Abstract
RNA modifications have recently emerged as critical posttranscriptional regulators of gene expression programs. They affect diverse eukaryotic biological processes, and the correct deposition of many of these modifications is required for normal development. Messenger RNA (mRNA) modifications regulate various aspects of mRNA metabolism. For example, N6-methyladenosine (m6A) affects the translation and stability of the modified transcripts, thus providing a mechanism to coordinate the regulation of groups of transcripts during cell state maintenance and transition. Similarly, some modifications in transfer RNAs are essential for RNA structure and function. Others are deposited in response to external cues and adapt global protein synthesis and gene-specific translation accordingly and thereby facilitate proper development.
Understanding normal tissue development and disease susceptibility requires knowledge of the various cellular mechanisms that control gene expression in multicellular organisms. Much work has focused on investigation of lineage-specific transcriptional networks that govern stem cell differentiation (1). Yet gene expression programs are dynamically regulated during development and require the coordination of both mRNA metabolism and protein synthesis. The deposition of chemical modifications onto RNA has emerged as a basic mechanism to modulate cellular transcriptomes and proteomes during lineage fate decisions in development.
Many of the more than 170 modifications present in RNA have been known for decades, but only in the past several years have sufficiently sensitive tools and high-resolution genome-wide techniques been developed to identify and quantify these modifications in low-abundance RNA species such as mRNA (2, 3). Some RNA modifications have been shown to affect normal development; these modifications can control the turnover and/or translation of transcripts during cell-state transitions and therefore play important roles during tissue development and homeostasis. In particular, the N6-methyladenosine (m6A) modification of mRNA is an essential regulator of mammalian gene expression (4, 5). Other modifications such as 5-methylcytosine (m5C) and N1- methyladenosine (m1A) are currently best described for their functional roles in noncoding RNAs but have also been studied in mRNA (4, 6).
We summarize here recent studies that elucidate the roles of RNA modifications in modulating gene expression throughout cell differentiation and animal development. Because of space limitations, we will focus on m6A in mRNA and m5C in tRNA as notable examples. RNA editing and RNA tail modifications, which have been comprehensively reviewed previously, will not be included.
Types of RNA modifications
Modifications in mRNA
In addition to the 5′ cap and 3′ polyadenylation, mRNAs contain numerous modified nucleosides, including base isomerization to produce pseudouridine (Ψ); methylation of the bases to produce m6A, m1A, and m5C; methylation of the ribose sugar to install 2′O-methylation (Nm, m6Am); and oxidation of m5C to 5-hydroxymethylcytosine (hm5C) (4). Of these, one of the most abundant and well-studied mRNA modifications is m6A. Of all transcripts encoded by mammalian cells, 20 to 40% are m6A methylated, and methylated mRNAs tend to contain multiple m6A per transcript (2, 3). m6A and other RNA modifications are also present in long noncoding and microRNAs.
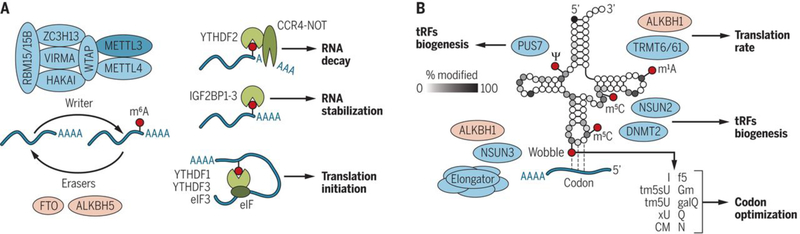
The biological functions of m6A are mediated by writer, eraser, and reader proteins (Fig. 1A) (4). m6A is installed by a multiprotein writer complex that consists of the METTL3 catalytic subunit, and many other accessory subunits (4). Two demethylases, FTO and ALKBH5, act as erasers (7, 8). m6A can both directly and indirectly affect the binding of reader proteins on methylated mRNAs to regulate the metabolism of these transcripts (4). For example, YTHDF2 binds to m6A in mRNA and targets the transcripts for degradation (4, 9), and YTHDF1, YTHDF3, and eIF3 promote translation of m6A-containing transcripts (4, 10, 11). The list of m6A readers that regulate mRNA homeostasis is still growing (12, 13), and the functions of m6A could depend on recognition by cell type–specific reader proteins. Reader and eraser proteins for other modifications are less well described.
Fig. 1. Regulation of gene expression by RNA modifications.
(A) m6A is installed by a multicomponent writer complex with the catalytic subunit METTL3 and removed by the demethylase enzymes FTO and ALKBH5. m6A reader proteins can specifically bind m6A transcripts and effect different outcomes for methylated mRNAs. (B) RNA modifications in human eukaryotic tRNAs according to Modomics (http://genesilico.pl/trnamodviz/jit_viz/select_tRNA). xU, other modified uracil (U); N, unknown modified. How often a base is modified is shown by the grayscale. Only examples of writers (TRMT6/61, DNMT2, NSUN2, NSUN3, PUS7, and Elongator) and erasers (ALKBH1) are shown and how they affect translation. Modifications at the wobble base are most diverse.
Modifications in tRNA and ribosomal RNA
In addition to mRNA, the faithful translation of the genetic code is orchestrated by at least two more types of RNAs, tRNA and ribosomal RNA (rRNA). Human rRNAs contain a set of chemical modifications that often cluster at functionally important sites of the ribosome, such as the peptidyltransferase center and the decoding site (14). Modification in tRNAs are the most diverse, with cytoplasmic and mitochondrial tRNAs carrying more than 100 different modifications (Fig. 1B). A human tRNA can contain between 11 and 13 different modifications that are deposited at different steps during its maturation and could directly affect translation (15). The modifications range from simple methylation and isomerization events—including m5C, m1A, Ψ, 5-methyluridine (m5U), 1- and 7-methylguanosine (m1G, m7G), and inosine (I)—to complex multiple-step chemical modifications (Fig. 1B) (15). The function of a modification depends on both its location in the tRNA and its chemical nature. For example, m5C is site-specifically deposited by at least three enzymes—NSUN2, NSUN3, and DNMT2 (Fig. 1B)—and all three enzymes influence tRNA metabolism differently. Modifications at the wobble position are the most diverse and often optimize codon usage during gene-specific translation (Fig. 1B) (16, 17).
RNA modifications in development
mRNA modifications in development
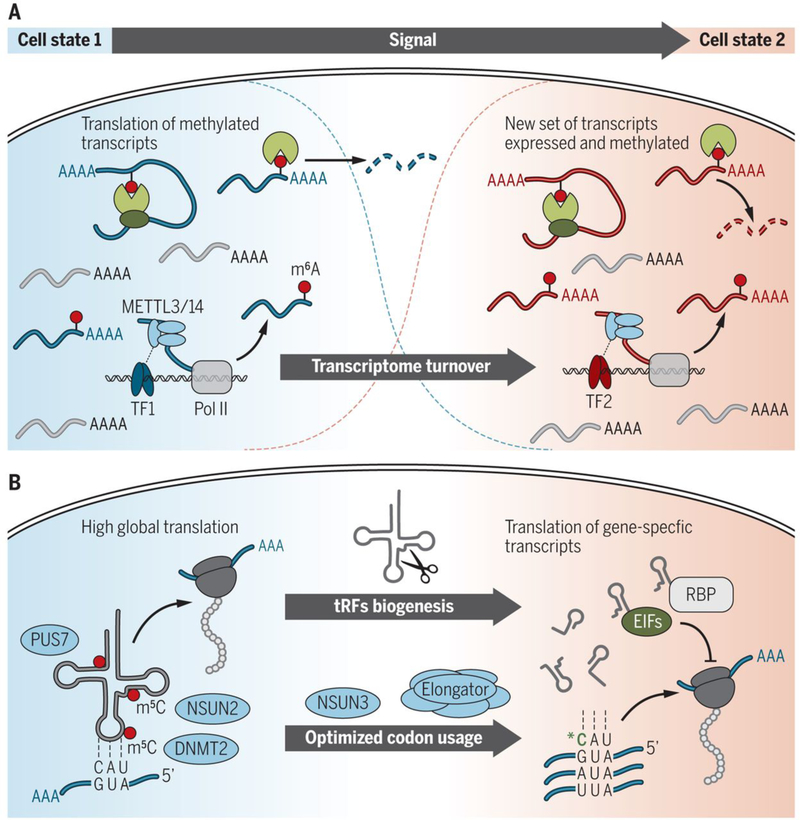
A wealth of recent studies identified an essential role for m6A during development, and many of them highlighted a role for m6A in the regulation of transcriptome switching during embryonic and adult stem cell differentiation (4). An early clue that m6A is essential for development was the observation that removal of the m6A writer enzyme Mettl3 is embryonic lethal in mice (5). Mettl3−/− embryos appear normal before implantation but begin to show defects after implantation and are absorbed by embryonic day 8.5. Examination of gene expression from these embryos and from embryonic stem cells (ESCs) depleted of Mettl3 suggested impaired exit from pluripotency because, for example, expression of the pluripotency factor Nanog was sustained (5, 18). Transcripts that encode certain pluripotency factors are methylated (5, 18, 19), which affects the turnover of these transcripts during differentiation. At least some of these transcripts are cotranscriptionally methylated through the recruitment of the m6A writer complex by cell-state specific transcription factors such as Smad2 and Smad3 (20). Therefore, m6A marks transcripts that encode important developmental regulators to facilitate their turnover during cell fate transitions and thereby enables cells to properly switch their transcriptomes from one cellular state to another (Fig. 2A).
Fig. 2. RNA modifications regulate cell differentiation and development.
(A) Model for the roles of m6A in cell differentiation. In the naïve, undifferentiated state, cell state–specific master transcription factors recruit the METTL3 complex to methylate transcripts that encode cell fate factors. Translation of these methylated factors may aid in the maintenance of cell state and prevent differentiation. When cells initiate differentiation and switch their transcriptional program, reader proteins mediate the turnover of the methylated transcripts to facilitate transcriptome switching. (B) Modification by NSUN2, DNMT2, and PUS7 protects tRNAs from cleavage and production of tRFs, which enables high global translation. In a different cell state, tRFs can affect global and gene-specific protein translation by displacing distinct RBPs and are therefore important players in stem cell differentiation. Wobble tRNA modifications—for example, by NSUN3 and Elongator—enhance the versatility of tRNA anticodon to recognize mRNA to optimize codon usage and translation of cytoplasmic and mitochondrial mRNAs during differentiation.
This paradigm has also been used to explain the differentiation of other cell types. Conditional knockout of Mettl3 in CD4+ T cells prevents the proliferation and differentiation of naïve T cells through stabilization of Socs family genes (21). Loss of Mettl14 (an essential component of the METTL3/14 methyltransferase complex) in the brain delays cortical neurogenesis and is associated with slower cell-cycle progression and impaired decay of transcripts that are involved in lineage specification of cortical neural stem cells (22). Similarly, deletion of Ythdf 2 delays mouse neuronal development through impaired proliferation and differentiation of neural stem and progenitor cells (23). m6A-mediated RNA decay also regulates various stages of zebrafish development. For example, during the maternal-to-zygotic transition, embryos that lack Ythdf 2 exhibit impaired clearance of maternal transcripts, delaying embryonic development (24). Loss of Mettl3 blocks the endothelial-to-hematopoietic transition in zebrafish because of loss of the Ythdf2- mediated decay of genes that specify endothelial cell fate, such as Notch1a and Rhoca (25).
Although these studies highlight the functional roles of the YTHDF2-mediated clearance of mRNAs, loss of Ythdf 2 only partially accounts for phenotypes associated with loss of Mettl3. For example, loss of Mettl3 impairs priming of mammalian ESCs, yet Ythdf2 knockout embryos are able to exit pluripotency (23, 26). Similarly, Mettl3 deletion in zebrafish is lethal owing to severe hematopoietic defects, but adult Ythdf2 knockout fish seem to be normal (24, 25). Work on gametogenesis highlights the importance of other m6A eraser and reader proteins in development because loss of Mettl3, Mettl14, Alkbh5, Ythdf2, and Ythdc2 are all associated with impaired fertility and defects in spermatogenesis and/or oogenesis (8, 26–32). These defects were associated with the altered abundance, translation efficiency, and splicing of methylated transcripts that encode regulators of gametogenesis. Work in Drosophila suggests important roles for m6A in mediating splicing because deletion of Ime4, the Mettl3 homolog, and other m6A writer complex subunits reduces viability of females owing to inappropriate splicing of Sex lethal (Sxl), an important regulator of dosage compensation and sex determination (33, 34).
Together, these studies demonstrate that the functional network that coordinates mRNA methylation is highly complex and high- light the requirement of m6A for the proper execution of stem cell differentiation programs (Fig. 2A). Transcripts that maintain a cell state are most likely cotranscriptionally decorated with m6A through the recruitment of the writer complex by cell state–specific transcription factors. Whereas m6A promotes the decay of these transcripts, active transcription may maintain them at steady-state levels, with other readers potentially aiding in mediating their processing and translation. Upon receiving the signal(s) for cells to differentiate and repress transcription of these factors, m6A coordinates the timely decay of these transcripts, which allows cells to differentiate. Although other posttranscriptional mechanisms aid in the promotion of cell-state switching, m6A writers and readers being required for many of these transitions suggests that m6A regulates gene expression in ways that cannot be substituted by other similar mechanisms.
tRNA modifications in development
Although RNA modifications are highly diverse and found in all RNA species, the recent discoveries underpin an emerging common theme: RNA modifications coordinate translation of transcripts that encode functionally related proteins when cells respond to differentiation or other cellular and environmental cues. Loss of tRNA modifying enzymes can delay stem cell differentiation, often only in distinct tissues. For instance, knockout of Nsun2 delays stem cell differentiation in the brain and skin (35, 36). Depletion of the pseudouridine synthase PUS7 impairs hematopoietic stem cell commitment, and loss of Dnmt2 delays endochondral ossification (37, 38). Knockout of Elp3, a core component of Elongator that modifies the tRNA wobble position, is embryonic lethal (39).
Several recent studies reveal that the dynamic deposition of tRNA modifications is a fast and efficient way for cells to adapt the protein translation machinery to external stimuli (38, 40–42). For example, self-renewing stem cells must be resilient to external differentiation cues and maintain protein synthesis at a low rate, yet their differentiation requires high levels of protein synthesis to produce committed progenitors (40, 43, 44). The deposition of RNA modifications into tRNAs represents an efficient way to adapt energy requirements to specific cell states.
Recent studies discovered that tRNA modifications regulate protein translation rates during development via tRNA-derived small noncoding RNA fragments (tRFs) (6, 38). Loss of NSUN2- mediated methylation at the variable loop increases the affinity to the endonuclease angiogenin, promotes cleavage of tRNAs into tRFs, and inhibits global protein synthesis (35, 40). Similarly, the Ψ writer PUS7 modifies tRNAs and thereby influences the formation of tRFs, which then target the translation initiation complex (38). Loss of DNMT2-mediated methylation at the anticodon loop (C38) causes both tRNA-specific fragmentation and codon-specific mistranslation (37). Thus, altered tRNA modification patterns shape tRF biogenesis and determine their intracellular abundances (Fig. 2B). tRFs could act on global and gene-specific protein translation by displacing distinct RNA-binding proteins (RBPs) and are therefore important players in stem cell differentiation (38, 40), sperm maturation (45), retrotransposon silencing (46), intergenerational transmission of paternally acquired metabolic disorders (47), and breast cancer metastasis (48).
Wobble tRNA modifications enhance the versatility of tRNA anticodons to recognize mRNA to optimize codon usage and translation of cytoplasmic and mitochondrial mRNAs (Fig. 2B) (16, 49–51). Mitochondria are crucial players in stem cell activation, fate decisions, tissue regeneration, aging, and diseases (52). Mitochondrial translation can be affected by mitochondrial tRNA and mRNA modifications. For example, mammalian mitochondria use folate-bound one- carbon (1C) units to methylate tRNA through the serine hydroxymethyl- transferase 2 (SHMT2). SHMT2 provides methyl donors to produce the taurinomethyluridine base at the wobble position of distinct mitochondrial tRNAs. Loss of the catalytic activity of SHMT2 impairs oxidative phosphorylation and mitochondrial translation (53).
Stem cell differentiation requires the constant and dynamic adaption of energy supply to fuel protein synthesis. A highly efficient and fast trigger to adapt global and gene-specific protein translation rates to external stimuli is the dynamic deposition and removal of modifications in tRNAs.
RNA modifications in disease
tRNA modifications in disease
Complex human pathologies that are directly linked to tRNA modifications include cancer, type 2 diabetes, neurological disorders, and mitochondrial-linked disorders (54). The human brain is particularly sensitive to defects in tRNA modifications (55), and the cellular defects are commonly caused by impaired translational efficiency and misfolded proteins, leading to a deleterious activation of the cellular stress response.
Similar to normal tissues, tumor cells are challenged by a changing microenvironment—for example, through hypoxia, inflammatory cell infiltration, and exposure to cytotoxic drug treatments (40). Thus, tumor cell populations rely on the correct deposition of tRNA modifications to switch their transcriptional and translation programs dynamically in response to external stimuli. For instance, mouse skin tumors that lack the NSUN2-mediated m5C modification repress global protein synthesis, leading to an enlarged undifferentiated tumor-initiating cell population (40). However, the up-regulation of NSUN2 and methylation of tRNAs is strictly required for cell survival in response to chemotherapeutic drug treatment, and NSUN2-negative tumors fail to regenerate after exposure to cytotoxic drug treatments (40). Thus, tumor-initiating cell populations require the tight control of protein synthesis for accurate cell responses and to maintain the bulk tumor.
Similarly, modifications found in other noncoding RNAs are likely to play important roles in their biogenesis and function. For instance, the biogenesis of rRNA is known to be substantially affected by various modifications, the defect of which could contribute to human ribosomopathies (56).
mRNA modifications in disease
mRNA modifications also contribute to the survival and growth of tumor cells, further high-lighting the importance of mRNA modifications in the regulation of cell fate decisions. The METTL3 and METTL14 subunits of the m6A writer complex are highly expressed in human hematopoietic stem and progenitor cells (HSPCs), and the expression of these two subunits declines during differentiation of HSPCs along the myeloid lineage (57, 58). Overexpression of METTL3 inhibits cell differentiation and increases cell growth (57, 58). Consistent with a role in maintaining self-renewal programs, METTL3 and METTL14 are overexpressed in acute myeloid leukemia (AML), and AML cells are sensitive to depletion of METTL3 and METTL14 (57–59). These effects could be mediated by changes in the methylation of cell state–specific transcripts such as MYC, MYB, BCL2, PTEN, and SP1 that help to maintain self-renewal and prevent differentiation (57–59). The stabilization of certain m6A methylated transcripts in AML cells may be mediated by the IGF2BP1–3 family of m6A reader proteins rather than the YTHDF1–3 family (13, 58).
An opposite role for m6A in leukemogenesis was found in certain subtypes of AML with increased expression of the demethylase FTO, resulting in decreased m6A and elevated levels of oncogene transcripts (60). Inhibition of FTO reduces AML cell proliferation and viability in these cell types (60, 61). The mechanisms and pathways for the writers and eraser to affect AML are likely distinct. Whereas elevated writer expression blocks differentiation of HPSCs to contribute to AML initiation and cell survival, elevated FTO mostly affects AML proliferation. This distinction is exemplified by the dual role of the oncometabolite R-2HG; its inhibition of TET2 contributes to AML initiation but also inhibits FTO in a subset of AML, leading to repressed proliferation (61). Decreased m6A is also associated with some solid tumors, likely promoting their proliferation. For example, in breast cancer, hypoxia was shown to induce the overexpression of ALKBH5, an m6A eraser, and ZNF217, a transcription factor that can inhibit METTL3, resulting in reduction of the m6A methylation and decay of transcripts such as Nanog (62, 63). Similarly, overexpression of ALKBH5 or down-regulation of METTL3 or METTL14 promotes the tumorigenicity of glioblastoma cells through stabilization of pro-proliferative transcripts such as FOXM1 (64, 65). In endometrial cancer, reduced m6A promotes cell proliferation through misregulation of transcripts encoding regulators of the AKT pathway (66). Additional mechanisms for how m6A alters gene expression to help drive cancer progression are likely to be discovered in the future.
Future perspectives
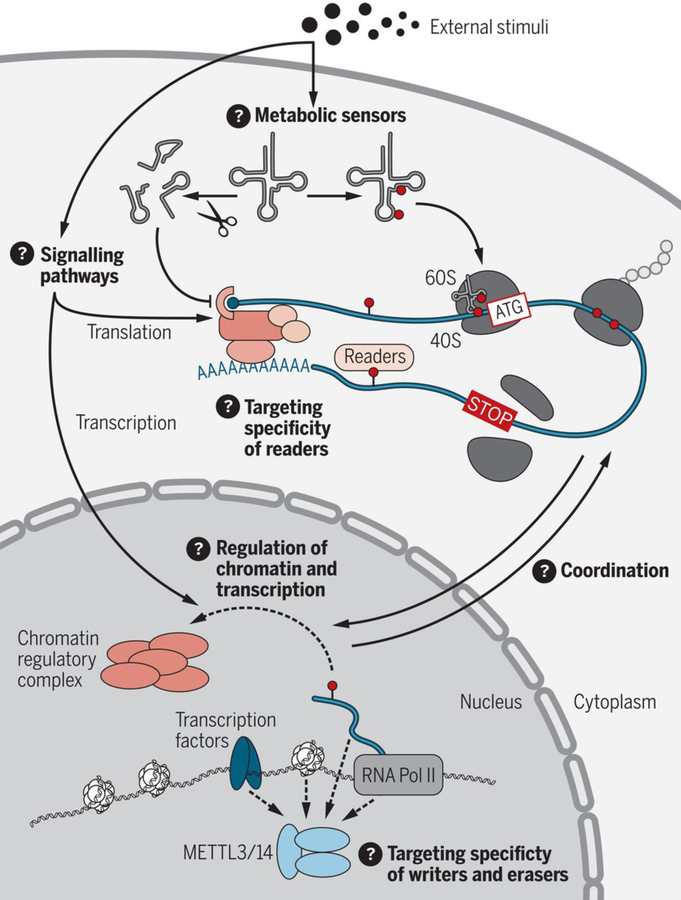
Although these studies demonstrate the roles of RNA modifications in various developmental processes, our understanding of how RNA modifications contribute to these processes remains incomplete, especially at the mechanistic level (Fig. 3). The development of new tools that can determine the transcriptome-wide distribution of RNA modifications at nucleotide resolution with quantitative information about the modification fraction would greatly help in these endeavors. Further, it will be essential to understand the intrinsic and extrinsic factors that determine the specificity of the RNA modification writers, readers, and erasers and how these proteins are regulated in different cell types across development. For many RNA modifications, there is only very little information available on how these modifications recruit or repel RBPs, yet this information is essential to understand how RNA modifications modulate the RNA processing or protein translation machineries. In addition, how cells adjust RNA modifications and adapt the protein synthesis machinery in response to metabolic requirements remains largely unclear—in particular, how these changes in translation could have cell type–specific effects. Last, recent studies have suggested that m6A could directly or indirectly influence chromatin state and transcription through regulation of chromatin regulatory complexes and long noncoding RNAs (67, 68). The potential roles of m6A and other RNA modifications in shaping chromatin states may provide additional mechanisms for explaining how these modifications contribute to gene regulation in development.
Fig. 3. Future directions for research into gene regulation by RNA modifications.
There are several unresolved questions in the field: Mechanistically, how do external stimuli regulate RNA modification to affect protein translation rates and transcription? How do RNA modifying enzymes act as metabolic sensors? How do RNA modifications directly or indirectly regulate chromatin regulatory complexes to affect chromatin state or transcription? What factors—such as transcription factors, chromatin, RNA, RBPs, or components of the RNA polymerase II complex— recruit m6A writer and eraser enzymes to their targets? What factors regulate and determine the target specificity of readers? How are the protein synthesis and transcription machineries coordinated by RNA modifications?
ACKNOWLEDGMENTS
Funding: B.T.H. is supported by National Cancer Institute fellowship F32 CA221007. C.H. is supported by the National Institutes of Health (HG008935 and GM071440). C.H. is an investigator of the Howard Hughes Medical Institute. M.F. is supported by a Cancer Research UK Senior Fellowship (C10701/A15181), the European Research Council (ERC; 310360), and the Medical Research Council UK (MR/M01939X/1). Part of this work was carried out in the framework of the European COST action EPITRAN 16120.
Footnotes
Competing interests: C.H. is a scientific founder of Accent Therapeutics and a member of its scientific advisory board. M.F. consults for Storm Therapeutics. All other authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Furlong EEM, Levine M, Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D et al. , Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD et al. , Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roundtree IA, Evans ME, Pan T, He C, Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geula S et al. , Science 347, 1002–1006 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Frye M, Blanco S, Development 143, 3871–3881 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jia G et al. , Nat. Chem. Biol 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng G et al. , Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X et al. , Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer KD et al. , Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X et al. , Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edupuganti RR et al. , Nat. Struct. Mol. Biol 24, 870–878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H et al. , Nat. Cell Biol 20, 285–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloan KE et al. , RNA Biol 14, 1138–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimmel P, Nat. Rev. Mol. Cell Biol 19, 45–58 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Schaffrath R, Leidel SA, RNA Biol 14, 1209–1222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson G, Coller J, Nat. Rev. Mol. Cell Biol 19, 20–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista PJ et al. , Cell Stem Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y et al. , Nat. Cell Biol 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertero A et al. , Nature 555, 256–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li HB et al. , Nature 548, 338–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon KJ et al. , Cell 171, 877–889.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M et al. , Genome Biol 19, 69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao BS et al. , Nature 542, 475–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C et al. , Nature 549, 273–276 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Ivanova I et al. , Mol. Cell 67, 1059–1067.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu K et al. , Cell Res 27, 1100–1114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z et al. , Cell Res 27, 1216–1230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu PJ et al. , Cell Res 27, 1115–1127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojtas MN et al. , Mol. Cell 68, 374–387.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Bailey AS et al. , eLife 6, e26116 (2017).29087293 [Google Scholar]
- 32.Jain D et al. , eLife 7, e30919 (2018).29360036 [Google Scholar]
- 33.Haussmann IU et al. , Nature 540, 301–304 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Lence T et al. , Nature 540, 242–247 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Blanco S et al. , EMBO J 33, 2020–2039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco S et al. , PLOS Genet 7, e1002403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuorto F et al. , EMBO J 34, 2350–2362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzzi N et al. , Cell 173, 1204–1216.e26 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Yoo H, Son D, Jang YJ, Hong K, Biochem. Biophys. Res. Commun 478, 631–636 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Blanco S et al. , Nature 534, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaunay S et al. , J. Exp. Med 213, 2503–2523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nedialkova DD, Leidel SA, Cell 161, 1606–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llorens-Bobadilla E et al. , Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Signer RA, Magee JA, Salic A, Morrison SJ, Nature 509, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma U et al. , Science 351, 391–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R, Cell 170, 61–71.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y et al. , Nat. Cell Biol 20, 535–540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodarzi H et al. , Cell 161, 790–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Haute L et al. , Nat. Commun 7, 12039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano S et al. , Nat. Chem. Biol 12, 546–551 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Haag S et al. , EMBO J 35, 2104–2119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Menzies KJ, Auwerx J, Development 145, dev143420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morscher RJ et al. , Nature 554, 128–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres AG, Batlle E, Ribas de Pouplana L, Trends Mol. Med 20, 306–314 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Bednářová A et al. , Front. Mol. Neurosci 10, 135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks MM et al. , Sci. Adv 4, eaao0665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vu LP et al. , Nat. Med 23, 1369–1376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weng H et al. , Cell Stem Cell 22, 191–205.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbieri I et al. , Nature 552, 126–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z et al. , Cancer Cell 31, 127–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su R et al. , Cell 172, 90–105.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C et al. , Proc. Natl. Acad. Sci. U.S.A 113, E2047–E2056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang C et al. , Oncotarget 7, 64527–64542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Q et al. , Cell Reports 18, 2622–2634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S et al. , Cancer Cell 31, 591–606.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J et al. , Nat. Cell Biol 20, 1074–1083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patil DP et al. , Nature 537, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y et al. , Nat. Neurosci 21, 195–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]