supported by grants from the Dirección General de Asuntos del Personal Académico de la UNAM/Mexico (grants IN203415 and IN202418) and from the Consejo Nacional de Ciencia y Tecnología (CONACYT)/Mexico (grant 254531) to V.H.B. C.Z-A. was supported by a master fellowship from CONACYT (grant 599442). M.M.B. was supported by a predoctoral fellowship from CONACYT (grant 403748).
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
We thank M. Fernández-Mora and F. J. Santana for technical assistance, L. C. Martínez for help during the early stages of this study, and I. Martínez-Flores for critical reading of this paper.
REFERENCES
- 1.Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 2.Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, Marks F. 2017. Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis 30:498–503. doi: 10.1097/QCO.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohl ME, Miller SI. 2001. Salmonella: a model for bacterial pathogenesis. Annu Rev Med 52:259–274. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 4.Fàbrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porwollik S, McClelland M. 2003. Lateral gene transfer in Salmonella. Microbes Infect 5:977–989. doi: 10.1016/S1286-4579(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 6.Bispham J, Tripathi BN, Watson PR, Wallis TS. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect Immun 69:367–377. doi: 10.1128/IAI.69.1.367-377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect Immun 73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A 107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 11.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. 2002. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 13.Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton JCD. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature 529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 15.Brown NF, Vallance BA, Coombes BK, Valdez Y, Coburn BA, Finlay BB. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog 1:e32. doi: 10.1371/journal.ppat.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao EA, Miller SI. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella Typhimurium. Proc Natl Acad Sci U S A 97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JCD. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Bustamante VH, Martínez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A 105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez LC, Banda MM, Fernández-Mora M, Santana FJ, Bustamante VH. 2014. HilD induces expression of Salmonella pathogenicity island 2 genes by displacing the global negative regulator H-NS from ssrAB. J Bacteriol 196:3746–3755. doi: 10.1128/JB.01799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol 65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 21.Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ. 2011. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem 286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomljenovic-Berube AM, Mulder DT, Whiteside MD, Brinkman FSL, Coombes BK. 2010. Identification of the regulatory logic controlling Salmonella pathoadaptation by the SsrA-SsrB two-component system. PLoS Genet 6:e1000875. doi: 10.1371/journal.pgen.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng X, Oropeza R, Kenney LJ. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol 48:1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee AK, Detweiler CS, Falkow S. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J Bacteriol 182:771–781. doi: 10.1128/JB.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng X, Walthers D, Oropeza R, Kenney LJ. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- 26.Bijlsma JJE, Groisman EA. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol 57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- 27.Linehan SA, Rytkönen A, Yu X-J, Liu M, Holden DW. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect Immun 73:4354–4362. doi: 10.1128/IAI.73.7.4354-4362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada N, Oi Y, Takeda-Shitaka M, Kanou K, Umeyama H, Haneda T, Miki T, Hosoya S, Danbara H. 2007. Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology 153:548–560. doi: 10.1099/mic.0.29259-0. [DOI] [PubMed] [Google Scholar]
- 29.Fass E, Groisman EA. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol 12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella Typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groisman EA, Chiao E, Lipps CJ, Heffron F. 1989. Salmonella Typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci U S A 86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellison DW, Miller VL. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol 9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 34.McVicker G, Sun L, Sohanpal BK, Gashi K, Williamson RA, Plumbridge J, Blomfield IC. 2011. SlyA protein activates fimB gene expression and type 1 fimbriation in Escherichia coli K-12. J Biol Chem 286:32026–32035. doi: 10.1074/jbc.M111.266619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyborn NR, Stapleton MR, Norte VA, Roberts RE, Grafton J, Green J. 2004. Regulation of Escherichia coli hemolysin E expression by H-NS and Salmonella SlyA. J Bacteriol 186:1620–1628. doi: 10.1128/JB.186.6.1620-1628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol 56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 37.Lithgow JK, Haider F, Roberts IS, Green J. 2007. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol 66:685–698. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez JC, Latifi T, Groisman EA. 2008. Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J Biol Chem 283:10773–10783. doi: 10.1074/jbc.M709843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorman CJ, Dorman MJ. 2017. Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium. Environ Microbiol 19:3834–3845. doi: 10.1111/1462-2920.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimada T, Takada H, Yamamoto K, Ishihama A. 2015. Expanded roles of two-component response regulator OmpR in Escherichia coli: genomic SELEX search for novel regulation targets. Genes Cells 20:915–931. doi: 10.1111/gtc.12282. [DOI] [PubMed] [Google Scholar]
- 41.Cordero-Alba M, Ramos-Morales F. 2014. Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium. J Bacteriol 196:3912–3922. doi: 10.1128/JB.02158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colgan AM, Kröger C, Diard M, Hardt W-D, Puente JL, Sivasankaran SK, Hokamp K, Hinton JCD. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith C, Stringer AM, Mao C, Palumbo MJ, Wade JT. 2016. Mapping the regulatory network for Salmonella enterica serovar Typhimurium invasion. mBio 7:e01024-16. doi: 10.1128/mBio.01024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banda MM, López C, Manzo R, Rico-Pérez G, García P, Rosales-Reyes R, De la Cruz MA, Soncini FC, García-del Portillo F, Bustamante VH. 2018. HilD and PhoP independently regulate the expression of grhD1, a novel gene required for Salmonella Typhimurium invasion of host cells. Sci Rep 8:4841. doi: 10.1038/s41598-018-23068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 47.Saini S, Ellermeier JR, Slauch JM, Rao CV. 2010. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog 6:e1001025. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrone BL, Stringer AM, Wade JT. 2014. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol 196:1094–1101. doi: 10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Main-Hester KL, Colpitts KM, Thomas GA, Fang FC, Libby SJ. 2008. Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect Immun 76:1024–1035. doi: 10.1128/IAI.01224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez-Flores I, Pérez-Morales D, Sánchez-Pérez M, Paredes CC, Collado-Vides J, Salgado H, Bustamante VH. 2016. In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Sci Rep 6:37858. doi: 10.1038/srep37858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer HM, Kühne C, Deditius JA, Hughes KT, Erhardt M. 2014. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol 196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 54.Duong N, Osborne S, Bustamante VH, Tomljenovic AM, Puente JL, Coombes BK. 2007. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J Biol Chem 282:34077–34084. doi: 10.1074/jbc.M707352200. [DOI] [PubMed] [Google Scholar]
- 55.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol 2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 56.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 57.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev 21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 58.Stoebel DM, Free A, Dorman CJ. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 59.Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog 5:e1000306. doi: 10.1371/journal.ppat.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol 263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 61.De la Cruz MÁ, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, Calva E. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol 66:727–743. doi: 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Morales D, Banda MM, Chau NYE, Salgado H, Martínez-Flores I, Ibarra JA, Ilyas B, Coombes BK, Bustamante VH. 2017. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathog 13:e1006497. doi: 10.1371/journal.ppat.1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt H, Hensel M. 2004. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baños RC, Vivero A, Aznar S, García J, Pons M, Madrid C, Juárez A. 2009. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet 5:e1000513. doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect 3:549–559. doi: 10.1016/S1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 66.Cameron ADS, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8:e1002615. doi: 10.1371/journal.pgen.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Norte VA, Stapleton MR, Green J. 2003. PhoP-responsive expression of the Salmonella enterica serovar Typhimurium slyA gene. J Bacteriol 185:3508–3514. doi: 10.1128/JB.185.12.3508-3514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mouslim C, Hughes KT. 2014. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog 10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browning DF, Busby SJW. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 70.Desai SK, Winardhi RS, Periasamy S, Dykas MM, Jie Y, Kenney LJ. 2016. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. Elife 5:e10747. doi: 10.7554/eLife.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayer MP. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41–46. doi: 10.1016/0378-1119(95)00389-N. [DOI] [PubMed] [Google Scholar]
- 72.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puente JL, Bieber D, Ramer SW, Murray W, Schoolnik GK. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol 20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 74.Hoiseth SK, Stocker BAD. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 75.Martínez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brosius J. 1984. Plasmid vectors for the selection of promoters. Gene 27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]