Abstract
Recent years have witnessed an explosion of interest in the human microbiota. Although commensal bacteria have dominated research efforts to date, mounting evidence suggests that endogenous viral populations (the ‘virome’) play key roles in basic human physiology. The most numerous constituents of the human virome are not eukaryotic viruses but rather bacteriophages, viruses that infect bacteria. Here, we review phages’ interactions with their immediate (prokaryotic) and extended (eukaryotic) hosts and with each other, with a particular emphasis on the temperate phages and prophages which dominate the human virome. We also discuss key outstanding questions in this emerging field and emphasize the urgent need for functional studies in animal models to complement previous in vitro work and current computational approaches.
Bacteriophages Dominate the Human Virome
Advances in metagenomics have led to significant progress in understanding the basic biology of the human virome (see Glossary). As with commensal bacteria, different viral communities inhabit the diverse microenvironments which collectively comprise the human host, but several paradigms are generalizable across niches. First, the healthy adult virome has relatively high interpersonal variation and relatively low intrapersonal variation [1–4]. Although a subset of viruses is broadly conserved across individuals [5–7], overall viral community composition varies considerably between individuals and remains largely stable over time. Second, phages greatly outnumber eukaryotic viruses throughout the body, including in the gut, mouth, and skin [1–4]. Most constituents of this ‘phageome’ are DNA viruses of the order Caudovirales (dsDNA) or the family Microviridae (ssDNA) and can be categorized by their lifecycle as either lytic or temperate (Figure 1). The lytic cycle is characterized by immediate viral protein production, inhibition of cellular processes, and host cell death. A temperate phage, by contrast, can also establish a long-term association with its host (lysogen) by forming an episome in the bacterial cytosol or by integrating directly into the bacterial chromosome [8,9]. In this latter scenario, called lysogeny, the mostly quiescent phage genome (prophage) is replicated by bacterial machinery and transmitted vertically upon binary fission. Many prophages are highly stable, but environmental stressors or stochastic fluctuations can trigger their induction (i.e., resumption of the lytic cycle), eventually culminating in the destruction of their host cell [10]. Lysogeny was previously believed to be evolutionarily favorable only at low host densities, but more recent evidence suggests that it is the dominant phage lifestyle in many natural ecosystems [11], including the human body [1–4]. Because induced prophages represent the primary source of extracellular phage particles in vivo [12], and integrated prophages actively modulate bacterial genotypes and phenotypes (reviewed in [13–16]), this review primarily emphasizes temperate phages within the broader context of phages’ interactions with their bacterial hosts, with their eukaryotic hosts, and with each other.
Figure 1. Lytic and Lysogenic Cycles of Phage Replication.
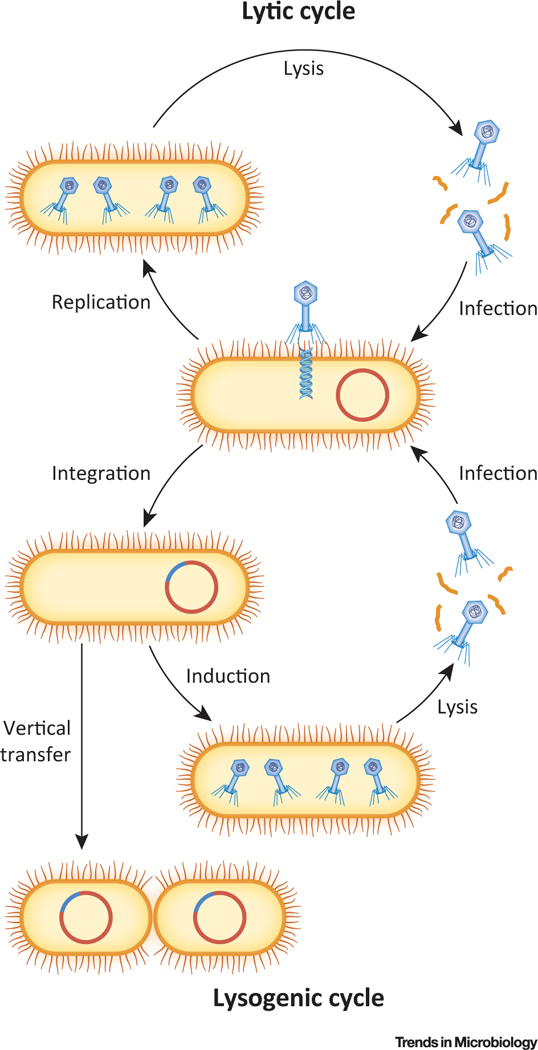
The lytic cycle (top) is characterized by the immediate transcription, replication, and translation of the newly injected phage genome, assembly of mature phage particles, and lysis of the bacterial cell. In the lysogenic cycle (bottom), the phage genome is not immediately replicated but instead integrated into the bacterial chromosome (or maintained episomally) as a prophage. Most phage genes – but not all – are transcriptionally repressed in prophages, which are replicated by bacterial machinery and segregated to daughter cells upon binary fission. DNA damage and other stimuli, along with stochastic fluctuations in gene expression, can result in prophage induction, in which the prophage genome is derepressed, excised, and replicated lytically.
Prophages Are Abundant in Bacterial Genomes
Phages are the most numerous biological entities on the planet [11], and over billions of years of coevolution, many bacterial genomes have become replete with prophages. Early studies, in which prophages were identified by similarity to known phage genes, estimated that roughly 70% of bacteria harbor prophages [17]. Less biased approaches, which discern prophages via general sequence characteristics (e.g., skewed G + C content and codon biases) rather than by specific gene identities, have established putative prophages in an even higher proportion (>80%) of bacterial genomes [18]. In a recent survey of the urinary microbiota, for example, 86% of bacterial isolates from the female bladder contained at least one prophage sequence [19]. Within the gut microbiota, prophages are considerably more common in Proteobacteria and Firmicutes than in Bacteroidetes or Actinobacteria [20], but for all phyla, prophage validation and characterization greatly lags behind identification by sequencing. Recent advances in bacterial culturomics [21], as well as ‘rebooting’ phage genomes in more tractable hosts [22,23], may facilitate the study of otherwise inaccessible phages from bacteria recalcitrant to culture.
Phages Modulate Bacterial Physiology
Phages interact with bacteria in ways which can benefit individual cells, entire communities, both, or neither. These interactions, which span the gamut from parasitic to commensal to mutualistic, include phage killing of bacteria, conveyance of novel bacterial phenotypes, and modulation of bacterial gene expression and evolution (reviewed in [13–16]). Importantly, however, many have not been demonstrated to occur in vivo (Table 1).
Table 1.
Key Prophage-associated Phenotypes and Their Reported Occurrencea
| In vitro prophage-associated phenotype | Reported to occur in vivo? | Reported to occur in commensals in vivo? |
|---|---|---|
| Killing of bacteria | ||
| Lytic replication | Yes [88,98] | Yes [88] |
| Prophage induction | Yes [48,92–94] | Yes [92,93] |
| Conveyance of novel phenotypes | ||
| Virulence | Yes [26,27,88] | No |
| Enhanced metabolic capacities | Yes [28] | No |
| Antibiotic resistance | Yes [29,30,51] (see also [32]) | Yes [29,30] (see also [32]) |
| Phage resistance | No | No |
| Stress tolerance | No | No |
| Biofilm formation | No | No |
| Killing of competitors | Yes [41,62,68] | Yes [62,68] |
| Modulation of bacterial gene expression and evolution | ||
| Generalized transduction | Yes [51,93] | Yes [93] |
| Phage-mediated transformation | No | No |
| Adaptive transposition | No | No |
| Phage repression of homologous bacterial sequences | No | No |
| Phage-encoded σ factors | No | No |
Prophages’ interactions with bacteria can be parasitic (e.g., lytic replication), mutualistic (e.g., protection from phage superinfection), or some combination thereof depending on whether individual bacteria or bacterial populations are considered (e.g., transposition via phage integration) (see text).
Unlike lytic replication, lysogeny does not immediately result in host cell death, but it can still compromise host fitness. Some temperate phages, such as Escherichia coli phage Mu, integrate randomly within the bacterial genome, thereby mutagenizing an infected population and eliminating those cells with insertions in essential genes [24]. Even if integration is successful, the addition of foreign DNA imposes a replicative burden on the host cell which can become nontrivial over evolutionary time [17]. On shorter timescales, however, the greatest threat of prophages to their hosts is the specter of induction. Prophages are genetic time bombs which can detonate during stressful conditions (e.g., upon DNA damage) or spontaneously [10,17], and induction results in the destruction of the bacterial cell and the release of viral progeny.
Nevertheless, prophages benefit evolutionarily by minimizing the detrimental effects of their parasitism. For example, many temperate phages insert into host tRNAs but, by carrying the corresponding tRNAs in their own genomes, complement the disrupted genes [25]. Furthermore, prophages often augment the fitness of their host lysogens and, in some cases, confer entirely novel phenotypes (lysogenic conversion). These traits include tolerance to various environmental stressors, auxiliary metabolic capabilities, antibiotic resistance, immunity to phage superinfection, and, most infamously, virulence [13–16]. Prophage-encoded exotoxins are the defining virulence factors of several prominent bacterial pathogens (reviewed in [26,27]), including Shigatoxin-encoding E. coli (STEC), Vibrio cholerae, Clostridium botulinum, and Corynebacterium diphtheriae, and phage-free strains are essentially avirulent. Prophages also endow Staphylococcus aureus, Streptococcus pyogenes, Salmonella enterica, and numerous other pathogens with toxins, enzymes, adhesins, and immune modulators [26–28]. Thecontributionsof prophages to bacterial commensals have not been similarly defined (Table 1), but endogenous phage communities may represent a reservoir of genes which enable the bacterial microbiota to recover from antibiotic exposure, dietary changes, and other perturbations [29–31]. Although a recent reanalysis has challenged the notion that phages frequently encode -antibiotic resistance genes [32], prophages need not carry any accessory genes to accelerate bacterial adaptation. Certain bacteriocins and the type VI secretion system, among others, likely arose from phage structural proteins which were retained and repurposed even as their progenitor prophages were eroded – and ultimately eliminated – by purifying selection [33–35].
Until recently, prophage induction was assumed to be inevitably detrimental (indeed, fatal) for an affected lysogen. In the intracellular pathogen Listeria monocytogenes, however, induction acts as a genetic switch which facilities the survival of that same lysogen [36]. The L. monocytogenes gene comK, encoding a master regulator of the Com (DNA uptake) machinery, is interrupted by an inserted prophage, ϕ10403S. The Com system is thus inactive for as long as ϕ10403S remains quiescent. After infecting a host cell, L. monocytogenes is engulfed into a phagosome, whereupon f10403S is induced by oxidative stress and precisely excised. Surprisingly, induction does not proceed to its typical endpoint, host cell death, because f10403S lysis proteins are not produced. Instead, the now-intact comK gene drives transcription of the Com machinery, which facilitates the escape of L. monocytogenes from the phagosome to the cytosol. This ‘active lysogeny’ (reviewed in [37]) has since been documented in other bacteria [38], as has binding of phage repressors to homologous bacterial sequences [39] and modulation of bacterial transcription by phage-encoded sigma factors [40]. Through these and other mechanisms yet to be characterized, prophages may play key roles in regulating gene expression across the microbiota.
Phages Modulate Bacterial Communities
Phage transposition and induction also have important functional implications for microbial communities. While these processes typically have neutral or negative consequences for individual lysogens, the line between antagonism and mutualism is blurred at the population level [15]. For example, mutagenesis from Mu-like phage transposition enhances the ability of Pseudomonas aeruginosa to adapt to a model of the cystic fibrosis lung [41]. Spontaneous prophage induction, followed by the release of bacterial DNA and other building blocks into the extracellular milieu, facilitates biofilm formation by remaining lysogens in diverse bacterial taxa [42,43]. Prophage induction also features prominently in intermicrobial warfare. When cocultured in vitro [44,45] or coinfected in vivo [46], prophage-bearing strains rapidly outcompete isogenic prophage-free strains as the latter are either killed or lysogenized by phages induced from the former. S. enterica produces colicin Ib, a pore-forming bacteriocin with activity against competing gut commensals, but lacks the requisite secretory machinery to export the protein itself. Remarkably, prophage induction in a subset of the population, rather than some form of ‘bacterial’ secretion, is the mechanism by which the colicin is released and deployed against niche competitors [47]. Similarly, prophage-borne Shiga toxin genes are among those repressed during lysogeny, and significant toxin production by STEC occurs only upon prophage activation [48,49]. These collective benefits of prophage induction may explain the retention of intact prophages, rather than the simple fixation of advantageous phage genes, in certain bacterial genomes, but their presence can be a double-edged sword. In the human nasopharynx, invasive Streptococcus pneumoniae displaces resident (and typically lysogenic) S. aureus by triggering prophage induction in nearby staphylococci [50].
Phages also alter the structure and function of microbial communities via horizontal gene transfer (HGT). Both lytic and temperate phages can mediate HGT by generalized transduction and by transformation [51–53]. Specialized transduction, by contrast, is specific to temperate phages [53]. All of these processes disseminate biomedically relevant genes, including determinants of antibiotic resistance and virulence, and are important contributors to the diversification of microbial genotypes and phenotypes [53]. However, HGT dynamics in vivo are poorly understood. Although the microbiota’s mobile genetic elements (MGEs) are beginning to be catalogued [54], other key aspects of phage-mediated HGT, including principal actors (efficient transducing phages, naturally competent bacteria, etc.) and relevant rates, have yet to be established for either steady or perturbed states. Emerging methodologies which allow the association of specific MGEs with specific hosts, including single-cell genomics and chromosome capture [55–58], may enhance our understanding of phages’ contributions to HGT within the human body.
Phages Modulate Viral Physiology
Phage particles greatly outnumber bacterial cells globally, and phage predation is a significant source of bacterial mortality in many natural environments [11]. Since prophages benefit from the survival and replication of their bacterial hosts, it is unsurprising that they enhance bacterial defenses against invading phages. Various prophages block almost every step of viral infection and replication, including adsorption, DNA injection, and transcription [59,60], and can, as a last resort, induce the altruistic suicide of their hosts [61]. As evidenced by the sheer magnitude of productive phage infections – 1023 per second globally, by some estimates [11] – none of these mechanisms are wholly effective, but they all reflect common actions by prophages to neutralize existential threats to their hosts and, by extension, to themselves.
Decision-making, even for viruses, does not occur in a vacuum, and temperate phages’ adoption of lysis or lysogeny is influenced by both environmental conditions and viral population dynamics. For example, the lysogenization rate of the model temperate coliphage λ is ∼0.1% when infecting rapidly growing bacteria, ∼50% when infecting starved cells, and ∼20% in the murine intestine [62]. Remarkably, it was recently demonstrated that this decision is, at least in certain Bacillus phages in vitro, mediated by chemical communication akin to quorum sensing [63]. Although this observation has not yet been documented in vivo, the high densities of microbes and microbial metabolites in the gut suggest considerable potential significance for phages and hosts alike.
Given the prevalence of polylysogeny, it is noteworthy that distinct prophage elements can mediate distinct phenotypes in a common host. The molecular dissection of nine cryptic prophages in E. coli BW25113 enabled the detection of prophage-specific phenotypes which were previouslyobscuredinaggregate[64]. Forinstance, the individual deletionofprophagese14, CPS-53, and CP4–57 all reduced E. coli’s acid tolerance in vitro, while CP4–6 and Qin deletion had the opposite effect. In E. coli double lysogens, within-host competition results in diminished replication of one or both phages [65], but Pseudomonas polylysogens outcompete nonlysogens more strongly than do single lysogens in an insect model of infection [66]. Cross-talk between seven prophage elements (pp1-pp7) in the pathobiont Enterococcus faecalis V583 has been mapped in even finer detail. In this strain, pp1, pp3, and pp5 block the excision of pp4 and pp6, while pp7, which lacks certain structural proteins and DNA packaging machinery, appropriates those of pp1 to form viable viral particles [67]. Phage production enhances E. faecalis competitiveness in vitro and in gnotobiotic mice [68], presumably by lysing phage-free strains. Prophage dynamics, already known to be affected by their hosts, are thus also affected by each other.
Phages Modulate Eukaryotic Physiology
As with the bacterial microbiota, a central finding from virome studies is that most viruses which inhabit the human body – including most eukaryotic viruses – do not cause overt disease, suggesting largely commensal or mutualistic interactions. Nevertheless, changes in community composition away from steady state have been associated with various pathologies, including Crohn’s disease and ulcerative colitis (expansion in phage richness) [69] and type I diabetes (diminished phage richness and diversity) [70]. Knockdown of the virome with a broad-spectrum antiviral cocktail also increased the severity of chemically induced colitis in mice [71]. In these and similar studies, however, it is unclear to what extent virome alterations actively contribute to – rather than simply reflect – broader microbiota dysbiosis and host pathology.
This difficulty inassessing correlationversus causation, letaloneestablishing specific mechanisms of action, reflects the dynamic and multifaceted nature of transkingdom interactions within the human body (reviewed in [72,73]). Given the prominence of prophages in bacterial physiology and of bacteria in human physiology, phages have obvious indirect implications for their eukaryotic hosts, but they may also have more direct activity. It was recently shown that coliphage PK1A2, which has high affinity for mammalian sialic acid, can bind, enter, and briefly persist within non-phagocyticcells in vitro, although the generalizability and in vivo implicationsof this observation are not yet clear [74]. Phages are omnipresent at mucosal surfaces, and it was recently proposed that bacteriophage adherence to mucus (BAM) constitutes a novel, non-host-derived form of innate immunity [75]. In this model, phage attachment to mucinglycoproteins enables their accumulation within mucus, where they reduce bacterial colonization and infiltration into underlying tissues. Phage lysis, including upon prophage induction, may further stimulate antibacterial innate immunity via the release of lipopolysaccharide and other bacterial immunogens [69]. Because dendritic cells actively phagocytose phage particles in vitro [76] and, along with manifold (M) cells, sample the contents of theintestinal lumen[77], it has been suggested that native phages mightalso prime antiviral immunity via endosomal Toll-like receptors and/or cytosolic nucleic acid sensors [78]. Neither the BAM model nor these types of phage immunomodulation, however, have been explicitly demonstrated to occur in a mammalian host. The ability of phages to translocate systemically from the lumen is similarly controversial. While a variety of model phages are efficiently transported across intestinal epithelial cells in vitro [79], dissemination of orally administered phages to the bloodstream is variable and may be species-specific [80]. Although the potential implications of this hypothesized ‘intrabody phageome’, including systemic antibacterial activity and immune stimulation, are far-reaching, considerably more work is required to establish its scope and significance in vivo (Figure 2, Key Figure).
Figure 2.
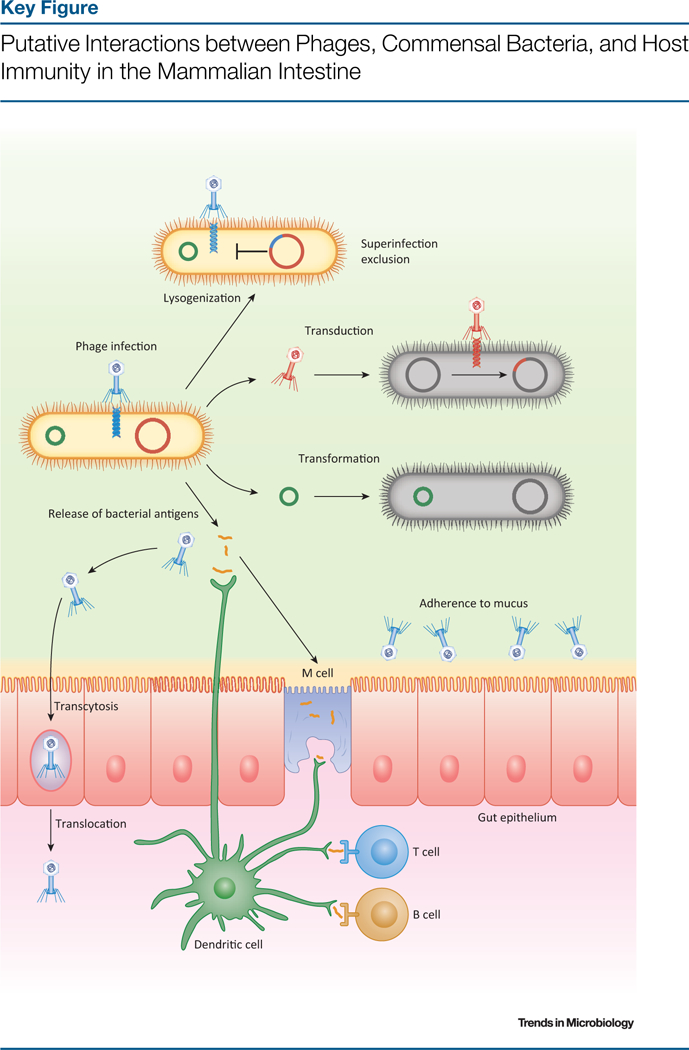
In this model, prophages block superinfection by other phages, and phage infection contributes to horizontal gene transfer via transduction and transformation. Phage particles adhere to epithelial mucus, where they reduce the densities of potentially invasive bacteria, and are occasionally translocated across the epithelium from the intestinal lumen. Phage antigens and bacterial debris released by lysis are transported through microfold (M) cells in Peyer’s patches and sampled by dendritic cells, which also acquire antigens directly from the intestinal lumen via cellular extensions between epithelial cells. These bacterial and phage antigens are presented to B and T cells in the Peyer’s patches and mesenteric lymph nodes, thereby driving a variety of immune responses not shown. Importantly, aspects of this model have yet to be explicitly demonstrated (see text).
Concluding Remarks and Future Perspectives
The first metagenomic analysis of the human virome, involving a single stool sample from a single individual, was published in 2003 [81]. In the intervening 15 years, the field of viral ecology has been transformed by novel methods and novel insights. Large-scale metagenomic studies have shed valuable light on how the composition of the virome develops over time [82], varies between body sites and individuals [1–7], and is affected by various perturbations and pathologies [29–31,69,70]. These efforts have defined the microbial constituents of human microenvironments in unprecedented detail, identified entirely new viruses [6] and revealed the unexpected abundance of others, and, perhaps most importantly, established the paradigm that the virome should not and cannot be examined exclusively through the lens of pathogenesis. While key challenges remain – most notably, a substantial majority (sometimes >90%) of sequencing reads from any given viral metagenome do not align to any known virus [83] – our knowledge of the viruses that inhabit our bodies has never been greater.
Despite this rapid progress, however, our understanding of the functional implications of the virome remains in its infancy. This is especially true for endogenous phage communities. Although an estimated 1015 phages inhabit the human body [84], fundamental aspects of their governing dynamics in vivo remain poorly understood, and their ultimate repercussions for human health are far from clear. Below, we highlight four foundational – and presently unresolved – questions at the intersection of phage, bacterial, and human biology with particular significance for this emerging field.
1. How Do Prophages Affect the Fitness of the Bacterial Microbiota?
As described above, prophage carriage is thought to augment bacterial fitness in a variety of contexts, but apart from virulence, many putative prophage-associated phenotypes have only been shown to occur in vitro, in a single bacterial species, or both (Table 1). Despite the prevalence of predicted prophages in the genomes of commensal bacteria [17–20], the evolutionary logic underlying mutualism [13–16], and an apparent correlation between bacterial doubling time and prophage abundance [85], it is unclear whether, how, and to what extent commensals benefit from prophage carriage. Few relevant details – which commensals and which prophages, via which mechanisms and under which conditions – have been established for any native microbiota. Nevertheless, simplified microbial consortia and gnotobiotic animals have begun to yield useful insights. For instance, a recent study compared the ability of E. coli MG1655 strains with and without λ to colonize the germ-free mouse gut [62]. Carriage of λ was detrimental upon monocolonization due to unexpectedly high rates of induction (50-fold higher than in vitro) but beneficial when lysogens were competed against wild-type MG1655. These findings reiterate that fitness is inherently context-dependent, even in highly artificial systems, and underscore the challenges of discerning prophage dynamics in their natural settings.
2. How Does Phage Predation Shape the Bacterial Microbiota?
Viral physiology is also inherently context-dependent. Given the nutrient and oxygen gradients in the mammalian intestine and the strong correlation between bacterial and phage growth rates, it is not surprising that phage replication kinetics are nonuniform in vivo and cannot be extrapolated from laboratory values [86–88]. Rates of replication, induction, mutation, and persistence, among other basic parameters, have not been established for the endogenous phageome, either for specific native phages or in aggregate. Consequently, the extent to which phage killing contributes to the turnover of the microbiota is unclear. Also unclear is how the microbiota fights back. Although a potpourri of bacterial antiphage defenses has been described, including CRISPR-Cas, restriction modification, toxin–antitoxin systems, and recently many others [89–91], it is not known which of these defenses, if any, are most important for commensals in vivo. Establishing these points is key to understanding the basic biology and functional significance of the phageome and to successfully leveraging phages to manipulate the composition of the microbiota.
3. What Mobilizes Prophages from the Bacterial Microbiota?
Prophages’ capacity for induction underlies many of their phenotypes most likely to be biomedically relevant, including bacterial killing and intermicrobial warfare, horizontal gene transfer, immunomodulation, and, in some cases, virulence factor production. However, surprisingly little is known about the factors which contribute to prophage induction in bacterial commensals. Numerous xenobiotics, most of which directly or indirectly damage DNA, efficiently induce prophages from various taxa in vitro [10,17], but other than certain antibiotics [48,92,93], the stimuli of induction in vivo are largely uncharacterized. Intriguingly, it was recently reported that pathogen-induced inflammation catalyzes prophage induction and transfer between Salmonella strains – and possibly within the microbiota at large – during infection [94]. Evaluating this model explicitly, as well as defining induction kinetics during steady state and upon various perturbations, may illuminate novel elements of pathogenesis and establish what is necessary to restore dysbiotic communities to equilibrium.
4. What Regulates Endogenous Phage Populations in vivo?
Despite phages’ abundance in vivo, our current understanding of immune control of phage dynamics – and of phage evasion of immune control – is rudimentary. Most efforts to explore phage persistence in animal models have focused on the pharmacodynamics of phage therapy [80,95,96], and although certain model phages stimulate antibody production in vivo when administered orally or intravenously [96,97], little is known about how native phage communities interface with host immunity. The extent to which the immune system (as opposed to bacterial availability) regulates the phageome is unclear, as is the relative importance of innate versus adaptive immunity at different body sites. It is not known which phage molecules are detected by which host pattern recognition receptors, which cytokines are produced by which cell types in response to phage exposure, or which phage motifs are displayed in antigen presentation. In addition, it is unclear whether specific lytic phages persist for extended periods in vivo and, if so, which structural features are the principal determinants of persistence. Given recent evidence that immune activity and phage killing are synergistic in combating bacterial infection [98], phage–immune system interactions may have important therapeutic implications along with their obvious significance for basic biology.
Addressing these and other outstanding questions will require a broad and versatile toolkit of methodologies (see Outstanding Questions). Though clearly valuable, neither in vitro studies nor traditional sequencing-based approaches are sufficient to fully unravel the simultaneous and overlapping interactions between phages, bacteria, and their human hosts. Instead, further breakthroughs will eventually necessitate the development and deployment of methods capable of addressing this intrinsic complexity, and hypotheses and assumptions derived from these approaches must be functionally tested in animal models. In some cases, this synergy is already illuminating new biology. For example, a virus’s host range is perhaps its most defining feature, yet neither in vitro assays nor conventional metagenomics can definitively establish the breadth of phage infection in any given environment [99]. To address these shortcomings, single-cell genomics and chromosome capture have been adapted to directly associate specific phage genomes with specific bacterial hosts within complex microbial communities, including seawater [55,56] and the native murine microbiota [58], respectively. Furthermore, phage host range was recently shown to expand more readily in vivo than in vitro because the microbiota harbors intermediate hosts which facilitate viral spillover to new niches [100].
Along with their immediate conclusions, these studies illustrate the ample opportunities for methodological innovation and foundational discoveries in this burgeoning field. Going forward, many more technological advances and functional insights, set against a backdrop of interdisciplinary collaboration and intellectual flexibility, will be necessary to fully unravel the intricacies at the intersection of viral, prokaryotic, and eukaryotic biology.
Highlights.
Bacteriophages (phages), viruses that infect bacteria, are the most abundant viruses in the human body. These phages influence their immediate (prokaryotic) hosts, their extended (eukaryotic) hosts, and each other.
Although phage–bacteria interactions have been extensively studied in vitro, phages’ roles in shaping the composition and function of the microbiota are not well understood. It remains broadly unclear whether, which, and how phages modulate the fitness of bacterial commensals in vivo.
Phage–phage and phage–eukaryote interactions, including phages’ repercussions for and regulation by the immune system, are even less established in vivo.
Despite recent advances, our understanding of endogenous phage communities (the ‘phageome’) is in its infancy. Foundational discoveries are waiting to be made.
Outstanding Questions.
Aberrant bacterial communities are associated with virtually all human diseases, but virome dysbiosis has been linked to fewer (and mostly gut-specific) pathologies to date. Does the virome and/or phageome influence a broader range of systemic phenotypes, such as susceptibility to infectious diseases and responsiveness to therapeutics and vaccines? If so, how?
Most virome and phageome proteins are of presently unknown function. How can this viral ‘dark matter’ best be characterized and harnessed for novel applications?
Phages are ubiquitous in nature and routinely ingested. Which factors determine whether a phage will successfully colonize and persist within a mammalian host?
Prophages are not distributed evenly across bacterial genomes. What evolutionary forces account for this variability?
Some phages do not employ canonical lytic replication or lysogeny. How prevalent are other life cycles, including pseudolysogeny and chronic infection, within the phageome?
Different phages exhibit different breadths of host range in vitro. To what extent do different phages’ host ranges overlap in vivo? Is the phageome largely comprised of generalists or specialists?
Acknowledgments
We thank Megan Baldridge, Alejandro Reyes, and members of the Dantas laboratory for valuable discussion and the Elsevier WebShop for illustration support. This work is supported in part by awards to G.D. through the Edward Mallinkcrodt Jr. Foundation (Scholar Award) and from the National Institute of Allergy and Infectious Diseases (NIAID: https://www.niaid.nih.gov/) and the Eunice Kennedy Shriver National Institute of Child Health & Human Development (https://www.nichd.nih.gov/) of the National Institutes of Health(NIH) underawardnumbers R01AI123394 and R01HD092414, respectively. ECKissupported by a National Science Foundation Graduate Research Fellowship(DGE-1143945). Thecontentissolely the responsibility ofthe authors and does not necessarily represent the official views of the funding agencies.
Glossary
- Cryptic prophage
a degraded, defective prophage which can no longer be induced or form functional particles.
- Generalized transduction
inadvertent packaging of random bacterial DNA into a phage particle during infection. Following lysis, subsequent infection delivers this DNA to a new host.
- Horizontal gene transfer
transfer of genetic material between members of a population by means other than by vertical descent (see mobile genetic elements).
- Human virome
the collective genome content of the acute, latent, and persistent viruses which infect eukaryotes, bacteria, and archaea in and on the human body (i.e., the viral fraction of the microbiome). Some conceptions of the virome also encompass plant viruses and endogenous retroviruses.
- Induction
excision and resumption of lytic replication (i.e., viral protein production) by a prophage. DNA damage and activation of the bacterial SOS response frequently triggers induction.
- Lysogen
a bacterial host cell harboring at least one prophage.
- Lysogenic conversion
acquisition of a novel phenotype (e.g., virulence) upon prophage integration.
- Lysogeny
a phage replication strategy in which a temperate phage genome is integrated into a bacterial chromosome or maintained as an extrachromosomal episome in the bacterial cytosol.
- Lytic cycle
a phage replication strategy characterized by immediate viral protein production, inhibition of cellular processes, and host cell death.
- Mobile genetic elements
self-mobilizable DNA sequences (e.g., transposons, prophages, and plasmids) which can move within and between genomes.
- Phageome
the collective genome content of endogenous phage populations within a defined environment such as the human body (see human virome).
- Polylysogeny
carriage of multiple prophages by a single bacterial lysogen.
- Prophage
a quasi-dormant but activatable (see induction) phage genome integrated into a bacterial chromosome or maintained as an extrachromosomal episome in the bacterial cytosol.
- Purifying selection
removal of deleterious genome content, which can include prophages.
- Specialized transduction
inadvertent excision and packaging of prophage-flanking bacterial sequences into phage particles upon induction. Following lysis, subsequent infection delivers this DNA to a new host.
- Temperate phage
a phage which can replicate via either the lytic cycle or lysogeny.
- Transformation
bacterial uptake and incorporation of exogenous chromosomal or plasmid DNA.
- Transkingdom interactions
biological interactions between multiple kingdoms of life inhabiting the same environment (e.g., regulation of enteric virus infectivity by gut bacteria).
References
- 1.Reyes A et al. (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minot S et al. (2011) The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannigan GD et al. (2015) The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 6, e01578–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeles SR et al. (2014) Human oral viruses are personal, persistent and gender-consistent. ISME J 8, 1753–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern A et al. (2012) CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res 22, 1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutilh BE et al. (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun 5, 4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manrique P et al. (2016) Healthy human gut phageome. Proc. Natl. Acad. Sci. U. S. A 113, 10400–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St-Pierre F and Endy D (2008) Determination of cell fate selection during phage lambda infection. Proc. Natl. Acad. Sci. U. S. A 105, 20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S et al. (2018) The developmental switch in bacteriophage l: a critical role of the Cro protein. J. Mol. Biol 430, 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanda AM et al. (2015) Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol 197, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles B et al. (2016) Lytic to temperate switching of viral communities. Nature 531, 466–470 [DOI] [PubMed] [Google Scholar]
- 12.Howe A et al. (2016) Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J 10, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bondy-Denomy J and Davidson AR (2014) When a virus is not a parasite: the beneficial effects of prophages on bacterial fitness. J. Microbiol 52, 235–242 [DOI] [PubMed] [Google Scholar]
- 14.Obeng N et al. (2016) The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol 24, 440–449 [DOI] [PubMed] [Google Scholar]
- 15.Harrison E and Brockhurst MA (2017) Ecological and evolutionary benefits of temperate phage: what does or doesn’t kill you makes you stronger. Bioessays Published online October 6, 2017 10.1002/bies.201700112 [DOI] [PubMed] [Google Scholar]
- 16.Argov T et al. (2017) Temperate bacteriophages as regulators of host behavior. Curr. Opin. Microbiol 38, 81–87 [DOI] [PubMed] [Google Scholar]
- 17.Canchaya C et al. (2003) Prophage genomics. Microbiol. Mol. Biol. Rev 67, 238–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HS et al. (2017) Prophage genomics reveals patterns in phage genome organization and replication. bioRxiv 114819
- 19.Miller-Ensminger T et al. (2018) Bacteriophages of the urinary microbiome. J. Bacteriol 200, e00738–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS and Bae JW (2018) Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J 12, 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagier JC et al. (2016) Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol 1, 16203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Kilcher S et al. (2018) Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl. Acad. Sci. U. S. A 115, 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando H et al. (2015) Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst 23, 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harshey RM et al. (2014) Transposable phage Mu. Microbiol. Spectr Published online October 2014. 10.1128/microbiolspec.MDNA3-0007-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell A (2003) Prophage insertion sites. Res. Microbiol 154, 277–282 [DOI] [PubMed] [Google Scholar]
- 26.Brussow H et al. (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev 68, 560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortier LC and Sekulovic O (2013) Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4, 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez CA et al. (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3, e00143–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modi SR et al. (2013) Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abeles SR et al. (2015) Effects of long-term antibiotic therapy on human oral and fecal viromes. PLoS One 10, e0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SW and Bae JW (2016) Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ. Microbiol 18, 1498–1510 [DOI] [PubMed] [Google Scholar]
- 32.Enault F et al. (2017) Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J 11, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobay LM et al. (2014) Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. U. S. A 111, 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholl D (2017) Phage tail-like bacteriocins. Annu. Rev. Virol 4, 453–467 [DOI] [PubMed] [Google Scholar]
- 35.Lopez J and Feldman MF (2018) Expanding the molecular weaponry of bacterial species. J. Biol. Chem 293, 1515–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabinovich L et al. (2012) Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 17, 792–802 [DOI] [PubMed] [Google Scholar]
- 37.Feiner R et al. (2015) A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol 13, 641–650 [DOI] [PubMed] [Google Scholar]
- 38.Abe K et al. (2014) Developmentally-regulated excision of the SPb prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet 10, e1004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y et al. (2005) Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol 3, e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuch R and Fischetti VA (2009) The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One 4, e6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies EV et al. (2016) Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc. Natl. Acad. Sci. U. S. A 113, 8266–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrolo M et al. (2010) Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One 5, e15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godeke J et al. (2011) Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J 5, 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bossi L et al. (2003) Prophage contributions to bacterial population dynamics. J. Bacteriol 185, 6467–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gama JA et al. (2013) Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One 8, e59043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies EV et al. (2016) Temperate phages enhance pathogen fitness in chronic lung infection. ISME J 10, 2553–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nedialkova N et al. (2016) Temperate phages promote colicin-dependent fitness of Salmonella enterica serovar Typhimurium. Environ. Microbiol 18, 1591–1603 [DOI] [PubMed] [Google Scholar]
- 48.Zhang X et al. (2000) Quinolone antibiotics induce Shiga toxinencoding bacteriophages, toxin production, and death in mice. J. Infect. Dis 181, 664–670 [DOI] [PubMed] [Google Scholar]
- 49.Tyler JS et al. (2013) Prophage induction is enhanced and required for renal disease and lethality in an EHEC mouse model. PLoS Pathog 9, e1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selva L et al. (2009) Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U. S. A 106, 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haaber J et al. (2016) Bacterial viruses enable their hosts to acquire antibiotic resistance genes from neighboring cells. Nat. Commun 7, 13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keen EC et al. (2017) Novel ‘superspreader’ bacteriophages promote horizontal gene transfer by transformation. mBio 8, e02115–e02116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balcázar JL (2018) How do bacteriophages promote antibiotic resistance in the environment? Clin. Microbiol. Infect 24, 447–449 [DOI] [PubMed] [Google Scholar]
- 54.Jiang X et al. (2017) Comprehensive analysis of mobile genetic elements in the gut microbiome reveals phylum-level niche-adaptive gene pools. bioRxiv 214213. [DOI] [PMC free article] [PubMed]
- 55.Roux S et al. (2014) Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 3, e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labonte J et al. (2015) Single-cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J 9, 2386–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beitel CW et al. (2014) Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ 2, e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marbouty M et al. (2017) Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci. Adv 3, e1602105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dedrick RM et al. (2017) Prophage-mediated defense against viral attack and viral counter-defense. Nat. Microbiol 9, 16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bondy-Denomy J et al. (2016) Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parma DH et al. (1992) The Rex system of bacteriophage lambda: tolerance and altruistic cell death. Genes Dev 6, 497–510 [DOI] [PubMed] [Google Scholar]
- 62.De Paepe M et al. (2016) Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet 12, e1005861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erez Z et al. (2017) Communication between viruses guides lysis–lysogeny decisions. Nature 541, 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X et al. (2010) Cryptic prophages help bacteria cope with adverse environments. Nat. Commun 1, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Refardt D (2011) Within-host competition determines reproductive success of temperate bacteriophages. ISME J 5, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burns N et al. (2014) Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol. Appl 8, 346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matos RC et al. (2013) Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet 9, e1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duerkop BA et al. (2012) A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. U. S. A 109, 17621–17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norman JM et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao G et al. (2017) Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. U. S. A 114, E6166–E6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang JY et al. (2016) Enteric viruses ameliorate gut inflammation via Toll-like receptor 3 and Toll-like receptor 7-mediated interferon-β production. Immunity 44, 889–900 [DOI] [PubMed] [Google Scholar]
- 72.Pfeiffer JK and Virgin HW (2016) Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mirzaei MK and Maurice CF (2017) Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol 15, 397–408 [DOI] [PubMed] [Google Scholar]
- 74.Lehti TA et al. (2017) Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun 8, 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barr JJ et al. (2013) Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U. S. A 110, 10771–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barfoot R et al. (1989) Some properties of dendritic macrophages from peripheral lymph. Immunology 68, 233. [PMC free article] [PubMed] [Google Scholar]
- 77.Rescigno M et al. (2001) Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol 2, 361–367 [DOI] [PubMed] [Google Scholar]
- 78.Duerkop BA and Hooper LV (2013) Resident viruses and their interactions with the immune system. Nat. Immunol 14, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen S et al. (2018) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. mBio 8, e01874–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miedzybrodzki R et al. (2017) Means to facilitate the overcoming of gastric juice barrier by a therapeutic staphylococcal bacteriophage A5/80. Front. Microbiol 23, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breitbart M et al. (2003) Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol 185, 6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim ES et al. (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med 21, 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aggarwala V et al. (2017) Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mob. DNA Published online October 3, 2017. 10.1186/s13100-017-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dalmasso M et al. (2014) Exploiting gut bacteriophages for human health. Trends Microbiol 22, 399–405 [DOI] [PubMed] [Google Scholar]
- 85.Touchon M et al. (2016) Genetic and life history traits associated with the distribution of prophages in bacteria. ISME J 10, 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weiss M et al. (2009) In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology 10, 16–23 [DOI] [PubMed] [Google Scholar]
- 87.Maura D (2012) Virulent bacteriophages can target O104:H4 enteroaggregative Escherichia coli in the mouse intestine. Antimicrob. Agents Chemother 56, 6235–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reyes A et al. (2013) Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. U. S. A 110, 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldfarb T et al. (2015) BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34, 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ofir G et al. (2018) DISARM is a widespread bacterial defense system with broad anti-phage activities. Nat. Microbiol 3, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doron S et al. (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science Published online 25 January 2018. 10.1126/science.aar4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Allen HK et al. (2011) Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2, e00260–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson TA et al. (2017) The in-feed antibiotic carbadox induces phage gene transcription in the swine gut microbiome. mBio 8, e00709–e00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diard M et al. (2017) Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355, 1211–1215 [DOI] [PubMed] [Google Scholar]
- 95.Merril CR et al. (1996) Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. U. S. A 93, 3188–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Majewksa J et al. (2015) Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 7, 4783–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ochs HD et al. (1971) Immunologic responses to bacteriophage phi-X 174 in immunodeficiency diseases. J. Clin. Invest 50, 2559–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roach DR et al. (2017) Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 12, 38–47 [DOI] [PubMed] [Google Scholar]
- 99.Edwards RA et al. (2016) Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol. Rev 40, 258–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Sordi L et al. (2017) The gut microbiota facilitates drifts in the genetic diversity and infectivity of bacterial viruses. Cell Host Microbe 22, 801–808 [DOI] [PubMed] [Google Scholar]


